Abstract
The proteasome is a key intracellular protease that regulates processes, such as signal transduction and protein quality control, through the selective degradation of specific proteins. Signals that target a protein for degradation, collectively known as degrons, have been defined for many proteins involved in cell signaling. However, the molecular signals involved in recognition and degradation of proteins damaged by oxidation have not been completely defined. The current study used biochemical and spectroscopic measurements to define the properties in calmodulin that initiate degradation by the 20S proteasome. Our experimental approach involved the generation of multiple calmodulin mutants with specific Met replaced by Leu. This strategy of site-directed mutagenesis permitted site-selective oxidation of Met to Met sulfoxide. We found that the oxidation-induced loss of secondary structure, as measured by circular dichroism, correlated with the rate of degradation for wild-type and mutants containing Leu substitutions in the C-terminus. However, no degradation was observed for mutants with Met to Leu substitution in the N-terminus, suggesting that oxidation-induced structural unfolding in the N-terminal region is essential for degradation by the 20S proteasome. Experiments comparing the thermodynamic stability of CaM mutants helped to further localize the critical site of oxidation-induced focal disruption to residues 51 and 72 in the N-terminal region. This work brings new biochemical and structural clarity to the concept of the degron, the portion of a protein that determines its susceptibility to degradation by the proteasome.
The selective degradation of proteins by the proteasome is a key mechanism for regulating processes essential for cell survival. The proteolytic core of the proteasome, known as the 20S proteasome, is composed of four stacked rings of seven subunits each. The catalytic sites are sequestered within the core’s interior, so proteins destined for degradation must first be unfolded to gain access to the catalytic sites. The functions of the 20S proteasome include both basal degradation of proteins containing an unfolded region inherent to their native conformation, e.g., p53, p21cip, and α-synuclein (1), and conditional degradation of proteins that contain an unfolded region induced by heat, stress, or mild oxidation (2, 3). The 26S proteasome, formed when the regulatory complex PA700 binds to the 20S catalytic core, recognizes and degrades both ubiquitin-modified (4–6) and some nonubiquitinated (7–11) protein substrates. Selective destruction of proteins involved in cell signaling, cell cycle progression, and regulation of apoptosis are examples of processes controlled by the 26S proteasome (5, 6).
The ATP-independent degradation of proteins by the 20S proteasome has been suggested as the primary mechanism for degrading oxidized proteins following an oxidative insult (3, 12). The signals that target a protein for degradation, such as specific amino acid sequences or structural motifs that are revealed by oxidation-induced posttranslational modifications, are collectively known as degrons. One well-described degron associated with the proteasome-dependent degradation of oxidized proteins is exposure of hydrophobic patches (13–15). However, this degron is not the universal signal for degradation of all oxidized proteins. For example, calmodulin (CaM)1 is one oxidized protein that does not utilize this degron (16), suggesting that other molecular signals trigger degradation of this oxidized protein.
CaM is a ubiquitously expressed calcium-sensing protein that binds to and regulates over 100 distinct proteins, including calcium channels and pumps, kinases, phosphatases, and transcription factors (17). Thus, regulation of cellular CaM levels is an important determinant in cell signaling. CaM’s ability to regulate large numbers of molecules can be attributed to its unique structure, which consists of N- and C-globular domains connected by a flexible linker. Different environmental conditions (e.g., calcium concentration, pH, ionic strength) can alter CaM’s conformation and change its affinity for select targets.
Previous work has demonstrated ubiquitin-independent degradation of CaM following in vitro oxidation or deamidation/isomerization of asparagines side chains by both the 20S and 26S proteasomes, respectively (10, 11, 16, 18, 19). In our previous work using hydrogen peroxide to selectively oxidize Met in CaM to Met sulfoxide, we showed that the extent of in vitro oxidation of CaM correlates with both the rate at which CaM was degraded by the 20S proteasome and the degree of CaM structural unfolding (16). Others have also reported a strong correlation between CaM global structural rearrangement induced by oxidation and degradation by the 20S proteasome (19). These results suggest that a loss of native structure induced by oxidation could expose a degron, i.e., a specific sequence or structural motif, which signals degradation by the proteasome. Additionally, it is possible that oxidation of specific Met residues is required for this signal to become solvent exposed, triggering proteasome proteolysis.
A limitation of using peroxide to oxidize Met to Met sulfoxide is that a mixture of multiple oxidized species of CaM is generated (16, 20). An additional limitation of our previous work (16) was that we could not distinguish the specific site of oxidation. To overcome these limitations, the current study used site-directed mutagenesis, facilitating site-selective oxidation, to define the Met residues that are critical for signaling proteasome degradation. We show that oxidation of N-terminal Met residues 51, 71, and 72 are essential for degradation by the 20S proteasome, suggesting the presence of a putative degron in CaM’s N-terminus.
EXPERIMENTAL PROCEDURES
Materials
Pigs were obtained from the University of Minnesota Experimental Farm and from Clemson University Research Farm Services. The QuickChange site-directed mutagenesis kit was purchased from Stratagene (LaJolla, CA). Fluorescamine was purchased from Sigma. 1-Anilinonaphthalene-8-sulfonate (ANS) was obtained from Molecular Probes (Eugene, OR). The bicinchoninic acid protein assay kit (BCA) was from Pierce (Rockford, IL). [3H]Ryanodine was purchased from PerkinElmer Life Sciences (Waltham, MA). Unlabeled ryanodine was obtained from Calbiochem (La Jolla, CA). Adenosine 5′-(β,γ-imido)triphosphate tetralithium salt (AMPPNP) was from Sigma (St. Louis, MO). All other reagents were of HPLC grade or better.
Purification of 20S Proteasome
The 20S proteasome was purified from frozen rat liver as previously described (16). Samples were stored in 10 mM Tris and 200 mM KCl (pH 7.5) at −80 °C. Protein concentrations were determined using the BCA assay with bovine serum albumin as the protein standard. The purity of 20S proteasome preparations, which includes HSP90, was similar to previous reports (16, 18).
Site-Directed Mutagenesis of CaM
Seven CaM mutants were generated, in which different combinations of Met were mutated to Leu. The nomenclature for the mutants indicates the number of Leu substitutions and their position within the protein (Figure 1). Mutants were constructed from the wild-type (WT) rat CaM cDNA by using primer-based site-directed mutagenesis following the protocol provided by Stratagene’s QuickChange site-directed mutagenesis kit. DNA sequence analysis, performed at the DNA Sequencing and Analysis Facility (University of Minnesota, Minneapolis, MN), confirmed the correct sequence for each mutant.
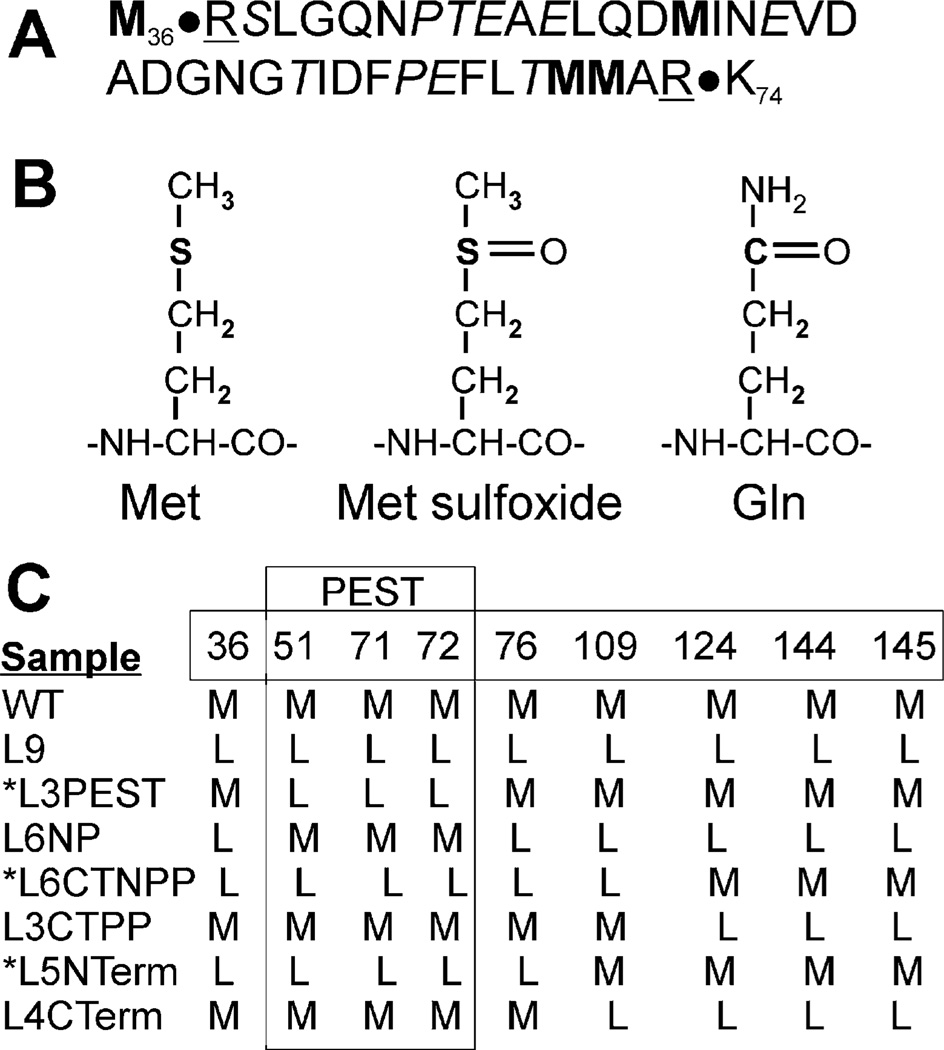
Figure 1.
Diagram of proposed PEST sequence and CaM mutants. (A) Putative PEST sequence in CaM (amino acids 37–73) includes Pro, Glu, Ser, or Thr (in italics) and is flanked by charged amino acids (underlined). (B) Structures of Met, Met sulfoxide, and glutamine. (C) CaM mutants; numbers indicate the positions of nine Met present in WT CaM. Amino acids 51, 71, and 72 are located within the putative PEST sequence. Mutant names reflect the number of Leu substitutions and the relative position of the mutation: all 9 Met mutated (L9), mutation of 51, 71, and 72 (L3 PEST), mutation of all non-PEST Met (L6 NP), and mutation of all Met in the N-terminus (L5 NTerm) or C-terminus (L4 CTerm). C-Terminal “pseudo-PEST” (L3 CTPP) indicates mutation of the C-terminal Met (124, 144, 145) that are analogous to the N-terminal PEST sequence. Mutation of all Met except 124, 144, and 145 is designated as C-terminal non-“pseudo-PEST” (L6 CTNPP). *PEST-region mutants contain Met to Leu substitutions for amino acids 51, 71, and 72.
Expression and Purification of WT and Mutant CaM
CaM was expressed in Escherichia coli (BL21(DE3)pLys5), purified via phenyl-Sepharose chromatography (21), and dialyzed overnight at 4 °C against 2 mM HEPES (pH 7.0). Protein concentration was determined using the Micro BCA protein assay using wild-type CaM as the protein standard. The concentration of the CaM standard was determined using the published molar extinction coefficient, ε277–320nm = 3029 M−1 cm−1 (22). The purity of the preparations was confirmed by a single protein band on Coomassie blue-stained gels that migrates at ~16 kDa.
Mass Spectrometric Analysis
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry was used to determine the masses of nonoxidized and oxidized mutants. Prior to analysis, samples were desalted using Millipore C4 ZipTips as instructed by the manufacturer’s protocol. Molecular mass was determined using a Bruker Biflex III mass spectrometer (Bruker Daltonics, Billerica, MA) and α-cyano-4-hydroxycinnamic acid as the matrix.
CaM Oxidation
Met in CaM were selectively oxidized by incubating 60 µM CaM (1 mg/mL) in 50 mM HOMOPIPES (pH 5.0), 0.1 M KCl, 1 mM MgCl2, and 0.1 mM CaCl2 with 50 mM H2O2 at 25 °C for 24 h (16). To stop the reaction, the sample was dialyzed (MWCO = 1000 kDa) overnight at 4 °C in distilled water buffered with 1 mM ammonium bicarbonate (pH 7.7).
Circular Dichroism (CD) Spectroscopy of CaM
CD spectra were recorded from 250 to 200 nm using a JASCO J-710 spectrophotometer and a temperature-jacketed spectral cell with a path length of 0.1 mm. Spectra were recorded at 1 nm intervals for CaM (50 µM) in 50 mM Tris (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM EGTA, and 600 µM CaCl2 at 37 °C. Two separate measurements were performed on each sample. CD spectra were analyzed based on the standard assumption that the CD spectrum is a linear combination of contributions from three discrete secondary structural populations
| (1) |
where E is the molar ellipticity per residue and xi and Ei(λ) are the mole fraction and molar ellipticity, respectively, of residues in α-helix, β-sheet, and random coil (23, 24). Specifically, at 208 and 222 nm (25)
| (2) |
But xB) 0 for CaM (26), so eq 2 reduces to
| (3) |
where R = E(208 nm)/E(222 nm) and xα is the α-helical content. Data are expressed as mean ± SD.
Thermal Denaturation Experiments
Thermal denaturation experiments were performed using a JASCO J-815 circular dichroism spectrophotometer with an automated temperature controller. Samples had a concentration of 10 µM in 50 mM Tris (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM EGTA, and 600 µM CaCl2 in a cuvette with a path length of 1.0 mm. Samples were heated from 10 to 90 °C at a rate of 2 °C/min and then cooled from 90 to 10 °C. Ellipticity was recorded at 222 nm at 1° increments. A minimum of two separate measurements was performed on each sample. Data were analyzed from only spectra that showed good reversibility with temperature.
Analysis of Hydrophobicity of Oxidized CaM
The surface hydrophobicity of CaM was evaluated from fluorescence enhancement of ANS. CaM (180 nM) was incubated at 37 °C in 50 mM HEPES, 100 mM KCl, 10 mM MgCl2, 1 mM EGTA, and 600 µM CaCl2 for 30 min with 16-fold molar excess of ANS. Fluorescence emission spectra, with excitation at 372 nm, were recorded from 450 to 600 nm with a Jobin-Evon Spex Fluoromax-2. Hydrophobicity was defined as the increase in integrated fluorescence intensity due to addition of CaM, normalized to WT. Four separate measurements were performed on each sample. Data are expressed as mean ± SD.
CaM Proteolysis by the 20S Proteasome
The rate of proteolysis using CaM as the substrate was determined using fluorescamine, which forms a fluorescent adduct with the amino termni of peptides released by proteasome cleavage of CaM (18). CaM (31.5 nM) was incubated at 37 °C in 50 mM HEPES, 100 mM KCl, 10 mM MgCl2, 1 mM EGTA, 600 µM CaCl2, and 1.5 nM 20S proteasome. Aliquots were removed every minute for 5 min. Measurement of the fluorescent adducts was performed in a Cytofluor 4000 multiwell plate reader (ex = 360 nm; em = 460 nm). The concentration of peptides hydrolyzed was determined by comparing the linear response of sample fluorescence with the fluorescence of a glycine standard curve. The rate of CaM degradation was linear with respect to proteasome concentration.
[3H]Ryanodine Binding to Skeletal Muscle Heavy Sarcoplasmic Reticulum (HSR)
Skeletal muscle HSR vesicles were prepared from porcine longissimus dorsi muscle, and [3H]ryanodine binding was performed as described previously (27). Ryanodine binds to the open RyR with high affinity and specificity and is therefore a sensitive indicator of channel activity (28, 29). HSR vesicles (0.2 mg/mL) were incubated for 90 min at 37 °C in medium containing 120 mM potassium propionate, 10 mM PIPES, pH 7.0, 1.5 mM AMPPNP, 100 nM [3H]ryanodine, and a Ca-EGTA buffer set to give the desired free Ca2+ concentration (30). HSR vesicles were then collected on Whatman GF/B filters and washed with 8 mL of ice-cold 100 mM KCl buffer. Radioactivity retained by the filters was determined by liquid scintillation counting. Estimates of maximal [3H]ryanodine binding capacity of each HSR vesicle preparation were determined in medium that in addition contained 500 mM KCl, 5 mM ATP, and 100 µM Ca2+. Nonspecific binding was measured in the presence of 20 µM nonradioactive ryanodine. [3H]Ryanodine binding is expressed as a percent of maximal [3H]ryanodine binding. The CaM concentration dependence of SR vesicle [3H]ryanodine binding was fit with a four-parameter Hill equation using SigmaPlot 9.0 (Systat Software, Point Richmond, CA). Data are presented as means ± SE.
Statistical Analysis
Degradation rates were compared for oxidized and nonoxidized samples using a Student’s t test. Linear regression analysis was performed to test the relationship between (i) α-helical content and the number of substitutions, (ii) degradation rate and α-helical content, or (iii) degradation rate and ANS binding. Student’s t test and linear regression were performed using the statistical software in Origin 7.5 (Microcal). Parameters derived from the Hill equation fit to [3H]ryanodine binding data were compared using a one-way analysis of variance with a Holm–Sidak multiple comparison as a post hoc test. Statistical analysis was performed using SigmaStat 3.1 (Systat Software, Point Richmond, CA). Statistical significance was set at p < 0.05. Results are expressed as mean ± SEM unless otherwise specified.
RESULTS
Description and Rationale for CaM Mutants
CaM contains nine Met that can be oxidized to Met sulfoxide. Previous work has shown that the extent of in vitro oxidation of CaM using hydrogen peroxide correlates with both the rate of CaM degradation by the 20S proteasome and the degree of CaM structural unfolding (16). These results suggest that loss of secondary structure induced by oxidation could unmask a specific sequence or structural motif that signals degradation by the proteasome. One well-characterized degradation signal, also known as a degron, is the PEST sequence. This degron is defined by the inclusion or exclusion of specific amino acids and the sequence hydrophobicity (31). The PEST sequence includes the amino acids Pro, Glu, Asp, Ser, and Thr that are flanked by positively charged amino acids; Lys and Arg are not permitted within the sequence. As determined using the PEST algorithm (https://emb1.bcc.univie.ac.at./toolbox/pestfind/), CaM contains a poor PEST sequence (score = −3.07, where positive scores indicate potential PEST sequences) within residues 37–73 (Figure 1A). Considering the structural and chemical similarities between Met sulfoxide and glutamine (Figure 1B), we substituted glutamine for Met (residues 51, 71, 72) and found that the PEST score improved to −0.33. It is likely that the presence of an oxygen on the side chain alters the hydrophobicity of the sequence, making it a stronger PEST degron. Thus, we hypothesize that the oxidation of Met within the putative N-terminal degron will cause conformational unmasking of a signal that elicits degradation by the proteasome.
A limitation of using peroxide to oxidize Met to Met sulfoxide is that a mixture of multiple oxidized species of CaM is generated (16), thus complicating identification of key Met. To overcome this limitation, site-directed mutagenesis and site-specific oxidation were used to define the Met residues that are critical for eliciting proteasome degradation. Figure 1C defines the site of Met to Leu substitutions for the seven mutants that were studied. (See also Figure 1 legend for explanation of the nomenclature. Note that the nomenclature utilizes “PEST” as an indication of the position of Met mutations in relation to residues 37–73 in the N-terminus. However, we are not inferring that this region contains a legitimate (confirmed) PEST sequence.) Of particular importance are the mutants containing Leu substitutions in positions 51, 71, and 72, which prevent oxidation in the putative PEST region of the protein. These mutants (L3 PEST, L6 CTNPP, L5 NTerm) are referred to as the PEST-region mutants. The reciprocal mutants (L6 NP, L3 CTPP, L4 CTerm) were generated for each PEST-region mutant as negative controls and contain oxidizable Met in the PEST region. The Met in positions 124, 144, and 145 are located within the C-terminal EF-hand at approximately the same position as Met 51, 71, and 72 that are within the N-terminal EF-hand. We refer to the sequence containing Met 124, 144, and 145 as the “pseudo-PEST” sequence because it is the positional correlate to the PEST sequence in the N-terminus. However, the “pseudo-PEST” sequence does not contain a putative PEST degron. Also included in the analysis are the two extremes: WT containing no substitutions and L9, where all nine Met are mutated to Leu.
Characterization of CaM Mutants
Analysis of the DNA sequence for each CaM mutant confirmed that the planned mutations were correctly generated (data not shown). Mass spectrometry was used to determine the molecular mass for WT and CaM mutants. WT CaM exhibits a single major peak at 16702 ± 1 Da, which is in close agreement with the experimental molecular mass for rat CaM (16706 Da) (Table 1). Each of the unoxidized CaM mutants exhibited a single major peak that closely matched the theoretical mass; the average percent difference was 0.04 ± 0.006%. Using experimental conditions where complete hydrogen peroxide-induced oxidation of Met to Met sulfoxide was achieved, there was a shift in the single major peak that corresponded to the addition of one oxygen (+16 Da) for each Met present in the sequence. For example, oxidation of WT CaM resulted in the addition of 144 Da. In contrast, replacement of all Met with Leu (L9) resulted in no oxidation-dependent change in mass. The average percent difference comparing theoretical with experimentally measured masses for oxidized samples was 0.03 ± 0.006%.
Table 1.
Characterization of CaM Mutants
| molecular massb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| nonoxidized | oxidized | α-helix contentc | hydrophobicityd | |||||||
| samplea | theor | exptl | er (%) | theor | exptl | er (%) | nonox | ox | dif (%) | ANS binding |
| WT | 16706 | 16702 | 0.03 | 63 ± 2 | 71 | 0.99 ± 0.02 | ||||
| ox WT | 16850 | 16846 | 0.02 | 18 ± 1 | 1.00 | |||||
| L9 | 16544 | 16552 | 0.05 | 16544 | 16558 | 0.08 | 52 ± 2 | 53 ± 1 | 0 | 1.05 ± 0.02 |
| L3 PESTe | 16652 | 16645 | 0.04 | 16748 | 16751 | 0.02 | 61 ± 1 | 43 ± 3 | 30 | 0.88 ± 0.03 |
| L6 NP | 16598 | 16589 | 0.05 | 16646 | 16645 | 0.01 | 58 ± 4 | 43 ± 1 | 41 | 0.96 ± 0.00 |
| L6 CTNPPe | 16598 | 16608 | 0.06 | 16646 | 16654 | 0.05 | 55 ± 1 | 48 ± 1 | 15 | 1.09 ± 0.06 |
| L3 CTPP | 16652 | 16666 | 0.08 | 16748 | 16754 | 0.04 | 65 ± 1 | 30 ± 1 | 54 | 0.94 ± 0.05 |
| L5 NTerme | 16616 | 16612 | 0.02 | 16680 | 16681 | 0.01 | 61 ± 1 | 47 ± 1 | 25 | 0.97 ± 0.02 |
| L4 CTerm | 16634 | 16628 | 0.04 | 16714 | 16711 | 0.02 | 60 ± 1 | 29 ± 1 | 52 | 0.96 ± 0.03 |
Nomenclature reflects the number and location of Met to Leu substitutions. See Figure 1 for description.
Theoretical molecular mass was based on the sequence for CaM from rat (P62161, Swiss Prot) and the loss of 18 Da for each Met to Leu substitution. Theoretical mass of oxidized samples includes the addition of one oxygen (+16 Da) for each Met. Experimental molecular mass was determined using MALDI-TOF mass spectrometry. The percent error was calculated by [(experimental MW − theoretical MW)/theoretical MW] × 100.
α-Helix content was calculated from the ratio of ellipticity at 208 and 222 nm, measured by CD spectroscopy. Values are mean ± SD of two separate measures for each sample. The percent difference (dif) in α-helix content due to oxidation was calculated as (1 – (oxidized/nonoxidized)) for each sample.
Hydrophobicity of oxidized samples was estimated from fluorescence of ANS binding. All values were normalized to ox WT for each trial. Values are mean ± SD of two separate measures for each sample.
PEST-region mutants.
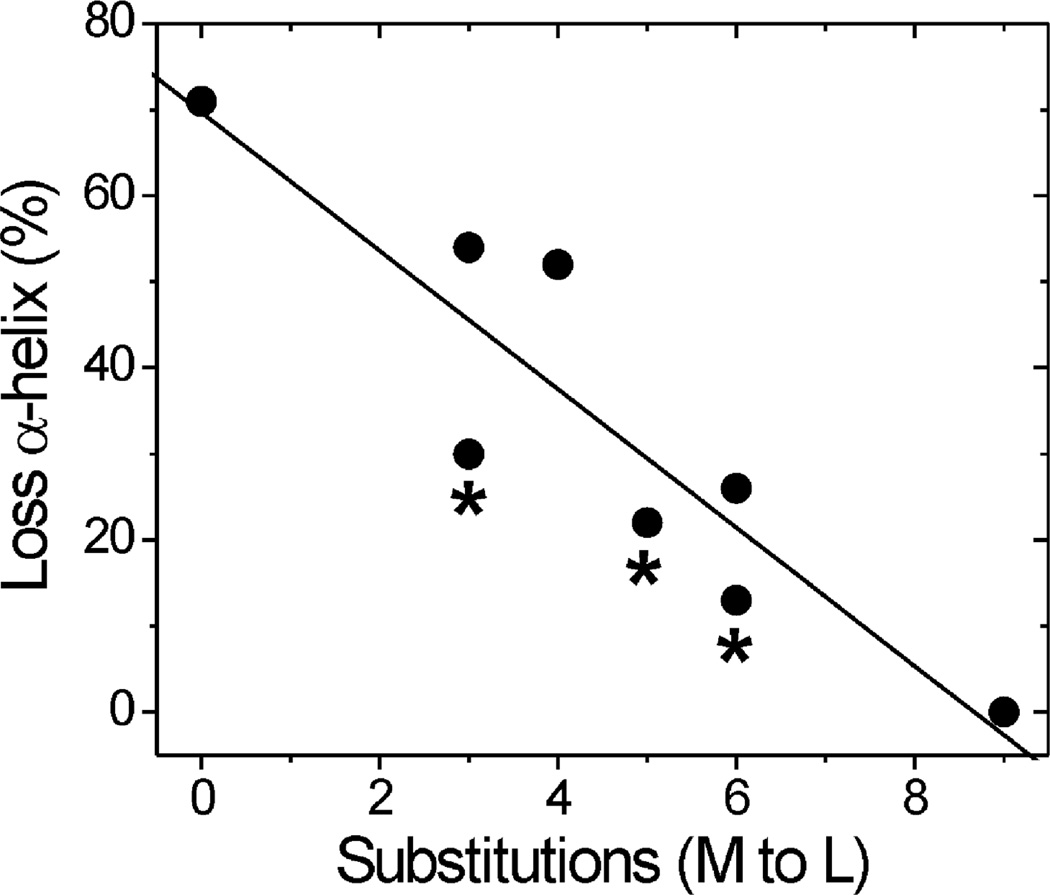
Circular dichroism (CD) spectroscopy was performed to evaluate changes in secondary structure due to either the Met to Leu substitutions or site-specific oxidation. Under our experimental conditions, WT CaM contains ~63% α-helix (Table 1). For the nonoxidized mutants, three to five substitutions had no effect on the α-helix content, whereas a minor loss in α-helix content was observed for mutants containing six or nine substitutions. With oxidation by hydrogen peroxide, all samples except L9 demonstrated a loss in secondary structure that correlated with the number of Met to Leu substitutions (Table 1, Figure 2). These results confirm that Met oxidation disrupts the secondary structure of CaM and that the decrease in α-helical content depends on the extent of Met oxidation. Of note, preventing oxidation of Met in the PEST region via their substitution to Leu, i.e., the PEST-region mutants, resulted in a loss in secondary structure that was similar to the reciprocal mutants containing an equivalent number of Met substitutions.
Figure 2.
Decreased α-helix content after oxidation correlates with the extent of Met substitutions. Linear regression analysis shows a significant negative relationship between the number of Met to Leu substitutions and the loss in α-helix content after oxidation (p = 0.002, R = 0.91, slope = −8.04). Secondary structure was evaluated by CD spectroscopy. α-Helical content was calculated from the ratio of ellipticity at 208 and 222 nm for nonoxidized and oxidized CaM samples. The percent loss in α-helical content following oxidation for each CaM mutant is provided in Table 1. * indicates PEST-region mutants.
PEST-Region Mutants Resist Degradation by the 20S Proteasome
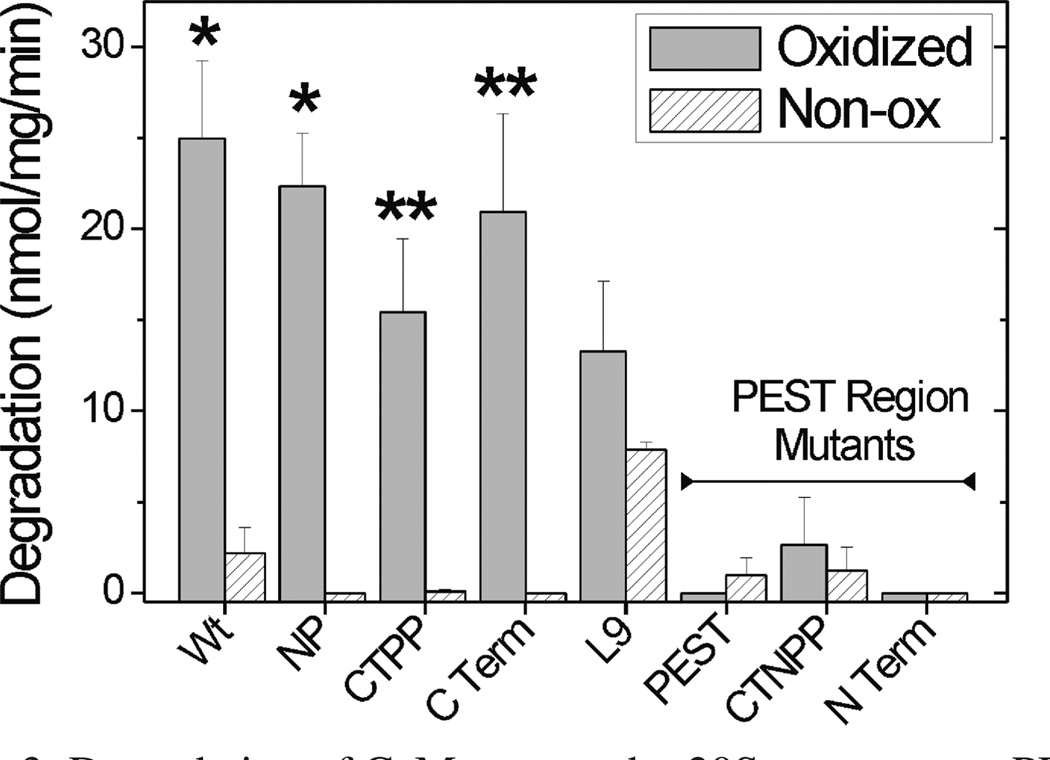
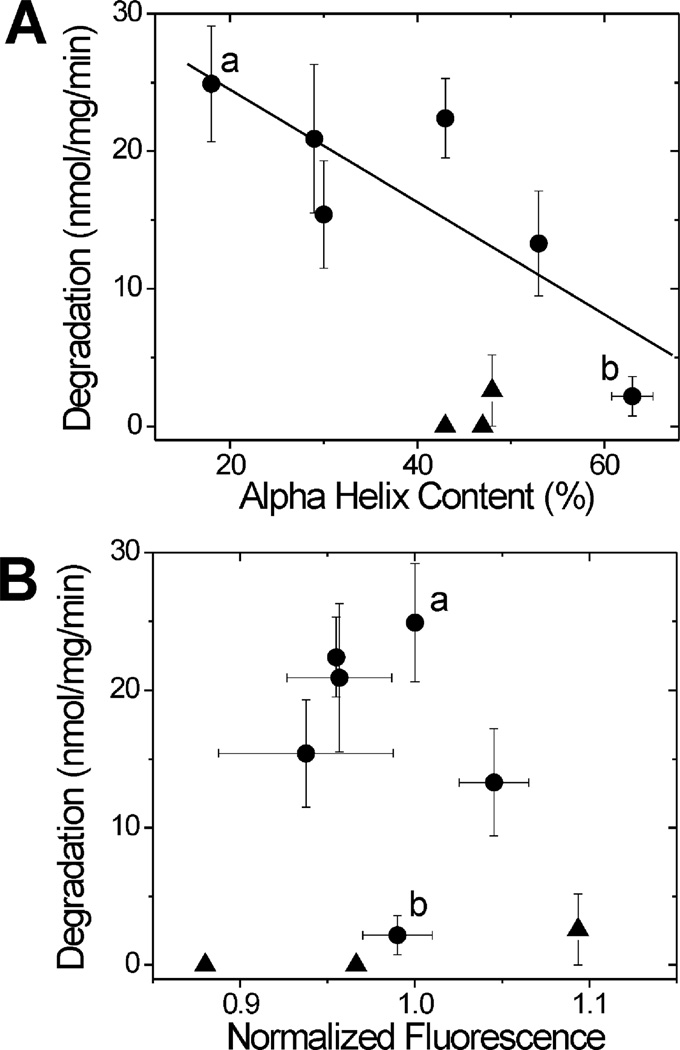
To determine whether oxidation of specific Met residues alters the proteolytic susceptibility of CaM, the rate of degradation for WT and CaM mutants by the 20S proteasome was compared (Figure 3). With the exception of L9, degradation of the nonoxidized proteins was negligible. For oxidized WT and mutants containing oxidizable Met in the PEST region (L6 NP, L3 CTPP, and L4 CTerm), a significant increase in degradation rate was observed. However, oxidized PEST-region mutants were resistant to degradation, even though the extent of secondary structure loss was similar to the reciprocal non-PEST mutants (Figure 2). In plotting the rates of degradation versus the α-helical content (Figure 4A), the dramatic differences between the PEST and non-PEST mutants are further highlighted. The loss of secondary structure observed for WT and oxidized non-PEST mutants demonstrates a significant correlation with their rate of degradation. In contrast, oxidation and the corresponding loss in secondary structure for PEST-region mutants had no effect on the rate of degradation by the proteasome.
Figure 3.
Degradation of CaM mutants by 20S proteasome: PEST-region mutants resist proteolysis. CaM degradation by the 20S proteasome was determined from the reaction of free amines generated by peptide bond cleavage with fluorescamine. A minimum of two to five separate measurements were performed in triplicate for each sample. *, p < 0.01; **, p < 0.06 comparing oxidized versus nonoxidized samples by t test analysis.
Figure 4.
Dependence of CaM degradation on secondary structure and hydrophobicity. (A) The rate of CaM degradation as a function of α-helical content demonstrates a significant linear relationship for WT and all non-PEST-region CaM mutants (p = 0.04, R = 0.83, slope = −0.41) (filled circles). PEST-region mutants are plotted as filled triangles. Calculation of α-helical content and CaM degradation are as described in Figures 2 and 3. Oxidized and nonoxidized WT are indicated by a and b, respectively. (B) The rate of CaM degradation showed no significant correlation with hydrophobicity (p = 0.65). Hydrophobicity was determined from the fluorescence due to binding of ANS, as described in Experimental Procedures. Normalized fluorescence is mean ± SD of two separate measures for each sample. Oxidized and nonoxidized WT are indicated by a and b, respectively.
Hydrophobicity Is Not Altered in PEST-Region Mutants
An oxidation-induced exposure of hydrophobic regions within proteins has been suggested as a signal for proteasome degradation (13–15). The relative hydrophobicity of WT and mutant CaM was measured to determine whether mutation of the PEST region preferentially altered the hydrophobicity of the mutant protein and therefore could explain their resistance to degradation. The fluorescence emission of 1-anilinonaphthalene-8-sulfonate (ANS) was used as an indicator of hydrophobicity; an increase in fluorescence is associated with binding of ANS to hydrophobic surfaces that become accessible to the probe (32). ANS binding to CaM is blue shifted and increased by binding of calcium to CaM, due to the exposure of hydrophobic pockets (33–35). To establish a benchmark for ANS fluorescence changes, WT CaM was incubated with ANS in a buffer containing either EGTA or high (700 µM) calcium. Calcium induced the expected blue shift in emission maximum along with a 1.9 ± 0.4-fold increase in integrated ANS fluorescence. By comparison, minimal differences in hydrophobicity were observed for oxidized CaM mutants relative to WT CaM; ANS fluorescence intensities ranged from 0.88 to 1.09 (Figure 4B) with no significant change in emission maximum (data not shown). These results indicate that altered hydrophobicity does not explain the resistance of PEST-region mutants to proteasome degradation.
Thermodynamic Stability
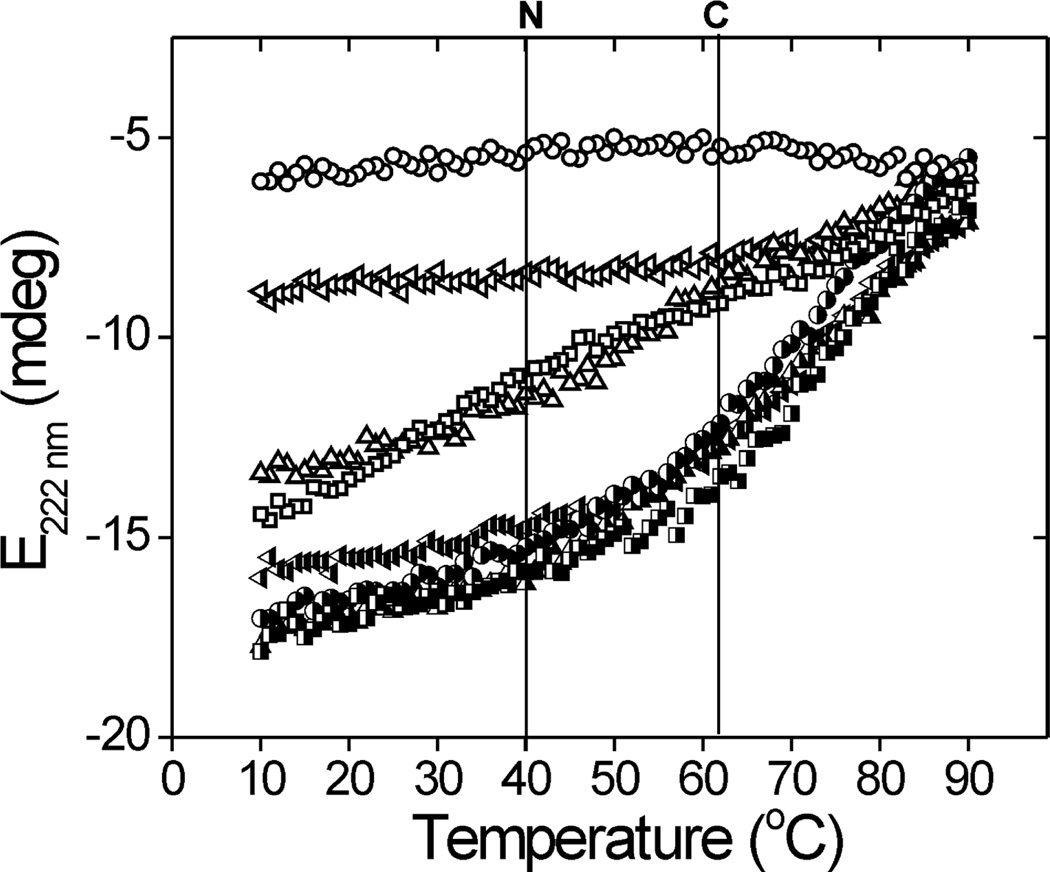
One possible mechanism explaining the difference in susceptibility to proteasome degradation is that oxidation of the N-terminal Met in the non-PEST mutants causes greater destabilization of CaM conformation. This destabilization could promote accelerated unfolding and subsequent rapid degradation by the proteasome. To test the conformational stability of select mutants that are oxidized in either the N- or C-terminus, we monitored the ellipticity at 222 nm using CD spectroscopy. Thermal denaturation and subsequent cooling were performed for temperatures between 10 and 90 °C. For all samples, ellipticity at each temperature with either heating or cooling was essentially identical, indicating that thermal unfolding was reversible (data not shown). Figure 5 reports the temperature-dependent changes in CD spectra, where the loss of ellipticity reflects CaM denaturation. The nonoxidized samples, including WT and mutants containing three to six Leu substitutions, displayed nearly identical patterns of unfolding (Figure 5), indicating that these Leu substitutions did not alter overall protein conformational stability. Two transitions of accelerated denaturation occurred at ~40 and ~62 °C. Previous thermal and chemical denaturation experiments with nonoxidized, calcium-bound CaM determined that the first and second transitions are associated with unfolding of CaM’s N- and C-domains, respectively (36).
Figure 5.
Thermal stability of WT, PEST-, and non-PEST-region mutants: WT (●), L3 PEST (▲), L4 CTerm (▼), and L6 NP (■). Oxidized and nonoxidized samples are represented by open and filled symbols, respectively. Vertical lines indicate C-terminal (C) and N-terminal (N) domain transitions. Thermal denaturation experiments were performed using CD spectroscopy and monitoring ellipticity (E) at 222 nm.
Spectra from oxidized WT CaM showed a significant decrease in signal amplitude that corresponds to a mainly unstructured protein (~18% α-helix at 37 °C, Table 1); no evidence of further unfolding occurred within the temperature range studied. Oxidation of the non-PEST mutant containing five N-terminal Met (L4 CTerm) also demonstrated a significant loss of signal amplitude compared with nonoxidized samples, indicating reduced secondary structure (~29% α-helix at 37 °C, Table 1). There is no evidence of the transition at 40 °C, suggesting the N-domain is significantly denatured by oxidation. A minor transition at 60 °C is consistent with denaturation of the C-domain. For the L3 PEST mutant, oxidation of five C-terminal Met and Met 36 in the N-terminus resulted in an intermediate loss of signal intensity and reduced secondary structure (~43% α-helix at 37 °C, Table 1). In contrast with L4 C-Term, the L3 PEST melting of the N-domain, suggesting that the C-domain is significantly denatured by oxidation. These results suggest that oxidation of five N-terminal Met (L4 CTerm) destabilizes CaM conformation more than oxidation of five C-terminal Met (L3 PEST), and thus decreased conformational stability may provide a partial explanation for the accelerated degradation of non-PEST mutants.
To test this idea further, we measured the thermodynamic stability of an additional non-PEST mutant, L6 NP. This mutant contains only three Met that are located in the putative PEST region of the N-terminus. Thus, the L6 NP mutant is the direct correlate to the L3 PEST mutant, where Leu replaces the three Met in the putative PEST region. Importantly, oxidized L6 NP is degraded as rapidly as oxidized L4 CTerm, which contains all five Met in the N-terminus (Figure 3). Spectra from oxidized L6 NP showed a loss of signal amplitude and reduced secondary structure (~43% α-helix at 37 °C, Table 1) that was similar to the thermal changes in ellipticity demonstrated for L3 PEST. Thus, decreased conformational stability is not an absolute requirement for eliciting proteasome degradation. Our results support the hypothesis that CaM degradation is triggered by oxidation-induced disruption of the specific region between Met 51 and Met 72 in the N-terminus. As a caveat, it is possible that limited sensitivity of CD measurements will not permit observing focal disruptions of CaM structure. However, these results help to localize critical points of disruption that trigger proteasome proteolysis.
Functional Consequences of PEST-Region Mutants
To determine the functional consequence of the site-specific mutagenesis and oxidation, we measured CaM regulation of the ryanodine receptor calcium release channel (RyR1) isolated from skeletal muscle. In vitro, calcium-activated CaM regulates RyR1 function by inhibiting channel opening, which is measured from the binding of [3H]ryanodine. Ryanodine is a neutral plant alkyloid that binds with high affinity and specificity to open RyR1 channels. Thus, changes in the amount of [3H] ryanodine bound by RyR1 indicate changes in the number of open release channels (28, 29).
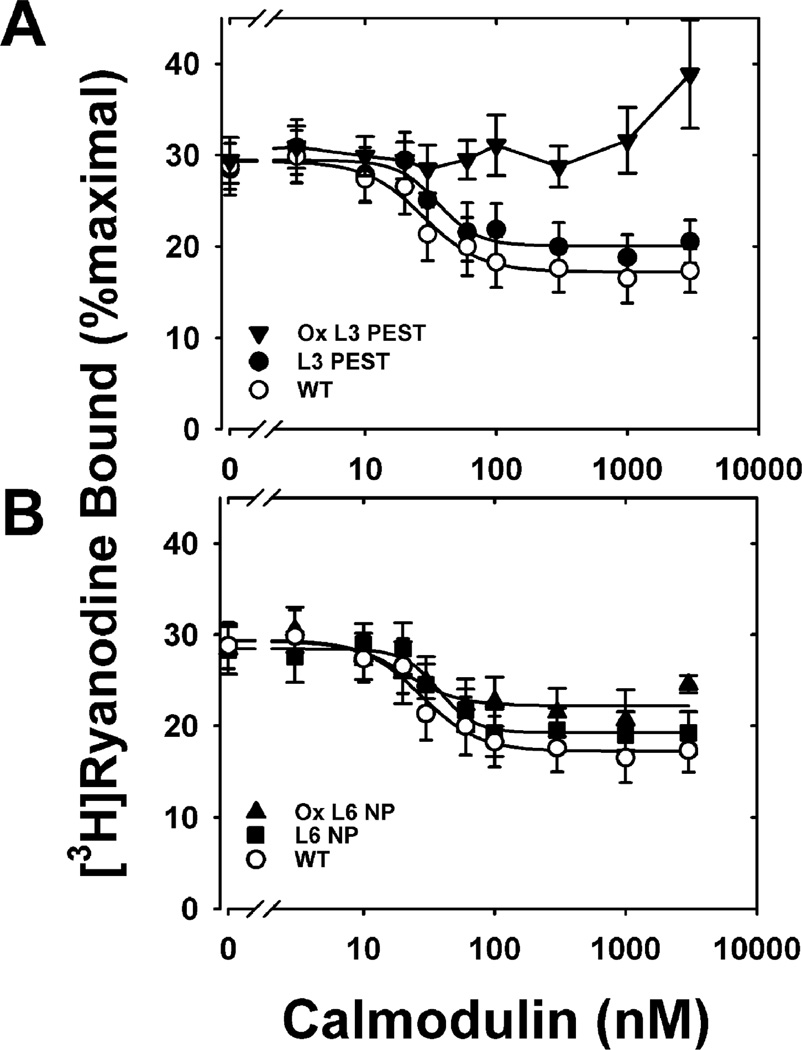
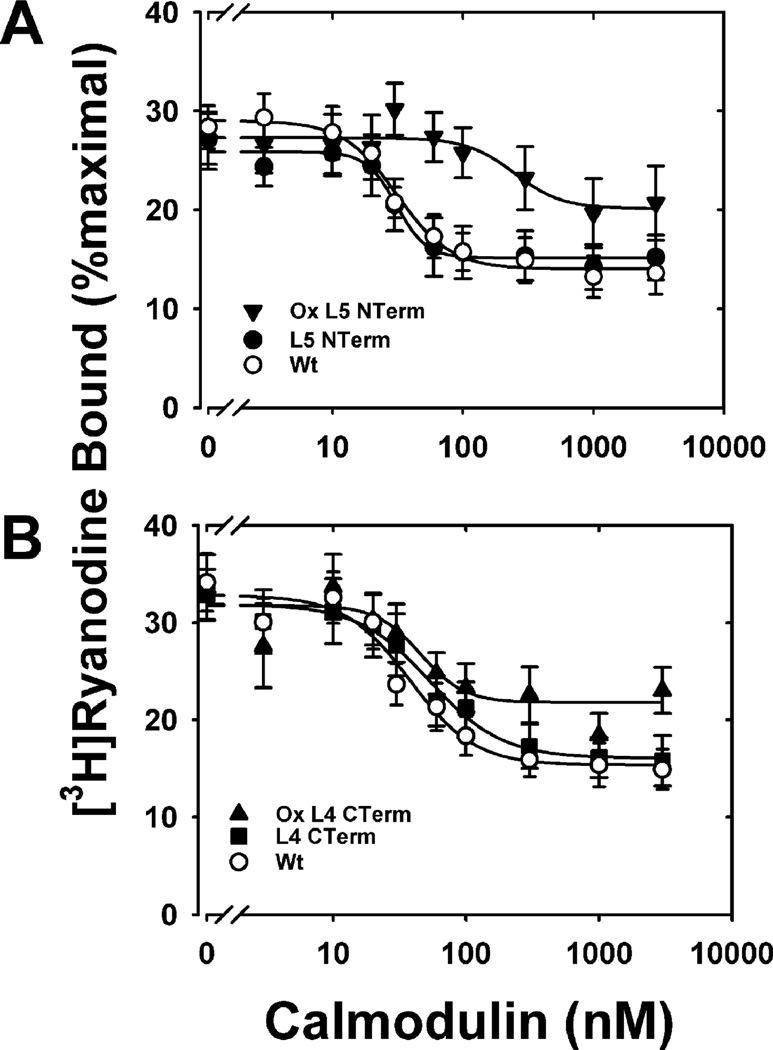
We have previously shown that oxidation of all nine Met residues abolished CaM regulation of RyR1 (24). Here we show that while the mutations had no or only minor functional effects on the extent of inhibition of ryanodine binding, the concentration of CaM required to achieve half-inhibition of ryanodine binding (IC50) or the Hill coefficient (ni) of ryanodine binding (Figures 6 and 7, Table 2), oxidation of the PEST-region mutants, L3 PEST and L5 NTerm, causes significant functional impairment for CaM regulation of RyR1. Oxidation of L3 PEST, containing three Met to Leu substitutions within the putative PEST sequence, completely abolished CaM inhibition of ryanodine binding (Figure 6). Oxidation of L5 NTerm, where all N-terminal Met are replaced by Leu and the C-terminal Met are oxidized, also exhibited a loss in function as demonstrated by the decreased extent of inhibition and an 8-fold increase in IC50 (Figure 7, Table 2). Compared to the effects of oxidizing PEST-region mutants, oxidizing non-PEST-region mutants, L6 NP and L4 CTerm, had relatively minor effects on CaM inhibition of RyR1. Specific oxidation of the Met residues within the PEST sequence (ox L6 NP) did not alter the extent of inhibition, the Hill coefficient, or the IC50 when compared to the unoxidized mutant (L6 NP) (Table 2, Figure 6). However, oxidation of L6 NP did cause a decrease in the extent of inhibition compared to WT CaM. Similarly, oxidation of all of the N-terminal Met (L4 CTerm) did not alter any of the Hill equation parameters when compared to the unoxidized mutant but did cause a significant decrease in the extent of inhibition compare to WT CaM. These results suggest that oxidation of the C-terminal Met residues (i.e., PEST-region mutants) has a greater negative impact on CaM regulation of the RyR1 than oxidation of Met residues in the N-terminus (i.e., non-PEST-region mutants). Since oxidation of C-terminal Met does not signal proteasome degradation (Figure 3), accumulation of this CaM oxiform within the cell has the potential to adversely affect CaM regulation of RyR1 and calcium homeostasis.
Figure 6.
Inhibition of skeletal muscle RyR1 [3H]ryanodine binding for WT and mutant L3PEST and L6NP CaM: (A) nonoxidized WT (○), nonoxidized L3PEST (●), and oxidized L3PEST (▼); (B) nonoxidized WT (○), nonoxidized L6 NP (■), and oxidized L6 NP (▲). RyR1 ryanodine binding was performed as described under Experimental Procedures in medium containing 700 µM Ca2+. Solid lines represent fits to a four-parameter Hill equation. Data are the mean ± SEM of six to ten experiments performed in duplicate. Table 2 reports the kinetic parameters derived from these fits, i.e., extent of inhibition of ryanodine binding, the concentration of CaM required to achieve half-inhibition of ryanodine binding (IC50), or the Hill coefficient (ni) of ryanodine binding for each fit.
Figure 7.
Inhibition of skeletal muscle RyR1 [3H]ryanodine binding for WT and mutant L5 NTerm and L4 CTerm CaM: (A) nonoxidized WT CaM (○), nonoxidized L5 NTerm CaM (●), and oxidized L5 NTerm CaM (▼); (B) nonoxidized WT CaM (○), nonoxidized L4 CTerm CaM (■), and oxidized L4 CTerm CaM (▲). RyR1 ryanodine binding was performed as described under Experimental Procedures in medium containing 700 µM Ca2+. Solid lines represent fits to a four-parameter Hill equation. Data are the mean ± SEM of six to ten experiments performed in duplicate. Table 2 reports the kinetic parameters derived from these fits, i.e., extent of inhibition of ryanodine binding, the concentration of CaM required to achieve half-inhibition of ryanodine binding (IC50), or the Hill coefficient (ni) of ryanodine binding for each fit.
Table 2.
Hill Equation Parameters for Fits to Calmodulin Concentration Dependence of Inhibition of Sarcoplasmic Reticulum Vesicle [3H]Ryanodine Binding
| wild type | L3 PEST | L6 NP | oxidized L6 NP | wild type | L5 NTerm | oxidized L5 NTerm | wild type | L4 CTerm | oxidized L4 CTerm | |
|---|---|---|---|---|---|---|---|---|---|---|
| inhibition (% maximal binding) | 12.1 ± 1.0 | 9.4 ± 1.2 | 9.2 ± 0.7 | 7.2 ± 1.4a | 15.0 ± 0.8 | 10.7 ± 0.8a | 7.2 ± 1.4a,b | 17.5 ± 1.7 | 15.8 ± 1.1 | 10.3 ± 2.3a |
| nic | −1.99 ± 0.53 | −2.84 ± 1.33 | −3.18 ± 0.93 | −2.12 ± 1.41 | −2.10 ± 0.38 | −3.77 ± 1.34 | −2.37 ± 2.09 | −1.78 ± 0.53 | −1.6 ± 0.34 | −2.39 ± 1.8 |
| IC50c(nM) | 27.8 ± 4.0 | 35.8 ± 7.5 | 37.8 ± 4.7 | 20.5 ± 6.8 | 31.3 ± 3.0 | 30.9 ± 3.0 | 248.3 ± 96.1a,b | 37.9 ± 7.4 | 53.7 ± 7.8 | 47.5 ± 18.8 |
| nc | 4 | 4 | 4 | 4 | 6 | 6 | 6 | 10 | 10 | 10 |
Significantly different (p < 0.05) from wild type.
Significantly different (p < 0.05) from unoxidized.
ni, the Hill coefficient of ryanodine binding; IC50, the concentration of CaM required to achieve half-inhibition of ryanodine binding; n, number of separate experiments performed.
DISCUSSION
System of Site-Specific Oxidation
The current study utilized site-directed mutagenesis coupled with site-selective oxidation of Met to identify residues that are critical for oxidation-induced degradation by the 20S proteasome. Mutation of Met to Leu is considered a conservative substitution based on data from evolution and the physicochemical properties of the two amino acids. Throughout evolution, Leu is the most common substitution for Met (37). For example, in comparing the CaM sequence from Saccharomyces cerevisiae and mammals, only three of the nine Met are retained in CaM from yeast; the remaining six residues contain Leu substitutions (Figure 8A). Considering the physicochemical properties, Leu is slightly more hydrophobic than Met, but both amino acids have similar volumes and propensity to form α-helices, which is important considering the abundance of α-helices in CaM (35, 38). Data from the current study is also consistent with the conservative nature of the Met to Leu substitutions; neither secondary structure nor hydrophobicity was substantially altered in CaM mutants containing three to five5 substitutions (Table 1).
Figure 8.
Alignment of CaM sequences. (A) Comparison of amino acids corresponding with rat CaM Met residues shows complete conservation of Met 36 and Met 72 among multiple species. Identity is the percent of amino acids in the entire sequence that are identical with rat CaM. In yeast, there was a Leu in position 51 and a Met in position 52. (B) Alignment of CaM sequences that correspond with the region containing the putative degron. Identical amino acids and conservative substitutions are highlighted in dark and light gray, respectively.
Model of Protein Degradation by the Proteasome
Recent evidence suggests that signals for substrate recognition by both the 26S and 20S proteasomes are often bipartite, involving binding of an unstructured region coupled with secondary interactions that tether the substrate to the proteasome (1, 39, 40). An additional element of the recognition signal is the presence of a sequence within the unstructured region, a degron, that is recognized by the proteasome (39). Since the degron is part of the structured region in native protein, it can only become accessible after a posttranslational modification, such as phosphorylation or oxidation, causes regional unfolding. Tethering the substrate to the proteasome can be accomplished via ubiquitin, which is recognized by subunits in PA700 (41). Other mechanisms for tethering substrates to either the 26S and 20S proteasomes are the ATPase subunits of PA700 and adaptor molecules that bind to the 20S, such as HSP90 or NAD(P)H quinone oxidoreductase 1, that recognize unfolded proteins or specific protein substrates (11, 42, 43). Recent studies suggest that, even in the absence of adaptor molecules, proteins containing disordered regions can bind to and be degraded by the 20S proteasome (44). Taken together, these studies suggest the critical element for initiating degradation is the engagement of an unstructured region of the substrate with either the 20S or 26S proteasome.
Model of CaM Degradation by the 20S Proteasome
Results from the current study show that preventing oxidation of three to five Met in the N-terminus via Leu substitutions produces mutants that are resistant to degradation (Figure 3, PEST-region mutants). Conversely, the unique oxidation of three to five N-terminal Met elicits rapid proteolysis by the 20S proteasome (Figure 3). These data suggest that the N-terminus contains a degradation signal that is revealed by site-specific oxidation of N-terminal Met. As a caveat, our in vitro results were obtained in the absence of the cadre of cellular proteins that could influence CaM proteolysis. Thus, these results may reflect an oversimplified view of CaM degradation by the proteasome. On the other hand, these in vitro experiments bring new biochemical and structural clarity to specific regions within CaM that contain a potential degron, thus providing important guidance for future experiments in cultured cells where the in vivo conditions are more closely aligned.
As a general model for CaM degradation, we propose that an essential step in eliciting digestion by the proteasome is regional unfolding of the N-terminus caused by events such as oxidation of N-terminal Met (Figure 9). The unstructured region, along with a cryptic sequence that becomes solvent exposed, serves as the recognition element for binding to the 20S proteasome and/or adaptor proteins, such as HSP90. Degradation of the N-terminus occurs following translocation of either an unstructured loop (endoproteolysis) or the free N-terminus into the catalytic chamber. As shown previously, the initial products of degradation are peptides derived from the N-terminus (16, 19, 20).
Figure 9.
Model for CaM degradation by the 20S proteasome. CaM ribbon structure is shown with Met residues indicated as gray balls. The 20S proteasome is represented as a cylinder of four seven-membered rings of subunits. CaM degradation is initiated by events, such as oxidation of N-terminal Met, that cause regional unfolding in the N-terminus (1). The degron, consisting of the disordered N-terminus plus a cryptic degradation sequence, is recognized and binds to the 20S and/or HSP90 (dark oval) (2). Degradation of the N-terminus occurs following translocation of either an unstructured loop (endoproteolysis) (3) or the free N-terminus (4) into the catalytic chamber. N-Terminal peptides are initially released (5).
In our preparations of proteasome from liver, HSP90 is tightly associated and copurifies with the 20S catalytic core (16, 18). Previous reports have demonstrated an essential role for HSP90 in the degradation of oxidized CaM by the 20S proteasome (42). Note that while we did not specifically test for mutant-specific differences in HSP90 interaction, this factor could potentially affect rates of degradation. Another factor that could influence the rate of degradation is conformational stability since less stable proteins may be more easily unfolded and rapidly degraded. We tested stability under conditions of heat denaturation (Figure 5) and found similar conformational stability for oxidized mutants containing either Leu substitutions for Met 51, 71, and 72 (L3 PEST) or Leu substitutions at all positions except Met 51, 71, and 72 (L6 NP). Oxidized L6 NP is rapidly degraded, but oxidized L3 PEST resists proteasome degradation. Taken together, these results suggest that altered conformational stability does not explain the difference in susceptibility to proteasome degradation. These results also support the hypothesis that oxidation-induced perturbation of the region localized to residues 51–72 triggers proteasome proteolysis.
Sequence alignment of CaM from multiple species shows complete conservation of Met 36 and 72 (Figure 8A) even between divergent species, i.e., rat, wheat, and Caenorhabditis elegans. With the exception of C. elegans, Met 51 is also highly conserved among different species. The evolutionary conservation suggests that these residues are involved in critical functions. In addition to the well-described function of Met in providing surface contact with target proteins (17), data from the current study suggest that oxidation of one or more of these residues may also be an essential component for triggering proteasome degradation.
Besides oxidation of N-terminal Met, mutations or oxidation of other sites in the protein that cause conformational rearrangement could also elicit proteasome proteolysis. In the current study, mutation of all nine Met to Leu (L9) resulted in a loss of secondary structure (Table 1) that correlated with increased proteasome degradation (Figures 3 and 4). These results are consistent with a previous report of increased susceptibility of the L9 CaM mutant to proteasome degradation (19). Notably, mutation or oxidation of a single Met in the C-terminus can also elicit rapid proteasome proteolysis. Squier and colleagues showed that either mutation or oxidation of Met145 caused global tertiary structural rearrangements that correlated with rapid proteolysis by the proteasome (19, 45). Thus, it is possible that the conformational changes associated with altering Met145 cause regional unfolding and exposure of the recognition elements in the N-terminus that initiate proteasome proteolysis.
The unstructured region is often part of a bipartite recognition element that forms in conjunction with other residues that occur nearby in the primary sequence. Some degrons have sequences that are unique to specific proteins. For example, the degron specific for ornithine decarboxylase contains a structural motif that consists of the Cys441-Ala442 recognition element plus an unstructured C-terminus (8). The degron for cFos includes the tripeptide motif Pro-Thr-Leu plus an additional undescribed structural element (9, 46). Notably, the “Pro-Thr-Leu” signal is unique to cFos since other protein family members contain a similar sequence that does not elicit degradation.
The PEST motif, which is rich in Pro, Glu, Asp, Ser, and Thr, is a well-known degron that is found in many proteins. While historically PEST was first described in proteins with short half-lives, our expanded understanding of protein degradation suggests that PEST is not limited to these proteins. For example, myosin heavy chain from rat heart has a half-life of 5.9 days (47) and contains a potential PEST sequence (PEST score +19.32) encompassing residues 369–382. Reported half-lives of CaM in rat muscle and brain are 10 and 18 h, respectively (48, 49). These half-lives are short compared with those of the sarcoplasmic reticulum Ca-ATPase (14 days) and plasma membrane Ca-ATPase (12 days) reported from rat muscle and brain (49, 50). Thus, CaM may qualify as a “short-lived protein” when compared with other calcium regulatory proteins in the same tissue.
Although analysis of the CaM sequence using the PEST algorithm indicated CaM residues 36–72 as a weak PEST signal (Figure 1), substitution of Gln for Met to mimic oxidation improved the likelihood of a PEST motif. Sequence alignment of CaM from divergent species shows considerable identity (Figure 8B), providing further support that this region contains a sequence that is part of the recognition element for proteasome degradation. It is also possible that a CaM-specific sequence in the N-terminus, such as the highly conserved residues 58DGNxxIDFxEF68, could serve as the degron.
One model of proteolytic digestion of substrates involves the translocation of the unstructured N- or C-termini into the catalytic chamber where the protein is fully degraded into peptides (46, 51). A second model involves endoproteolysis, where a disordered polypeptide loop is translocated into the catalytic chamber, resulting in partial digestion of the substrate (45, 52). This is the putative mechanism utilized for activation of transcription factors, such as the processing of the active nuclear factor κ B transcription factor p50 from the inactive precursor p105 (52). Another factor that influences the extent and rate of degradation includes the secondary structure of the region directly adjacent to the degradation signal. For example, proteins are readily digested when the degradation signal is next to a region containing either α-helices or irregular loops (53). Conversely, regions containing highly stable secondary structure, such as β-sheet, resist degradation (53).
With CaM, the N-terminal globular domain is followed by a flexible linker region (residues 75–86) consisting of an eight-turn α-helix that tethers the two opposing domains (17). Biophysical measures of the solution structure of CaM report that the central linker exhibits considerable flexible and can adopt a nonhelical conformation upon binding many of its target proteins (17). Thus, the presence of a favorable secondary structure adjacent to the putative degron that is exposed upon oxidation of the N-terminal Met may promote rapid CaM digestion. Additionally, the flexible central linker region could permit endoproteolysis of CaM when conditions cause disorder in this region. Indeed, peptides generated from proteolysis of oxidized WT CaM by the 20S proteasome are consistent with both endoproteolysis and N-terminal degradation. Supporting endoproteolysis of oxidized CaM are the release of large N-terminal domain fragments (A1-F92 and A1-K30) (19, 20) and multiple peptides originating from the flexible linker region (20). Consistent with proteolytic digestion of the free N-terminus, data from our previous work (16) showed numerous peptides originating mainly from the N-terminal domain were produced after 1 h digestion of oxidized WT CaM. These differences in peptides released by the 20S proteasome could depend on the region and extent of CaM structural unfolding or could be due to differences in the adaptor molecules that copurify with the 20S proteasome. Nonetheless, these data support the hypothesis that the site of recognition and initial degradation is in CaM’s N-terminus.
Physiological Relevance of CaM Oxidation and Proteasome Proteolysis
The binding of up to four calcium ions to CaM elicits significant conformational changes that increase the exposure of hydrophobic residues, e.g., Met, that serve as binding sites for target proteins (17). This conformational flexibility allows CaM to regulate over 100 different proteins in a wide variety of configurations (17). Additionally, the extent of calcium (Ca2+) saturation and location of bound Ca2+ ions also influence CaM’s binding to targets. For example, the Ca2+-loaded N-lobe of CaM binds to a peptide of the Ca2+-activated small conductance potassium channel while the Ca2+-free C-lobe binds to a noncontiguous region of the target (54). Conversely, CaM binds anthrax adenylyl cylcase endotoxin via a Ca2+-loaded C-lobe and a Ca2+-free N-lobe (55). Thus, CaM binding to specific targets can alter the solvent exposure of select Met residues. Because solvent accessibility is a major determinant of susceptibility to oxidation, the specific Met residues subjected to oxidative modification may vary depending on the cellular conditions that determine intracellular calcium levels and, subsequently, CaM binding partners.
Data from the current study (Figures 6 and 7) and others (56–58) have shown that oxidation of specific Met can significantly alter CaM’s ability to regulate protein targets. Therefore, efficient removal of oxidized CaM is critical for maintaining cell signaling. In the case of RyR1, oxidation of CaM’s C-terminal Met was more deleterious to regulation of the channel than oxidation of CaM’s N-terminal Met (Figures 6 and 7). Unless C-terminal oxidized CaM is removed from the cell, cell signaling would be significantly impaired.
In the current study, data support the presence of a degron in the N-terminus that is revealed by oxidation of N-terminal Met (Figure 3). The presence of an additional degron in CaM’s C-terminus has also been proposed (19). Squier and colleagues (19) reported that oxidation of Met145 induced an alteration in CaM’s tertiary structure that triggered degradation. In the present study, oxidation of multiple C-terminal Met (L6 CTNPP), including Met 145, did not increase the rate of CaM degradation (Figure 3). It is possible that under our experimental conditions, i.e., oxidation of multiple C-terminal Met and a lower free calcium concentration (~10 versus 100 µM), the C-terminal degron is not exposed. Therefore, we cannot rule out the possibility that CaM contains degrons in both the N- and C-terminus. Multiple degrons would be advantageous considering the potential for site-selective oxidation that occurs when CaM is complexed with specific proteins.
The extent of CaM methionine oxidation in vivo has been estimated at ~two Met sulfoxide per CaM in both young and aged rat muscle (59) and in the brain of aged rats (60). In brain, the in vivo sites of modification were defined by HPLC separation of trypsin-generated peptides followed by mass spectrometry (60). In both young and aged brains, Met sulfoxide at position 76 was detected, suggesting that this site is readily oxidized in vivo and is likely not a signal for degradation. In aged brain, there was a general increase in oxidation of Met based on the appearance of multiple tryptic peptides containing Met sulfoxide. However, abundantly present in aged brain were two tryptic peptides containing a single added oxygen that corresponded with oxidation of Met 36 and oxidation of either Met 51, 71, or 72. The presence of oxidized Met in the putative CaM degron coupled with decreased proteasome activity reported in aged rat brain (61) suggests aging is accompanied by defects in proteasome-dependent removal of damaged CaM, which could lead to an in vivo loss in CaM signaling. In support of this idea, the age-dependent increase in Met sulfoxide content correlated with a reduction in the ability of CaM isolated from aged brain to activate the plasma membrane Ca-ATPase, an in vivo CaM target (60). Hence, even limited Met oxidation can alter the ability of CaM to regulate target proteins, further supporting an essential role for proteasome in maintaining cell signaling via the efficient removal of oxidatively damaged proteins.
CONCLUSION
We have used site-directed Met oxidation, followed by measurement of proteasome susceptibility and circular dichroism, to define both the chemical and structural features of the degron, the portion of CaM that is involved in targeting proteasome degradation. These results are important not only in providing the molecular details of CaM as a proteasome substrate but also in furthering our fundamental understanding of proteasome action on oxidized protein substrates.
ACKNOWLEDGMENT
We acknowledge the Mass Spectrometry Consortium for the Life Sciences, University of Minnesota, for the assistance in acquisition and interpretation of mass spectral data.
Footnotes
The work was supported by National Institutes of Health Grants EY013623, AG032391 (D.A.F.), and AG025392 (D.D.T.), the Fesler-Lampert Foundation, and an unrestricted grant to the Department of Ophthalmology from the Research to Prevent Blindness Foundation.
Abbreviations: CaM, calmodulin; ANS, 1-anilinonaphthalene-8-sulfonate; BCA, bicinchoninic acid; AMPPNP, adenosine 5′-(β,γ-imido)triphosphate tetralithium salt; HPLC, high-performance liquid chromatography; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; Tris, 2-amino-2-(hydroxymethyl)propane-1,3-diol; WT, wild-type CaM; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; MALDI-TOF, matrix-assisted laser desorption ionization time of flight; HOMOPIPES, homopiperazine-1,4-bis(2-ethanesulfonic acid); HSR, heavy sarcoplasmic reticulum; PIPES, 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Asher G, Reuven N, Shaul Y. 20S proteasomes and protein degradation “by default”. BioEssays. 2006;28:844–849. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 3.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 5.Wang T. The 26S proteasome system in the signaling pathways of TGF-beta superfamily. Front. Biosci. 2003;8:d1109–d1127. doi: 10.2741/1057. [DOI] [PubMed] [Google Scholar]
- 6.Jesenberger V, Jentsch S. Deadly encounter: ubiquitin meets apoptosis. Nat. Rev. Mol. Cell. Biol. 2002;3:112–121. doi: 10.1038/nrm731. [DOI] [PubMed] [Google Scholar]
- 7.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi J, Chen H, Hoyt MA, Coffino P. Structural elements of the ubiquitin-independent proteasome degron of ornithine decarboxylase. Biochem. J. 2008;410:401–407. doi: 10.1042/BJ20071239. [DOI] [PubMed] [Google Scholar]
- 9.Acquaviva C, Brockly F, Ferrara P, Bossis G, Salvat C, Jariel-Encontre I, Piechaczyk M. Identification of a C-terminal tripeptide motif involved in the control of rapid proteasomal degradation of c-Fos proto-oncoprotein during the G(0)-to-S phase transition. Oncogene. 2001;20:7563–7572. doi: 10.1038/sj.onc.1204880. [DOI] [PubMed] [Google Scholar]
- 10.Tarcsa E, Szymanska G, Lecker S, O’Connor CM, Goldberg AL. Ca2+-free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26 S proteasomes without ubiquitination. J. Biol. Chem. 2000;275:20295–20301. doi: 10.1074/jbc.M001555200. [DOI] [PubMed] [Google Scholar]
- 11.Benaroudj N, Tarcsa E, Cascio P, Goldberg AL. The unfolding of substrates and ubiquitin-independent protein degradation by proteasomes. Biochimie. 2001;83:311–318. doi: 10.1016/s0300-9084(01)01244-5. [DOI] [PubMed] [Google Scholar]
- 12.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem. J. 1998;335(Part 3):637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacifici RE, Kono Y, Davies KJ. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1993;268:15405–15411. [PubMed] [Google Scholar]
- 14.Giulivi C, Pacifici RE, Davies KJ. Exposure of hydrophobic moieties promotes the selective degradation of hydrogen peroxide-modified hemoglobin by the multicatalytic proteinase complex, proteasome. Arch. Biochem. Biophys. 1994;311:329–341. doi: 10.1006/abbi.1994.1245. [DOI] [PubMed] [Google Scholar]
- 15.Friguet B, Szweda LI, Stadtman ER. Susceptibility of glucose-6-phosphate dehydrogenase modified by 4-hydroxy-2-nonenal and metal-catalyzed oxidation to proteolysis by the multicatalytic protease. Arch. Biochem. Biophys. 1994;311:168–173. doi: 10.1006/abbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- 16.Ferrington DA, Sun H, Murray KK, Costa J, Williams TD, Bigelow DJ, Squier TC. Selective degradation of oxidized calmodulin by the 20 S proteasome. J. Biol. Chem. 2001;276:937–943. doi: 10.1074/jbc.M005356200. [DOI] [PubMed] [Google Scholar]
- 17.Yamniuk AP, Vogel HJ. Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 18.Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- 19.Sacksteder CA, Whittier JE, Xiong Y, Li J, Galeva NA, Jacoby ME, Purvine SO, Williams TD, Rechsteiner MC, Bigelow DJ, Squier TC. Tertiary structural rearrangements upon oxidation of methionine145 in calmodulin promotes targeted proteasomal degradation. Biophys. J. 2006;91:1480–1493. doi: 10.1529/biophysj.106.086033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strosova M, Voss P, Engels M, Horakova L, Grune T. Limited degradation of oxidized calmodulin by proteasome: formation of peptides. Arch. Biochem. Biophys. 2008;475:50–54. doi: 10.1016/j.abb.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Gopalakrishna R, Anderson WB. Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem. Biophys. Res. Commun. 1982;104:830–836. doi: 10.1016/0006-291x(82)90712-4. [DOI] [PubMed] [Google Scholar]
- 22.Strasburg GM, Hogan M, Birmachu W, Thomas DD, Louis CF. Site-specific derivatives of wheat germ calmodulin. Interactions with troponin and sarcoplasmic reticulum. J. Biol. Chem. 1988;263:542–548. [PubMed] [Google Scholar]
- 23.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 24.Saxena VP, Wetlaufer DB. A new basis for interpreting the circular dichroic spectra of proteins. Proc. Natl. Acad. Sci. U.S.A. 1971;68:969–972. doi: 10.1073/pnas.68.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenfield N, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 26.Kretsinger RH, Rudnick SE, Weissman LJ. Crystal structure of calmodulin. J. Inorg. Biochem. 1986;28:289–302. doi: 10.1016/0162-0134(86)80093-9. [DOI] [PubMed] [Google Scholar]
- 27.Balog EM, Norton LE, Bloomquist RA, Cornea RL, Black DJ, Louis CF, Thomas DD, Fruen BR. Calmodulin oxidation and methionine to glutamine substitutions reveal methionine residues critical for functional interaction with ryanodine receptor-1. J. Biol. Chem. 2003;278:15615–15621. doi: 10.1074/jbc.M209180200. [DOI] [PubMed] [Google Scholar]
- 28.Chu A, Diaz-Munoz M, Hawkes MJ, Brush K, Hamilton SL. Ryanodine as a probe for the functional state of the skeletal muscle sarcoplasmic reticulum calcium release channel. Mol. Pharmacol. 1990;37:735–741. [PubMed] [Google Scholar]
- 29.Meissner G, el-Hashem A. Ryanodine as a functional probe of the skeletal muscle sarcoplasmic reticulum Ca2+ release channel. Mol. Cell. Biochem. 1992;114:119–123. doi: 10.1007/BF00240306. [DOI] [PubMed] [Google Scholar]
- 30.Brooks SP, Storey KB. Bound and determined: a computer program for making buffers of defined ion concentrations. Anal. Biochem. 1992;201:119–126. doi: 10.1016/0003-2697(92)90183-8. [DOI] [PubMed] [Google Scholar]
- 31.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 32.Cardamone M, Puri NK. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 1992;282(Part 2):589–593. doi: 10.1042/bj2820589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Neil KT, DeGrado WF. A predicted structure of calmodulin suggests an electrostatic basis for its function. Proc. Natl. Acad. Sci. U.S.A. 1985;82:4954–4958. doi: 10.1073/pnas.82.15.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Neil KT, DeGrado WF. How calmodulin binds its targets: sequence independent recognition of amphiphilic alphahelices. Trends Biochem. Sci. 1990;15:59–64. doi: 10.1016/0968-0004(90)90177-d. [DOI] [PubMed] [Google Scholar]
- 35.Gellman SH. On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry. 1991;30:6633–6636. doi: 10.1021/bi00241a001. [DOI] [PubMed] [Google Scholar]
- 36.Masino L, Martin SR, Bayley PM. Ligand binding and thermodynamic stability of a multidomain protein, calmodulin. Protein Sci. 2000;9:1519–1529. doi: 10.1110/ps.9.8.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dayhoff MO. Atlas of Protein Sequence and Structure. Vol. 5. Washington, DC: National Biomedical Research Foundation; 1978. [Google Scholar]
- 38.Sharp KA, Nicholls A, Friedman R, Honig B. Extracting hydrophobic free energies from experimental data: relationship to protein folding and theoretical models. Biochemistry. 1991;30:9686–9697. doi: 10.1021/bi00104a017. [DOI] [PubMed] [Google Scholar]
- 39.Kenniston JA, Sauer RT. Signaling degradation. Nat. Struct. Mol. Biol. 2004;11:800–802. doi: 10.1038/nsmb0904-800. [DOI] [PubMed] [Google Scholar]
- 40.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 41.Janse DM, Crosas B, Finley D, Church GM. Localization to the proteasome is sufficient for degradation. J. Biol. Chem. 2004;279:21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 42.Whittier JE, Xiong Y, Rechsteiner MC, Squier TC. Hsp90 enhances degradation of oxidized calmodulin by the 20 S proteasome. J. Biol. Chem. 2004;279:46135–46142. doi: 10.1074/jbc.M406048200. [DOI] [PubMed] [Google Scholar]
- 43.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin D, Sun H, Ferrington DA, Squier TC. Closer proximity between opposing domains of vertebrate calmodulin following deletion of Met(145)-Lys(148) Biochemistry. 2000;39:10255–10268. doi: 10.1021/bi000949y. [DOI] [PubMed] [Google Scholar]
- 46.Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol. Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 47.Zak R, Martin AF, Prior G, Rabinowitz M. Comparison of turnover of several myofibrillar proteins and critical evaluation of double isotope method. J. Biol. Chem. 1977;252:3430–3435. [PubMed] [Google Scholar]
- 48.Peterson-von Gehr JaJ, HP Abstr. Commun. 12 Int. Congr. Biochem; 1982. [Google Scholar]
- 49.Ferrington DA, Chen X, Krainev AG, Michaelis EK, Bigelow DJ. Protein half-lives of calmodulin and the plasma membrane Ca-ATPase in rat brain. Biochem. Biophys. Res. Commun. 1997;237:163–165. doi: 10.1006/bbrc.1997.7105. [DOI] [PubMed] [Google Scholar]
- 50.Ferrington DA, Krainev AG, Bigelow DJ. Altered turnover of calcium regulatory proteins of the sarcoplasmic reticulum in aged skeletal muscle. J. Biol. Chem. 1998;273:5885–5891. doi: 10.1074/jbc.273.10.5885. [DOI] [PubMed] [Google Scholar]
- 51.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 52.Rape M, Jentsch S. Taking a bite: proteasomal protein processing. Nat. Cell Biol. 2002;4:E113–E116. doi: 10.1038/ncb0502-e113. [DOI] [PubMed] [Google Scholar]
- 53.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 54.Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 55.Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, Grabarek Z, Bohm A, Tang WJ. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 56.Balog EM, Norton LE, Thomas DD, Fruen BR. Role of calmodulin methionine residues in mediating productive association with cardiac ryanodine receptors. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H794–H799. doi: 10.1152/ajpheart.00706.2005. [DOI] [PubMed] [Google Scholar]
- 57.Yao Y, Yin D, Jas GS, Kuczer K, Williams TD, Schoneich C, Squier TC. Oxidative modification of a carboxylterminal vicinal methionine in calmodulin by hydrogen peroxide inhibits calmodulin-dependent activation of the plasma membrane Ca-ATPase. Biochemistry. 1996;35:2767–2787. doi: 10.1021/bi951712i. [DOI] [PubMed] [Google Scholar]
- 58.Bartlett RK, Bieber Urbauer RJ, Anbanandam A, Smallwood HS, Urbauer JL, Squier TC. Oxidation of Met144 and Met145 in calmodulin blocks calmodulin dependent activation of the plasma membrane Ca-ATPase. Biochemistry. 2003;42:3231–3238. doi: 10.1021/bi026956z. [DOI] [PubMed] [Google Scholar]
- 59.Boschek CB, Jones TE, Smallwood HS, Squier TC, Bigelow DJ. Loss of the calmodulin-dependent inhibition of the RyR1 calcium release channel upon oxidation of methionines in calmodulin. Biochemistry. 2008;47:131–142. doi: 10.1021/bi701352w. [DOI] [PubMed] [Google Scholar]
- 60.Gao J, Yin D, Yao Y, Williams TD, Squier TC. Progressive decline in the ability of calmodulin isolated from aged brain to activate the plasma membrane Ca-ATPase. Biochemistry. 1998;37:9536–9548. doi: 10.1021/bi9803877. [DOI] [PubMed] [Google Scholar]
- 61.Keller JN, Hanni KB, Markesbery WR. Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech. Ageing Dev. 2000;113:61–70. doi: 10.1016/s0047-6374(99)00101-3. [DOI] [PubMed] [Google Scholar]