Abstract
Autophagy (also known as macroautophagy) captures intracellular components in autophagosomes and delivers them to lysosomes, where they are degraded and recycled. Autophagy can have two functions in cancer. It can be tumour suppressive through the elimination of oncogenic protein substrates, toxic unfolded proteins and damaged organelles. Alternatively, it can be tumour promoting in established cancers through autophagy-mediated intracellular recycling that provides substrates for metabolism and that maintains the functional pool of mitochondria. Therefore, defining the context-specific role for autophagy in cancer and the mechanisms involved will be important to guide autophagy-based therapeutic intervention.
Autophagy is a process by which cells capture intracellular proteins, lipids and organelles, and deliver them to the lysosomal compartment where they are degraded1,2. The products of autophagic degradation of intracellular material are exported from lysosomes into the cytoplasm where they are recycled3. The intracellular recycling function of autophagy has a varying impact on cellular and organismal physiology that depends on the circumstances. There is a long-term need for autophagy to prevent tissue damage and disease, and there is also an acute requirement for autophagy to sustain homeostasis in stressful environments.
Autophagy occurs at a low basal level, but it can be induced and can implement selective or non-selective bulk degradation. Thus, the regulation of autophagy is crucial in controlling the level, timing and specificity of cargo elimination. Under normal conditions, the low level of basal autophagy acts as needed to remove unfolded proteins and damaged or superfluous organelles. This prevents their gradual accumulation over time, and maintains protein and organelle ‘quality control’. Autophagy is dramatically induced by starvation and stress. Bulk intracellular degradation by autophagy during starvation recycles cellular components to supply the building blocks for metabolic pathways and for specific biomass generation to synthesize stress-response proteins3. This allows synthetic pathways and energy homeostasis to be maintained. For example, the generation of amino acids from protein degradation can support new protein synthesis and essential metabolic pathways, including tricarboxylic acid (TCA) cycle function, and so compensates for the absence of external nutrients3,4.
Selective autophagy recognizes, captures and eliminates stress-induced unfolded proteins and damaged organelles to ensure the preservation of normal and essential cellular components. To ensure the identification and selective elimination of such proteins and organelles, they are tagged by ubiquitin modification and are recognized by autophagy receptors that link the cargo to the autophagosome machinery5. This process is essential in stressful conditions, as the accumulation of damaged proteins and organelles is toxic to the cell.
The ability of autophagy to capture, degrade, eliminate and recycle intracellular components affects metabolism, enables host defence, remodels the proteome, regulates trafficking, alters signalling and influences cellular interactions. We are only beginning to understand the role of autophagy in normal and disease states. In this Review, I focus on the context-specific role of autophagy in suppressing and promoting cancer.
Consequences of autophagy disruption
Autophagy-related ATG genes encode the intracellular machinery that controls the initiation of autophagosome formation, cargo collection and trafficking to the lysosomal compartment, and most of these genes are conserved between yeast and humans2,6. Tissues from mutant mice with defects in autophagy accumulate ubiquitylated protein aggregates, abnormal organelles, particularly mitochondria, as well as excess peroxisomes, endoplasmic reticulum (ER), ribosomes and lipid droplets7. The functional consequences of this failure of protein and organelle quality control are not entirely clear, but they are associated with the production of reactive oxygen species (ROS), metabolic insufficiency and increased proteotoxicty. This can promote cellular damage, reduce stress tolerance and compromise survival. Indeed, mice with systemic mosaic whole-body and tissue-specific autophagy defects display neurodegeneration, chronic inflammation, steatohepatitis and muscle damage, and have an increased susceptibility to tumour development and infection by pathogens7,8. These animals also show an accumulation of autophagy substrates, including polyubiquitylated proteins, autophagy receptors such as p62 and NBR1, chaperones and other stress-response regulators, as well as aggregation-prone proteins9,10.
In stressful conditions or in situations in which the expression of mutant proteins with increased protein misfolding and aggregation, autophagy has an important role in eliminating protein aggregates. Failure to eliminate protein aggregates through autophagy causes their accumulation as Mallory–Denk bodies in the liver, ubiquitylated and mutant protein aggregates in the brain, and as α1-antitrypsin aggregates in the lung, liver and other tissues10–13. Although the proteaseome pathway can degrade individual, soluble proteins, autophagy is required to degrade aggregated proteins, and may supplement proteasome-mediated degradation in times of stress. Autophagy is stimulated by proteasome pathway inhibition to which autophagy-defective cells are highly sensitized, providing evidence that there is a partial, complementary interaction and interdependence between the two main mechanisms of protein degradation10,14.
Tumour suppression by autophagy
Suppression of liver tumour initiation by autophagy
The demonstration that autophagy could have a role in tumour suppression came from the examination of mice with allelic loss of the essential autophagy gene beclin 1 (Becn1; also known as Atg6). These mice are partially defective for autophagy and develop hepatocellular carcinomas with advancing age15,16. Liver tumours arising from allelic loss of Becn1 do not undergo loss of heterozygosity, suggesting obligate haploinsufficiency, and that tumour cells cannot tolerate the complete loss of Becn1 and autophagy. Cancers in other tissue types have been reported in these mice15,16; however, mosaic deletion of the essential autophagy gene Atg5 in mice produces hepatomas only, suggesting that tumour suppression by autophagy is liver-specific17.
Allelic loss of BECN1 has been reported in some human cancers18,19; however, a comprehensive analysis of copy number variation, mutation frequencies and expression levels of essential autophagy genes using cancer genome and gene expression data is necessary to validate these findings. This is important because BECN1 has autophagy-independent functions20.
Autophagy and antioxidant defence
An important subtype of autophagy is the specific autophagy of mitochondria that is known as mitophagy. Dysfunctional mitochondria lose membrane potential, triggering the activation of PTEN-induced putative kinase 1 (PINK1). PINK1 activates the E3 ligase parkin (PARK2) to ubiquitylate mitochondrial outer membrane proteins, providing an ‘eat me’ signal for recognition by the autophagy machinery21. This causes the selective elimination of damaged mitochondria, thereby maintaining mitochondrial quality control. A reduction in mitochondrial number through mitophagy, and particularly the selective elimination of damaged mitochondria, is a potential means to reduce ROS and oxidative stress. The physiological settings in which mitophagy is important are only now beginning to emerge, but inactivating mutations in PINK1 and PARK2 are found in Parkinson’s disease21. PARK2 is also a tumour-suppressor gene22, and Park2 deletion in mice causes hepatocellular carcinoma that is similar to allelic loss of Becn1 (REF. 23). It is likely that toxic consequences of failed mitophagy and mismanagement of oxidative stress contribute to neuro-degenerative diseases, such as Parkinson’s disease, and also contribute to tumour promotion when autophagy is defective.
In cells with an intact autophagy pathway that are not subject to stress, levels of p62 (a protein targeted by autophagy (BOX 1)) are low. p62 is a crucial activator of nuclear factor erythroid 2-related factor 2 (NRF2)12,24. Cells protect themselves from oxidative stress not only by eliminating the mitochondrial ROS source through mitophagy, but also by turning on the transcription of antioxidant-defence genes. NRF2 is a transcription factor that is responsible for activating this response.
Box 1. Regulation of signalling and autophagy by p62.
p62 is an adaptor protein that possesses many binding motifs that allow it to bring together proteins and assemble them into protein complexes that regulate signal transduction96. p62 binds both polyubiquitin on autophagy cargo via its ubiquitinassociated (UBA) domain and the autophagosome protein light chain 3 (LC3; also known as MAP1LC3A) via its LC3-interacting region (LIR) domain, which directs cargo to autophagosomes for degradation. The PB1 domain of p62 also interacts with itself, promoting self-aggregation, and with the autophagy receptor neighbour of BRCA1 (NBR1), to promote packaging of cargo and delivery to the autophagy pathway. As p62 is an autophagy substrate, autophagy defects cause accumulation of p62, which perturbs signal transduction in multiple pathways. Through its PB1 domain, p62 can bind ERK1, MAP2K5, MAP3K3 and atypical protein kinase C, and thereby may influence nuclear factor-κB (NF-κB) activation, cell growth and polarity. The ZZ domain of p62 interacts with RIP1 and TRAF6, and complex formation can promote NF-κB activation, leading to cell growth, survival, inflammation and promotion of antioxidant defence. p62 interacts with kelch-like ECH-associated protein 1 (KEAP1) through its KEAP1-interacting region (KIR domain), which releases NRF2 and enables it to induce genes involved in antioxidant defence. p62 also interacts with regulatory-associated protein of mTOR (RAPTOR), thereby promoting nutrient sensing and cell growth by mTOR complex 1. By virtue of these interactions, p62 has a role in cancer, differentiation, inflammation, metabolism and cell growth. Autophagy defects increase p62 levels and promote some of these pathways, however, accumulation of p62 to high levels causes p62 aggregation that sequesters and inactivates p62-interacting proteins to inhibit some of these pathways.
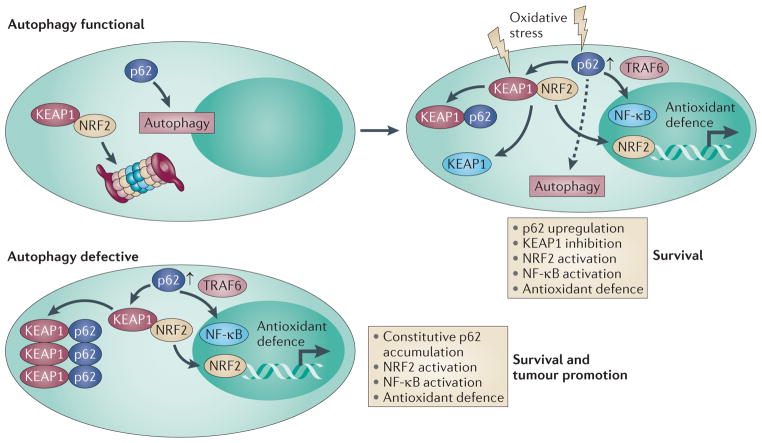
NRF2 transcriptional activity is suppressed under normal conditions by direct binding to its inhibitor, kelch-like ECH-associated protein 1 (KEAP1), a component of the cullin 3 (CUL3)–RBX1 E3-ligase complex (FIG. 1). In the absence of oxidative stress, NRF2 is bound to KEAP1–CUL3–RBX1 and is degraded, and antioxidant-defence genes are not activated. Oxidative stress causes the modification of KEAP1 that releases NRF2, or causes the upregulation of SQSTM1 (the gene that encodes p62). The binding of p62 to KEAP1 competitively displaces NRF2, which is then free to translocate to the nucleus where it turns on the expression of numerous ROS-detoxification genes, promoting cell survival25 (FIG. 1).
Figure 1. Regulation of NRF2 by autophagy, KEAP1 and p62.
Under normal conditions, nuclear factor erythroid 2-related factor 2 (NRF2) is bound to kelch-like ECH-associated protein 1 (KEAP1) and is inactivated as a transcription factor for antioxidant-defence genes by proteasome-mediated degradation. p62 is degraded through autophagy under normal conditions. In the presence of oxidative stress, KEAP1 is either modified so that it can no longer bind NRF2 or it is sequestered by p62, the expression of which is increased in response to oxidative stress. This displaces KEAP1 from NRF2 so that NRF2 can activate antioxidant-defence genes and promote survival. Oxidative stress also activates nuclear factor-κB (NF-κB) as a result of p62 upregulation and tumour necrosis factor receptor-associated factor 6 (TRAF6) complex formation, or by other mechanisms, to turn on antioxidant-defence gene expression. p62 is still subject to autophagy in cells experiencing cellular stress (dashed arrow). In autophagy-defective cells and tissues, the autophagy substrate p62 is not degraded, and so accumulates to high levels. p62 binds and sequesters KEAP1 in aggregates, resulting in the constitutive activation of NRF2 and antioxidant defence. This p62 induction can also activate NF-κB, antioxidant defence and survival.
NRF2 is also regulated by autophagy because p62 is a prominent autophagy substrate12,24,26. Although in normal cells the activation of the KEAP1–NRF2 pathway can be tumour suppressive through the induction of antioxidant-defence systems, in cells that have a disrupted autophagy pathway, NRF2 may be protumorigenic. In autophagy-defective cells and mice, p62 accumulates to high levels because it is not degraded, generating more p62 that is available to bind to and inhibit KEAP1 and sequester it in aggregates, which promotes the activation of NRF2 (FIG.1). Thus, autophagy defects can enable cell survival by preventing p62 degradation and by promoting NRF2 activation and the induction of antioxidant defence. Indeed, p62 deficiency impairs the tumorigenicity of genetically altered cell lines and the development of lung tumours in mice after RAS activation10,27,28. Moreover, liver-specific deletion of the essential autophagy gene Atg7 causes p62 accumulation, nuclear localization of NRF2, upregulation of NRF2-target genes and tumorigenesis, which is blocked by p62 deficiency12,13,17.
Interestingly, deficiency in p62 or in NRF2 greatly suppresses the development of oncogenic RAS-driven non-small-cell lung cancer in mouse models27,29. It will, therefore, be important to test whether NRF2 deficiency prevents tumorigenesis that is induced by autophagy deficiency and p62 accumulation.
There are activating mutations in NRF2 and inactivating mutations in KEAP1 in human cancers, indicating that NRF2 can function as an oncogene and KEAP1 as a tumour suppressor gene25,30. One prediction is that p62 upregulation by autophagy deficiency, or perhaps by activating mutations or gene amplification, is a major mechanism of tumour promotion by the NRF2 pathway. Another prediction is that autophagy suppression leading to upregulation of p62 and activation of NRF2-mediated survival might provide an alternative interpretation for the phenomenon of autophagic cell death. Increased cell survival with knockdown of essential autophagy genes has been attributed to the loss of autophagic cell death31. For example, the killing of cancer cells by some agents, and of a normal cell line through excessive RAS activation, can be decreased by autophagy inhibition32,33. It will be important to test whether this is instead due to the induction of p62 and the activation of NRF2 (or of nuclear factor-κB (NF-κB))-dependent survival when autophagy is inhibited (discussed below). To rule out this possibility, p62 elimination will need to have no effect on survival. If the activation of survival pathways by autophagy inhibition is not a factor, it will be interesting to explore the potential mechanism of cell killing by autophagy activation.
Although it is clear that degrading p62 by autophagy and suppressing NRF2 activity is a crucial tumour suppressive mechanism, there may be additional consequences to deregulation of p62 through activation of additional oncogenic signalling pathways. p62 is an important signalling adaptor protein that interacts with tumour necrosis factor receptor-associated factor 6 (TRAF6) and promotes NF-κB activation34, and autophagy defects result in NF-κB induction in the liver10. Recent evidence suggests that p62 interacts with mTOR, RAPTOR and RAG proteins to promote mTOR signalling35. It will be important to identify the role and function of p62 in human cancers, and when and how p62 deregulation is related to autophagy suppression and tumour promotion.
The importance of tumour suppression by autophagy
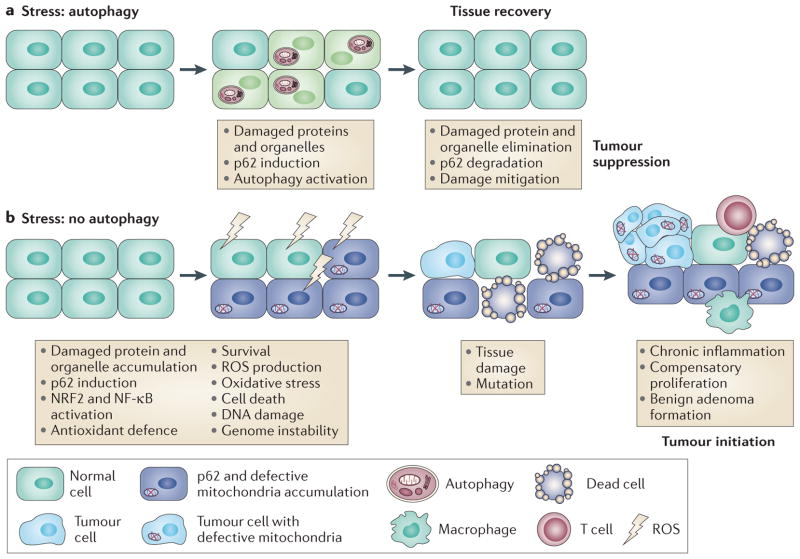
The gradual picture that is emerging is that autophagy is required to suppress p62 accumulation and inappropriate activation of NRF2, and perhaps other oncogenic signalling pathways, which can promote survival and tumorigenesis. Autophagy defects cause the accumulation of abnormal mitochondria that are a potential source of ROS. NRF2 activation may not be sufficient for a complete and sustained suppression of ROS when autophagy is blocked, which might eventually overwhelm the NRF2 antioxidant-defence system (FIG. 2). Autophagy defects cause the activation of the DNA damage response, DNA copy number variations and genetic instability, which is consistent with the eventual failure of cellular protection by NRF2 and the acquisition of genome mutations that drive tumorigenesis17,36,37 (FIG. 2). This situation of chronic tissue damage also provokes an inflammatory response that can further promote tumour growth through cytokine and chemokine production (FIG. 2). Chronic inflammation is known to contribute to liver cancer38,39, and activation of inflammation is also observed with allelic loss of Becn1 in the liver10, in tumour allografts with autophagy defects40 and following expression of a hypomorphic allele of Atg16l1 in the gut in a model of Crohn’s disease41. Thus, tumour promotion conferred by autophagy defects may result from both mutagenesis and the creation of an inflammatory environment (FIG. 2). Alternatively, autophagy may facilitate oncogene-induced senescence to limit tumorigenesis in some settings42. Going forwards, it will be interesting to examine these different roles for autophagy in cancer models and human tumours to determine their importance and contribution.
Figure 2. Mechanism of autophagy-mediated tumour suppression.
a| Autophagy, either basal or stress-induced, prevents the accumulation of oncogenic proteins such as p62, as well as damaged proteins and organelles. b | In autophagydefective tissues, p62 and damaged proteins and organelles accumulate. This is associated with the activation of oncogenic signalling pathways (nuclear factor erythroid 2-related factor 2 (NRF2) and nuclear factor-κB (NF-κB)) that promote survival but that are probably eventually overwhelmed by sustained oxidative stress. This leads to reactive oxygen species (ROS) production, chronic tissue damage, inflammation and genome instability, creating a tumour-initiating and tumour-promoting environment.
Autophagy stimulation for cancer prevention
Autophagy limits inflammation, tissue damage and genome instability that can promote cancer initiation, suggesting that stimulating autophagy may be beneficial for cancer prevention17,36,37,40,43,44. The aberrant accumulation of the autophagy substrate p62 in autophagy-deficient liver promotes liver damage and tumorigenesis10,13,17, and deficiency in p62 impairs tumorigenesis10,27, suggesting that inhibiting p62 may be valuable for cancer prevention and treatment. The damaging consequence of aggregation-prone mutant protein expression (such as mutant α1-antitrypsin Z), which predisposes to cancer, is also mitigated by autophagy stimulation11. The added burden of mutant protein expression may also compromise the effectiveness of autophagy, and further increase cancer risk by a feedforward mechanism.
Increased inflammation is associated with cancer when autophagy is suppressed10,40,41,45,46, suggesting that anti-inflammatory agents and autophagy promoters might be effective in cancer prevention. There is also evidence that the suppression of cancer owing to calorific restriction47 and the health benefits of exercise48,49 may be attributed to autophagy. Thus, by clearing away cellular waste, particularly mutated aggregationprone proteins and damaged mitochondria, and p62, autophagy may contribute to tumour suppression in some settings. Thus, functional autophagy status may be a predictor of cancer predisposition and the cancer prevention effectiveness of calorie restriction and exercise.
Tumour promotion by autophagy
Autophagy enables survival during starvation
Autophagy was originally found to be induced in yeast in response to starvation; autophagy supports the survival of yeast under starvation conditions by preserving amino acid levels and by upregulating starvation-response genes and mitochondrial function50–52. Similarly, deletion of Atg5 in mice revealed that autophagy was required for mammalian survival during the interval between placental separation and suckling53. Atg5-deficient mice die shortly after birth, which coincides with a period of physiological starvation during which autophagy is upregulated. Tissues from these mice have lower amino acid and ATP levels, which are suggestive of a metabolic crisis. Autophagy is also induced by the fertilization of mouse embryos and is required for proteome remodelling, as well as preimplantation development and survival54. Thus, recycling to support metabolism, and protein and organelle quality control — the main functions for autophagy — are highly conserved.
Autophagy is a cancer cell survival pathway
Autophagy is robustly activated in tumour cells by a multitude of stressors, including starvation, growth factor deprivation, hypoxia, damaging stimuli and proteasome inhibition. In the vast majority of cases, autophagy induction promotes survival in response to stress. Survival by autophagy in response to growth factor withdrawal or metabolic stress is particularly dramatic when apoptosis is disabled, which results in dormancy or quiescence, survival for weeks and then the resumption of cell growth on return to normal conditions40,55. Ischaemia (that is, glucose deprivation and hypoxia), a common physiological stress in the tumour microenvironment, upregulates autophagy in cancer cells and enables survival in vitro36,37,40. Autophagosomes are most prominent in tumour cells that are located in hypoxic tumour regions, and deletion of essential autophagy genes results in tumour cell death specifically in these hypoxic regions36,37,40. This was the first indication that tumours can commandeer the survival function of autophagy to promote tumorigenesis.
High basal autophagy and autophagy addiction in cancer
Normal cells and tissues have low levels of basal autophagy, but autophagy is dramatically induced by stress and starvation. This is most evident in autophagy-reporter transgenic mice in which starvation increases autophagosome numbers in many different tissues56,57. Thus, in normal cells and tissues, minimal autophagy is required in the absence of stress, but upregulation of autophagy in response to stress is crucial for survival. By contrast, many cancer cell lines have high basal levels of autophagy even under fed conditions that do not increase much further under stress. Moreover, activation of oncogenic RAS, which can induce tumour growth, is sufficient to upregulate basal autophagy28,58,59, and these cells have a limited ability to further increase autophagy levels, which diminishes their adaptation to stress. This suggests that oncogene activation may be a stress that requires autophagy induction to maintain homeostasis, and, therefore, that RAS activation restricts stress adaptation. Indeed, many cancer cell lines with activated RAS are highly dependent on autophagy for survival under basal, but especially stress, conditions.
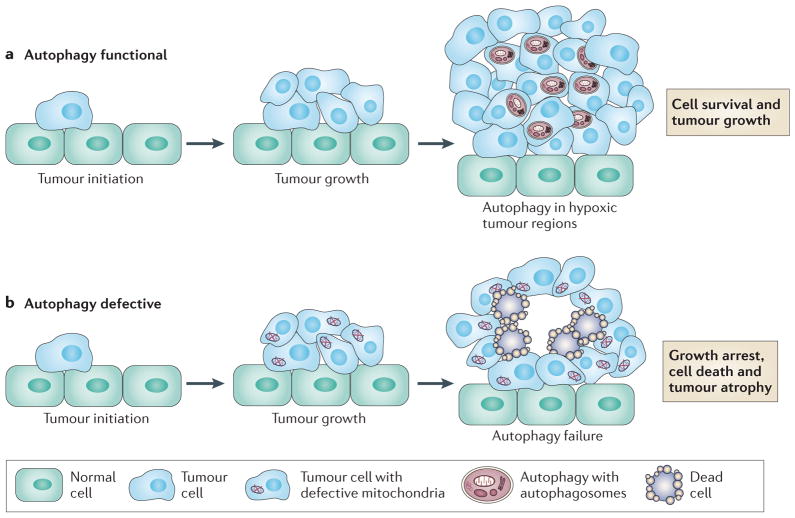
Autophagy deficiency almost abrogates the tumorigenicity of oncogenic RAS-expressing human and mouse cancer cell line models, suggesting that RAS activation and autophagy deficiency are synthetically lethal28,59 (FIG. 3). These observations may not be limited to RAS, as inactivation of Fip200, the mouse homologue of yeast Atg17 (also known as Rb1cc1) in the polyoma middle T (PyMT) mouse mammary cancer model impairs tumour growth46, and deletion of Atg5 or Atg7 in the liver causes hepatoma formation without progression to hepatocellular carcinoma17. These findings suggest that autophagy could be required to support the growth of aggressive cancers (FIG. 3). However, the mechanism by which autophagy supports tumorigenesis is not yet clear.
Figure 3. The role of autophagy in supporting the growth of aggressive cancers.
a | Autophagy is upregulated in RAS-driven cancers and is also induced in hypoxic tumour regions where it supports tumour cell survival. b | Autophagydeficient tumour cells accumulate defective mitochondria and are prone to cell death in hypoxic regions. This can lead to impairment of the growth of RAS-driven cancers and perhaps other cancers.
Autophagy is required to maintain mitochondrial function
Inactivation of autophagy in tumours results in the accumulation of morphologically abnormal mitochondria17,28,46,59. A comparison of autophagy wild-type and deficient cancer cell lines that were used to generate these tumours revealed defective mitochondrial respiration28,59, which is also observed in autophagy-deficient mouse muscle and yeast50,60. Thus, maintaining the pool of respiring mitochondria is a function of autophagy that is conserved from yeast to mammals.
The route cause of mitochondrial defects needs to be addressed. Failure to remove damaged mitochondria can explain their accumulation in autophagy-defective cells; however, autophagy might have a direct role in regulating mitochondrial function by supplying substrates in the form of amino acids and fatty acids. The absence of these autophagy-supplied substrates might cause mitochondrial dysfunction, leading to toxic ROS production and mitochondrial damage, with the accumulation of damaged mitochondria amplified by the failure to purge them from the population3. As mitochondrial respiration is required for tumorigenesis that is induced by oncogenic RAS61, the deterioration of mitochondrial function in autophagy-defective tumours could explain their impaired tumorigenicity17,28,46,59,62. However, the precise mechanisms by which autophagy supports mitochondrial function are currently unknown.
Autophagy-supplied substrates support mitochondrial metabolism
The collection, degradation and recycling of intracellular material in starvation is a main function of autophagy. Mitochondria are important for ATP production, energy homeostasis, the generation of building blocks from cataplerosis (such as the use of citrate for fatty acid synthesis and membrane biogenesis) and the production of ROS to activate signal transduction pathways. All of these functions are likely to be contributors to tumour-promoting functions of autophagy. Autophagy defects in tumour cells cause a deficit in ATP levels and energy charge, depletion of key TCA cycle intermediates such as citrate and aberrant ROS production (either high toxic or deficient ROS levels, depending on acute versus chronic autophagy inhibition)10,28,36,37,59.
Autophagy can supply substrates to replenish TCA cycle intermediates through anaplerotic reactions to sustain mitochondrial function in stress and starvation. Degradation of proteins by autophagy generates amino acids that can feed into the TCA cycle at multiple points to sustain mitochondrial metabolism (FIG. 4). Lipids harvested from lipid droplets by lipophagy63, or from organelle membrane degradation by autophagy, can be used to produce acetyl-CoA to maintain TCA cycle function (FIG. 4). It is also possible that autophagy-recycled sugars can generate pyruvate and acetyl-CoA from glycolysis to feed the TCA cycle. The challenge is to determine which of these mechanisms are crucial for autophagy-mediated tumour survival.
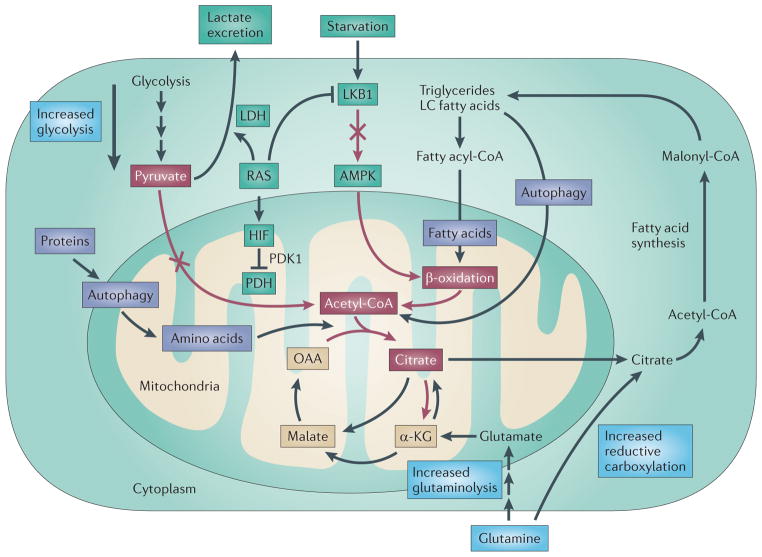
Figure 4. Mechanism of autophagy addiction of RAS-driven cancers.
The generation and use of acetyl CoA (depicted by red arrows and red boxes) is an essential component of the tricarboxylic acid (TCA) cycle. There are three known mechanisms by which RAS diminishes the pool of acetyl-CoA (shown in the green boxes). First, RAS can activate lactate dehydrogenase (LDH) that converts pyruvate to lactate, which is excreted. Second, RAS can activate hypoxia-inducible factor (HIF), inhibiting pyruvate dehydrogenase (PDH) and the conversion of pyruvate to acetyl-CoA. Third, RAS inhibits liver kinase B1 (LKB1), blocking AMP kinase (AMPK) and β-oxidation. Autophagy defects result in reduced citrate levels, impaired TCA cycle function and loss of mitochondrial respiration28,59. Autophagy can potentially compensate for the metabolic reprogramming by RAS by degrading proteins and lipids that provide amino acid and fatty acid substrates, producing acetyl-CoA (purple boxes). Tumour cells might also compensate for autophagy impairment by upregulating glycolysis, glutaminolysis or reductive carboxylation of α-ketoglutarate (α-KG) from glutamine (blue boxes). LC, long chain; OAA, oxaloacetate; PDK1, pyruvate dehydrogenase kinase 1.
Why RAS-driven cancers are autophagy-dependent
RAS impairs acetyl-CoA production through several mechanisms: by stimulating lactate dehydrogenase (LDH), which depletes pyruvate; by activating hypoxia-inducible factors (HIFs) and pyruvate dehydrogenase kinase 1 (PDK1), which inhibits pyruvate dehydrogenase (PDH)64,65; and by inhibiting liver kinase B1 (LKB1) and blocking AMP-activated protein kinase (AMPK) activation, preventing the mobilization of lipid stores and β-oxidation66,67. Thus, RAS potentially leaves cells dependent on autophagy to provide substrates, such as amino acids and fatty acids, for acetyl-CoA biosynthesis (FIG. 4). Experiments to verify this are ongoing. RAS signalling may amplify this problem by rendering cancer cells dependent on autophagy to maintain functional mitochondria. One prediction is that RAS-driven cancers would be addicted not only to autophagy but also to glutamine for glutaminolysis to compensate for curtailed acetyl-CoA production. Thus, glutamine-derived α-ketoglutarate can promote the TCA cycle when there are reduced levels of acetyl-CoA that are normally supplied by pyruvate and β-oxidation (FIG. 4).
RAS may also starve mitochondria of TCA cycle substrates and shorten their lifespan, which is exacerbated by RAS-dependent HIF activation, as HIF1 activation impairs mitochondrial biogenesis68. Thus, autophagy addiction of RAS-driven cancers may arise as a specific adaptation to, and compensation for, the metabolic reprogramming by RAS. If this is the case, the specific oncogenic events that are inherent to cancers may dictate whether autophagy is required, and the particular aspect of autophagy that is important.
Adaptation strategies of cancer cells to autophagy inhibition
As autophagy defects compromise mitochondrial function, an intriguing issue is whether cancer cells can adapt and circumvent this problem by altering their metabolism. Autophagy deficiency in both normal and cancer cells can upregulate glycolysis as a potential compensatory mechanism to cope with defective mitochondria58,60 (FIG. 4). Cancer cells may get by with a partially defective TCA cycle by relying on glutaminolysis to bypass citrate depletion and flux through that part of the TCA cycle (FIG. 4). One prediction from this is the glutamine dependence of autophagy-deficient cells. Glutamine-derived α-ketoglutarate can be converted to citrate for cataplerotic reactions by reductive carboxylation (FIG. 4). Indeed, cancer cells with defective mitochondria owing to mutational defects in electron transport chain complex III do exactly that69. Another way to at least partially compensate for autophagy deficiency is to upregulate proteasome-mediated protein degradation or chaperone-mediated autophagy. Both pathways provide ways to derive amino acid substrates for anaplerosis from soluble proteins, use the same targeting mechanism of ubiquitin substrate modification, and their inhibition potentiates cell death with autophagy inhibition and can compromise cancer cell growth5,10,14,28,70. It will be interesting to determine the extent and mechanisms by which cancer cells adapt to autophagy inhibition.
Autophagy in cancer therapy
Since the realization that autophagy is a survival pathway for tumour cells, there has been great interest in inhibiting autophagy for cancer therapy44,71–73. Although small-molecule autophagy inhibitors are in development, the lysosomotropic and anti-malarial agent hydroxychloroquine (HCQ), which blocks the degradation of the products of autophagy by inhibiting lysosome function, is being actively assessed in the clinic71,73. Whether HCQ will be an effective autophagy inhibitor in human tumours, how patients who would benefit will be identified and their tumours assessed, and how to determine the best drugs to combine with HCQ, have yet to be resolved. It will also be important to establish whether any anticancer activity of HCQ is due to autophagy impairment, as it may act by other mechanisms74. Our current understanding of the role of autophagy in cancer has provided a number of insights.
Stress augmentation
Autophagy inhibition may augment ambient or therapy-induced stress to promote cancer cell death. As autophagy promotes the survival of tumour cells that reside in hypoxic tumour regions, and because this subpopulation is commonly more refractory to cell death, combining autophagy inhibitors with radiation or chemotherapy would be expected to increase death among this resistant subpopulation. Increasing tumour metabolic stress by interfering with the tumour blood supply following surgery or with the use of angiogenesis inhibitors may increase the tumour cell subpopulation that is susceptible to autophagy inhibition. Dietary manipulation may also improve therapy in conjunction with autophagy inhibition. Starvation for leucine fails to activate autophagy in a mouse melanoma model, thereby creating sensitivity to autophagy inhibition that impairs tumorigenesis75. This suggests that disengaging nutrient sensing from catabolism creates metabolic vulnerability. Compounding ER stress and proteotoxicity with autophagy inhibition is another option. Concurrent autophagy and proteasome inhibition, especially in diseases such as multiple myeloma, where there is an inherent burden of unfolded protein owing to immunoglobulin secretion, is also being explored14. Alternatively, blocking the ER stress response while inhibiting autophagy might promote tumour cell killing.
Potentiation of DNA damage
Autophagy defects can activate the DNA damage response and promote genome damage, potentially taxing DNA repair mechanisms36,37. Many agents that either damage DNA or that inhibit DNA repair are successfully used in the clinic, and their activity may be augmented by autophagy inhibition. It will be interesting to determine whether autophagy inhibition induces genome damage by promoting toxic ROS production or whether there is direct regulation of the DNA repair machinery by autophagy that might dictate the optimal approach.
Blocking autophagy-mediated survival by targeted therapies
Activation of the PI3K pathway is a ubiquitous feature of human cancers, and many pathway component-specific inhibitors are currently in development or in the clinic76. These include mTOR, PI3K and AKT inhibitors, all of which potently activate autophagy. As these agents often do not produce a durable response in patients, there is interest in increasing their efficacy. Autophagy activation by these agents is potentially counterproductive. In preclinical models, combining PI3K pathway inhibitors with HCQ or genetic ablation of autophagy has demonstrated increased tumour regression59,75,77– 90. Clinical trials to test this concept in patients are ongoing71,73. The identification of susceptible patients and the mechanisms behind any antitumour activity needs to be determined to guide clinical application.
Susceptible cancer subtypes
A major limitation of our current knowledge is the ability to identify patients who would most benefit from autophagy inhibition. Good candidates are RAS-driven cancers, particularly pancreatic cancer in which the majority of patients have activating mutations in KRAS, and for which there is evidence for autophagy addiction from preclinical models59. The ability to assess human tumour tissue for high autophagic flux rather than only static levels of autophagosomes would be a potential means to direct therapy. Determining the effectiveness of autophagy inhibitors in clinical samples is also necessary. The accumulation of autophagy substrates such as p62 can help to assess autophagy inhibition, although a reliable panel of markers requires development. Autophagy is induced during cancer cell detachment (anoikis) and promotes survival, suggesting a possible role in facilitating metastasis91. As most cancer patients die from disseminated disease, this may be another important application of autophagy inhibitors.
Collateral damage
Autophagy inhibition, especially in combination therapy, may pose cytotoxicity issues, such as to the liver and brain. Stem cells may also be more sensitive to autophagy inhibition than differentiated tissue92. It is hoped is that the metabolic and growth alterations inherent to most cancers will provide a sufficient therapeutic window. The role of autophagy in regulating the immune response to tumours is still an open question. Indeed, autophagy in tumour cells may be required to supply extracellular ATP to increase the antitumour immune response, as has been implied by the study of tumour xenografts treated with cytotoxic chemotherapy93.
In summary
Autophagy can both suppress cancer initiation and promote the growth of established cancers, and we are at the early stages of using this information to benefit patients. These findings have raised new issues. For example, are autophagy genes cancer susceptibility loci and are they commonly altered in human cancer? Which autophagy-supplied substrates are important for sustaining metabolism? This is important to determine because blocking this process would be expected to kill autophagy-addicted cancer cells. Will metabolic flux analysis identify oncogenic anabolic and catabolic pathways and exploitable vulnerabilities? Is autophagy an escape mechanism for therapies that target metabolic pathways? Will autophagy modulation be valuable in cancer prevention or therapy? Can autophagy addiction be exploited as a liability in cancer? Does autophagy diminish the efficacy of targeted therapies, and, if so, how? This may be most relevant to mTOR inhibitors for which the activation of autophagy is direct, and improving efficacy is desirable76. What is the mechanism by which high basal autophagy in cancers overrides autophagy inhibition by active mTOR? Is ammonia derived from glutaminolysis, which has been shown to potently activate autophagy, one possible mechanism94,95? How do autophagy defects promote inflammation? Is there a role of autophagy in tumour–stroma interactions? How does autophagy affect the immune response to cancer? What can we learn about the role of autophagy from genetically engineered mouse models for cancer? The answers to these and many other interesting questions seem set to maintain the current interest in autophagy and its links to tumour development and metabolism, with the promise of using this information to improve cancer treatment.
At a glance.
Autophagy is a cellular self-cannibalization process that captures and digests cellular proteins and organelles in lysosomes.
Autophagy levels are normally low but are dramatically induced by starvation and stress.
Recycling of cellular material by autophagy sustains cellular and mammalian metabolism necessary for survival in starvation.
The elimination of damaged proteins and organelles by autophagy is required for cellular homeostasis.
Autophagy can be tumour suppressive by preventing chronic tissue damage and cancer initiation.
Autophagy is induced in and required for the survival of tumour cells in hypoxic tumour regions.
Many cancer cells upregulate autophagy that is required to support metabolism, tumorigenesis and survival to therapy.
In aggressive cancers, autophagy inhibition may be therapeutically advantageous.
Acknowledgments
I thank members of the White laboratory for helpful discussions and advice. This work was supported by grants from the US National Institutes of Health (R37 CA53370, RO1 CA130893 and RC1 147961), DOD (W81XWH06-1-0514 and W81XWH05) and the V Foundation.
Glossary
- Tricarboxylic acid (TCA) cycle
Also known as the citric acid cycle or the Krebs cycle, the TCA cycle is a series of chemical reactions that generate energy and building blocks through the oxidation of acetate derived from carbohydrates, fats and proteins into carbon dioxide and water
- Reactive oxygen species (ROS)
Chemically reactive molecules containing oxygen that cause cellular damage or that activate signalling
- Steatohepatitis
A pathological condition, also known as fatty liver disease, characterized by inflammation and fat accumulation in the liver
- Proteasome
A large protein complex responsible for the degradation of soluble proteins
- NF-κB
A transcription factor that controls the immune response to damage and infection
- mTOR
A serine/threonine protein kinase that regulates cell growth, cell proliferation, cell survival, protein synthesis and transcription in response to nutrient and growth factor availability
- Hypoxia
A pathological condition in which the body as a whole or a region of the body, such as a tumour, is deprived of an adequate oxygen supply
- Cataplerosis
The process by which metabolic intermediates are removed from metabolic pathways
- Glycolysis
The metabolic pathway that converts glucose into pyruvate and in the process produces ATP and reduced NADH
- Glutaminolysis
A series of biochemical reactions in which the amino acid glutamine is degraded to glutamate then to α-ketoglutarate for further metabolism in the tricarboxylic acid cycle
- Anaplerosis
The process of replenishment of depleted metabolic cycle or pathway intermediates
- ER stress
A stress adaptation pathway activated by the accumulation of unfolded proteins in the endoplasmic reticulum (ER)
Footnotes
Competing interests statement
The author declares no competing financial interests.
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21:113–119. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. Proteomics were used to identify proteins that accumulate in autophagy-defective tumour cells, one of which is p62 that is required for tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidvegi T, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z. and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. This paper identifies p62 accumulation in autophagy-defective cells and KEAP1 interaction that activates NRF2. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. This paper identifies that p62 accumulation in autophagy-defective mouse liver is required for inclusion body formation and hepatic toxicity. [DOI] [PubMed] [Google Scholar]
- 14.Ding WX, et al. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol Cancer Ther. 2009;8:2036–2045. doi: 10.1158/1535-7163.MCT-08-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. This paper describes the requirement for Becn1 in mouse development and that alleic loss renders mice tumour prone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. This paper describes the requirement for Becn1 in mouse development and that alleic loss renders mice tumour prone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. This paper demonstrates that Atg5 or Atg7 deficiency in mice causes hepatoma development that requires p62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 19.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. This paper reports a role for beclin 1 in autophagy and in inhibiting tumorigenesis when overexpressed. [DOI] [PubMed] [Google Scholar]
- 20.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature Rev Mol Cell Biol. 2010;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesari R, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25–q27. Proc Natl Acad Sci USA. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujiwara M, et al. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- 24.Lau A, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. This paper demonstrates that p62 binds and inhibits KEAP1 thereby pronoting NRF2 activation in autophagy-defective cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2–Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2010;13:1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 27.Duran A, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. This paper demonstrates that p62 deficiency prevents KRAS lung tumorigenesis. [DOI] [PubMed] [Google Scholar]
- 28.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. This paper reports autophagy addiction of RAS-driven cancers, specifically that RAS activation upregulates basal autophagy that is required for mitochondrial function and tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. This paper demonstrates that NRF2 is required for KRAS and BRAF lung tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Shen S, Kepp O, Kroemer G. The end of autophagic cell death? Autophagy. 2012;8:1–3. doi: 10.4161/auto.8.1.16618. [DOI] [PubMed] [Google Scholar]
- 32.Turcotte S, et al. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin–1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Moscat J, Diaz-Meco M. T p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. This paper describes that autophagy prevents genome damage and promotes tumour cell survival in a model of mammary cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. Autophagy defects cause activation of the DNA damage response, DNA copy number variations and an elevated mutation rate, suggesting that autophagy suppresses genome instability to limit tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakurai T, et al. Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14:156–165. doi: 10.1016/j.ccr.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun B, Karin M. NF-kappaB signaling, liver disease and hepatoprotective agents. Oncogene. 2008;27:6228–6244. doi: 10.1038/onc.2008.300. [DOI] [PubMed] [Google Scholar]
- 40.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. Autophagy is induced in hypoxic tumour regions and is required for tumour cell survival and for limiting inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. Atg16l1 is required in Paneth cells to prevent an injury response and may have a role in Crohn’s disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen HY, White E. Role of autophagy in cancer prevention. Cancer Prev Res (Phila) 2011;4:973–983. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nature Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei H, Gan B, Wu X, Guan JL. Inactivation of FIP200 leads to inflammatory skin disorder, but not tumorigenesis, in conditional knock-out mouse models. J Biol Chem. 2009;284:6004–6013. doi: 10.1074/jbc.M806375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei H, et al. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blagosklonny MV. Linking calorie restriction to longevity through sirtuins and autophagy: any role for TOR. Cell Death Dis. 2010;1:e12. doi: 10.1038/cddis.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS ONE. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. Autophagy is required in yeast to prevent mitochondrial inpairment and cell death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 52.Kamada Y, Sekito T, Ohsumi Y. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol. 2004;279:73–84. doi: 10.1007/978-3-642-18930-2_5. [DOI] [PubMed] [Google Scholar]
- 53.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. Autophagy promotes survival of mice during neonatal starvation. [DOI] [PubMed] [Google Scholar]
- 54.Tsukamoto S, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 55.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. Autophagy enables long-term survival of lymphoid cells to growth factor deprivation. [DOI] [PubMed] [Google Scholar]
- 56.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol. 2009;452:13–23. doi: 10.1016/S0076-6879(08)03602-1. [DOI] [PubMed] [Google Scholar]
- 57.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lock R, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2010;22:165–178. doi: 10.1091/mbc.E10-06-0500. RAS upregulates autophagy that promotes transformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. This paper reports that pancreatic cancers display autophagy addiction with high basal autophagy that is required for growth, survival and tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu JJ, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valentin-Vega YA, et al. Mitochondrial dysfunction in ataxia telangiectasia. Blood. 2011;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chun SY, et al. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1alpha and HIF-2alpha target genes. Mol Cancer. 2010;9:293. doi: 10.1186/1476-4598-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 67.Zheng B, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell. 2009;33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 69.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kon M, et al. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garber K. Inducing indigestion: companies embrace autophagy inhibitors. J Natl Cancer Inst. 2011;103:708–710. doi: 10.1093/jnci/djr168. [DOI] [PubMed] [Google Scholar]
- 73.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maycotte P, et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200–212. doi: 10.4161/auto.8.2.18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19:613–628. doi: 10.1016/j.ccr.2011.03.012. Autophagy induction by leucine starvation is required for the survival of melanomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feldman ME, Shokat KM. New inhibitors of the PI3K-Akt-mTOR pathway: insights into mTOR signaling from a new generation of Tor Kinase Domain Inhibitors (TORKinibs) Curr Top Microbiol Immunol. 2010;347:241–262. doi: 10.1007/82_2010_64. [DOI] [PubMed] [Google Scholar]
- 77.Altman BJ, et al. Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis. Oncogene. 2011;30:1855–1867. doi: 10.1038/onc.2010.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Mycinduced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bellodi C, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carew JS, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Degtyarev M, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan QW, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han W, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS ONE. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma XH, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17:3478–3489. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan Y, et al. Targeting autophagy augments in vitro and in vivo antimyeloma activity of DNA-damaging chemotherapy. Clin Cancer Res. 2011;17:3248–3258. doi: 10.1158/1078-0432.CCR-10-0890. [DOI] [PubMed] [Google Scholar]
- 87.Parkhitko A, et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci USA. 2011;108:12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saleem A, et al. Effect of dual inhibition of apoptosis and autophagy in prostate cancer. Prostate. 2012 Jan 12; doi: 10.1002/pros.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shi YH, et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159–1172. doi: 10.4161/auto.7.10.16818. [DOI] [PubMed] [Google Scholar]
- 90.Wu Z, et al. Autophagy blockade sensitizes prostate cancer cells towards Src family kinase inhibitors. Genes Cancer. 2010;1:40–49. doi: 10.1177/1947601909358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mortensen M, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Michaud M, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. Autophagy is required to promote antitumour immunity in response to cytotoxic chemotherapy. [DOI] [PubMed] [Google Scholar]
- 94.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31. doi: 10.1126/scisignal.2000911. Ammonia produced from glutaminolysis induces autophagy. [DOI] [PubMed] [Google Scholar]
- 95.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moscat J, Diaz-Meco MT. p62: a versatile multitasker takes on cancer. Trends Biochem Sci. 2012 Mar 14; doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]