Abstract
Type 2 diabetes is highly prevalent and is the major cause of progressive chronic kidney disease in American Indians. Genome wide association studies identified several loci associated with diabetes but their impact on susceptibility to diabetic complications is unknown. To measure this we studied the association of 18 type 2 diabetes genome wide association single nucleotide polymorphisms (SNPs) with the estimated glomerular filtration rate (eGFR) (MDRD equation) and urine albumin to creatinine ratio in 6,958 individuals in the Strong Heart Study family and cohort participants. Center specific residuals of eGFR and the log urine albumin to creatinine ratio, obtained from linear regression models adjusted for age, sex and body mass index, were regressed onto SNP dosage using variance component in family data and linear regression models in unrelated individuals. Estimates were then combined across centers. Four diabetic loci were associated with eGFR and one locus with the urine albumin to creatinine ratio. A SNP in the WFS1 gene (rs10010131) was associated with higher eGFR in younger individuals and with increased albuminuria. The SNPs of the FTO, KCNJ11 and TCF7L2 genes were associated with lower eGFR, not albuminuria, and were not significant in prospective analyses. Our findings suggest a shared genetic risk for type 2 diabetes, its kidney complications, and a potential role for WFS1 in early onset diabetic nephropathy in American Indian populations.
Introduction
American Indians have a high prevalence of obesity, type 2 diabetes (T2D) and metabolic syndrome. 1, 2 These conditions, together with hypertension, hyperlipidemia 3, 4 and smoking 5, 6 may contribute to an increased burden of chronic kidney disease (CKD), defined either by increased albuminuria or by decreased glomerular filtration rate (GFR).7-9 CKD is associated with increased cardiovascular disease events and death in American Indians. 10, 11 Therefore, understanding the pathways leading to CKD is relevant to public health.
T2D is one of the most common conditions associated with CKD and kidney failure in Western populations. 12 Diabetic nephropathy most typically progresses through an early phase in which microalbuminuria, detected by a spot urine albumin to creatinine ratio (UACR) ≥ 30 mg/g, is associated with supranormal glomerular filtration rate (GFR), i.e., glomerular hyperfiltration 13-15. These initial abnormalities may be reversible but then may progress to macroalbuminuria (UACR ≥300 mg/g) and an irreversible decline in GFR. Alternatively, in some T2D individuals, impaired GFR can occur without substantial albuminuria. 16 In the Strong Heart Study (SHS), American Indians with T2D often have micro or macroalbuminuria associated with decreased GFR. 17 Similarly, young Pima Indians with T2D, aged 5 to 19 years, frequently have microalbuminuria which is a strong predictor of macroalbuminuria at follow up. 18
Diabetic nephropathy occurs more often among family members 19 suggesting a genetic susceptibility to this complication. The heritability of kidney traits in American Indians has been documented for estimated GFR (eGFR) and UACR, a marker of kidney damage. 20-23 Genome-wide association studies (GWAS) have identified several loci associated with T2D (reviewed in 24). There is some evidence for associations of T2D loci with diabetic nephropathy including reported variants in PPARG in Chinese 25 and KCNQ1 in Japanese individuals. 26 In addition, TCF7L2 has been associated with diabetic nephropathy in individuals with early onset T2D 27 and among middle-aged individuals. 28 Given the large number of newly identified loci for T2D, the effects of genes associated with T2D on kidney function and damage merit exploration in American Indian populations.
We recently genotyped several GWAS validated single nucleotide polymorphisms (SNPs) in American Indians in the Genetic Epidemiology of Causal Variants Across the Life Course (CALiCo) Consortium as part of the Population Architecture using Genomics and Epidemiology (PAGE) study. 29 We describe associations of T2D SNPs with prevalent eGFR and UACR in over 6,900 American Indians from the Strong Heart Family Study (SHFS) and SHS cohort. We also explored the evidence for SNP associations with incident CKD and new-onset microalbuminuria using longitudinal data from the SHS participants.
Results
Baseline characteristics of the SHFS and SHS cohort American Indian samples are displayed in Table 1. By design, the mean age of SHFS participants was lower than that of the SHS cohort participants. The prevalence of T2D and impaired fasting glucose varied across centers but the mean body mass index (BMI) was similar when comparing cohort to family members for a same recruiting center. Cohort participants had lower mean eGFR and higher median UACR than family study participants. Eighteen SNPs representing 15 loci were examined. Minor allele frequencies (MAF) per center are shown in Supplemental Table 1. Two SNPs in each the FTO and the TCF7L2 genes were highly correlated in American Indians (r2>0.95 in the overall sample and within each center).
Table 1. Baseline characteristics of American Indians of the Strong Heart Family Study (1998-2003) and Strong Heart Study (1989-1991).
| Strong Heart Family Study | Strong Heart Study | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Dakota | Arizona | Oklahoma | Dakota | Arizona | Oklahoma | |
| Number | 1176 | 1165 | 1167 | 1188 | 1147 | 1115 |
| Mean age (SD), years | 38.4 (16.8) | 36.2 (15.5) | 43.4 (17.3) | 56.3 (8.1) | 55.8 (7.9) | 56.8 (8.4) |
| Female sex % | 58.7 | 62.1 | 59.3 | 56.8 | 63.8 | 57.9 |
| Type 2 diabetes % | 13.4 | 31.4 | 19.8 | 33.1 | 63.7 | 36.1 |
| Impaired fasting glucose % | 7.9 | 8.1 | 6.2 | 14.7 | 10.7 | 14.5 |
| Mean BMI (SD), kg/m2 | 30.1 (6.8) | 35.5 (8.8) | 31.1 (6.9) | 29.3 (5.5) | 32.5 (7.1) | 30.7 (6.0) |
| Mean eGFR (SD), ml/min/1.73 m2* | 96.6 (23.8) | 114.5 (28.9) | 93.6 (22.9) | 81.3 (20.3) | 86.1 (23.6) | 78.6 (17.5) |
| Median UACR (25-75%), mg/g | 7.1 (1.7-7,805.9) | 10.4 (1.7-9,756.1) | 7.1 (1.1-9,645.7) | 7.5 (0.1-7,493) | 27.1 (0.1-65,000.0) | 8.5 (0.04-12,419.8) |
SD, standard deviation; BMI, body mass index; eGFR, estimated glomerular filtration rate; UACR, urine albumin-to-creatinine ratio.
eGFR was estimated using the MDRD equation
Cross-sectional associations with eGFR and UACR in the SHS and SHFS samples
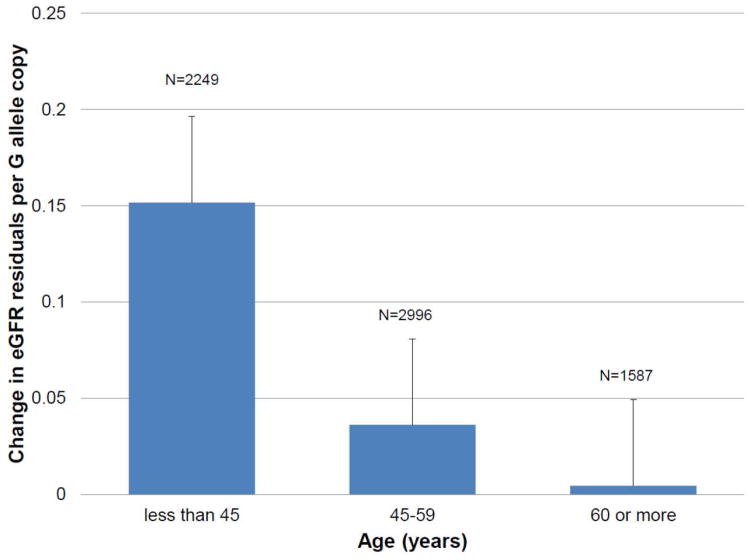
We identified significant associations of SNPs in FTO, TCF7L2, and KCNJ11 with eGFR (p<0.003) (Table 2). The FTO rs8050136 A allele and TCF7L2 rs7901695 C allele (both associated with increased risk of T2D) were associated with a 0.08 and 0.09 reduction of eGFR residuals per allele copy, respectively. The KCNJ11 rs5219 T allele, previously associated with increased T2D risk, was associated with 0.05 reductions in eGFR residuals per allele copy. We also identified nominal associations of WFS1 with eGFR and UACR. The WFS1 rs10010131 G allele was associated with increased eGFR and UACR but we noted a significant between study heterogeneity for the association with eGFR (p for heterogeneity=0.07). In center-specific analyses, we noted the lowest p-values in analyses of the SHFS compared to the older (>45 years) SHS participants (Table 2 footnote). We thus hypothesized age-specific effects on eGFR. We then performed analyses using age categories (<45, 45 to 59 and 60 years or more). The magnitude of effect of WFS1 rs10010131 G allele on eGFR was larger in American Indians <45 years-old (p=0.0003) compared to those 45 to 59 or ≥60 years, in whom the SNP effect was greatly attenuated (Figure 1, Supplemental Table 2, p for heterogeneity=0.05, I2 = 67.5%).
Table 2. Meta-analyses findings for type 2 diabetes-SNP associations for eGFR and log UACR residuals.
| Trait | SNP | Loci/gene | Coded Allele | Other Allele | β | SE | P-value | Number | P-value for heterogeneity | Direction of effect for coded allele |
|---|---|---|---|---|---|---|---|---|---|---|
| eGFR | rs8050136* | FTO | A | C | -0.078 | 0.0253 | 0.002 | 6835 | 0.13 | ↑BMI and T2D |
| rs9939609* | FTO | A | T | -0.0751 | 0.0252 | 0.003 | 6802 | 0.18 | ↑BMI and T2D | |
| rs5219 | KCNJ11 | T | C | -0.0543 | 0.0178 | 0.002 | 6807 | 0.91 | ↑T2D | |
| rs7901695* | TCF7L2 | C | T | -0.0884 | 0.0265 | 0.0008 | 6834 | 0.24 | ↑T2D | |
| rs7903146* | TCF7L2 | T | C | -0.0758 | 0.0269 | 0.005 | 6837 | 0.17 | ↑T2D | |
| rs10010131† | WFS1 | G | A | 0.0704 | 0.0252 | 0.005 | 6837 | 0.07 | ↑T2D | |
| UACR | ||||||||||
| rs10010131 | WFS1 | G | A | 0.0684 | 0.0263 | 0.009 | 6784 | 0.34 | ↑T2D |
SNP, single nucelotide polymorphism; SE, standard deviation; eGFR, estimated glomerular filtration rate; log UACR (urine albumin-to-creatinine ratio); T2D, type 2 diabetes
FTO variants (rs8050136 and rs9939609) and TCF7L2 variants (rs7901695 and rs7903146) are in linkage disequilibrium: r2>0.95 in the overall sample and within each center.
Estimates for the family study are: beta (se) = 0.12 (0.04), p=0.0008) and for cohort study beta (se) = -0.02 (0.04), p=0.63.
Figure 1.
Age-specific effects of WFS1 rs1001013 G allele on eGFR residuals. Numbers are 2249, 2996 and 1587 for age<45, 45 to 59 and 60 years or older.
To examine if these effects were mediated by diabetes, we also performed diabetes-stratified analyses (Supplemental Table 3) and analyses adjusting for T2D. Interestingly, TCF7L2 SNP association with eGFR was unchanged when adjusting for T2D. The overall T2D-adjusted associations of WFS1(p=0.08), KCNJ11 (p=0.72) and FTO (p=0.03) with eGFR were no longer significant but the age-specific association with eGFR for the WFS1SNP was mostly unchanged (p=0.0003 to 0.0008, n=2249, for association among individuals 45 years or younger). The association of rs10010131 with UACR was no longer significant when adjusting for T2D.
Longitudinal associations with CKD and microalbuminuria in the SHS cohort
In prospective data of SHS participants, incident (stage 3) CKD events were defined by the new occurrence of an eGFR of 60 ml/min/1.73 m2 or lower, or by self-report of maintenance dialysis or kidney transplantation among individuals with eGFR > 60 ml/min/1.73 m2. Approximately 477 CKD events occurred among 3,160 individuals at a median follow-up of 8 years. Of the 3,918 SHS participants with baseline eGFR ≥ 60 ml/min/1.73 m2, 1,035 with albuminuria at baseline were excluded for the analysis of incident albuminuria (approximately 499 events). A FTO SNP (rs9939609) was significantly associated with incident CKD in the Dakotas but not in the analyses of all three centers (Supplemental Table 4) and none of the loci was associated with new-onset microalbuminuria (Supplemental Table 5). However, the direction of the effect of the association with microalbuminuria was consistent with the findings for continuous UACR.
Discussion
CKD affects minorities disproportionally, who also progress more often to end-stage renal disease (ESRD) than individuals of European ancestry. 8, 9 American Indians have a high prevalence of obesity and T2D, which contribute to the large burden of cardiovascular and kidney diseases in this population. 7-9, 30 We investigated the genetic contribution of recently discovered T2D genetic variants to kidney function and markers of kidney damage in American Indians. The selected SNPs were initially identified in European and Asian ancestry studies and some of these variants have also been replicated for T2D associations in American Indian participants of the SHS (Haiman et al., Diabetes, under review). Several loci (FTO, KCNJ11, TCF7L2 and WFS1) were associated with eGFR and a SNP in WFS1 was associated with albuminuria. WFS1 product is a transmembrane protein localized to the endoplasmic reticulum (ER) 31 which is involved in ER stress signaling pathways. 32 The WFS1 SNP T2D risk allele was associated with increased albuminuria and eGFR, with a large effect on eGFR in American Indians younger than 45 years-old compared to those 45 to 59 years or older than 60 years, for whom the SNP effect was greatly attenuated. In analyses adjusted for both BMI and T2D, the association with eGFR was slightly changed in the younger age category but was no longer significant for UACR. These are interesting findings as American Indians develop diabetic nephropathy at earlier ages than individuals of other ancestry. 33 Early stages in the pathogenesis of the diabetic nephropathy are characterized by microalbuminuria and hyperfiltration. Our findings of increasing eGFR and albuminuria for WFS1, although modest, are consistent with the early development of nephropathy and suggest that the genetic effect is partially mediated by diabetes. Our findings will require further confirmation in American Indians and other populations, and in studies using direct measures of kidney function to detect higher ranges of GFR.
In contrast, the alleles associated with T2D risk for FTO, KCNJ11, TCF7L2 were associated with reduced eGFR in American Indians and were not associated with albuminuria. FTO is known to confer risk for T2D through its association with obesity 34 in both children and adults. 34, 35 Variants in this gene have been recently reported to be associated with end-stage renal disease in two case-control studies 36. Of interest, the associations with low eGFR in American Indians occurred in models adjusting for BMI suggesting an effect independent of body size, although the mean BMI in American Indians in our study was high and in the range of overweight to obese. The KCNJ11 gene was identified in T2D associations in individuals of European and Asian ancestry. 37-39 It impairs glucose-induced insulin release and it has also been associated with increased BMI 40, 41. It is intriguing that variants in both these loci are associated with lower eGFR in the context of obesity, which is a risk factor for CKD. 42 However, for both loci, the associations seem to be mediated by diabetes as they are no longer significant in analyses adjusted for T2D.
TCF7L2 has shown consistent associations with T2D across ancestry groups 43, 44 and, in American Indians, it has been associated with glucose homeostasis traits (unpublished data). TCF7L2 is a key component of the Wnt signaling pathway involved in the regulation of pancreatic beta-cell proliferation, differentiation and insulin secretion 45 but also important in kidney embryogenesis and disease. 46 The rs7903146 T allele was significantly associated with lower eGFR in American Indians but not with incident CKD in contrast to a previous report in a population-based study of middle-aged individuals. 28 Interestingly, a recent large study of T2D showed association of the rs7903146 T allele with diabetic nephropathy, with larger effect in individuals with early onset T2D.27 In our study, this genetic variant shows no evidence of heterogeneity across age categories. In addition, the association was slightly attenuated when adjusting for diabetes. Therefore, our findings suggest that at least four T2D loci are associated with kidney traits in American Indians of which two may have effects that are partially independent of diabetes. These findings may have important implications in prevention efforts for CKD in this population.
Our prospective associations with incident CKD and new onset-albuminuria were restricted to the SHS cohort study which has long-term longitudinal data. Although we noted significant associations for the FTO rs9939609 A allele in the Dakotas, our samples were limited. In addition, we did not identify associations with new-onset albuminuria for tested SNPs.
This study used kidney function estimates from an equation derived from CKD individuals. Calibrated serum creatinine concentrations are currently not available in the SHS. The SNPs investigated were identified in individuals of European ancestry. However, the FTO, WFS1 and TCF7L2 SNPs associations have replicated in American Indians for T2D or glycemic traits.
In summary, we identified four T2D loci associated with kidney function in American Indians including two previously reported associations with kidney traits in individuals of European ancestry. The WFS1 variant was associated with increased albuminuria and eGFR and it showed age-specific effects on eGFR. These findings suggest shared genetic risk factors for T2D and its kidney complications in this population.
Concise Methods
Population and phenotypes
The SHS is a population-based cohort study funded by the National Heart, Lung, and Blood Institute (NHLBI) of 4,549 American Indian men and women, ages 45-74 years and recruited from 13 tribes located in Arizona, North and South Dakota, and Oklahoma, who underwent a baseline (1989-1991) and then two follow-up (1993-1995 and 1996-1999) examinations. 47 The SHFS started in 1998 to study the genetics of cardiovascular disease among American Indian populations and it recruited family members from the original cohort study. 48 Over 3,600 American Indians aged 14 to 93 years and in 94 multigenerational families were examined, of whom 540 also are SHS cohort members (and were included in the family data only for analyses). The SHS and SHFS protocols were approved by the Indian Health Service Institutional Review Board, by the Institutional Review Boards of the participating Institutions, and by the Indian tribes participating in these studies. 47, 48 All participants gave informed consent for genetic testing.
Demographic data and medical history were obtained by a personal interview during a clinical exam. Anthropometric measures of body weight (kg) and height (m) were used to estimate BMI (kg/m2). Blood was collected after an overnight fast and plasma or serum samples were stored at −80°C until analyzed. T2D was defined as a fasting blood glucose of 126 mg/dl or higher, history of diabetes or use of diabetic medications. 49 Impaired fasting glucose and impaired glucose tolerance were defined by the World Health Organization criteria based on fasting plasma glucose and 75 gram oral glucose tolerance test results. Serum and urine creatinine were assayed by a kinetic alkaline picrate method and urine albumin by a sensitive nephelometric method, both on the Hitachi 717 platform (Roche Diagnostics, Indianapolis, IN). Urine albumin excretion was estimated as the UACR (mg/g) and eGFR was calculated using the Modified Diet and Renal Disease (MDRD) equation. 50
In prospective data of SHS participants, incident (stage 3) CKD events were defined by the new occurrence of an eGFR of 60 ml/min/1.73 m2 or lower, or by self-report of maintenance dialysis or kidney transplantation. CKD events included events that occurred during a median of 8 years follow-up between the baseline and visit 3 examinations in those individuals without a prior history of CKD (n=516 with CKD at baseline were excluded). New onset albuminuria was defined either as: 1. UACR ≥ 30 mg/g at follow-up among individuals with UACR< 30 mg/g at baseline or as 2. UACR ≥ 30 mg/g at follow-up among individuals with UACR< 30 mg/g at baseline and with a baseline eGFR ≥60 ml/min/1.73 m2. Of 4,549 SHS participants, 1,035 with albuminuria at baseline were excluded for the analysis of incident albuminuria by definition 1. Of 3,918 SHS participants with baseline eGFR ≥ 60 ml/min/1.73 m2, 1,313 with albuminuria at baseline were excluded for the incident albuminuria defined by definition 2. Because results were unchanged using definition 1 or 2, we reported only findings using definition 1.
SNP selection, genotyping and quality control
SNPs were selected from T2D GWAS publications prior to January 2009 and our analysis is thus restricted to these loci. Genotyping was performed at the University of Texas Health Science Center – Houston using Taqman assays (Applied Biosystems). Replica samples were included as controls. Quality control performed included sample call rates (>95%), concordance of blinded replicates (>98%), and deviation from Hardy-Weinberg equilibrium among founders (p<0.01). Individuals with more than 10% of missing genotypes (n=64) were excluded. Eighteen SNPs on 15 loci passed quality control and were included in these analyses (Supplemental Table 1). SNPs that failed genotyping were rs4430796 (HNF1b), rs2074196 (KCNQ1) and rs1326634 (SLC30A8). We estimated the linkage disequilibrium between SNPs in the same loci using Haploview.
Statistical Analysis
Cross-sectional analyses
Quantitative traits with non-normal distribution were natural log-transformed for analysis. We obtained center-specific residuals of eGFR and log UACR (LACR) using linear regression models adjusted for age, age2, sex, age and sex interactions and BMI. These residuals were then regressed onto SNP dosage using variance component models for family data to account for family relatedness and population history 51 and using linear regression for analysis of unrelated individuals (SHS). For SHS analyses, we also adjusted for self-reported blood quantum to account for ancestry. To account for age-specific effects, analyses were performed within categories of age (<45, 45 to 59 and 60 or older). We performed a quantitative transmission disequilibrium test 52 in the SHFS and found no evidence of population substructure within centers but significant evidence of population stratification when combining centers. Therefore analyses were stratified by center.
Summary estimates were combined across age-categories and centers using a weighted average of point estimates meta-analyses. We also tested for evidence of between-study heterogeneity 53 for age- and center-estimates by estimating the between-study variance and the I2 metric, which is a measure of the percentage of the total variation across strata due to heterogeneity rather than chance 54. Unadjusted p-values are reported. A Bonferroni correction for multiple testing p=0.003 (α=0.05/15 independent tests) was considered significant. In sensitivity analyses, we excluded 333 individuals younger than 18 years (range 15 to 17) from eGFR analyses and age-specific results were unchanged. We also performed analyses adjusting for T2D and stratified analyses by diabetes.
Longitudinal analyses
For significant SNP associations in cross-sectional analysis, we evaluated associations with incident CKD and increased albuminuria in the SHS using center-stratified logistic regression models adjusted for age, age2, sex and BMI. Odds ratio (OR) and 95% confidence intervals (CI) are reported. A Bonferroni correction for multiple testing p=0.01 (α=0.05/4 independent tests) was considered significant.
Supplementary Material
Acknowledgments
Funding: The Population Architecture Using Genomics and Epidemiology (PAGE) program is funded by the National Human Genome Research Institute (NHGRI), supported by U01HG004803 (CALiCo), U01HG004798 (EAGLE), U01HG004802 (MEC), U01HG004790 (WHI), and U01HG004801 (Coordinating Center). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The complete list of PAGE members can be found at http://www.pagestudy.org.
Funding support for the Genetic Epidemiology of Causal Variants Across the Life Course (CALiCo) program was provided through the NHGRI PAGE program (U01HG004803). The Strong Heart Study (SHS) is supported by NHLBI grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654, and U01 HL65521. The opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the Indian Health Service.
Footnotes
Disclosures: The authors report absence of any interest to disclose.
References
- 1.Kurella M, Lo JC, Chertow GM. Metabolic syndrome and the risk for chronic kidney disease among nondiabetic adults. J Am Soc Nephrol. 2005;16:2134–2140. doi: 10.1681/ASN.2005010106. [DOI] [PubMed] [Google Scholar]
- 2.Lucove J, Vupputuri S, Heiss G, et al. Metabolic syndrome and the development of CKD in American Indians: the Strong Heart Study. Am J Kidney Dis. 2008;51:21–28. doi: 10.1053/j.ajkd.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Fried LF, Orchard TJ, Kasiske BL. Effect of lipid reduction on the progression of renal disease: a meta-analysis. Kidney Int. 2001;59:260–269. doi: 10.1046/j.1523-1755.2001.00487.x. [DOI] [PubMed] [Google Scholar]
- 4.Fagot-Campagna A, Nelson RG, Knowler WC, et al. Plasma lipoproteins and the incidence of abnormal excretion of albumin in diabetic American Indians: the Strong Heart Study. Diabetologia. 1998;41:1002–1009. doi: 10.1007/s001250051023. [DOI] [PubMed] [Google Scholar]
- 5.Gambaro G, Verlato F, Budakovic A, et al. Renal impairment in chronic cigarette smokers. J Am Soc Nephrol. 1998;9:562–567. doi: 10.1681/ASN.V94562. [DOI] [PubMed] [Google Scholar]
- 6.Pinto-Sietsma SJ, Mulder J, Janssen WM, et al. Smoking is related to albuminuria and abnormal renal function in nondiabetic persons. Ann Intern Med. 2000;133:585–591. doi: 10.7326/0003-4819-133-8-200010170-00008. [DOI] [PubMed] [Google Scholar]
- 7.Robbins DC, Knowler WC, Lee ET, et al. Regional differences in albuminuria among American Indians: an epidemic of renal disease. Kidney Int. 1996;49:557–563. doi: 10.1038/ki.1996.79. [DOI] [PubMed] [Google Scholar]
- 8.Scavini M, Stidley CA, Paine SS, et al. The burden of chronic kidney disease among the Zuni Indians: the Zuni Kidney Project. Clin J Am Soc Nephrol. 2007;2:509–516. doi: 10.2215/CJN.02780806. [DOI] [PubMed] [Google Scholar]
- 9.Luo F, Wang Y, Wang X, et al. A functional variant of NEDD4L is associated with hypertension, antihypertensive response, and orthostatic hypotension. Hypertension. 2009;54:796–801. doi: 10.1161/HYPERTENSIONAHA.109.135103. [DOI] [PubMed] [Google Scholar]
- 10.Shara NM, Resnick HE, Lu L, et al. Decreased GFR estimated by MDRD or Cockcroft-Gault equation predicts incident CVD: the strong heart study. J Nephrol. 2009;22:373–380. [PMC free article] [PubMed] [Google Scholar]
- 11.Shara NM, Wang H, Valaitis E, et al. Comparison of estimated glomerular filtration rates and albuminuria in predicting risk of coronary heart disease in a population with high prevalence of diabetes mellitus and renal disease. Am J Cardiol. 107:399–405. doi: 10.1016/j.amjcard.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.US Renal Data System: Excerpts from the USRDS 2006 Annual Data Report. Am J Kidney Dis. 2006;49(suppl 291):S1–S296. [Google Scholar]
- 13.Silveiro SP, Friedman R, de Azevedo MJ, et al. Five-year prospective study of glomerular filtration rate and albumin excretion rate in normofiltering and hyperfiltering normoalbuminuric NIDDM patients. Diabetes Care. 1996;19:171–174. doi: 10.2337/diacare.19.2.171. [DOI] [PubMed] [Google Scholar]
- 14.Pruijm M, Wuerzner G, Maillard M, et al. Glomerular hyperfiltration and increased proximal sodium reabsorption in subjects with type 2 diabetes or impaired fasting glucose in a population of the African region. Nephrol Dial Transplant. 2010;25:2225–2231. doi: 10.1093/ndt/gfq008. [DOI] [PubMed] [Google Scholar]
- 15.Brenner BM, Lawler EV, Mackenzie HS. The hyperfiltration theory: a paradigm shift in nephrology. Kidney Int. 1996;49:1774–1777. doi: 10.1038/ki.1996.265. [DOI] [PubMed] [Google Scholar]
- 16.Kramer HJ, Nguyen QD, Curhan G, et al. Renal insufficiency in the absence of albuminuria and retinopathy among adults with type 2 diabetes mellitus. JAMA. 2003;289:3273–3277. doi: 10.1001/jama.289.24.3273. [DOI] [PubMed] [Google Scholar]
- 17.Franceschini N, Voruganti VS, Haack K, et al. The association of the MYH9 gene and kidney outcomes in American Indians: the Strong Heart Family Study. Hum Genet. 127:295–301. doi: 10.1007/s00439-009-0769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim NH, Pavkov ME, Knowler WC, et al. Predictive value of albuminuria in American Indian youth with or without type 2 diabetes. Pediatrics. 125:e844–851. doi: 10.1542/peds.2009-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleyer AJ, Sedor JR, Freedman BI, et al. Risk factors for development and progression of diabetic kidney disease and treatment patterns among diabetic siblings of patients with diabetic kidney disease. Am J Kidney Dis. 2008;51:29–37. doi: 10.1053/j.ajkd.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 20.Imperatore G, Knowler WC, Nelson RG, et al. Genetics of diabetic nephropathy in the Pima Indians. Curr Diab Rep. 2001;1:275–281. doi: 10.1007/s11892-001-0046-2. [DOI] [PubMed] [Google Scholar]
- 21.MacCluer JW, Scavini M, Shah VO, et al. Heritability of measures of kidney disease among Zuni Indians: the Zuni Kidney Project. Am J Kidney Dis. 56:289–302. doi: 10.1053/j.ajkd.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mottl AK, Vupputuri S, Cole SA, et al. Linkage analysis of albuminuria. J Am Soc Nephrol. 2009;20:1597–1606. doi: 10.1681/ASN.2008080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mottl AK, Vupputuri S, Cole SA, et al. Linkage analysis of glomerular filtration rate in American Indians. Kidney Int. 2008;74:1185–1191. doi: 10.1038/ki.2008.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfs MG, Hofker MH, Wijmenga C, et al. Type 2 Diabetes Mellitus: New Genetic Insights will Lead to New Therapeutics. Curr Genomics. 2009;10:110–118. doi: 10.2174/138920209787847023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Zheng T, Wang F, et al. Pro12Ala polymorphism in the PPARG gene contributes to the development of diabetic nephropathy in Chinese type 2 diabetic patients. Diabetes Care. 33:144–149. doi: 10.2337/dc09-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohshige T, Tanaka Y, Araki S, et al. A single nucleotide polymorphism in KCNQ1 is associated with susceptibility to diabetic nephropathy in japanese subjects with type 2 diabetes. Diabetes Care. 33:842–846. doi: 10.2337/dc09-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buraczynska M, Swatowski A, Markowska-Gosik D, et al. Transcription factor 7-like 2 (TCF7L2) gene polymorphism and complication/comorbidity profile in type 2 diabetes patients. Diabetes Res Clin Pract. doi: 10.1016/j.diabres.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Kottgen A, Hwang SJ, Rampersaud E, et al. TCF7L2 variants associate with CKD progression and renal function in population-based cohorts. J Am Soc Nephrol. 2008;19:1989–1999. doi: 10.1681/ASN.2007121291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matise TC, Ambite J, Buyske S, et al. Population Architecture using Genetics and Epidemiology. Am J Epidemiol. 2011 doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard BV, Lee ET, Cowan LD, et al. Coronary heart disease prevalence and its relation to risk factors in American Indians. The Strong Heart Study. Am J Epidemiol. 1995;142:254–268. doi: 10.1093/oxfordjournals.aje.a117632. [DOI] [PubMed] [Google Scholar]
- 31.Takeda K, Inoue H, Tanizawa Y, et al. WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet. 2001;10:477–484. doi: 10.1093/hmg/10.5.477. [DOI] [PubMed] [Google Scholar]
- 32.Ueda K, Kawano J, Takeda K, et al. Endoplasmic reticulum stress induces Wfs1 gene expression in pancreatic beta-cells via transcriptional activation. Eur J Endocrinol. 2005;153:167–176. doi: 10.1530/eje.1.01945. [DOI] [PubMed] [Google Scholar]
- 33.Knowler WC, Pettitt DJ, Saad MF, et al. Diabetes mellitus in the Pima Indians: incidence, risk factors and pathogenesis. Diabetes Metab Rev. 1990;6:1–27. doi: 10.1002/dmr.5610060101. [DOI] [PubMed] [Google Scholar]
- 34.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elks CE, Loos RJ, Sharp SJ, et al. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med. 7:e1000284. doi: 10.1371/journal.pmed.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubacek JA, Viklicky O, Dlouha D, et al. The FTO gene polymorphism is associated with end-stage renal disease: two large independent case-control studies in a general population. Nephrol Dial Transplant. doi: 10.1093/ndt/gfr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omori S, Tanaka Y, Takahashi A, et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57:791–795. doi: 10.2337/db07-0979. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Han XY, Ren Q, et al. Effect of genetic variants in KCNJ11, ABCC8, PPARG and HNF4A loci on the susceptibility of type 2 diabetes in Chinese Han population. Chin Med J (Engl) 2009;122:2477–2482. [PubMed] [Google Scholar]
- 40.Nielsen EM, Hansen L, Carstensen B, et al. The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes. 2003;52:573–577. doi: 10.2337/diabetes.52.2.573. [DOI] [PubMed] [Google Scholar]
- 41.Yang L, Zhou X, Luo Y, et al. Association between KCNJ11 gene polymorphisms and risk of type 2 diabetes mellitus in East Asian populations: a meta-analysis in 42,573 individuals. Mol Biol Rep. doi: 10.1007/s11033-011-0782-6. [DOI] [PubMed] [Google Scholar]
- 42.Hsu CY, McCulloch CE, Iribarren C, et al. Body mass index and risk for end-stage renal disease. Ann Intern Med. 2006;144:21–28. doi: 10.7326/0003-4819-144-1-200601030-00006. [DOI] [PubMed] [Google Scholar]
- 43.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 44.Palmer ND, Hester JM, An SS, et al. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes. 60:662–668. doi: 10.2337/db10-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson ER. Translating TCF7L2: from gene to function. Diabetologia. 2009;52:1227–1230. doi: 10.1007/s00125-009-1356-1. [DOI] [PubMed] [Google Scholar]
- 46.Lancaster MA, Gleeson JG. Cystic kidney disease: the role of Wnt signaling. Trends Mol Med. 16:349–360. doi: 10.1016/j.molmed.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee ET, Welty TK, Fabsitz R, et al. The Strong Heart Study A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 48.North KE, Howard BV, Welty TK, et al. Genetic and environmental contributions to cardiovascular disease risk in American Indians: the strong heart family study. Am J Epidemiol. 2003;157:303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 49.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 50.Levey AS, Bosch JP, Lewis JB, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 51.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 52.Havill LM, Dyer TD, Richardson DK, et al. The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification. BMC Genet. 2005;6(1):S91. doi: 10.1186/1471-2156-6-S1-S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.