SUMMARY
V(D)J recombination is essential for generating a diverse array of B and T cell receptors that can recognize and combat foreign antigen. As with any recombination event, tight control is essential to prevent the occurrence of genetic anomalies that drive cellular transformation. One important aspect of regulation is directed targeting of the RAG recombinase. Indeed, RAG accumulates at the 3’ end of individual antigen receptor loci poised for rearrangement, however, it is not known whether focal binding is involved in regulating cleavage, and what mechanisms lead to enrichment of RAG in this region. Here we show that mono-allelic looping out of the 3’ end of Tcra, coupled with transcription and increased chromatin/nuclear accessibility, are linked to focal RAG binding and ATM-mediated regulated mono-allelic cleavage on looped out 3’ regions. Our data identify higher order loop formation as a key determinant of directed RAG targeting and the maintenance of genome stability.
INTRODUCTION
Epigenetic regulation of gene expression involves dynamic changes in transcription factor (TF) binding, chromatin packaging and nuclear organization. In recent years we have learned a great deal about the genome wide binding patterns of TFs and chromatin marks that determine the ‘on’ or ‘off’ state of target genes in a given lineage and developmental stage. These data sets are beginning to be integrated with genome wide chromosome conformation capture analyses to determine how binding patterns translate into changes in nuclear organization. The combined information from these experiments indicates that co-regulated active and repressed genes contact each other in activator or repressor enriched regions of the nucleus, respectively (Cheutin and Cavalli, 2012; Schoenfelder et al., 2010). Aggregation of co-regulated genes in a population of cells implies the interaction of multiple different loci. However, individual chromosomes occupy discrete territories in interphase nuclei and bearing in mind these physical constraints, it is not clear how genes on different chromosomes meet up with each other in the nucleus to form hubs. Indeed, live imaging studies indicate that chromosome territories move very little after mitosis, thus, unless chromosome territories are neighboring, gene interactions must rely on the mobility of individual loci, which are known to be more dynamic (Muller et al., 2010). Higher-order looping facilitates movement of genes away from their chromosome territories, and this is often correlated with open chromatin and an active transcriptional status, while silent genes are positioned more internally (Fraser and Bickmore, 2007; Heard and Bickmore, 2007; Kalhor et al., 2012; Splinter et al., 2011). However, while higher-order looping has been shown to facilitate stochastic interactions between genes on different chromosomes (Kalhor et al., 2012), we know little about whether higher-order looping contributes to co-ordinated regulation of genes in trans.
The antigen-receptor loci have afforded a rich model for studying the epigenetic mechanisms underlying regulation of programmed recombination in developing lymphocytes. It has been shown that intra-locus loop formation (locus contraction), changes in chromatin landscape and dynamic association of antigen-receptor loci with repressive pericentromeric heterochromatin (PCH) contribute (Chaumeil and Skok, 2012; Hewitt et al., 2010). However, we do not know how higher-order nuclear organization of antigen receptor loci is integrated with these various aspects of regulation and what role it plays in controlling this complex process in individual cells. This is particularly pertinent in the light of recent data showing that the lymphoid-specific recombinase proteins, RAG1 and RAG2, bind to active chromatin and localize to the J segments at the 3’ end of each antigen receptor locus in rearranging cells (Ji et al., 2010), but it is not known how focal RAG binding alters higher-order nuclear organization and the regulation of cleavage.
V(D)J recombination of immunoglobulin (Ig) and T cell receptor (Tcr) loci occurs in developing B and T cells, respectively. Tight control is exerted at multiple levels to ensure lineage and stage specific rearrangement of variable (V), diversity (D) and joining (J) gene segments within the individual loci. Furthermore, recombination is regulated at the level of the individual allele (allelic exclusion) to ensure that each B and T cell expresses an antigen receptor with limited specificity. The RAG proteins recognize highly conserved recombination signal sequences (RSSs) bordering each V, D and J gene segment (Helmink and Sleckman, 2012; Schatz and Ji, 2011)}. RAG binding mediates synapsis of a pair of compatible RSSs (that have conserved conserved heptamer and nonamer sequences separated by a 12 and 23 base pair space) prior to cleavage, and following the introduction of double strand breaks (DSBs) the recombinase secures the four broken ends in a postcleavage complex. Repair of these recombination intermediates occurs through the combined action of the DNA damage response pathway (which involves the factors ATM, γH2AX, 53BP1, and the MRN complex) as well as the ubiquitous non-homologous end joining (NHEJ) machinery (Helmink and Sleckman, 2012; Puebla-Osorio and Zhu, 2008). This gives rise to a new antigen receptor gene and a signal joint, which is frequently excised during cell division.
Productive rearrangement of the different Tcr loci gives rise to two distinct T cell lineages: recombination of Tcrg/Tcrd and Tcrb/Tcra leads to γδ and αβ T cells, respectively (Figure 1A) (Ciofani and Zuniga-Pflucker, 2010; Krangel, 2009). Rearrangement of the Tcrg, Tcrd and Tcrb loci occurs early on in development in CD4−CD8− double negative DN2/3 cells, while Tcra recombination occurs later at the double positive (DP) stage (Livak et al., 1999). Because the Tcrd locus is embedded within the Tcra locus (Figure 1B), regulation of Tcrd and Tcra recombination is especially complex. Furthermore, the Tcrd and Tcra loci are not stringently subjected to allelic exclusion and productively assembled genes are frequently bi-allelically expressed. Thus recombination can occur on both alleles. Moreover, individual Tcra alleles typically undergo numerous recombination events in each DP cell. Given the large number of recombination events that can occur during Tcra assembly, tight control at the DP cell stage is particularly important. Here we have investigated the role of transcription, higher-order looping and nuclear organization in regulating these processes to safeguard genome stability.
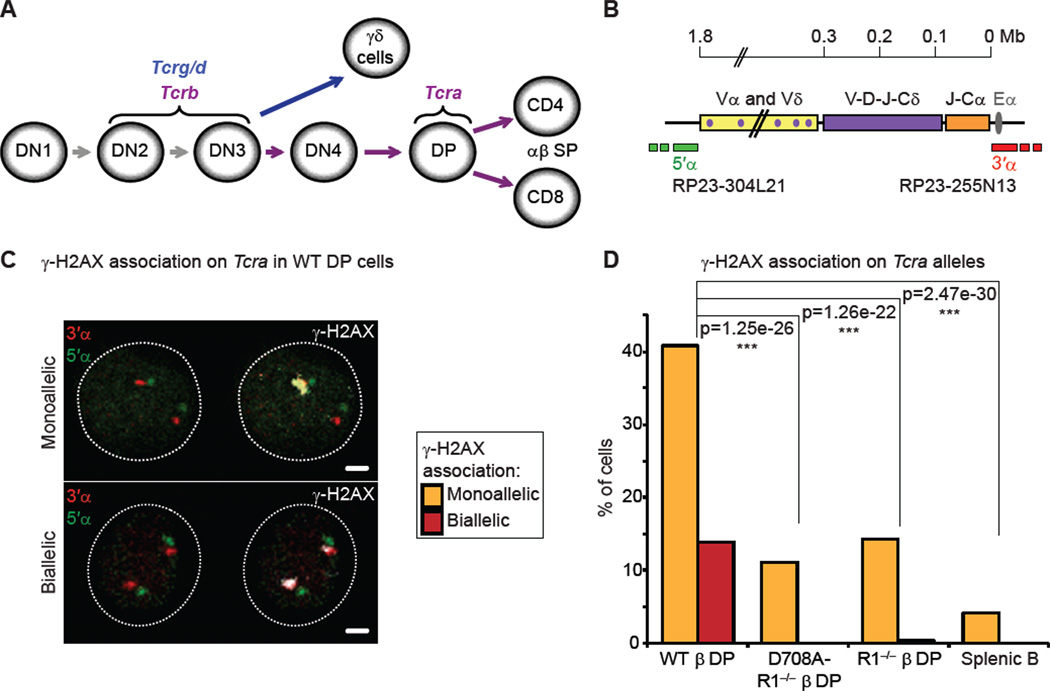
Figure 1. Recombination of Tcra occurs on one allele at a time in DP cells.
(A) Recombination of the 4 Tcr loci at different stages of T cell development.
(B) Scheme representing the Tcra/d locus. Probes for 3D-DNA FISH were generated from BACs hybridizing outside of the Vα segments (5’α, green) and of the Cα region (3’α, red).
(C) Confocal sections showing examples of γ-H2AX association (in white) on Tcra alleles (3’α in red and 5’α in green) in WT DP cells: mono-allelic association on the top panel, and bi-allelic association with the two foci of different size and intensity, on the bottom panel. Scale bars = 1 µm.
(D) Frequency of γ-H2AX association on Tcra in WT, RAG1-mutant DP cells and splenic B cells. The p values calculated using a two-tail Fisher exact test are shown as: -ns- no significance (p ≥ 5.00e-2), -*- significant (5.00e-2 > p ≥ 1.00e-2), -**- very significant (1.00e-2 > p ≥ 1.00e-3), -***- highly significant (p < 1.00e-3). Graphs and p values represent a combination of two independent and representative experiments (individual data sets and full statistical analyses in Table S1). See also Figure S1.
RESULTS
Recombination of Tcra occurs on one allele at a time in DP cells
As the Tcra locus is not subject to allelic exclusion, we first asked whether both Tcra alleles are targeted for recombination at the same time. For this we examined RAG-mediated cleavage of Tcra in individual DP cells by 3D immuno-DNA FISH, using two differentially labeled BAC probes that hybridize to the 3’α and 5’α ends of the locus (which in the germline form are 1.8Mb apart) (Skok et al., 2007) in combination with an antibody that detects the phosphorylated form of the histone variant H2AX (γ-H2AX) as a read out for DSBs (Chen et al., 2000; Rogakou et al., 1998) (Figure 1B,C). For these studies we sorted DP cells from RAG1 deficient and control wild-type mice, known as R1−/− β and WT β, respectively. Both mice carry a functionally rearranged Tcrb transgene (β) that allows T cell development to proceed to the DP stage in the absence of Tcrb rearrangement. As a complementary control we also included DP cells from D708A-R1−/− β mice, which in addition to the Tcrb transgene carry another transgene expressing a catalytically inactive version of the RAG1 protein that has a D708A active site substitution preventing DNA cleavage without impairing binding and synapsis of RSSs by the RAG complex (Fugmann et al., 2000; Kim et al., 1999; Landree et al., 1999).
As expected, we found that γ-H2AX was associated with Tcra at very low levels in control splenic B cells, while in WT or WT β DP cells, consistent with a previous study (Chen et al., 2000), foci of γ-H2AX were colocalized with one Tcra allele in around 40% of cells and with both alleles in around 14% of the cells (Figure 1C,D and Table S1). We noticed however, that in more than 80% of WT DP cells harboring bi-allelic breaks, one γ-H2AX focus was substantially smaller in size and intensity than the other, even though the two DNA FISH signals were equivalent (see surface plots in Figure S1A). Furthermore, in RAG-deficient cells mono-allelic γ-H2AX association on Tcra alleles occurred at a similar frequency as the bi-allelic breaks in WT cells (around 14%), with similar small and faint foci as those found on the second allele in WT cells (Figure 1D and Figure S1A,B). These data indicate that only the large γ-H2AX foci in WT cells are specific to RAG-mediated cleavage, whereas the small RAG-independent foci may result from other cellular processes such as transcription. Indeed the difference between the level of γ-H2AX association on Tcra alleles in RAG deficient and splenic B cells suggests that transcription could be a factor. Thus, despite the absence of allelic exclusion, RAG-mediated cleavage of Tcra occurs on one allele at a time in DP cells.
Mono-allelic higher-order looping out of the Tcra 3’ end coincides with mono-allelic RAG-mediated cleavage
To investigate whether differential organization of Tcra alleles relative to their chromosome territories could be involved in asynchronous RAG cleavage on the two Tcra alleles, we performed 3D DNA FISH for Tcra combined with a chromosome 14 paint. For our analysis we classified the location of Tcra relative to the chromosome territory into the following four categories: i) outside - looping out of the bulk of the chromosome territory (defined here as higher-order loops), ii) outer edge - adjacent to the territory edge, iii) edge - spanning the territory edge, and iv) inside the territory (See Extended Experimental Procedures section for details). Interestingly, we found that Tcra was more likely to adopt an external location and form large higher-order loops outside of the chromosome territory in the presence of WT or catalytically inactive RAG1 (Figure 2A, Figure S2A and Table S2). In contrast, in splenic B cells where Tcra is transcriptionally inactive and RAG is not expressed, the alleles adopted a more internal location compared to RAG1-mutant DP cells, in which Tcra is active. Thus, consistent with previous published reports (for review see (Heard and Bickmore, 2007)), transcription may be linked to looping out. Here however, we have also uncovered a novel aspect of control: higher-order loop formation is dependent on trans acting factors (in this case the RAG recombinase).
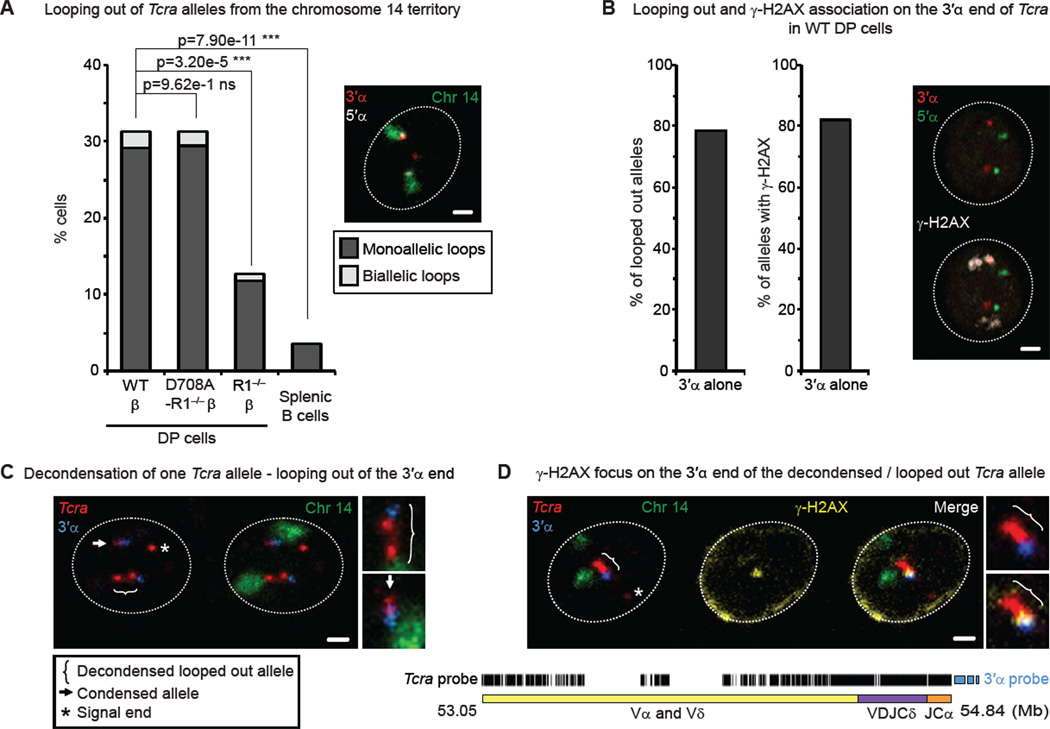
Figure 2. Mono-allelic higher-order looping of the 3’ end of Tcra coincides with mono-allelic RAG-mediated cleavage.
(A) Frequency of looping out of Tcra alleles from their chromosome 14 territories in WT, RAG1-mutant DP cells and splenic B cells. p values were calculated using a two-tail Fisher exact test. Confocal section shows an example of mono-allelic looping out of a decondensed Tcra allele: The 3’α end (red) loops out while the 5’α (white) end remains inside of the chromosome 14 territory (green). Scale bar = 1 µm.
(B) Proportion of looped out alleles (left) and alleles associated with γ-H2AX (right) that involve only the 3’α end of Tcra. Confocal sections show an example of γ-H2AX association (white) on the 3’α end (red) of the decondensed Tcra allele (5’α in green). Scale bar = 1 µm.
(C, D) Confocal examples of mono-allelic higher-order looping and decondensation of Tcra (C), correlated with γ-H2AX foci on the looped out 3’α end (D). The scheme of the mix of oligonucleotide probes for the entire Tcra/d locus is shown below. Tcra in red, 3’α in blue, chromosome 14 paint in green and γ-H2AX in yellow. Scale bars = 1 µm.
See also Figure S2 and Table S2 for details and full statistical analyses.
Analysis of looped out alleles showed that the majority had a decondensed conformation (separation of > 0.5 µm between the 3’α and 5’α ends), with the 3’α end positioned away from the chromosome 14 territory, while the 5’α end remained at the edge of the territory (Figure 2A,B (left) and Table S2). (Additional points of discussion clarifying the differences between decondensation and decontraction can be found in Extended Results section and Figure S2B.) Intriguingly, these higher-order loops were mono-allelic more than 90% of the time (example in Figure 2A and Table S2). In a similar manner, γ-H2AX foci, that are predominantly mono-allelic (Figure 1D), were mostly associated with decondensed Tcra alleles, and in the majority of cases these foci were located on the 3’α end alone and never on the 5’α end alone (Figure 2B (right), Figure S2C and Table S2). Furthermore, when breaks were detected in cells with one condensed and one decondensed allele, γ-H2AX foci were preferentially associated with the 3’ end of the latter (see example in Figure 2B).
To investigate higher-order looping in more depth we designed a probe comprised of oligonucleotides that cover the entire Tcra locus with the exception of the most repetitive regions (see scheme below Figure 2C,D, Table S2 and Extended experimental procedures for details). (The use of complex oligonucleotide-based probes for FISH analysis was first described by Boyle et al. (Boyle et al., 2011).) With this probe we could also detect deleted signal ends, (a by-product of recombination) that were separated from the rest of the locus that was marked by the presence of the 3’α BAC (see asterisks in Figure 2C,D). (For additional points of discussion about signal ends see Extended Results section.) The combined use of this Tcra probe with the 3’α probe and the chromosome 14 paint showed that indeed looped out alleles had a more elongated, decondensed conformation (with the 3’ region being the most external) compared to alleles that remained associated with the chromosome 14 territory (Figure 2C). Furthermore, immuno-DNA FISH confirmed that γ-H2AX foci were predominantly associated with the 3’ end of the looped out decondensed alleles (Figure 2D).
Taken together our data indicate that mono-allelic higher-order looping of Tcra, which separates the 3’ end from the chromosome territory, occurs in a RAG-dependent, cleavage independent manner, indicating that this change in nuclear conformation occurs prior to the introduction of a break. Moreover, the link between mono-allelic looping out of the 3’α end of Tcra and mono-allelic association of γ-H2AX foci in this 3’α region, suggest that these changes in nuclear organization could participate in the regulation of recombination.
Looping out of the 3’ end of Tcra is linked to transcription, increased accessibility and mono-allelic cleavage
To further understand the mechanism underlying the formation of mono-allelic higher-order loop formation we performed sequential RNA/DNA FISH, with nascent RNA detected by the large Tcra probe (see Figure 2C,D) followed by DNA FISH with the 3’α probe and the chromosome 14 paint in WT DP cells. Briefly, RNA FISH was imaged by confocal microscopy and coordinates of the fields of nuclei were recorded in order to track the nuclei after the DNA FISH part (See the sequential RNA/DNA FISH section of Extended Experimental procedures in the Supplementary Information section for details). RNA signals were detected on both alleles in 26.3% of cells and on one allele in 30% of cells (in total 56% of cells had RNA signals) (Table S3). It is of note that although the large Tcra probe was used for this analysis, RNA signals were predominantly only found associated with the 3’ end of the locus (see example in Figure 3A). This is likely due to limitations in detection of transcription below a certain level using this RNA FISH assay. Interestingly, transcription was correlated with the formation of higher-order loops: more looped out Tcra alleles were transcribed compared to alleles that were positioned at the outer edge or edge of the territory (Figure 3B and Table S3). Furthermore, no RNA signals were found on alleles that were located inside the chromosome territory. Taken together our data show that the formation of higher-order mono-allelic Tcra loops is dependent on the presence of RAG as well as correlated with transcription.
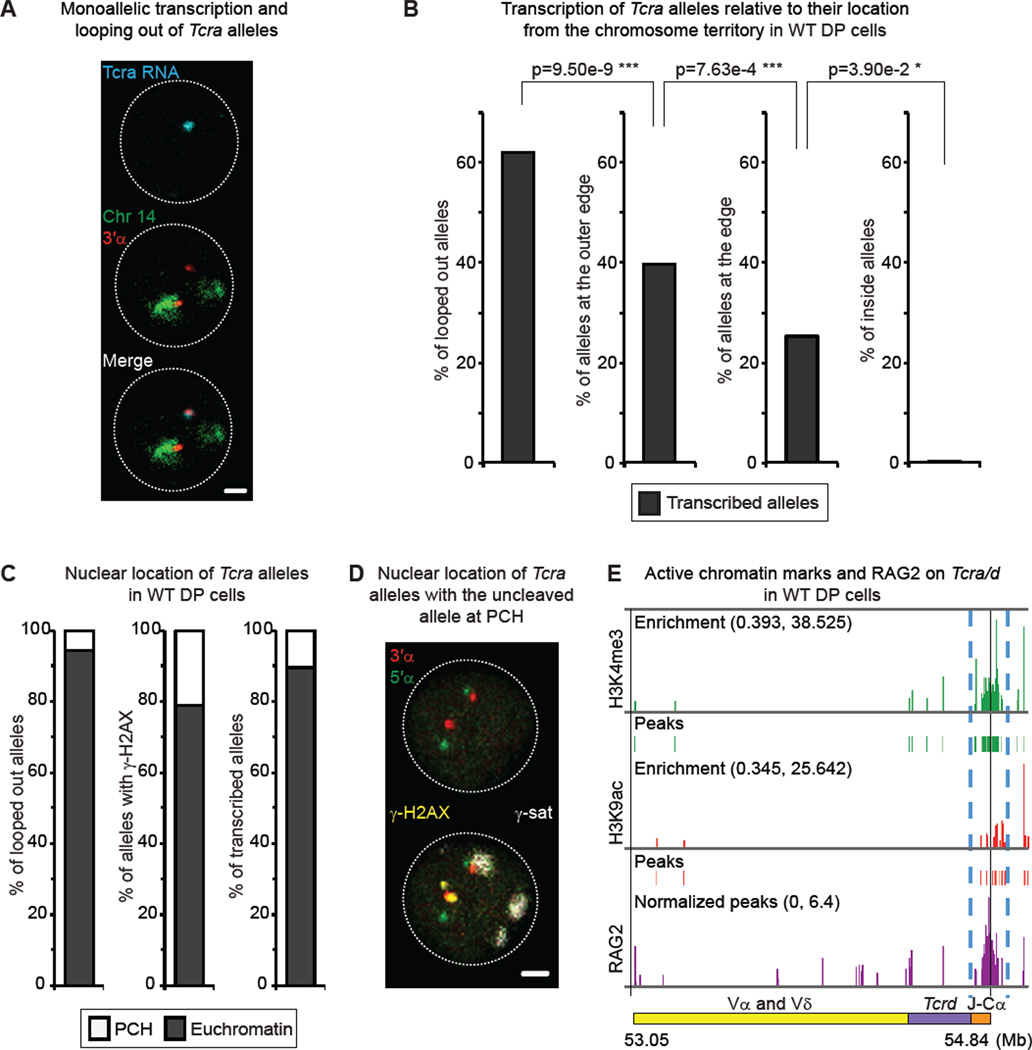
Figure 3. Looping out of the 3’ end of Tcra is linked to transcription, increased accessibility and mono-allelic cleavage.
(A) Confocal example shows mono-allelic transcription of looped out Tcra allele. Merge was created subsequently based on the DAPI signals of the RNA FISH and DNA FISH images. Tcra nascent transcript in blue, 3’α in red, chromosome 14 paint in green. Scale bar = 1 µm.
(B) Percentage of Tcra alleles located outside (loops), at the outer edge, at the edge or inside the chromosome 14 territory which are transcribed in WT DP cells. p values were calculated using a two-tail Fisher exact test.
(C) Nuclear location of looped out alleles (left), alleles associated with γ-H2AX (middle), and transcribed alleles (right) within euchromatin or heterochromatin (PCH).
(D) Confocal sections show an example of the uncleaved Tcra allele (no γ-H2AX) being at PCH while the γ-H2AX-associated one remains in euchromatin. 3’α in red, 5’α in green, γ-H2AX in yellow, and γ-satellite (PCH) in white. Scale bar = 1 µm.
(E) Alignment of ChIP-seq data at the Tcra locus showing levels of enrichment and peaks of H3K4me3 (green) and H3K9ac (red), as well as RAG2 binding (purple) (Ji et al., 2010). Blue dashed lines highlight the focal enrichment in active marks and RAG2in the Tcra 3’α region.
See also Table S3 for details and full statistical analyses.
We next asked whether looping of the 3’ end of the locus is linked to changes in accessibility and enrichment of RAG, which preferentially binds regions of active chromatin (Ji et al., 2010; Liu et al., 2007; Matthews et al., 2007; Shimazaki et al., 2009). To determine this we examined the position of alleles relative to euchromatin (active chromatin) or to pericentromeric heterochromatin (PCH; repressive chromatin that can be detected with a γ-satellite probe (Brown et al., 1999)). Our analyses indicate that the vast majority of alleles that were (i) looped out, (ii) associated with γ-H2AX or (iii) transcribed were positioned in euchromatic regions (Figure 3C and Table S3). Importantly, in cells with mono-allelic γ-H2AX, the cleaved allele remained euchromatic while the other uncleaved allele was predominantly positioned at PCH (example in Figure 3D). Further, ChIP-seq analyses of histone modifications (histone H3 lysine 4 trimethylation (H3K4me3) and histone H3 lysine 9 acetylation (H3K9ac)) confirmed that the 3’ end of Tcra was enriched for these active marks (Figure 3E and Table S3). Moreover, these marks line up with RAG2 enrichment in DP cells (data obtained from (Ji et al., 2010)) (Figure 3E). Together, these data suggest that mono-allelic looping out of the 3’ end of Tcra in euchromatic regions as well as increased accessibility (active chromatin marks coupled with transcription) are linked to focal RAG binding and mono-allelic cleavage in this region.
Homologous pairing of Tcra alleles is linked to transcription, increased accessibility and mono-allelic cleavage
During the course of our analyses, we discovered that looped-out alleles were often in close proximity to their partner homologues. Moreover, we recently showed that homologous pairing and regulated asynchronous cleavage of Ig homologues are linked to allelic exclusion in developing B cells (Hewitt et al., 2009). We thus looked at whether Tcra alleles associate with one another in cells undergoing recombination. Our analyses of WT DP and control splenic B cells indicate that Tcra alleles were preferentially associated in T lineage cells (Figure 4A, Figure S3A and Table S4). Sliding Fisher exact tests comparing Tcra interallelic distances in WT β and R1−/− β DP cells showed that the lowest p values were found for distances under 1 µm (Chaumeil et al., 2012). Thus, consistent with previous studies (Augui et al., 2007; Bacher et al., 2006; Hewitt et al., 2009), we defined homologous pairing when the two alleles were separated by less than 1 µm from each other (Figure 4A, Figure S3A; see Extended experimental procedures section for details). Within the DP subset we found that the frequency of homologous Tcra pairing was higher in WT β (or WT) compared to RAG1-deficient cells; however, the presence of the D708A catalytically inactive RAG1 protein was able to restore association to WT levels (Figure 4A and Table S4). Thus, the elevated level of pairing of Tcra alleles in DP cells depends on the presence of RAG1 but not on RAG-mediated cleavage, suggesting that pairing occurs prior to cleavage. This is in analogy to what we observed for pairing of Ig loci in B cells (Hewitt et al., 2009). However, it is likely that other factors are also involved because pairing occurs at a higher frequency in RAG1-deficient DP cells compared to splenic B cells (Figure 4A). Since Tcra is transcribed in DP cells but not in splenic B cells, it is possible that transcriptional activity, in addition to the presence of RAG, promotes pairing. Indeed, the proportion of RNA signals in cells with paired alleles was significantly increased compared to cells with unpaired alleles (Figure 4B and Table S4). Thus, as with higher-order loop formation, pairing of homologues is dependent on the presence of RAG as well as correlated with transcription. As for total alleles, we found that the formation of mono-allelic higher-order loops involving the 3’ end of Tcra, mono-allelic cleavage on the looped out 3’ end and association of the uncleaved allele with PCH also occurred on paired alleles (Figure 4C, Figure S3B,C and Table S4).
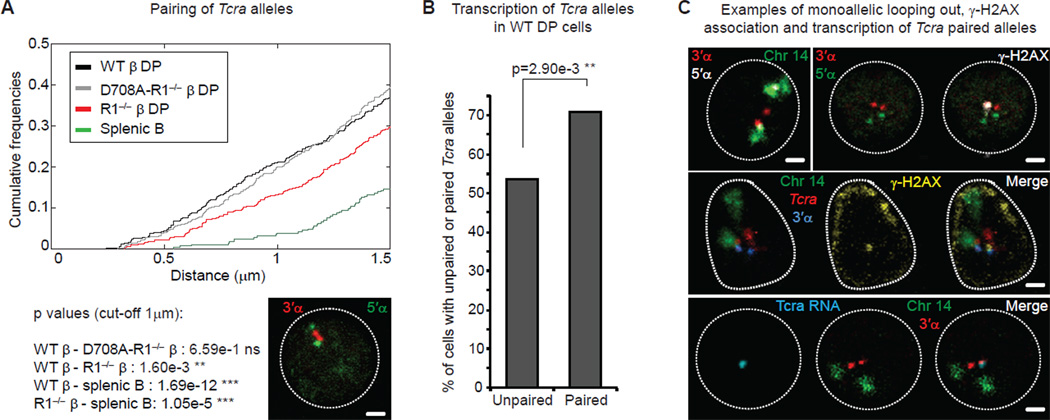
Figure 4. Homologous pairing of Tcra alleles is linked to transcription, increased accessibility and mono-allelic cleavage.
(A) Cumulative frequency curves of Tcra inter-allelic distances in WT, RAG1-mutant DP cells and splenic B cells (cut-off at 1.5 µm; p values for a cut-off at 1 µm). A left shift indicates closer association. Confocal section shows paired Tcra alleles (3’α in red and 5’α in green). Scale bar = 1 µm.
(B) Frequency of transcription of unpaired and paired Tcra alleles assessed by RNA FISH detecting nascent transcripts in WT DP cells.
(C) Confocal sections show examples of mono-allelic decondensation, looping out, association with γ-H2AX and transcription in Tcra pairs in WT DP cells. Top left: 3’α in red, 5’α in white and chromosome 14 paint in green. Top right: 3’α in red, 5’α in green and γ-H2AX in white. Middle: Tcra in red, 3’α in blue, chromosome 14 paint in green and γ-H2AX in yellow. Bottom: Tcra nascent transcript in blue, 3’α in red, chromosome 14 paint in green. Scale bars = 1 µm.
All p values were calculated using a two-tail Fisher exact test. See also Figure S3 and Table S4 for details and full statistical analyses.
In summary, these data indicate that RAG1 enhances the propensity of one allele of Tcra to adopt a decondensed conformation, with the 3’α end of the locus looped away from its chromosome territory. Higher-order looping of the 3’α end coincides with transcription, focal RAG binding and enrichment of active histone marks (Ji et al., 2010), as well as cleavage in this region, suggesting that these events are linked. We speculate that the formation of these higher order loops could serve to bring RAG enriched regions together in the nucleus, thereby increasing the concentration of RAG in localized recombination centers. The local concentration of RAG may also be a determinant of cleavage, in the sense that the more RAG that is present, the more likely the occurrence of a DSB. Furthermore, close proximity of RAG-enriched homologues provides a mechanism for trans regulation of mono-allelic cleavage.
ATM is involved in ensuring mono-allelic RAG-mediated cleavage on Tcra alleles
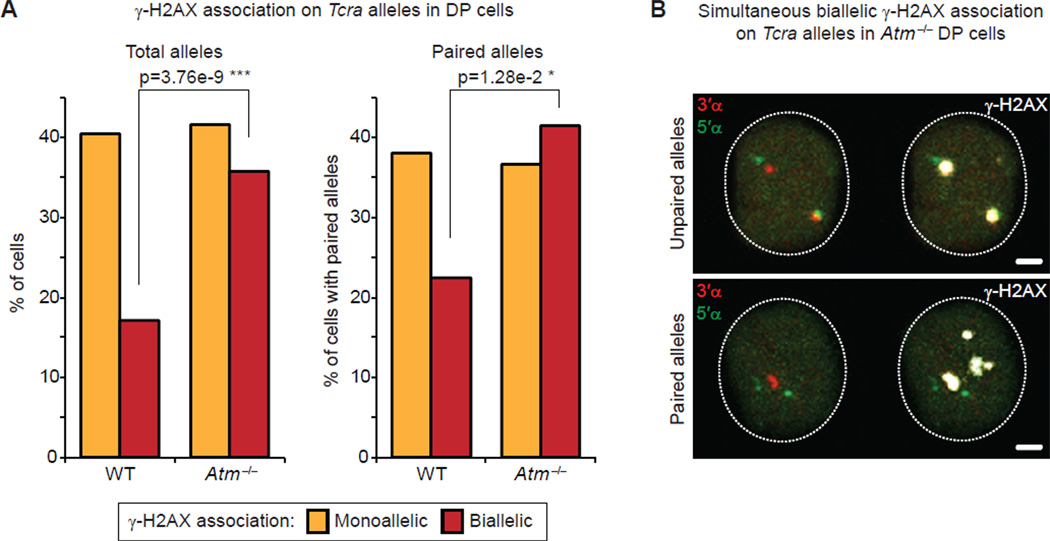
In developing B cells, RAG enhances homologous pairing of both Igh and Igk and repositioning of one allele to repressive pericentromeric heterochromatin (PCH) (Hewitt et al., 2009). Furthermore, the DNA damage factor ATM is implicated in regulating mono-allelic recombination in trans through repositioning of unrearranged Ig alleles to PCH: in the absence of ATM both Ig alleles remain euchromatic and there is a significant increase in bi-allelic breaks and translocations (Hewitt et al., 2009). Similarly, in DP cells we detected a significant increase in the frequency of γ-H2AX foci associated with both Tcra alleles in Atm−/− cells but no significant change in mono-allelic γ-H2AX association (Figure 5A and Table S5). Moreover, in Atm−/− cells γ-H2AX formed two large foci of a similar size and intensity, which is in contrast to what we observed in WT cells with bi-allelic breaks (Figure 5B and Figure S4A,B).
Figure 5. ATM is involved in ensuring mono-allelic RAG-mediated cleavage on Tcra alleles.
(A) Frequency of γ-H2AX association on total (left) or paired (right) Tcra alleles in WT and Atm−/− DP cells. p values were calculated using a two-tail Fisher exact test.
(B) Confocal sections show examples of simultaneous bi-allelic γ-H2AX association on unpaired (top) and paired (bottom) Tcra alleles in Atm−/− DP cells. 3’α in red, 5’α in green and γ-H2AX in white. Scale bars = 1 µm.
See also Figure S4 and Table S5 for details and full statistical analyses.
One important aspect of ATM’s regulation is to co-ordinate DSB repair with the progression of cell cycle through activation of proteins that regulate cell cycle checkpoints, including Chk2 and p53 (Helmink and Sleckman, 2012). Thus, in the absence of ATM it is possible that unrepaired breaks could accrue as damaged alleles in dividing cells. However, since we only analyze intact alleles (see section below and Figure 6D,E for details on categories of damage) in DP cells and these are non-cycling cells, the increase in bi-allelic breaks cannot be attributed to this aspect of control. Moreover, the increase in bi-allelic breaks is also not simply a result of a defect in repair because in the absence of another DNA sensing factor, 53BP1, we did not observe the same phenotype. Rather, we found a significant increase in the frequency of mono-allelic γ-H2AX foci associated with Tcra suggesting that in these cells, cleavage of the second allele cannot occur prior to repair of the first allele (Figure S4C and Table S5). These findings are in agreement with what we observed in Artemis−/− cells, which, due to an absence of this NHEJ factor, accumulate unrepaired RAG-mediated breaks: in the absence of Artemis we observed breaks on only one allele, while in Artemis−/− Atm−/− cells breaks were found on both alleles (Hewitt et al., 2009). These data are consistent with the notion that RAG-mediated cleavage of Tcra is regulated to occur on one allele at a time through the action of ATM.
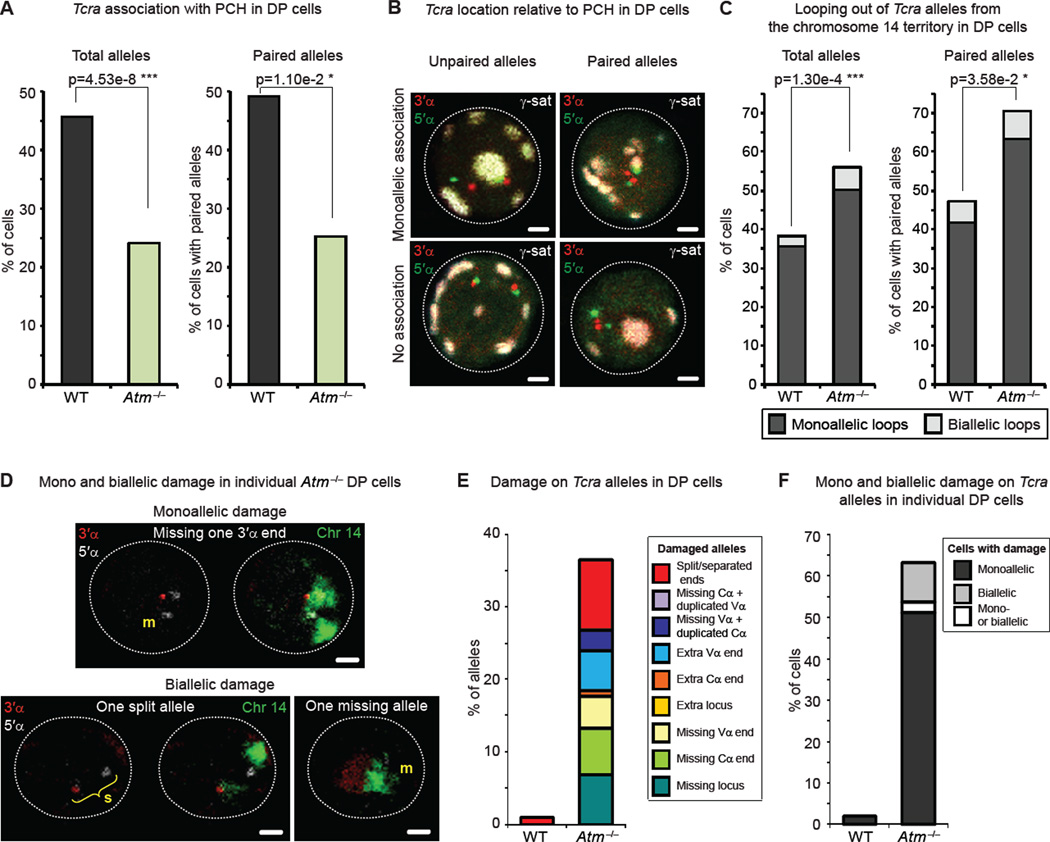
Figure 6. ATM-mediated suppression of bi-allelic cleavage and preservation of genome integrity involves modulation of nuclear accessibility and higher-order looping.
(A) Frequency of PCH association of total (left) and paired (right) Tcra alleles in WT and Atm−/− DP cells.
(B) Confocal sections show representative examples of unpaired (left) or paired (right) Tcra alleles associated with PCH (top) or not associated (bottom). 3’α in red, 5’α in green and γ-satellite (PCH) in white. Scale bars = 1 µm.
(C) Frequency of looping out of total (left) or paired (right) Tcra alleles from their chromosome 14 territories in WT and Atm−/− DP cells.
(D) Confocal sections show examples of mono and bi-allelic damage on Tcra alleles. 3’α in red, 5’α in white, and chromosome 14 paint in green. m = missing end or locus; s = split locus. Scale bars = 1 µm.
(E,F) Frequency of damaged Tcra alleles (E) and frequency of cells with mono or bi-allelic Tcra damage (F) in WT and Atm−/− DP cells.
All p values were calculated using a two-tail Fisher exact test. See also Figure S5 and Table S6 for details and full statistical analyses.
ATM-mediated suppression of bi-allelic cleavage and preservation of genome integrity involves modulation of nuclear accessibility and higher-order looping
To determine whether higher-order nuclear organization participates in coordinated ATM mediated regulation of recombination on individual Tcra alleles we simultaneously examined higher-order looping, homologous pairing and association with PCH. Although ATM does not regulate association of Tcra homologues we found that both paired and unpaired Tcra alleles were more frequently located in euchromatic regions of the nucleus in ATM-deficient versus control WT DP cells (Figure 6A,B, Figure S5A and Table S6). It is of note that in 53bp1−/− DP cells we observed no significant change in homologous pairing or PCH association (Figure S5B,C and Table S6). Thus, repositioning of Tcra to PCH is dependent on ATM, and this function is not shared by all DNA damage sensing factors.
Importantly, we found that decreased association of Tcra with PCH in Atm−/− DP cells was linked to a significant increase in higher-order looping out of paired and unpaired Tcra alleles (Figure 6C, Figure S5D and Table S6). These data indicate that trans regulation of mono-allelic cleavage by ATM occurs by modulating nuclear accessibility of Tcra through changes in higher-order loop formation and PCH association. Recall that in cells with mono-allelic γ-H2AX, the cleaved allele remained euchromatic while the uncleaved allele was predominantly positioned at PCH (Figure 3D). Thus ATM, recruited to the site of a RAG-mediated break on one allele, (directly or indirectly) acts in trans on the uncleaved allele, repositioning it to PCH and inhibiting loop formation.
To determine how ATM-mediated regulation of bi-allelic cleavage and changes in nuclear accessibility are linked with maintenance of genome stability we next analyzed mono and bi-allelic damage in Atm−/− interphase DP cells. For this, we performed a DNA FISH experiment using BAC probes that hybridize outside the 3′ and 5′ ends of Tcra (See scheme Fig. 1B) in combination with a chromosome 14 paint. Damage assessed included split alleles (> 1.5µm in between the two ends) and duplicated or missing signals (see examples in Figure 6D). Whereas only 1% of WT cells carried a Tcra allele with split ends, Atm−/− DP cells showed a whole range of damage on as much as 37.2% of Tcra alleles (which represents 62.4% of cells carrying at least one damaged allele) (Figure 6D,E and Table S6). Importantly, in 9.6% of Atm−/− DP cells, both Tcra alleles were damaged in the same cell (Figure 6D,F and Table S6), which is consistent with previous analyses (Matei et al., 2007). In contrast, only mono-allelic damage could be found in 53bp1−/− DP cells, consistent with the increase in mono-allelic γ-H2AX association (Figure S4C, S5E,F and Table S6). In sum, ATM-mediated changes in nuclear organization (repositioning to PCH and higher-order looping) are linked with maintenance of genome stability. We suggest that these chromosomal movements could reduce accessibility to RAG proteins, preventing cleavage on the second allele, which could lead to genomic instability.
DISCUSSION
Although regulation of the Tcra/d locus is uniquely complicated, we have found that recombination occurs on only one allele at a time. As with Ig recombination, regulation of mono-allelic cleavage occurs in trans through the action of ATM (Hewitt et al., 2009). To understand how recombination between different alleles is coordinated, we analyzed a novel aspect of nuclear organization: the formation of higher order loops. Here we show that higher-order mono-allelic looping connects allelic interactions, transcription, active chromatin, nuclear accessibility and focal RAG binding with mono-allelic cleavage at the 3’ end of individual loci. These studies provide a plausible explanation for the mechanisms underlying directed mono-allelic RAG targeting and regulated cleavage in localized recombination centers (Ji et al., 2010).
Previous studies have shown that dynamic movements of genes away from their chromosome territory are linked with their transcriptional status. For example, sequential looping out / decondensation of the Hox genes has been shown to be linked to expression (Chambeyron and Bickmore, 2004; Morey et al., 2009). Furthermore, X-linked genes are reorganized during X-chromosome inactivation, with internalization of repressed genes and looping out of escapees (Chaumeil et al., 2006). However, there have been no studies that connect gene interactions and regulation in trans with looping out of loci from their chromosome territories. Furthermore, higher-order looping was previously not known to play a role in recombination. Looping correlates with nuclear accessibility of the Tcra locus, as judged by transcription and its location in euchromatic regions of the nucleus. Moreover, in line with the notion that transcription can alter chromatin conformation and movement away from the chromosome territory, we found that the majority of looped out Tcra alleles were ‘decondensed’, with the 3’ end separated from the 5’ end (which remained embedded in the territory), and RAG mediated breaks were preferentially associated with the 3’ end of these alleles. Thus we propose that transcription and focal binding of RAG at the 3’ end of the locus induce mono-allelic looping/chromatin decondensation, which directs cleavage on one allele. Our data indicate that allelic differences in transcription could also contribute to the regulation of mono-allelic cleavage since DSBs are preferentially introduced on looped out decondensed alleles, and the formation of the latter occur predominantly on transcribed alleles. Thus allelic differences in transcription levels could determine which allele will be cleaved. Whether allelic differences are stochastic or the result of epigenetic differences linked to for example, differences in chromatin packaging and replication timing (Farago et al., 2012; Goldmit et al., 2005; Mostoslavsky et al., 2001) remains to be determined.
Our data indicate that ATM mediates repositioning of the uncleaved allele to PCH and movement to this repressive compartment is known to be accompanied by changes in chromatin and transcriptional repression (Su et al., 2004). Since active chromatin marks and transcription at the 3’ end of Tcra are both linked to higher-order looping out of the 3’ end of the locus, it is possible that inhibition of looping occurs on the uncleaved pericentric allele to prevent the introduction of further breaks in trans. This notion is supported by our finding that the vast majority of transcribed alleles were positioned in euchromatic regions. However, we do not conclude that repositioning to PCH is accompanied by complete transcriptional silencing on Tcra as it is likely our RNA FISH assay can only detect transcription occurring above a certain level. Thus, transcription could continue on repositioned loci, albeit at a reduced level (perhaps across a localized region) that could impact on higher-order loop formation. In addition, since we know that ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks (Shanbhag et al., 2010), we cannot exclude that inhibition of looping out could also occur on the cleaved allele.
The experiments we have performed here provide additional insight into ATM-mediated regulation of RAG cleavage in trans. We know from our previous work that ATM (recruited to the site of a mono-allelic Tcra break) helps reposition uncleaved alleles to repressive pericentromeric heterochromatin (Hewitt et al., 2009). Here we now show that ATM also functions to reduce accessibility by inhibiting higher-order loop formation. These changes in location and conformation correlate with inhibition of cleavage on the second allele. The temporal regulation of higher-order looping and PCH association are likely to be controlled by signaling pathways that impact on accessibility and transcription. Rapid release of alleles from heterochromatic regions and externalization from the territory could kick-start another round of recombination.
Our studies indicate that RAG-mediated looping away from the territory facilitates RAG-mediated interactions with homologous antigen receptor alleles providing a mechanism for regulation of cleavage in trans. However, the changes in cleavage and accessibility that we observe on paired alleles are always mirrored at the level of the overall population, so we cannot rule out that regulation in trans is equally effective whether alleles are associated or separated, as shown in the two versions of the Model (Figure S6). Nonetheless, we favor the first version in which ATM regulates nuclear accessibility and RAG-mediated cleavage of genes in the neighborhood of a DNA break (Figure S6). We propose that homologous alleles come together in localized recombination centers for coordinated regulation of RAG cleavage. Separation of alleles could occur during the repair phase of the break, which would account for the equivalent frequency of γ-H2AX association on paired and unpaired alleles. Of course, in the absence of live cell imaging studies that trace the dynamics of cleavage and repair at these loci, this is impossible to prove. Nonetheless, it is clear that closely associated uncleaved alleles will have immediate access to a high concentration of activated ATM and its downstream targets that accumulate at the site of the RAG break. Either way though, whether the mode of trans regulation occurs on paired or unpaired alleles it is apparent that an ATM-mediated negative feedback loop provides a mechanism for limiting the number of RAG-mediated breaks that are introduced on antigen receptor alleles in each cell. Furthermore, this mode of regulation is linked to maintenance of genome stability in ATM deficient mice (Callen et al., 2007; Deriano et al., 2011; Liyanage et al., 2000; Zha et al., 2010) and Ataxia Telangiectasia (AT) patients (Kobayashi et al., 1991).
Interestingly, our model for ATM-mediated feedback control of RAG cleavage at the site of a recombination break is remarkably similar to what has recently been proposed for ATM mediated feedback regulation of recombination in meiosis (Lange et al., 2011). Activation of ATM by DSBs triggers a negative feedback loop that leads to the inhibition of closely associated Spo-11p mediated DSB formation. In this context it has also been proposed that feedback control operates at a local level to halt the introduction of further DSBs. Our data indicate that different programmed DNA recombination events share common regulatory mechanisms. As in meiosis, feedback control during V(D)J recombination could also be important for inhibiting further RAG cleavage in cis after the initial breaks are introduced. These studies provide new insight into the mechanisms underlying the regulation of recombination and the phenotype of the AT disease.
EXPERIMENTAL PROCEDURES
Mouse strains
Appropriate wild-type mice (C57BL/6 CBA, 129 or WT β) or litter mates were used as control mice for the mutant phenotypes. WT, Rag1−/− and Rag1−/−-Rag1D708A mice carrying a functionally rearranged Tcrb transgene (WT β, R1−/− β and D708A-R1−/− β) were provided by Yanhong Ji and David Schatz. The Atm−/− mice generated through the interbreeding of 129SvEv Atm+/− mice, were given by Craig Bassing, Ludovic Deriano and David Roth and genotyped as described (Barlow et al., 1996). The 53BP1−/− mice were provided by Davide Robbiani and Michel Nussenzweig (Ward et al., 2003). Animal care was approved by NYU School of Medicine Animal Care and Use Committee of (protocol number 090505-02)
T cell flow cytometry sorting
Flow cytometry cell sorting was performed on a MoFlo or Reflection sorter. Antibodies were as follows: Thy1.2 PE-Cy7 (CD90.2, clone 53-2.1, eBioscience, 1:1000 dilution), TCRβ APC-eFluor780 (clone H57-597, eBioscience, 1:500 dilution), CD4 APC (L3T4, RM4-5, BD Biosciences, 1:500 dilution), CD8a FITC (clone 53-6.7, BD Biosciences, 1:500 dilution), CD25 PE (PC61, BD Biosciences, 1:500 dilution). Sorting was done as Thy1.2+/TCRβint/CD4+/CD8+ for DP cells and as Thy1.2+/TCRβlo/CD4−/CD8−/CD25+ for DN2/3 cells.
Splenic B cell purification
B cells were purified from spleens of 6- to 10-week-old mice. Cells were suspended to 108 cells per 500 µl, and labeled with magnetic microbeads coupled to anti-CD43 (Miltenyi Biotech, Auburn, CA) to remove T cells and macrophages. Cells were passed over a LS+ column following the manufacturer’s instructions, and flow-through B cells (CD43−) were collected.
Probes for 3-D DNA FISH
For details see Supplementary Information section
3-D DNA FISH and immuno-FISH
3D-DNA FISH and combined DNA FISH-immunofluorescence for γ-H2AX (immuno-FISH) were carried out on T or splenic B cells adhered to poly-L lysine coated coverslips or slides as previously described for immuno-FISH (Hewitt et al., 2009). DNA FISH and immuno-FISH combined with the chromosome 14 paint were carried out as above except that the cells were denatured in 50% formamide / 2× SSC (pH 7–7.4) for 30 minutes at 80°C before overnight hybridization, and the 3 washes were performed in 50% formamide / 2 × SSC (pH 7–7.4) and 3 times in 2 × SSC at 37°C. For details see Supplementary Information section.
Sequential RNA/DNA FISH
Sequential RNA/DNA FISH was performed as previously described (Chaumeil et al., 2008). For details see Supplementary Information section.
Confocal microscopy and analysis
For details see Supplementary Information section
Statistical analysis
For details see Supplementary Information section
ChIP-seq
Cells cross-linking, preparation of mononucleosomes-containing chromatin, chromatin immunoprecipitation and ChIP-Seq library preparation and sequencing were carried out as previously described using H3K9ac (Abcam) and H3K4me3 (Active Motif) antibodies (Ntziachristos et al., 2012). See Supplementary Information section for details on ChIP-seq alignment and ChIP-seq analysis.
4C-seq
1×107 splenic B cells were processed as previously described (Simonis et al., 2006). Gene specific PCR primers for the Tcra Eα enhancer bait were HindIII AGACAGACCCTGCGAAGCTT and DpnII TAAGACTGGACCCACAGAAC, The Illumina specified adapters for llumina GAIIx sequencing were included at the 5′ end of each primer. The 4C library was sequenced on an Illumina GAIIx single-read 72-cycle run. For details on analysis see the Supplementary Information section.
Supplementary Material
HIGHLIGHTS.
-
-
RAG dependent monoallelic loop formation is linked to monoallelic RAG cleavage
-
-
RAG enrichment, cleavage and higher-order loop formation occur at the 3’ end of Tcra
-
-
Looping out is a determinant of directed RAG targeting
-
-
ATM mediated control of looping out is linked to the maintenance of genome stability
ACKNOWLEDGEMENTS
The authors thank M. Nussenzweig and D. Robbiani for providing the 53BP1−/− mice, and C. Bassing, D. Roth and L. Deriano for providing the Atm−/− mice. We would like to thank Jeanne Allinne, Susannah Hewitt, Pedro Rocha, Thomas Trimarchi and MacLean Sellars for their help with 4C-seq experiments; Fabio Parisi and Francesco Strino for their help with statistical analyses; and members of the Skok lab for discussions and comments on the study. This work is supported by the National Institute of Health: R01GM086852 (J.A.S.), R37AI32524 (D.G.S.), RO1CA133379, RO1CA105129, RO1CA149655, and RO1GM088847 (I.A.). J.A.S. is a Leukemia & Lymphoma Society (LLS) scholar. J.C. is an Irvington Institute Fellow of the Cancer Research Institute. M.M. was supported by an NSF IGERT 0333389. L.D. was a LLS Fellow. E.N. was supported by the French Ministry of Research and the Association pour la Recherche sur le Cancer (France). I.A. is also supported by the Leukemia & Lymphoma Society (TRP grant), The V Foundation for Cancer Research, and the Dana Foundation. D.G.S. is an Investigator and I.A. an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare that they have no competing financial interests.
REFERENCES
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2011;19:901–909. doi: 10.1007/s10577-011-9245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Augui S, Chow JC, Heard E. Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol Biol. 2008;463:297–308. doi: 10.1007/978-1-59745-406-3_18. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Micsinai M, Skok JA. Combined Immunofluorescence And DNA FISH on 3D-preserved Interphase Nuclei to Study Changes in 3D Nuclear Organization. J Vis Exp. 2012:e50087. doi: 10.3791/50087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Skok JA. The role of CTCF in regulating V(D)J recombination. Curr Opin Immunol. 2012;24:153–159. doi: 10.1016/j.coi.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, et al. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, Cavalli G. Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion. PLoS Genet. 2012;8:e1002465. doi: 10.1371/journal.pgen.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Zuniga-Pflucker JC. Determining gammadelta versus alphass T cell development. Nat Rev Immunol. 2010;10:657–663. doi: 10.1038/nri2820. [DOI] [PubMed] [Google Scholar]
- Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV, Chang S, Skok JA, Roth DB. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago M, Rosenbluh C, Tevlin M, Fraenkel S, Schlesinger S, Masika H, Gouzman M, Teng G, Schatz D, Rais Y, et al. Clonal allelic predetermination of immunoglobulin-kappa rearrangement. Nature. 2012;490:561–565. doi: 10.1038/nature11496. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SL, Chaumeil J, Skok JA. Chromosome dynamics and the regulation of V(D)J recombination. Immunol Rev. 2010;237:43–54. doi: 10.1111/j.1600-065X.2010.00931.x. [DOI] [PubMed] [Google Scholar]
- Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nature biotechnology. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Tycko B, Soreng AL, Sklar J. Transrearrangements between antigen receptor genes in normal human lymphoid tissues and in ataxia telangiectasia. J Immunol. 1991;147:3201–3209. [PubMed] [Google Scholar]
- Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak F, Tourigny M, Schatz DG, Petrie HT. Characterization of TCR gene rearrangements during adult murine T cell development. J Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- Liyanage M, Weaver Z, Barlow C, Coleman A, Pankratz DG, Anderson S, Wynshaw-Boris A, Ried T. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]
- Matei IR, Gladdy RA, Nutter LM, Canty A, Guidos CJ, Danska JS. ATM deficiency disrupts Tcra locus integrity and the maturation of CD4+CD8+ thymocytes. Blood. 2007;109:1887–1896. doi: 10.1182/blood-2006-05-020917. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome research. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- Muller I, Boyle S, Singer RH, Bickmore WA, Chubb JR. Stable morphology, but dynamic internal reorganisation, of interphase human chromosomes in living cells. PLoS One. 2010;5:e11560. doi: 10.1371/journal.pone.0011560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla-Osorio N, Zhu C. DNA damage and repair during lymphoid development: antigen receptor diversity, genomic integrity and lymphomagenesis. Immunol Res. 2008;41:103–122. doi: 10.1007/s12026-008-8015-3. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Dynamic assembly of silent chromatin during thymocyte maturation. Nat Genet. 2004;36:502–506. doi: 10.1038/ng1351. [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Bassing CH, Sanda T, Brush JW, Patel H, Goff PH, Murphy MM, Tepsuporn S, Gatti RA, Look AT, et al. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. J Exp Med. 2010;207:1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.