Abstract
Background
Postinfluenza Staphylococcus aureus pneumonias are increasingly recognized as a major form of life-threatening infections.
Methods
A mouse model of postinfluenza S. aureus pneumonia was developed. Mice were intranasally infected with bacteria alone or bacteria plus virus. Infection was assessed by mouse survival, lung histopathology, bacterial density in the lungs, and cellular response to infection.
Results
Mice infected with both influenza virus and S. aureus showed higher mortality, greater lung parenchymal damage, and greater bacterial density at metastatic tissue sites than mice infected with only S. aureus. At 4 h, more polymorphonuclear leukocytes and fewer CD11c+ cells were found in lung samples from mice infected with virus and bacteria than in those from mice infected with bacteria. α-Hemolysin and protein A were maximally expressed 4 h after infection, and Panton-Valentine leukocidin was maximally expressed 72 h after infection, with higher levels of α-hemolysin expression in mice infected with bacteria alone. Interferon γ expression was higher in tissue collected from mice infected with virus plus bacteria than in those from bacteria-infected mice.
Conclusions
The results from this model demonstrate diverse effects caused by antecedent influenza virus infection, which have a profound influence on the morbidity and mortality associated with S. aureus pneumonia.
Infections caused by community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains have spread throughout the United States [1, 2]. Although most of these infections involve the skin and soft tissues, the less frequent systemic ones, including sepsis, necrotizing fasciitis, osteomyelitis, and pneumonia, are often characterized by fulminant onset, rapid progression, and, in a subset of patients, a fatal outcome [3]. In the United States many of these infections have been due to the CA-MRSA strains USA300 or USA400. Among the invasive infections, necrotizing S. aureus pneumonia has emerged as one of the most lethal [4, 5]. Many of these pneumonias have been preceded by an influenza virus infection [6, 7]. This association of necrotizing bacterial pneumonia with antecedent influenza virus infection is well recognized. Recent studies have noted the importance of bacterial infections due to Streptococcus pneumoniae, Haemophilus influenzae, and S. aureus in the fatal cases associated with the 1918 influenza pandemic [8-10]. The increased influenza-associated mortality from coinfections with S. aureus among the pediatric age group resulted in a 2008 health advisory for the Centers for Disease Control and Prevention (CDCHAN-00268-2008-01-30-ADV-N) [11]. Although the association of influenza virus with S. pneumoniae respiratory infections has been investigated, few studies have investigated the association of influenza virus with S. aureus infections [12-14]. We have developed a murine model of postinfluenza S. aureus pneumonia to examine the factors associated with this life-threatening infection.
METHODS
Influenza virus strain and preparation
The mouse-adapted influenza virus A/Puerto Rico/8/34(H1N1) was grown on Madin-Darby canine kidney cells (American Type Culture Collection) [15], aliquoted, and stored, and the viral density was determined by viral plaque assay, as described elsewhere [12, 16]. Viral aliquots were diluted in phosphate-buffered saline (PBS) supplemented with 0.1% bovine serum albumin and adjusted to the final concentration.
Bacterial strains and preparation
The S. aureus strains used in this study included clinical isolates from subjects with S. aureus pneumonia and isogenic mutants defective in Panton-Valentine leukocidin (PVL) production (Table 1). Bacteria were grown to the stationary phase at 37 °C in Todd-Hewitt broth (Becton Dickinson). The bacteria were then harvested by centrifugation, washed, resuspended in PBS, and adjusted to a final concentration.
Table 1. Descriptions of Staphylococcus aureus Isolates Used in the Study.
| Strain | USA type | Description | Source |
|---|---|---|---|
| 933 | 300 | Pediatric pneumonia isolate | New York (present study) |
| 204 | 500 | Clinical isolate | K. Taylor, Nabi Pharmaceuticals |
| LAC, LACΔpvl | 300 | Isogenic pair, wild type and lukF/S-PVL knockout | Voyich et al [17], F. DeLeo |
| MW2, MW2Δpvl | 400 | Isogenic pair, wild type and lukF/S-PVL knockout | Voyich et al [17], F. DeLeo |
Pneumonia model
The model was a modification of that described by McCullers for S. pneumoniae [18]. Six-week-old female BALB/c mice (Charles River Laboratories) were challenged intranasally with influenza virus (50 μL/mouse). Seventy-two hours after viral infection, the mice were intranasally infected with S. aureus (50 μL/mouse). Different doses of influenza virus (low dose, 2.0 × 104 plaque-forming units [PFU]/mL or 1 × 103 PFU/mouse; high dose, 1.0 × 106 PFU/mL or 5 × 104 PFU/mouse) and/or S. aureus (low dose, 1.8 × 108 colony-forming units [CFU]/mL or 9 × 106 CFU/mouse; high dose, 2.4 × 109 or 1.2 × 108 CFU/mouse) were used in separate studies. The higher dose was used only in the mouse survival studies; the lower dose was used in the survival studies, as well as those assessing bacterial density in the lungs and other tissues. After infection, mice demonstrating signs of distress were killed. In the survival studies, these mice were included with mice for the nearest survival time point.
In separate experiments mice were killed at different time points after bacterial infection (low dose) to determine the bacterial density in the lung, kidney, liver, and spleen. Tissue samples were weighed, homogenized, and plated into mannitol salt agar (Becton Dickinson) and incubated for 48 h at 37 °C for bacterial colony counts. Results were expressed as the number of CFUs per gram of tissue. The animal protocol was reviewed and approved by the Columbia University Institutional Animal Care and Use Committee.
Histology
Harvested lungs were fixed in 10% formaldehyde and embedded in paraffin. Sections (6 μm) were then prepared and stained with either hematoxylin-eosin or Gram stain. A pulmonary pathologist (A.B.) performed a blinded examination of histological sections prepared from mice at 4, 24, or 72 h. Samples from ≥10 mice were examined for each time point. Sections from uninfected mice, mice infected with virus or bacteria alone, and mice infected with both virus and bacteria were evaluated for the degree of inflammation, necrosis, and type of cellular infiltrate.
Bronchoalveolar lavage
Bronchoalveolar lavage (BAL) was performed immediately after death in mice infected with the low doses of either virus plus bacteria or bacteria alone. A blunt 2.5-cm-long 21-gauge needle was used to perform lavage with PBS. The lavage fluid was mixed with RNAprotect Bacteria Reagent (Qiagen) and frozen until used.
RNA isolation
Prokaryotic RNA was isolated from BAL fluid obtained from mice infected with low doses of virus or virus and bacteria, using a method provided by Craig Rubens and Donald Chaffin (personal communication, 2008). RNA from S. aureus cultures at both logarithmic and stationary growth phases were obtained with the RNeasy kit (Qiagen) and used as in vitro controls for gene expression. Eukaryotic RNA was isolated from separate groups of infected mice with the same kit, according to the manufacturer’s instructions.
Quantitative real-time reverse-transcriptase polymerase chain reaction determination of S. aureus gene expression
RNA samples were analyzed by reverse-transcriptase polymerase chain reaction (RT-PCR) on a SmartCycler system (Cepheid) using TaqMan probes (Integrated DNA Technologies), primers (Table 2), and the SuperScript III Platinum One-Step Quantitative RT-PCR system (Invitrogen) to measure expression of lukS, hla, and spa. In vitro RNA samples were adjusted to an RNA concentration similar to that of the in vivo samples. Relative target expression was calculated according to the 2-ΔΔCT method of Livak and Schmittgen [19] and expressed as the fold change of the different genes compared with the housekeeping gene gyrB.
Table 2. Primer and Probe Sequences for Quantitative Real-Time Reverse-Transcriptase Polymerase Chain Reaction.
| Gene | Primer (5′→3′) | Probe | |
|---|---|---|---|
| Forward | Reverse | ||
| gyrB | AACGGACGTGGTATCCCAGTTGAT | TTGTATCCGCCACCGCCGAATTTA | 56-FAMAAATGGGACGTCCAGCTGTCGAAGTT- 3IABlkFQ |
| lukS | ATTGTCGTTAGGAATAATCACT | CCCACTTATCGCTACTTGTATCTTCTG | Cy5-TTGGTGATGGCGCTGAGGTAGTCAAA- 3IAbRQSp |
| spa | TTTGTCAGCAGTAGTGCCGTTTGC | GGCAACAAGCCTGGCAAAGAAGAT | 56-FAMCCAGGTTTAACGACATGTACTCCGTTACC- 3IABLFQ |
| hla | CCGGTACTACAGATATAGGAAGCAATA | GCGCCTTCTTCGCTATAAACTC | 5HEX-ACGAAAGGTACCATTGCTGGTCAGT- 3IABLFQ |
| GAPDH | ACCACAGTCCATGCCATCAC | TCCACCACCCTGTTGCTGTA | SYBR Green |
| Ifng | GACATGAAAATCCTGCAGAGC | TGAGCTCATTGAATGCTTGG | SYBR Green |
Cellular response to respiratory infection
BAL fluid was obtained from the infected mice, as outlined above. Cellular components were collected by centrifugation at 162 g for 10 min at 4 °C. Single cell suspensions from harvested lungs were obtained by filtration through 40-μm nylon mesh filters to create lung tissue suspensions. Erythrocyte lysis was performed on the lung tissue suspensions and BAL cells by incubating the suspensions for 5 min at room temperature in ammonium chloride lysing solution. The samples were then treated with mouse serum and mouse Fc Block (BD Biosciences) before being labeled with the appropriate fluorophore-conjugated antibodies. Lung and BAL samples were analyzed separately.
Cell types were determined by incubation for 30 min at 4 °C with the appropriate isotype controls or with Ly6G/Ly6C-fluorescein isothiocyanate for polymorphonuclear (PMN) leukocytes (BD Biosciences), CD45–phycoerythrin (PE) for the total leukocyte population (Invitrogen), and CD11c-PE–cyanine 5.5 for phagocytic cells (eBioscience). After incubation, cells were washed twice with PBS plus 1% fetal calf serum and fixed in 1% paraformaldehyde. Samples were analyzed with a BD FACS-Calibur flow cytometer using CellQuest software (version 3.3; BD Biosciences). Samples were gated based on their forward and side scatter characteristics as well as their expression of CD45, using the open-source software WinMDI (Scripps Institute). Data are presented as the percentages of CD45+ cells expressing the indicated markers.
BAL fluid was also subjected to keratinocyte-derived chemokine (KC) enzyme-linked immunosorbent assay, following the manufacturer’s instructions (DuoSet; R&D Systems). All samples were assayed in triplicate.
Quantitative RT-PCR determination of interferon γ expression
To measure expression of interferon γ (IFN-γ) (Table 2), complementary DNA (cDNA) of isolated host RNA (obtained from lung homogenates) from mice infected with low-dose virus and/or bacteria was generated using the iScript cDNA synthesis kit (Bio-Rad Laboratories). This was followed by real-time PCR using the Power SYBR Green PCR Master Mix (Applied Biosystems) with the StepOnePlus Real-Time PCR System (Applied Biosystems). Results are presented as fold changes compared with the housekeeping gene GAPDH, using the 2−ΔΔCT method of Livak and Schmittgen [19] as described above.
Statistical analyses
Lung bacterial and viral densities were normalized for analysis using logarithmic transformation. Logrank tests were used to determine significant differences among the survival assay groups. Normally distributed samples were analyzed using the Student t test. Samples that did not follow a normal distribution were analyzed using the nonparametric Mann-Whitney test. For multiple comparisons, statistical significance was determined by analysis of variance.
RESULTS
Mouse survival with and without antecedent influenza virus infection
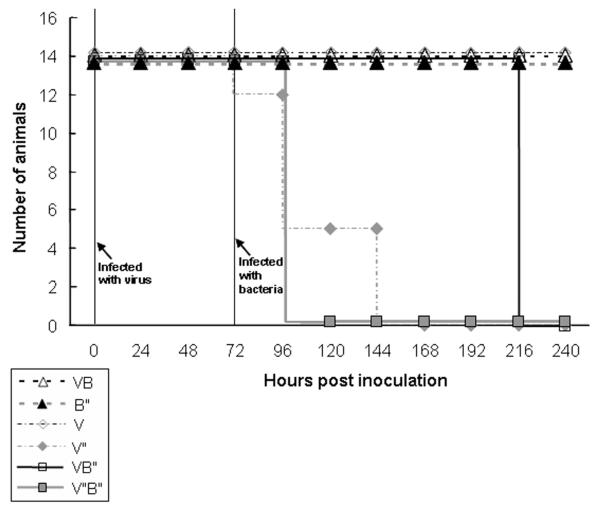
The survival of 6 treatment groups after infection was compared: (1) high dose of influenza virus alone, (2) low dose of virus, (3) high dose of S. aureus bacteria, (4) low dose of bacteria, (5) high doses of virus and bacteria, and (6) low dose of virus plus high dose of bacteria (Figure 1). All mice survived in the low-dose virus, low-dose bacteria, and high dose bacteria groups; there were significant differences in survival found among the remaining treatment groups (χ2 = 119.16; df = 5; P < .001). Mice infected solely with the high dose of influenza virus started to die 4 days after viral infection. There were no survivors 7 days after infection. All mice infected with the high dose of S. aureus survived, but all mice infected with an antecedent high viral dose plus a high bacterial dose died 2 days after bacterial challenge. In contrast, all mice challenged with the low viral dose and high S. aureus dose were dead 7 days after the bacterial challenge, but all mice infected with low doses of virus or both virus and bacteria survived 7 days. In summary, the antecedent viral infection appeared to accelerate death in the mice infected with bacteria.
Figure 1.
Survival assay with mice infected with different inocula of influenza virus (V, low dose; V“, high dose) and Staphylococcus aureus (B, low dose; B”, high dose). Mice were infected as outlined in Methods, with bacterial challenge 72 h after viral inoculation.
Lung histopathology
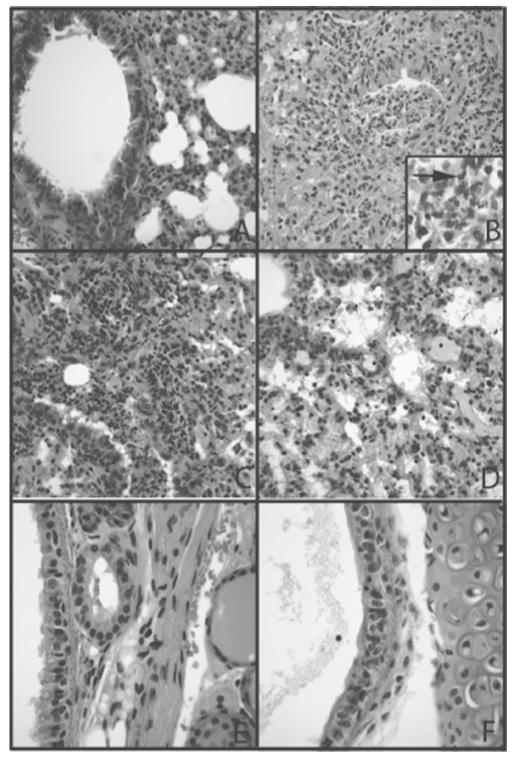
There was evidence of increased acute inflammation and injury in the lungs of mice infected with both influenza and S. aureus compared with lungs from mice challenged with either PBS or S. aureus only (Figure 2A and 2B). These differences were most apparent at the 4-h time point after bacterial infection, compared with 24 h (Figure 2C) and 72 h (Figure 2D). Sections from the later time points revealed increased organization of the pneumonic process and increased lymphocytic infiltration (Figure 2D). Gram staining of the tissue seldom revealed bacteria. However, S. aureus were detected at 4 h in animals infected with both virus and bacteria (Figure 2B, inset). Mice infected solely with influenza virus showed abnormalities that were localized to the trachea, where there was evidence of cellular regeneration and patchy loss of cilia (Figure 2F).
Figure 2.
Histopathology of mouse lungs infected with low viral and bacterial doses. A, Lung tissue after instillation of phosphate-buffered saline showing unremarkable bronchiole and lung parenchyma without acute or chronic inflammation. B, Lung tissue 72 h after virus instillation and 4 h after bacterial infection showing acute suppurative bronchiolitis with extension into alveoli adjacent to the airway. Inset, Tissue Gram stain showing gram-positive cocci (arrow) in areas of suppurative inflammation (original magnification, ×600). C, Presence of acute pneumonia with neutrophils in the distal bronchiole and in alveolar spaces 24 h after bacterial instillation. Mild chronic inflammation is also present in the interstitium and in perivascular locations. D, Reduced acute inflammation 72 h after bacterial instillation. Inflammation is still seen as neutrophils and neutrophil debris throughout the lung interstitium with congestion and intraalveolar fluid. E, Section of trachea after vehicle instillation showing normal pseudostratified ciliated epithelium and lamina propria accessory glands. F, Sections of trachea 72 h after viral instillation showing disarray of epithelium consistent with regeneration and large reactive basal epithelial cells with loss of ciliated cell polarity. A–F, hematoxylineosin stain; original magnification, ×200.
Bacterial density in the lung and in tissue homogenates
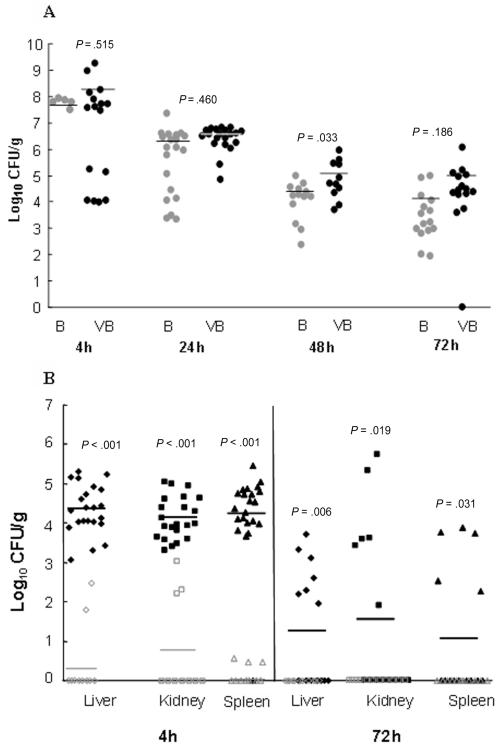
Bacterial densities were compared between lungs from mice infected with combined low doses of virus and bacteria and lungs from those infected with low doses of bacteria alone (Figure 3A). Although bacterial density was higher in the mice infected with both virus and bacteria at each of the time points, the differences were not significant. There were also no significant differences in bacterial density between mice infected with the isogenic PVL USA300 or USA400 strain (data not shown). In contrast, mice infected with both virus and bacteria showed significantly higher numbers of S. aureus in the blood (spleen), kidneys, and liver than the mice infected with bacteria only at both 4 and 72 h after bacterial infection (Figure 3B).
Figure 3.
Bacterial density in the lung, blood (spleen), and other tissue sites. A, Lung bacterial burden in mice infected with low-dose bacteria at different time points with or without antecedent low dose of virus. B, Bacterial counts in the livers, kidneys, and spleens of mice after infection with low doses of either virus or bacteria. B, bacteria; CFU, colony-forming units; VB, virus plus bacteria.
Cellular response to influenza virus and S. aureus infection
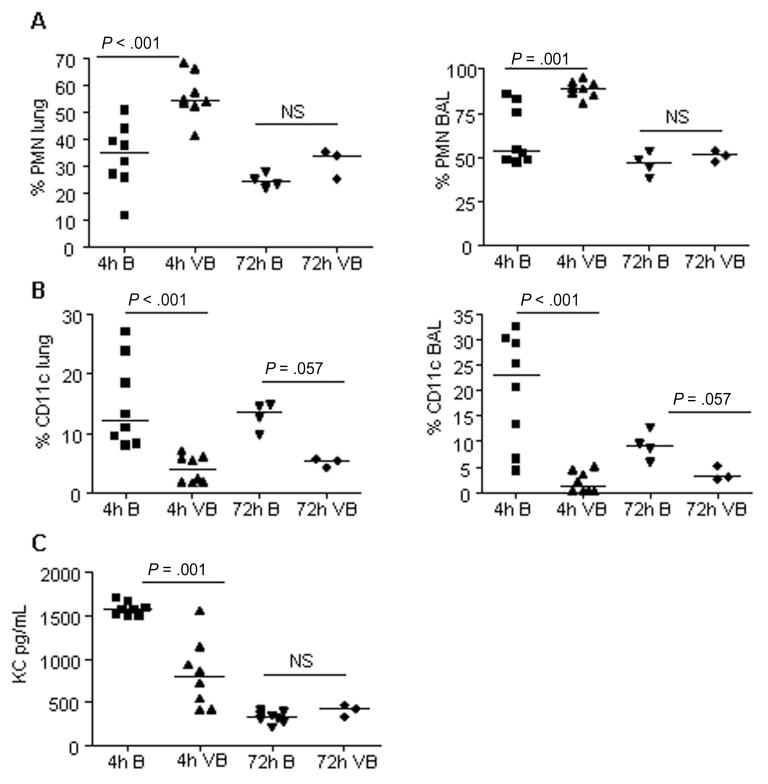
The cellular response to the sequential infections was assessed by comparison of cells harvested from the lungs and BAL of mice infected with low doses of bacteria only or both virus and bacteria (Figure 4). Four hours after bacterial exposure, mice infected with virus plus bacteria showed significantly fewer CD11c+ cells (P < .05) but significantly more PMN leukocytes in lung homogenates and BAL than mice infected with only bacteria. The latter group had significantly higher amounts of the major PMN chemoattractant KC in their BAL fluids than mice infected with virus plus bacteria. Thus, the presence of KC did not appear to be responsible for the increased levels of leukocytes present in the mice infected with virus and bacteria.
Figure 4.
Flow cytometry and enzyme-linked immunosorbent assay measurement of cellular responses in mice infected with influenza virus plus Staphylococcus aureus or S. aureus only. A, Polymorphonuclear (PMN) leukocytes (Ly6G/CD45+). B, Dendritic cells and alveolar macrophages (CD11c+/CD45+). Data are percentages of total CD45+ population. C, Levels of keratinocyte-derived chemokine (KC) in bronchoalveolar lavage (BAL) fluid of infected mice. B, bacteria only; NS, not significant; VB, virus followed by bacteria.
In vivo gene expression of S. aureus protein A, α-hemolysin, and PVL
RNA samples isolated from BAL fluid at 4 and 72 h after bacterial infection were used to compare in vivo gene expression of lukS, hla, and spa. RNA samples from in vitro cultures of S. aureus were used to measure expression levels of protein A, α-hemolysin, and PVL.
The greatest increase in expression levels of protein A occurred in the mice infected with virus plus bacteria at 4 versus 72 h after bacterial infection (mean fold change, 22.28 ± 14.88 vs 4.49 ± 5.15; P = .036). α-Hemolysin expression was also highest at 4 versus 72 h after infection with bacteria alone (mean fold change, 27.37 ± 2.19 vs 6.59 ± 8.67; P = .003), but there was no difference in levels between the 2 time points for the mice infected with virus plus bacteria (mean fold change, 47.42 ± 5.71 vs 3.44 ± 5.27; P = .28). In contrast, PVL levels were highest in both treatment groups at 72 h (mean fold change, 0.29 ± 0.56 for virus plus bacteria and 0.69 ± 0.53 for bacteria only), but only the group infected with bacteria alone had expression significantly different from that at the 4-h time point (mean fold change, 0.07 ± 0.09 for virus plus bacteria and 0.03 ± 0.03 for bacteria only; P = .048). When levels of expression were compared between the infection groups, the level of α-hemolysin was significantly reduced in the virus plus bacteria groups at the 4-h time point (mean fold change, 7.42 ± 5.71 for virus plus bacteria vs 27.37 ± 2.19 for bacteria only; P < .001).
In vivo gene expression of IFN-γ
IFN-γ expression was measured using RNA harvested from mice infected with PBS, virus or bacteria alone, or virus plus bacteria. Although there was no difference between the virus and bacteria groups, there was a significant difference between the virus-plus-bacteria group (fold change, 26.22 ± 19.04) and either the bacteria-only group (fold change, 1.72 ± 1.32) or the virus-only group (fold change, 0.74 ± 0.36) (P < .05).
DISCUSSION
A mouse model of postinfluenza CA-MRSA pneumonia was developed for this study. Our findings highlight a number of differences in the respiratory infection and its outcome on the basis of whether there was antecedent influenza virus infection. Most noteworthy was the increased mortality in mice infected with virus plus bacteria. This increased mortality was associated with evidence of greater pathological damage to the lung parenchyma, a higher-grade bacteremia, increased metastatic seeding to the liver and kidney, and an altered cellular response to infection in the infected mice. In contrast there were minimal differences in virulence between different strains of CA-MRSA. Of interest, the pattern of PVL expression was different from those of both protein A and α-hemolysin, regardless of time point or whether there was antecedent viral infection.
The importance of postinfluenza bacterial pneumonias as a primary cause of death in the 1918 influenza pandemic has recently been reemphasized [9, 10, 20]. Commenting on the mortality associated with the 1918 pandemic, investigators noted that most influenza pneumonias were self-limited and that the supervening bacterial superinfections accounted for most of the fatalities. This interpretation is supported by an analysis of the timeline of infections and reviews of firsthand accounts as well as the histopathological material available from the epidemic [9]. This concern regarding the primary cause of death has taken on increased importance with the emergence of the highly pathogenic H5N1 influenza strain that appears genetically similar to the H1N1 1918 strain, as well as the recently detected “swine flu” H1N1 strain [20, 21].
Investigators have recently described several models of CA-MRSA pneumonia [22-24]. These studies examined the virulence of different strains of CA-MRSA. However, the role of antecedent viral infections in these different models was not examined. Montgomery et al [24] reported that USA300 isolates were more virulent than USA400 strains in a rat model of pneumonia. These investigators also examined the in vitro expression of S. aureus virulence determinants, including PVL and α-hemolysin, finding higher levels of virulence gene expression in the USA300 strains. A subsequent study using the same model failed to identify differences in the host inflammatory and cytokine response in mice infected with the PVL isogenic pair [25]. In contrast, Labandeira-Rey et al [22] reported that the PVL toxin was primarily responsible for the parenchymal damage. Other investigators have failed to confirm these findings and presented data suggesting that α-hemolysin was the primary factor [26]. Our studies failed to find a difference in virulence between the isogenic strains USA300 and USA400 [17, 27]. The role of PVL in CA-MRSA pneumonia therefore remains uncertain, perhaps in part because of the use of different strains and/or different animal species. In our study, PVL levels were low overall and maximally expressed only 72 h after infection; in contrast, for α-hemolysin higher levels were observed 4 h after infection, perhaps suggesting a lesser role for PVL.
Our histopathological findings demonstrated (as have findings of others) that influenza virus infection alone primarily damages the tracheobronchial epithelium [28]. This damage may contribute to the inability to clear secretions efficiently and may lead to increased accumulation of debris in the alveoli. Damage at the 4-h time point was increased in mice previously infected with influenza virus, with a pronounced inflammatory response and extensive parenchymal infiltration. This difference was not evident at the later time points.
In addition to the pathological changes that undoubtedly contributed to the severity of these infections, immunological effects caused by the viral infection contributed to the progression of subsequent bacterial infections. Several studies have recently demonstrated a role for the IFN-γ signaling pathway as a mediator of the inflammatory—and more specifically neutrophil—response to S. aureus infection [29, 30]. Shornick et al [31] reported that the innate immune response due to activation of the IFN signaling protein Stat1 is impaired during influenza virus infection. This may result from the effects of the influenza nonstructural protein NS1 [32]. More recently, Sun and Metzger [33] and Shahangian et al [14] reported an impaired antibacterial response to S. pneumoniae infection mediated by the IFN-γ produced during influenza virus infection. S. aureus was also demonstrated by Martin et al [34] to induce type I IFN. These observations are further supported by our results showing higher levels of expression of IFN-γ, as well as an increased PMN cellular response in mice infected with both virus and bacteria compared with those infected with bacteria alone. The failure to contain the infection in the lungs, as evidenced by seeding of other tissues, also suggests an impaired host response, possibly caused by the defects in alveolar macrophage function described elsewhere or the demonstrated parenchymal damage. These results highlight the potential importance of the impaired immune response in the observed pathological damage and the resultant inability to contain the bacterial infection.
The in vivo RNA levels were higher for α-hemolysin in the bacteria-infected group than in the mice infected with bacteria plus influenza virus. The basis for this observation, though uncertain, is of interest. The finding suggests that influenza virus may cause alterations in the level of selected bacterial gene expression, perhaps via effects on staphylococcal regulatory genes such as agr, which up-regulates both α-hemolysin and PVL expression [35]. Though a preliminary observation, it is worth further investigation.
This study has a number of limitations. The mouse model, though in many ways reflecting the clinical setting of necrotizing CA-MRSA pneumonia, does not necessarily reflect the host response in humans. Specifically, mouse white blood cells appear to be less susceptible to staphylococcal leukotoxins than human cells [36]. This effect may have diminished the pathological findings in the lungs. Only a limited number of S. aureus isolates were used in this study. Results may vary when additional strains are used. Despite the limitations, these findings demonstrate the importance of antecedent influenza infections in the outcome of possible subsequent S. aureus infections.
Our model not only demonstrated the differences between bacterial and dual viral and bacterial respiratory tract infection, but it also provides a possible rationale for the enhanced morbidity and mortality encountered in these dual infections. Future studies can now address the pathogenesis and therapy of these life-threatening infections in greater detail.
Acknowledgments
We gratefully acknowledge the technical advice of Peter Palese, Gina Conenello, Craig Rubens, and Donald Chaffin.
Financial support: Centers for Disease Control and Prevention (grant CCR223380); Pfizer.
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Chambers HF. The changing epidemiology of Staphylococcus aureus. Emerg Infect Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 3.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 4.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 5.Francis JS, Doherty MC, Lopatin U, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–107. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 6.Hageman JC, Uyeki TM, Francis JS, et al. Severe community-acquired pneumonia due to Staphylococcus aureus, 2003-04 influenza season. Emerg Infect Dis. 2006;12:894–899. doi: 10.3201/eid1206.051141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallen AJ, Hageman J, Gorwitz R, Beekmann SE, Polgreen PM. Characteristics of Staphylococcus aureus community-acquired pneumonia during the 2006-2007 influenza season. Clin Infect Dis. 2007;45:1655. doi: 10.1086/523721. [DOI] [PubMed] [Google Scholar]
- 8.Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis. 2006;6:303–312. doi: 10.1016/S1473-3099(06)70466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brundage JF, Shanks GD. Deaths from bacterial pneumonia during 1918-19 influenza pandemic. Emerg Infect Dis. 2008;14:1193–1199. doi: 10.3201/eid1408.071313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finelli L, Fiore A, Dhara R, et al. Influenza-associated pediatric mortality in the United States: increase of Staphylococcus aureus coinfection. Pediatrics. 2008;122:805–811. doi: 10.1542/peds.2008-1336. [DOI] [PubMed] [Google Scholar]
- 12.McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis. 2002;186:341–350. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- 13.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shahangian A, Chow EK, Tian X, et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaush CR, Smith TF. Replication and plaque assay of influenza virus in an established line of canine kidney cells. Appl Microbiol. 1968;16:588–594. doi: 10.1128/am.16.4.588-594.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Sastre A, Egorov A, Matassov D, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 17.Voyich JM, Otto M, Mathema B, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 18.McCullers JA, Bartmess KC. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis. 2003;187:1000–1009. doi: 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 21.Outbreak of swine-origin influenza A (H1N1) virus infection: Mexico, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:467–470. [PubMed] [Google Scholar]
- 22.Labandeira-Rey M, Couzon F, Boisset S, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 23.Wardenburg JB, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–1044. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montgomery CP, Boyle-Vavra S, Adem PV, et al. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery CP, Daum RS. Transcription of inflammatory genes in the lung after infection with community-associated methicillin-resistant Staphylococcus aureus: a role for Panton-Valentine leukocidin? Infect Immun. 2009;77:2159–2167. doi: 10.1128/IAI.00021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wardenburg JB, Bae T, Otto M, DeLeo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 27.Diep BA, Palazzolo-Ballance AM, Tattevin P, et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS ONE. 2008;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hers JF, Masurel N, Mulder J. Bacteriology and histopathology of the respiratory tract and lungs in fatal Asian influenza. Lancet. 1958;2:1141–1143. doi: 10.1016/s0140-6736(58)92404-8. [DOI] [PubMed] [Google Scholar]
- 29.McLoughlin RM, Lee JC, Kasper DL, Tzianabos AO. IFN-gamma regulated chemokine production determines the outcome of Staphylococcus aureus infection. J Immunol. 2008;181:1323–1332. doi: 10.4049/jimmunol.181.2.1323. [DOI] [PubMed] [Google Scholar]
- 30.Muralimohan G, Rossi RJ, Guernsey LA, Thrall RS, Vella AT. Inhalation of Staphylococcus aureus enterotoxin A induces IFN-gamma and CD8 T cell-dependent airway and interstitial lung pathology in mice. J Immunol. 2008;181:3698–3705. doi: 10.4049/jimmunol.181.5.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shornick LP, Wells AG, Zhang Y, et al. Airway epithelial versus immune cell Stat1 function for innate defense against respiratory viral infection. J Immunol. 2008;180:3319–3328. doi: 10.4049/jimmunol.180.5.3319. [DOI] [PubMed] [Google Scholar]
- 32.Geiss GK, Salvatore M, Tumpey TM, et al. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci U S A. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat Med. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 34.Martin FJ, Gomez MI, Wetzel DM, et al. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J Clin Invest. 2009;119:1931–1939. doi: 10.1172/JCI35879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 36.Szmigielski S, Prevost G, Monteil H, Colin DA, Jeljaszewicz J. Leukocidal toxins of staphylococci. Zentralbl Bakteriol. 1999;289:185–201. doi: 10.1016/s0934-8840(99)80105-4. [DOI] [PubMed] [Google Scholar]