Abstract
Self-reinforcing negative feedback loops are commonly observed in biological systems. RNA-mediated negative feedback loops have been described in the formation of heterochromatin at centromeres in fission yeast and the inactive X chromosome in mammalian cells. The telomere repeat-containing RNA (TERRA) has also been implicated in the formation of telomeric heterochromatin through a self-reinforcing negative feedback loop. In cells derived from human ICF syndrome, TERRA levels are abnormally elevated and telomeres are abnormally shortened. We now show that telomere heterochromatin is also abnormal in ICF cells. We propose that ICF cells fail to reinforce the TERRA-dependent negative feedback loop as a result of the inability to establish heterochromatin at subtelomeres. This failure is likely due to the lack of DNMT3b and DNA methylation, which is a genetic lesion associated with ICF syndrome. Failure of this feedback mechanism leads to catastrophic telomere dysfunction and chromosome instability.
Keywords: TERRA, TRF2, CpG methylation, heterochromatin, ICF syndrome
Introduction
A remarkable recent discovery has been the finding that mammals and yeast express telomere repeat-containing noncoding RNA referred to as TERRA (also TelRNAs).1,2 The function and regulation of TERRA has been the focus of much recent interest and several recent reviews.3–6 Previous to these reports, it was generally assumed that telomeres were heterochromatic nucleoprotein structures that function exclusively in the protection of chromosome ends. The mechanism of end protection involved formation of heterochromatin and transcriptional silencing of genes in close proximity to the telomere repeat tracts. However, like centromeres and the inactive X-chromosome, telomeres are paradoxically transcribed despite being silenced by heterochromatin.
TERRA has been implicated in several telomere and non-telomere related functions. In vitro, TERRA containing oligonucleotides can antagonize telomerase activity by competitive hybridization to the catalytic RNA of telomerase enzyme.2 This mode of action is consistent with the observations that TERRA expression varies inversely with telomerase activity during cellular development and tumorigenesis. However, there are exceptions to this pattern since TERRA RNA levels diminish in aging fibroblasts which have no detectable telomerase activity and shortened telomere repeat tracts.7 Like other non-coding RNAs, TERRA has also been implicated in the establishment of heterochromatin. TERRA physically associates with telomere foci, but can also be found at other subcellular locations, including the sex chromosomes in stem cells.8 The mechanisms regulating TERRA localization, expression and function remain important questions in telomere and chromatin biology, but have, as yet, only partial answers.
TERRA levels are regulated by several mechanisms, including cellular differentiation and telomeric histone modifications.2,7 TERRA is transcribed primarily by RNA polymerase II (RNAPII), and the transcription is further facilitated by shelterin protein TRF1.2 TERRA transcription is also dependent on histone H3 K4 di/trimethylation at subtelomeres and the histone methyltransferase MLL.7 TERRA levels can be induced with treatment of histone deacetylase inhibitor Trichostatin A (TSA) in human cells,3 or by mutations in the histone H3 K9 and H4 K20 methyltransferase in mouse cells.2 In yeast, TERRA levels were upregulated by deletion of Rat1p, an RNA 5'-3' exonuclease and downregulated by deletion of Pap1, responsible for polyadenylation of mRNA.9
TERRA localization is also subject to complex regulation that depends on multiple factors including cellular differentiation and proliferative capacity.1,2 TERRA localization at telomeres is controlled, in part, by factors involved in the non-sense mediated RNA decay pathway (NMD), including UPF1 and SMG1.1 A mechanism for TERRA localization to mammalian telomeres has been proposed recently.10 In our recent study, we showed that TRF2 and TRF1 bind directly to TERRA using RNA-ChIP and RNA-affinity chromatography. The TRF2 amino-terminal basic domain bound directly to TERRA RNA in vitro, and overexpression of a TRF2ΔB mutant caused the delocalization of TERRA from telomere foci. Interaction of TERRA with shelterin proteins, especially TRF1 and TRF2, could account for its colocalization with telomeres.
A function for TERRA in telomere heterochromatin formation was also proposed.10 TERRA was found to interact with several heterochromatin-associated proteins, including methyl CpG binding protein (MeCP2) and heterochromatin protein 1 (HP1), and heterochromatic histone modifications, especially H3 K9me3. The interaction with these heterochromatin factors, including histone H3 K9 methyltransferases, may be sufficient to nucleate heterochromatin complexes. TERRA was also implicated in the recruitment of the Origin Recognition Complex (ORC) to telomeres. In an earlier study, the TRF2 N-terminal domain was shown to bind ORC1 in an RNA-dependent manner.11 ORC has well-established functions in DNA replication initiation, but has also been implicated in heterochromatin formation. Depletion of ORC subunits caused defects in telomere heterochromatin and structural stability. Similarly, siRNA depletion of TERRA caused a similar loss of telomere heterochromatin and structural stability.10 These studies suggest that TERRA mediates an interaction between TRF2 and ORC, HP1 and H3K9me3 to promote heterochromatin formation at telomeres. However, it remains unclear whether these few players are sufficient to maintain stable telomeric heterochromatin and how this structure remains sufficiently dynamic to maintain telomere length in proliferating cells.
Recent studies into Immunodeficiency, Centromeric region instability, and Facial anomalies syndrome (ICF) may help to provide an answer. ICF is an autosomal recessive disease that involves spontaneous chromosome instability and immunodeficiency12,13 (reviewed in ref. 14). The molecular basis for ICF Syndrome has been linked to the mutations in DNA methyltransferase 3b (DNMT3b).15–17 A recent study revealed that ICF cells express an abnormally high level of TERRA and sustain telomere DNA structural abnormalities.12 Here, we examine the status of telomere repeat binding factors and histone modifications at telomeres in ICF cells relative to the parental control cells. We conclude that DNA methylation is essential for maintaining a stable negative feedback loop through which low levels of TERRA can promote heterochromatin formation at telomeres.
Results and Discussion
Loss of CpG methylation from subtelomeres of ICF cells
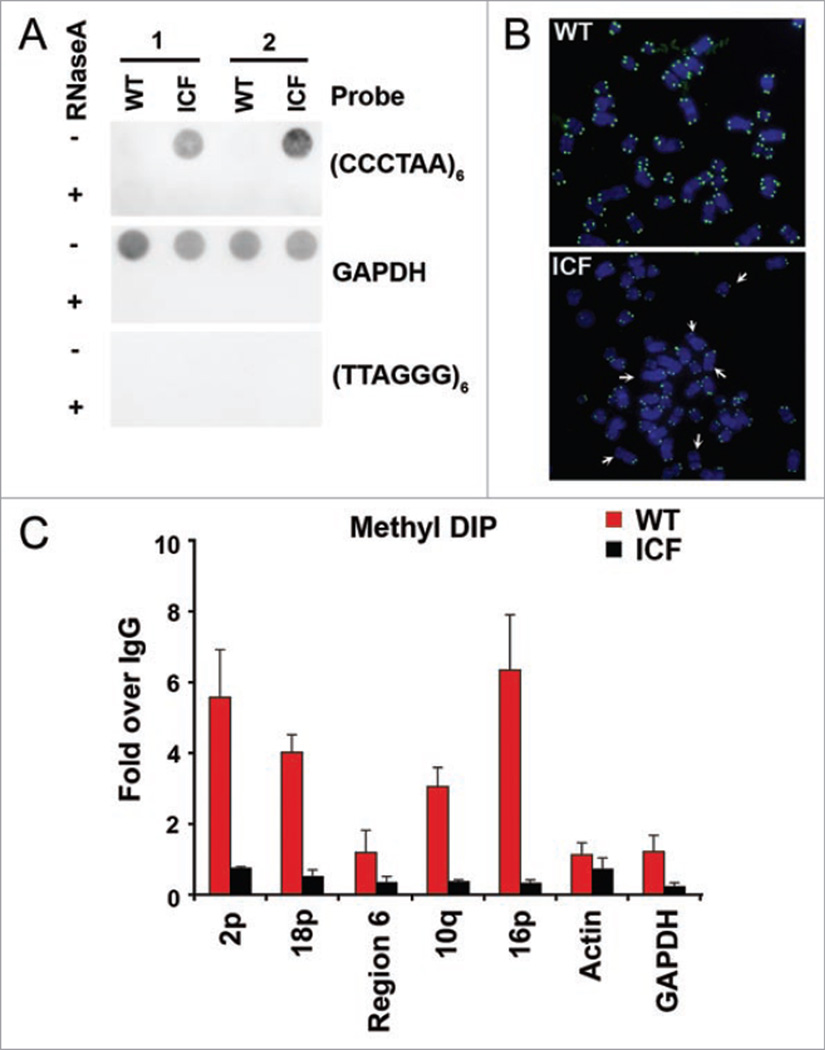
Previous studies have shown that ICF cells have elevated TERRA expression and telomere abnormalities.12 To ensure that the ICF cells used in our experiments have similar properties, we compared lymphoblastoid cells derived from ICF and genetically matched normal individual for TERRA expression using RNA dot blotting methods. Consistent with published studies, we found that TERRA levels were elevated 6–10 fold in ICF cells relative to normal (wt) cells (Fig. 1A). We also confirmed that metaphase telomeres were highly abnormal, with a high prevalence of telomere signal loss (Fig. 1B). To assess the level of cytosine methylation in the subtelomeres, we used the methyl DNA IP (MeDIP) method to compare ICF and wt cells at several different subtelomeric locations (Fig. 1C). The MeDIP was quantified relative to IgG control for each location. We found that CpG methylation was reduced in ICF cells relative to wt cells at all locations assayed, including β-actin and GAPDH loci. This is consistent with the finding that ICF cells lack DNMT3b activity. Interestingly, we found that CpG methylation at subtelomeres was affected more significantly than housekeeping genes, like β-actin and GAPDH. While CpG methylation at β-actin was reduced ~2 fold, the reduction in CpG methylation at subtelomere 2p and 16p were reduced more than 10 fold. These findings indicate that ICF cells have a dramatic loss of CpG methylation at subtelomeres, and may suggest that loss of DNA methylation may be selectively enriched at subtelomeres. This was supported by the findings that repetitive sequences are primary targets of DNMT3b.18
Figure 1.
Loss of telomeric heterochromatin formation in ICF cells. (A) RNA dot-blot analysis of total RNA isolated from ICF LCLs (GM08714) or WT control LCLs from the father (GM08729) in two independent experiments. RNA was detected by hybridization with probes containing (CCCTAA)6, GAPDH, or (TTAGGG)6, as indicated. RNase A treatment (+) was used as an RNA specificity control. (B) Representative telomere DNA FISH analysis on metaphase spreads for telomere defects in WT and ICF LCLs. Arrows indicate ends with telomeric signals loss. (C) Methylated DNA Immuoprecipitation (MeDIP) analysis of Cytosine methylation in WT (red) or ICF (black) LCLs. Real-time PCR was used to quantify methylated DNA signals by primers specific for indicated subtelomeric regions (2p, 18p, region 6, 10q or 16p) or controls β-actin and GAPDH loci. The bar graph represents the value of MeDIP relative to IgG IP.
TRF2 binding to subtelomeres is reduced in ICF cells
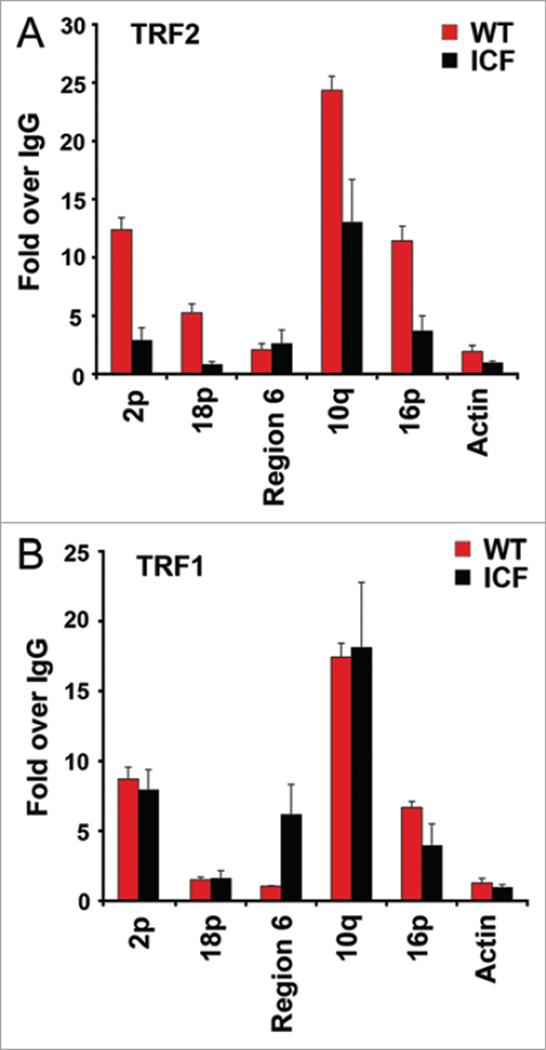
To investigate further the mechanism of telomere dysfunction in ICF cells, we assayed the binding of TRF2 and TRF1 to the same subtelomeric regions that were found to have substantially reduced CpG DNA methylation using ChIP assays (Fig. 2). We found that TRF2 binding was reduced ~4 fold at several subtelomeres (e.g., 2p and 16p) where CpG methylation was reduced in ICF cells relative to wt cells (Fig. 2A). In contrast, TRF1 binding to subtelomeres was not reduced in ICF cells relative to wt cells (Fig. 2B), and may even increase at subtelomere region 6. It is interesting to note that subtelomere region 6 consists of TTAGGG tracts located >2–100 kb from telomere termini in multiple chromosomes. The binding of TRFs to subtelomeres is likely to be both direct and indirect, since consensus and degenerate telomere repeats are found in the subtelomeres, and since DNA-looping from terminal repeats to subtelomeres is also known to form.
Figure 2.
Decreased association of TR F2 with subtelomeres in ICF cells. (A) ChIP analysis by TR F2 antibody at indicated subtelomeric regions or at the control Actin loci in WT or ICF LCLs. ChIP DNA was assayed by real-time PCR with specific primers as indicated. TRF2 binding was first normalized to corresponding input DNA and then shown as fold change relative to IgG. (B) Same as in (A), except that TRF1 antibody was used for ChIP analysis.
Subtelomeric heterochromatin is disrupted in ICF cells
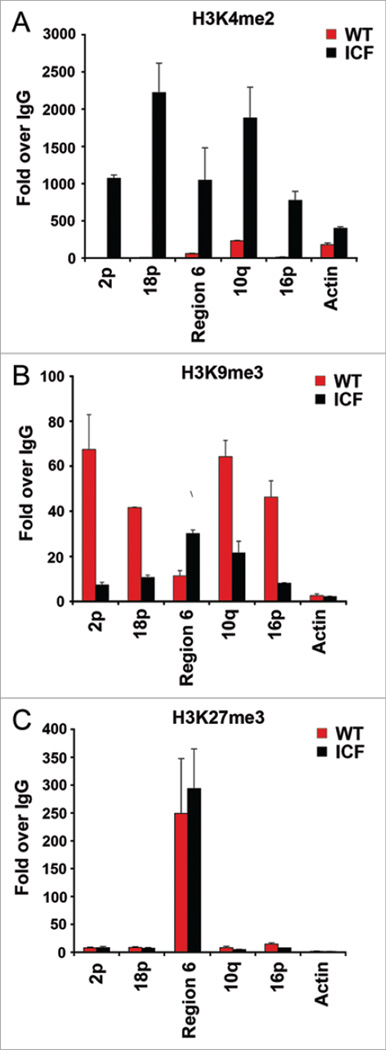
Previous studies have shown that telomeres and subtelomeres are enriched in heterochromatin-associated histone modifications, especially histone H3 K9me3.19 TERRA transcription has been shown to be regulated by telomeric heterochromain2 and be dependent on euchromatic mark of histone H3 K4me2/3 mediated by MLL histone methyltransferase.7 Others have also found evidence for polycomb associated H3 K27me3 modifications in subtelomeric heterochromatin.20 To investigate the effect of ICF cells on subtelomere histone modifications, we assayed H3 K4me2, H3 K9me3 and H3 K27me3 for changes in ICF cells relative to wt cells (Fig. 3). We found that subtelomeres of ICF cells had a dramatic enrichment of H3 K4me2 relative to wt cells (Fig. 3A). The average increase was greater than 100 fold at the subtelomeres, while β-actin was only increased by 2 fold. A reciprocal loss of H3 K9me3 was also observed at almost every subtelomeres examined in ICF relative to wt cells, with the exception of subtelomere region 6 (Fig. 3B). The average loss of H3 K9me3 was ~5 fold. The increase in H3 K9me3 at region 6 may correlate with the increase in TRF1 binding at this region, indicating that it behaves differently from other subtelomere regions that are proximal to telomeres. No significant change in H3 K27me3 was detected (Fig. 3C). The change in histone modifications at subtelomeres is striking given that a normal pattern of histone modifications has been shown on the inactive X chromosome of ICF cells relative to the parental cells.21 We conclude that histone modifications associated with heterochromatin formation are substantially altered at subtelomeres in ICF cells. The increase in H3 K4me2 and decrease in H3 K9me3 is also consistent with the large increase in TERRA transcription in ICF cells.
Figure 3.
DNA hypomethylation correlates with increased H3K4 methylation and reduced H3K9 methylation at subtelomeres of ICF cells. (A–C) ChIP analysis of subtelomeric histone modifications with antibodies specific for histone H3 K4 di-methylation (H3 K4me2), H3 K9 tri-methylation (H3 K9me3), or H3 K27 tri-methylation (H3 K27me3) in WT (red) or ICF (black) LCLs, respectively.
Conclusions
Our findings indicate the ICF cells have multiple defects at subtelomeres, including a loss of CpG methylation (Fig. 1C), loss of TRF2 binding (Fig. 2A), and a loss of heterochromatic histone mark H3 K9me3 (Fig. 3B). The most remarkable finding is that euchromatic histone modification H3 K4me2 is increased by several orders of magnitude. Since ICF cells are known to have a deficiency in DNMT3b, it is likely that all of these changes in telomere structure, TERRA RNA expression, and histone modifications are an indirect consequence of the loss of CpG methylation. However, in contrast the high levels of TERRA transcription and telomere shortening seen in ICF cells, mouse ES cells deficient in DNMT3b (DNMT3b−/−) show a slight reduction of TERRA levels and a dramatic elongation of telomere length.2 Also, no apparent change of TRF2 binding and histone modifications was observed in telomeric and subtelomeric regions of DNMT3b−/− ES cells.22 Therefore, precisely how CpG methylation mediates these downstream events is not entirely clear.
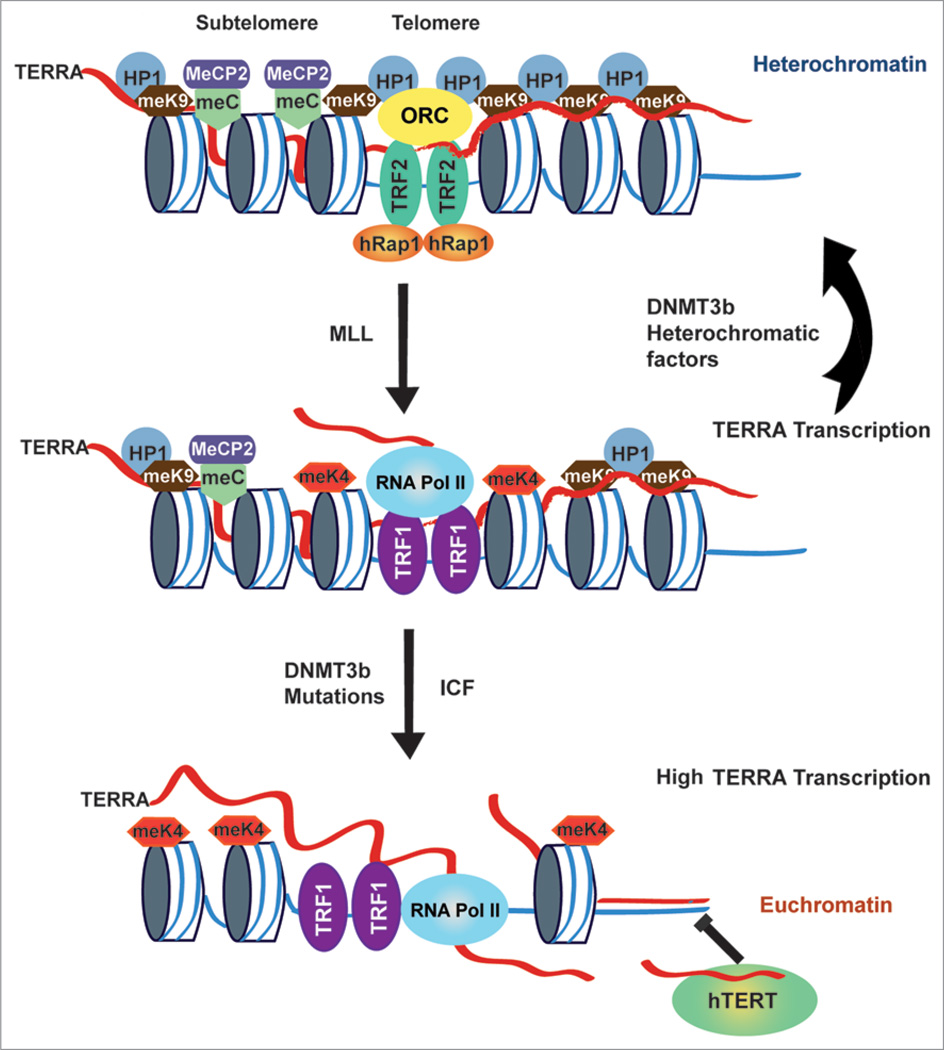
In a previous study, we found that TERRA RNA could interact with MeCP2.10 It is possible that MeCP2 binding to methyl CpG at subtelomeres is essential for recruiting TERRA and establishing heterochromatin at subtelomeres. We also observed that TRF2 binding to subtelomeres is reduced in ICF cells. The loss of TRF2 binding is not easily explained as a consequence of loss of CpG methylation, since there is no evidence that CpG methylated DNA alters TRF2 DNA binding directly. However, we have shown that TRF2 can bind to TERRA RNA, and it is possible that TRF2-TERRA binding may facilitate interactions of the terminal repeats with the subtelomeric regions. This potential telomere-subtelomere interaction must also be dependent on CpG DNA methylation. Alternatively, the loss of CpG methylation is likely to allow transcription factor assembly and RNA polymerase II unimpeded access to subtelomeres. This is likely to result in high levels of TERRA transcription and consequently high levels of H3 K4 me2. In this case, loss of TRF2 binding and elevated histone H3 K4me2 may be an indirect consequence of deregulated RNA polymerase II transcription through the subtelomere and telomere repeats. In normal cells, it is likely that TERRA promotes heterochromatin through recruitment of DNMT3b and generation of CpG methylation at subtelomeres (Fig. 4).
Figure 4.
Schematic model for dynamic changes of subtelomeric and telomeric heterochromatin regulated by TERRA RNA transcription in ICF cells. In normal cells, TERRA facilitates heterochromatin formation through a negative feedback looping mechanism, which is likely mediated by the recruitment of DNMT3b and the generation of CpG methylation at subtelomeres. TERRA RNA further stabilizes the telomeric heterochromatin through physical interactions with ORC, MeCP2, and heterochromatin factors including HP1. In ICF cells, loss of CpG methylation at subtelomeres due to DNMT3b mutations results in an irreversible chromatin state, as well as a loss of negative feedback looping by TERRA. This leads to an “open” chromatin state enriched with H3 K4 dimethylation and allows transcription factor assembly and RNA polymerase II unimpeded access to subtelomeres. Eventually, highly induced TERRA levels may contribute to telomere dysfunction in ICF cells either by inhibiting telomerase activity, forming RNA-DNA hybrids with telomeres, or by interactions with telomere binding proteins.
The mechanism by which TERRA promotes CpG methylation remains elusive since there is no direct evidence for an interaction between TERRA and DNMT3b. Though DNMTs have been shown to bind to XIST RNA, along with polycomb factors,23,24 we did not find any evidence for polycomb-associated histone H3 K27me3 modifications at subtelomeres. This certainly does not rule out that a similar, or related process of RNA-dependent heterochromatin formation is occurring at telomeres as it does at the inactive X-chromosome or at centromeres in fission yeast. It is also possible that ICF cells have additional mutations to DNMT3b that further disrupt telomere heterochromatin formation. This would partially explain why ICF cells behave differently than mouse cells with a targeted disruption in DNMT3b. Future studies will be necessary to determine what additional factors are required for TERRA-dependent heterochromatin formation, as well as potential other functions of TERRA in regulating telomere length and structural stability. Along these lines, it is interesting to note that TERRA RNA can be processed into small pi-like RNAs (tel-sRNAs) containing a 2'-O-methyl group at the 3' terminus.25 PiRNAs have been implicated in silencing retrotransposons through DNA methylation.26,27 Thus, it will be important to determine if tel-sRNAs provide a mechanism for DNA methylation at subtelomeres.
Acknowledgements
We thank Dr. Kuzuko Nishikura (The Wistar Institute) for ICF cells, Dr. Harold Riethman (The Wistar Institute) for sequence information of subtelomeric DNA, Andreas Wiedmer for superb technical assistance, and the Wistar Core Facilities in Microscopy, Flow Cytometry and Genomics. This work was funded in part by grants from NIH (CA CA093606) to P.M.L. and the Leukemia-Lymphoma Society to Z.D.
References
- 1.Azzalin CM, Reichenback P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance ractors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 2.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 3.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 4.Schoeftner S, Blasco MA. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res. 2008;122:194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- 7.Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol. 2009;29:4519–4526. doi: 10.1128/MCB.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang LF, Ogawa Y, Ahn JY, Namekawa SH, Silva SS, Lee JT. Telomeric RNAs mark sex chromosomes in stem cells. Genetics. 2009;182:685–698. doi: 10.1534/genetics.109.103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5' to 3' exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, Lieberman PM. RNA-dependent recruitment of the origin recognition complex. EMBO J. 2008;27:3024–3035. doi: 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Human Mol Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 13.Tuck-Muller CM, Narayan A, Tsien F, Smeets DF, Sawyer J, Fiala ES, et al. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet Cell Genet. 2000;89:121–128. doi: 10.1159/000015590. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich M, Jackson K, Weemaes C. Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF) Orphanet J Rare Dis. 2006;1:2. doi: 10.1186/1750-1172-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrlich M, Buchanan KL, Tsien F, Jiang G, Sun B, Uicker W, et al. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Human Mol Genet. 2001;10:2917–2931. doi: 10.1093/hmg/10.25.2917. [DOI] [PubMed] [Google Scholar]
- 16.Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 18.Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 19.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 20.Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gartler SM, Varadarajan KR, Luo P, Canfield TK, Traynor J, Francke U, et al. Normal histone modifications on the inactive X chromosome in ICF and Rett syndrome cells: implications for methyl-CpG binding proteins. BMC Biol. 2004;2:21. doi: 10.1186/1741-7007-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 23.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 25.Cao F, Li X, Hiew S, Brady H, Liu Y, Dou Y. Dicer independent small RNAs associate with telomeric heterochromatin. RNA. 2009;15:1274–1281. doi: 10.1261/rna.1423309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]