Multiple self-healing squamous epithelioma (MSSE, OMIM132800) also known as Ferguson-Smith disease (FSD) is a rare inherited skin cancer syndrome characterized by multiple invasive keratoacanthoma-like skin tumors that regress spontaneously leaving pitted scars (Ferguson-Smith, 1934; Goudie et al., 1993). The majority of the first-identified affected families have a shared Scottish ancestry, but MSSE has also been described in families of non-Scottish origin (D’Alessandro et al., 2007). Recently, the causative gene for MSSE was identified as TGFBR1, encoding a transmembrane serine/threonine kinase receptor involved in TGF-β signaling (Goudie et al., 2011). TGFBR1 mutations in MSSE are functionally null implying a tumor suppressor action in this disease, whereas mis sense mutations in the same gene can lead to Marfan syndrome-related disorders (Goudie et al., 2011; Loeys et al., 2005). TGFBR1 had previously been excluded as a MSSE candidate because the gene lies outside the original shared at-risk haplotype (SRH) in the Scottish families (Bose et al., 2006). However using large scale sequencing technology, Goudie et al. (2011) identified TGFBR1 germline mutations in a total of 18 MSSE families including 12 Scottish families. Of 9 Scottish families with the SRH, 7 families shared the same TGFBR1 mutation (p.G52R), importantly, however 2 families had different causative mutations (p.N45S and p.R414X). This suggests that unidentified rare variants associated with MSSE pathogenesis may exist in the non-TGFBR1 SRH region that has been conserved in 9 families. The fact that this relatively large conserved haplotype had not undergone recombination over many generations in these 9 affected Scottish MSSE families suggested that manifestation of the disease may require both the causative TGFBR1 mutation and additional variant(s) located within the SRH region.
To search for these SRH variants we used the Agilent SureSelect Target Enrichment system followed by sequencing on the Illumina GAII platform to sequence all of the target genes between the markers D9S197 and D9S1809 on 9q22 31 (D’Alessandro et al., 2007). 13 families including 7 Scottish families with the SRH were available (Table S1). Family numbers hereafter correspond to the study by Goudie et al. 2011. We performed targeted sequencing in 5 families with the SRH and 5 families without the SRH, along with 6 CEPH controls. Coding regions of the 15 then-known genes were sequenced previously (D’Alessandro et al., 2007), but based on NCBI36 (hg18), there are now 29 known genes in the targeted region (Table S5), encompassing ~2.2-Mb in total, and this entire genomic region of these genes was sequenced. After filtering out common variants included in dbSNP and shared with CEPH controls, we identified 9 non-coding variants shared by all 5 families with the SRH. None of the 9 rare variants were detected in MSSE families without the SRH. These 9 variants are located in intronic regions of 5 different genes: FAM120A, PHF2, C9orf3, FANCC, and PTCH1 (Table 1). Sanger dideoxy sequencing confirmed these 9 variants in all 5 families plus 2 additional families (Table S1, Figure S1). Thus, all 9 variants were conserved over this ~2.2Mb region in 7 Scottish families with the SRH, although TGFBR1 mutations were different in families 2 and 18 (Figure 1a). In 231 unrelated healthy controls, including 118 Scottish individuals, the minor allele frequency (MAF) of the 9 variants was rare, ranging from 0 to 0.022 and the association of these variants with the MSSE phenotype was highly significant (Table 1). This suggests that these 9 variants are MSSE-associated and segregated in individuals with this rare skin malignancy condition. Interestingly, these variants are located at either end of the ~2.2-Mb target region leaving a ~1.4-Mb central region where 24 genes are densely located (Figure 1a). Further analysis of MSSE families lacking the SRH identified distinct MSSE-associated rare variants in two Scottish families (Table S2-3). In family 17, we identified 8 distinct variants that were detected in 4 affected family members (Table S3). Interestingly, these 8 variants were all clustered in a ~1.4-Mb central region of the SRH that excluded the 9 MSSE associated variants discussed above (Figure 1a). This family harbored the most complex TGFBR1 mutation (c.1059_1062delACTGinsCAATAA) that was not observed in other families. All of these variants are non-coding and not frequently found in 162 healthy Scottish controls tested.
Table 1.
9 rare non-coding variants identified in 7 Scottish MSSE families with the SRH proximal to the causative TGFBR1 locus.
| Positions on Chr 9 | Gene | W | V | Controls (N=231) |
MAF2 | P value4 | |||
|---|---|---|---|---|---|---|---|---|---|
| (NCBI36/hg18) | (GRCh37/hg19) | W/W | W/V | V/V | |||||
| 95319014 | 96279193 | FAM120A | C | ins A | 222 | 0 | 0 | 0 | 6.13E-33 |
| 95323192 | 96283371 | FAM120A | G | A1 | 211 | 1 | 0 | 0.0023 | 1.23E-29 |
| 95393569 | 96353748 | PHF2 | G | C | 198 | 0 | 0 | 0 | 2.74E-28 |
| 95447694 | 96407873 | PHF2 | C | T | 203 | 0 | 0 | 0 | 2.37E-30 |
| 96870898 | 97831077 | C9orf3 | G | T1 | 208 | 2 | 0 | 0.005 | 5.89E-26 |
| 97001607 | 97961786 | FANCC | T | C1 | 214 | 1 | 0 | 0.002 | 2.13E-42 |
| 97289083 | 98249262 | PTCH1 | G | ins A | 223 | 5 | 0 | 0.011 | 2.84E-25 |
| 97296872 | 98257051 | PTCH1 | T | C1 | 209 | 6 | 0 | 0.014 | 1.14E-22 |
| 97309311 | 98269490 | PTCH1 | G | C1 | 195 | 9 | 0 | 0.022 | 8.75E-18 |
Abbreviations: Chr, chromosome; W, wild-type; V, variant; MAF, minor allele frequency; ins A, A insertion
Variants are reported in 1000 Genomes (http://www.1000genomes.org).
The minor allele frequency was calculated using Haploview 4.2. Individuals with >50% missing genotypes excluded.
rs117519070, MAF for European (EUR) population is 0.007 sourced from 1000 Genomes.
P value calculated by Chi-square test in Haploview 4.2 using all MSSE patients analyzed in this study and normal controls
Figure 1.
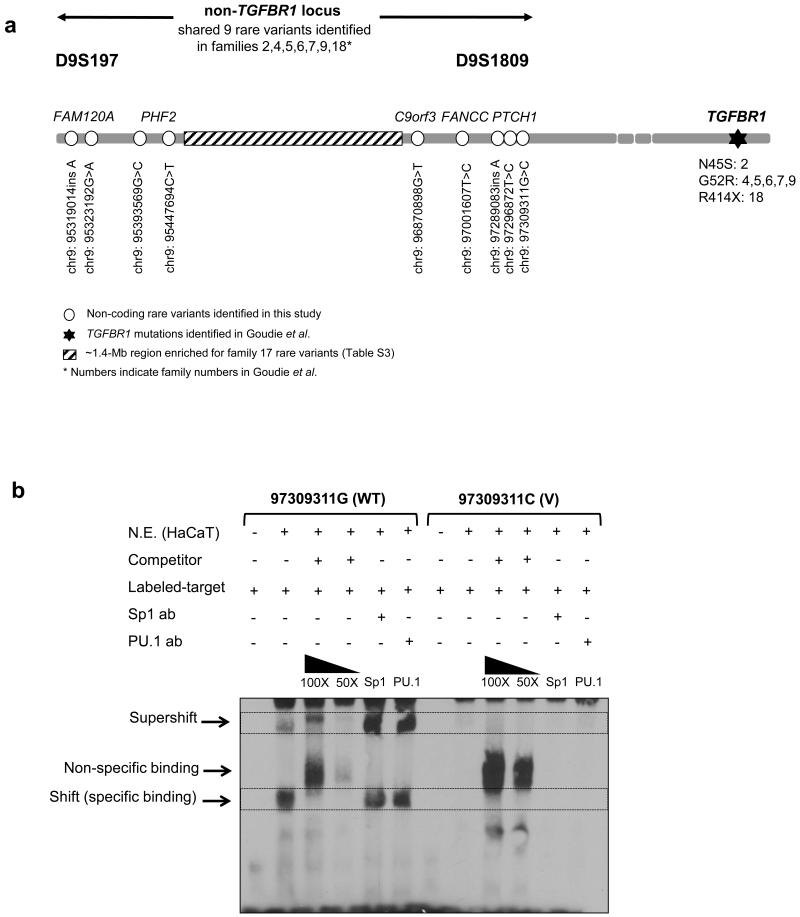
Schematic summary of the 9 rare variants identified in this study and functional effect (a) 7 Scottish MSSE families shared all the 9 non-coding non-TGFBR1 variants identified in this study, but the TGFBR1 mutations are different amongst these families (as described by Goudie et al. 2011). The 9 variants are located at either end of a non-TGFBR1 locus spanning ~2.2-Mb leaving a ~1.4-Mb central region where family 17 has diverged. In this family, 8 distinct rare variants were identified in the ~1.4-Mb central region. Family numbers follow Goudie et al., 2011. (b) One of PTCH1 variants, 97309311G>C was assayed by EMSA to see the effect of the variant on nuclear protein binding. While the major allele (97309311G) bound to SP1 and PU.1 transcription factors in nuclear extracts from HaCaT immortalized human keratinocyte cells, the minor allele (97309311C) showed disruption of this binding. WT for wild-type; V for variant; ab for antibody
Three of the 9 variants found were located in intronic regions of the PTCH1 gene, of which germline loss of function mutations are responsible for nevoid basal cell carcinoma syndrome (NBCCS) (Hahn et al., 1996; Johnson et al., 1996). Our previous study using mouse models suggested that a gain of function polymorphic variant in the mouse Ptch1 gene conferred susceptibility to squamous cell carcinoma (SCC) development (Wakabayashi et al., 2007). We considered the possibility that these rare PTCH1 variants may play a role in predisposition to SCC development in MSSE patients. We carried out a computational analysis of the possible functional significance of all variants using the program AliBaba2.1. This analysis identified the 97309311G>C PTCH1 variant as having possible functional significance. An electromobility shift assay (EMSA) was performed on the major 97309311G>C variant, which is reported to bind to multiple transcription factors including SP1 and PU.1 (http://genome.ucsc.edu/ENCODE/). While the major allele (97309311G) could bind to SP1 and PU.1 transcription factors in nuclear extracts from HaCaT immortalized human keratinocyte cells, the minor allele (97309311C) showed complete disruption of this binding, indicating that the MSSE-associated rare variant perturbs normal interaction between this binding site and transcription factors (Figure 1b).
In our study, we identified a rare MSSE-associated haplotype comprising 9 variants spanning a 2.2Mb region lying 1.5Mb proximal to the causative TGFBR1 gene. Several of the genes in this region have been implicated in cancer development, and regulatory polymorphisms may affect tumor susceptibility (Sinha et al., 2008). The co inheritance of these variants in the majority of Scottish families with MSSE suggests that these variants, or other as yet unknown linked variants (or their combinations), may affect the expression of the skin cancer phenotype induced by the mutations in TGFBR1. Their locations at opposite ends of the conserved haplotype may suggest that they modify chromatin loop structure to influence expression of genes within the region. Alternatively, the variants may directly affect expression of relevant genes, particularly the PTCH1 gene, which is implicated in susceptibility to different forms of skin cancer and carries three intronic variants, one of which affects transcription factor binding. Functional analysis of the possible roles of these and other variants in determining the MSSE phenotype, for example by gene expression or ChIP-seq analysis to assess differential transcription factor binding, will require further studies using keratinocytes or cultured primary tumor cells from MSSE affected patients. Finally, it is possible that since loss of function germline mutations in TGFBR1 are very rare, the conserved haplotype may act as a modifier to increase survival of individuals carrying strong mutations in this potent developmental regulator. This mechanism is supported by the identification of unlinked variant genetic modifiers that are preferentially inherited in mice haploinsufficient for Tgfb1 (Benzinou et al. 2012).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Program Project Grant (PPG) 5P01AR050440 from the US NIAMS (National Institute Of Arthritis And Musculoskeletal And Skin Diseases), National Institutes of Health (NIH). AB is supported by grants from the NCI MMHCC (National Cancer Institute Mouse Models of Human Cancer Consortium) 2U01 CA08422-06. We thank Malcolm Dunlop for providing normal control samples used in the study.
Abbreviations
- MSSE
multiple self-healing squamous epitheliomas
- FSD
Ferguson-Smith Disease
- TGFBR1
transforming growth factor beta receptor 1
- GA II
Genome Analyzer II
- CEPH
Centre d’ Etude du Polymorphisme Humain
- MAF
minor allele frequency
- BCC
basal cell carcinomas
- SCC
squamous cell carcinomas
- SRH
shared at-risk haplotype
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest.
REFERENCES
- Benzinou M, Clermont FF, Letteboer TG, et al. Mouse and human strategies identify PTPN14 as a modifier of angiogenesis and hereditary haemorrhagic telangiectasia. Nat Commun. 2012;10(3):616. doi: 10.1038/ncomms1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Morgan LJ, Booth DR, et al. The elusive multiple self healing squamous epithelioma (MSSE) gene: further mapping, analysis of candidates, and loss of heterozygosity. Oncogene. 2006;25:806–12. doi: 10.1038/sj.onc.1209092. [DOI] [PubMed] [Google Scholar]
- D’Alessandro M, Coats SE, Morley SM, et al. Multiple self healing squamous epithelioma in different ethnic groups: more than a founder mutation disorder? J Invest Dermatol. 2007;127:2336–44. doi: 10.1038/sj.jid.5700914. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith J. A case of multiple primary squamous-celled carcinomata in a young man, with spontaneous healing. Br J Dermatol. 1934;46:267. [Google Scholar]
- Goudie DR, Yuille MA, Leversha MA, et al. Multiple self-healing squamous epitheliomata (ESS1) mapped to chromosome 9q22-q31 in families with common ancestry. Nat Genet. 1993;3:165–9. doi: 10.1038/ng0293-165. [DOI] [PubMed] [Google Scholar]
- Goudie DR, D’Alessandro M, Merriman B, et al. Multiple self-healing squamous epithelioma is caused by a disease-specific spectrum of mutations in TGFBR1. Nat Genet. 2011;43:365–9. doi: 10.1038/ng.780. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–51. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–71. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Chen J, Neptune ER, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37:275–81. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- Sinha S, Singh RK, Alam N, et al. Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at chromosomal 9q22.3 region: Pathological significance in early- and late-onset breast carcinoma. Mol Canc. 2008;7:84. doi: 10.1186/1476-4598-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Mao JH, Brown K, et al. Promotion of H-ras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature. 2007;445:761–65. doi: 10.1038/nature05489. [DOI] [PubMed] [Google Scholar]
- Alibaba2 http://www.gene-regulation.com/pub/programs/alibaba2/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.