Abstract
Background
The genus Camellia, belonging to the family Theaceae, is economically important group in flowering plants. Frequent interspecific hybridization together with polyploidization has made them become taxonomically “difficult taxa”. The DNA content is often used to measure genome size variation and has largely advanced our understanding of plant evolution and genome variation. The goals of this study were to investigate patterns of interspecific and intraspecific variation of DNA contents and further explore genome size evolution in a phylogenetic context of the genus.
Methodology/Principal Findings
The DNA amount in the genus was determined by using propidium iodide flow cytometry analysis for a total of 139 individual plants representing almost all sections of the two subgenera, Camellia and Thea. An improved WPB buffer was proven to be suitable for the Camellia species, which was able to counteract the negative effects of secondary metabolite and generated high-quality results with low coefficient of variation values (CV) <5%. Our results showed trivial effects on different tissues of flowers, leaves and buds as well as cytosolic compounds on the estimation of DNA amount. The DNA content of C. sinensis var. assamica was estimated to be 1C = 3.01 pg by flow cytometric analysis, which is equal to a genome size of about 2940 Mb.
Conclusion
Intraspecific and interspecific variations were observed in the genus Camellia, and as expected, the latter was larger than the former. Our study suggests a directional trend of increasing genome size in the genus Camellia probably owing to the frequent polyploidization events.
Introduction
The genome size is the amount of DNA in an unreplicated, basic, gametic chromosome set [1]. The study on genome size variation often provides a strong unifying element in biology with practical and predictive uses. Myriad organismal and ecological traits are frequently associated with the variation in genome size [2], [3], [4]. Therefore, the measurement of the DNA content and genome size is often employed to better understand plant evolution and enhance comparative analyses of genome evolution [5].
Genome size variation among angiosperms nearly 2400-fold, ranging from 1C = 0.06 pg in Genlisea margaretae to 1C = 152.23 pg in the Paris japonica [6], with an extensive variation occurring even within groups. The average within-genus size variation is 3-fold, with an upper bound of more than 63-fold [7]. Indeed, intraspecific variation in genome size has also been observed in many plants [8], [9]. The observed 37% variation in DNA content was found to be correlated with the number and size of heterochromatic knobs in Zea mays [10]. Another example is DNA content of flax, Linum usitatissimum, which may vary within a single generation when the plants are grown under specific environmental conditions [11]. However, Greilhuber [12] suggested that earlier numerous reports of genome size variation below the species level were dismissed by inaccurate methods which lead to the unreliable measurement results, as clearly shown in studies on endogenous staining inhibitors [13], [14], [15]. Moreover, a great stability of the nuclear genome size has been reported in geographically isolated populations of Sesleria albicans [16], different species of Settaria [17], Cistus [18], Capsicum [19], and diverse cultivars of pea and onion [20], [21]. Nevertheless, these findings should instantly provoke the question whether it is a real variation in DNA amount or simply an artifact of intraspecific variation in genome size.
The relative frequency of increases and decreases in DNA content still remains unresolved in angiosperm phylogeny [22]. Besides polyploidization, genome size is primarily influenced by the proportion of non-genic repetitive DNA, much of which originates from transposable elements [23], [24]. In particular, copy number of retrotransposons may dramatically vary from one to another genome [25], [26]. An increase in genome size may result from the amplification and accumulation of retrotransposons. Nevertheless, the decrease in genome size can be caused by a higher overall rate of deletions than insertions, selection against transposable elements, unequal crossing over, and illegitimate recombination [27]. The occurrence and extent of genome size variation among and within plant species as well as evolutionary mechanisms behind still remain controversial and more investigations are fairly needed.
The genus Camellia has been long attracted considerable attention due to its greatly economic values, broadly geographic distribution and remarkable species diversity. The main economic value of Camellia is the production of tea made from the young leaves of C. sinensis var. sinensis and C. sinensis var. assamica. In addition, C. oleifera has been primarily used for cooking oil extracted from seeds [28]. Besides, Camellia species are of great ornamental values especially represented by C. japonica, C. reticulata and C. sasanqua. The genus is taxonomically ranked as one of the most challengingly difficult taxa in plants, whose complexity is primarily governed by frequent hybridization, accompanied by polyploidization and subsequent stabilization of novel forms by clonal growth [29]. The classification of species using a morphology-based system is often changeable and also disputed based on chromosome pairing behavior of hybrids [30]. As a result, the boundaries between taxa of various ranks are still a subject of dispute. According to Chang et al. [31], Camellia was classified into a total of 18 sections of four subgenera, which approximately comprised 361 species. However, Min et al. [28] taxonomically classified the genus into 14 sections of two subgenera, consisting of only about 120 species. The available sequence-based phylogeny of this genus is necessarily limited, and many controversies have long existed with regard to their taxonomical classification. The nuclear DNA content is in some cases useful as a supportive marker for a reliable delineation of problematic taxa and possesses a predictive value to infer evolutionary relationships [32]. Unfortunately, the lack of nuclear DNA contents apparently prevents us from understanding the diversification and evolution of the Camellia species. The knowledge of interspecific and intraspecific patterns of genome size variation may help to enlighten the evolution and particularly the involved evolutionary events such as hybridization and polyploidization in the genus. In the present study, we estimated genome size of C. sinensis var. assamica by using flow cytometric analysis. In the hope of better understanding the diversification and evolution in the genus Camellia, we extensively investigated interspecific and intraspecific patterns of DNA content variation in representative sections and species. The data presented here are intended to fill a gap that exists in the current genomic knowledge base of Camellia and take nuclear DNA content variation as a useful marker to predict and infer evolutionary relationships in such problematic taxa.
Materials and Methods
Plant materials
Materials of the Camellia plants used in this study were kindly provided by Kunming Institute of Botany (Chinese Academy of Sciences), Tea Research Institute (Yunnan Academy of Agricultural Sciences, China) and International Camellia Species Garden (Jinhua, Zhejiang, China) from May to July of 2010. All necessary permits were obtained for the described field studies; names of the persons or authority who issued the permission for each location are as below: Wei-bang Sun, Kunming Botanical Garden, Chinese Academy of Sciences; Ming-zhi Liang, Tea Research Institute, Yunnan Agricultural Academy of Sciences, Yunnan, China; Ji-yuan Li, International Camellia Species Garden, Jinhua, Zhejiang, China. We collected flowers, leaves and buds from field-growing trees, which were either analyzed immediately or maintained in a refrigerator on moistened paper for a maximum of two days until use. Considering many controversies of the genus Camellia, the collected plant materials were classified and analyzed by using two taxonomical treatments (Min taxonomic system: MTS; Chang and Ren taxonomic system: CRTS) [28], [31] in hope of the delineation of problematic taxa based on nuclear DNA contents.
Sample preparation
Approximately 40–50 mg of flowers, leaves and buds were separately used for the sample preparation. Nuclei suspensions were improved according to Galbraith et al. [33] and WPB isolation buffers [34], including 0.2 mM Tris.HCl, 4 mM MgCl2.6H2O, 2 mM EDTA Na2.2H2O, 86 mM NaCl, 2.0 mM dithiothreitol (Sigma-Aldrich CHIEMIE Gmbh, Steinheim, Germany), 1% (w/v) PVP-10, 1% (v/v) Triton X-100, (pH 7.5). For each case, 1 mL of ice-cold nuclei suspensions was added to a Petri dish containing the plant tissue, which was chopped using a sharp razor blade. The resulting homogenate was filtered through a 50-µm nylon filter to remove cell fragments and large debris. Nuclei were treated with 50 µg mL−1 RNase (Fluka, Buchs, Switzerland) and stained with 50 µg mL−1 propidium iodide (PI) (Sigma, St. Louis, MO, USA). The samples were kept on ice until further uses. Maize (Z. mays L. cv. B73) with a DNA content of 1C = 2.35 pg, namely 2300 Mb [35], was employed as a standard.
Flow cytometry measurements
Nuclear samples were analyzed by using a BD FACSCalibur (USA) flow cytometer. The instrument was equipped with an air-cooled argon-ion laser tuned at 15 mW and operating at 488 nm. PI fluorescence was collected through a 645-nm dichroic long-pass filter and a 620-nm band-pass filter. The amplifier system was set to a constant voltage and gained throughout the experiments. Usually, 10,000 nuclei were analyzed for each sample. The results of flow cytometry were further analyzed by using the Cellquest software and gated to selectively visualize all cells of interest which gather densely in dotplot map while eliminating results from unwanted particles. Here, CV = D/M×l00%, D is the standard deviation of the cell distribution and M is the average of cell distribution. The average of coefficient of variation values (CV) was used to evaluate the results with which CV<5% were considered as reliable. Nuclear DNA content was calculated as a linear relationship between the ratio of 2C-value peaks of the sample and standard.
Tests for inhibitors
To determine the impact of secondary metabolites on the fluorescence of nuclei, we tested the unidentified compounds in leaves of C. sinensis var. assamica cv. yunkangshihao that reduce PI fluorescence of maize (Z. mays L. cv. B73) nuclei as follows. Treatment A consisted of PI-stained nuclei from the independently processed and stained 20–25 mg leaves of C. sinensis var. assamica and Z. mays, respectively. C. sinensis var. assamica and Z. mays were simultaneously processed (co-chopped) and stained with PI, called as treatment B. After staining, these samples were individually measured for mean PI fluorescence, and the experiment was replicated for a total of three times. The fluorescence of nuclei from leaves of the marker simultaneously processed with C. sinensis var. assamica materials was compared with that from independently processed leaves of the marker and gave evidence of inhibitors.
Statistical analyses
Differences and correlations among variables between the Camellia species as well as different tissues were statistically tested using one-way ANOVA implemented with the software SPSS (SigmaStat for Windows Version 3.1, SPSS Inc., Richmond, CA, USA).
Results
Optimization of DNA flow cytometry for the Camellia species
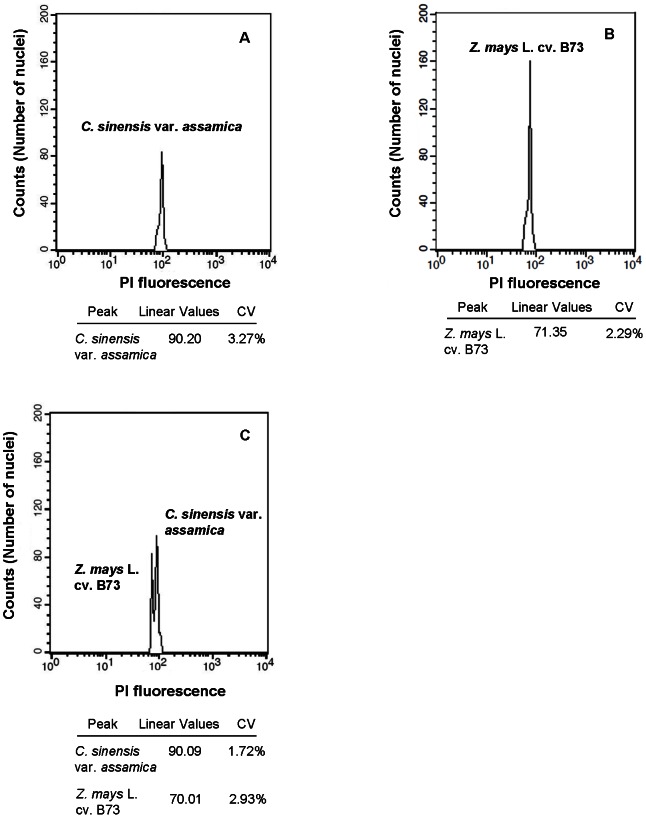
In this study, a total of five nuclear isolation buffers were compared, which included Galbraith [33], LB01 [36], Otto [37], [38], Tris.MgCl2 [39], and WPB [34] (data not shown). An improved WPB isolation buffer was finally chosen and employed in the flow cytometry, which was able to counteract the negative effects of tannic acid better than the other four buffers [40], [41]. The optimization of DNA flow cytometry generated high-quality results with low CV<5% in the present study. To determine a suitable plant tissue for the flow cytometry analysis for the Camellia species, we sampled and detected a total of three tissues, including flowers, leaves and buds from the eight species, representing up to five sections of the genus, C. oleifera, C. pyxidiacea var. rubituberculata, C. impressinervis, C. grijsii var. grijsii, C. reticulata (cv. honghuayoucha and cv. zipao), C. editha, and C. japonica (cv. feilipu). The nuclear DNA contents of Camellia species were presented as picograms and the variability of 2C-values among different tissues from a single specie was tested using one-way ANOVA (Table 1). Our results showed that 2C-values of the three tissues from a single Camellia plant had no significant differences between each other (P>0.05). The estimation of 2C-values, taking C. impressinervis for example, were 4.56±0.167, 4.59±0.138 and 4.61±0.161 pg for flower, leave and bud, respectively (P = 0.925>0.05). The largest discrepancy (0.13 pg/2C) between 2C-values of the three tissues were observed in C. editha, with 2C-value of 5.65±0.123 pg in flowers and 5.52±0.409 pg in leaves, respectively (P = 0.782>0.05). The standard deviation (SD) of 2C-value of three tissues from a single plant was more evident in the species with a large genome than the species with a small genome. For example, C. oleifera with the highest SD (0.691) in flowers and buds had the average 2C-value of 17.47 pg, while 2C-values of the three tissues of C. oleifera had no significant differences between each other (P>0.05). In addition, results showed that the flower color pigments had no obvious influence on staining results (Table 1). In order to test the impact of cytosolic compounds on the fluorescence of Camellia nuclei, we further measured and compared two filtrates of C. sinensis var. assamica cv. yunkangshihao (Fig. 1a) and Z. mays L. cv. B73 (Fig. 1b) which were treated individually, with a mixed filtrate which was co-chopped together (Fig. 1c). The PI fluorescence (linear values) of C. sinensis var. assamica and Z. mays was 90.20 and 71.35, respectively, when they were individually treated. In the co-chopped treatment, the PI fluorescence (linear values) for these two species was 90.09 and 70.01, respectively, with lower intensity peaks compared with the former. There existed 0.11 and 1.34 differences of PI fluorescence between samples treated individually and simultaneously. The average of CV were 3.27% and 2.29% for C. sinensis var. assamica (Fig. 1a) and Z. mays (Fig. 1b) alone, while the average of CV were 1.72% and 2.93% for them (Fig. 1c), respectively, which were simultaneously processed and stained.
Table 1. Comparisons of nuclear DNA amount (2C, pg) estimated with flow cytometry in different tissues of the eight species in the genus Camellia.
| Species (Min et al. 2010) [28] | Species (Chang and Ren, 1998) [31] | Flower | SD | Leaf | SD | Bud | SD | Flower colors | P |
| sect. Paracamellia | sect. Oleifera | ||||||||
| C. oleifera | C. oleifera | 17.53 | 0.691 | 17.46 | 0.213 | 17.41 | 0.691 | White | 0.968 |
| sect. Tuberculata | sect. Tuberculata | ||||||||
| C. pyxidiacea var. rubituberculata | C. rubituberculata | 4.56 | 0.245 | 4.57 | 0.112 | 4.63 | 0.113 | Red | 0.863 |
| sect. Archecamellia | sect. Chrysantha | ||||||||
| C. impressinervis | C. impressinervis | 4.56 | 0.167 | 4.59 | 0.138 | 4.61 | 0.161 | Yellow | 0.925 |
| sect. Paracamellia | sect. Paracamellia | ||||||||
| C. grijsii var. grijsii | C. yuhsienensis | 15.24 | 0.530 | 15.22 | 0.27 | 15.21 | 0.330 | White | 0.996 |
| sect. Camellia | sect. Camellia | ||||||||
| C. reticulata cv. honghuayoucha | C. reticulata cv. honghuayoucha | 15.31 | 0.339 | 15.37 | 0.015 | 15.33 | 0.550 | Light red | 0.980 |
| sect. Camellia | sect. Camellia | ||||||||
| C. reticulata cv. zipao | C. reticulata cv. zipao | 15.04 | 0.254 | 15.14 | 0.285 | 15.13 | 0.381 | Dark red | 0.911 |
| sect. Camellia | sect. Camellia | ||||||||
| C. edithae | C. edithae | 5.65 | 0.123 | 5.52 | 0.409 | 5.64 | 0.037 | Red | 0.782 |
| sect. Camellia | sect. Camellia | ||||||||
| C. japonica cv. feilipu | C. japonica cv. feilipu | 5.72 | 0.023 | 5.81 | 0.264 | 5.75 | 0.155 | Pink | 0.824 |
Z. mays L. cv. B73 was employed as a standard. The colors of flowers are given in the Table. All materials were collected from Kunming Institute of Botany, Chinese Academy of Sciences (KIBCAS).
Figure 1. Cytogram of fluorescence intensity of C. sinensis var. assamica and Z. mays L. cv. B73 nuclei isolated with an improved WPB buffer.
Leaves of C. sinensis var. assamica and Z. mays that were treated individually (a, b) or simultaneously processed (co-chopped) (c), and stained with PI. X: Relative fluorescence; Y: Number of nuclei.
Intraspecific genome size variation within C. sinensis var. assamica
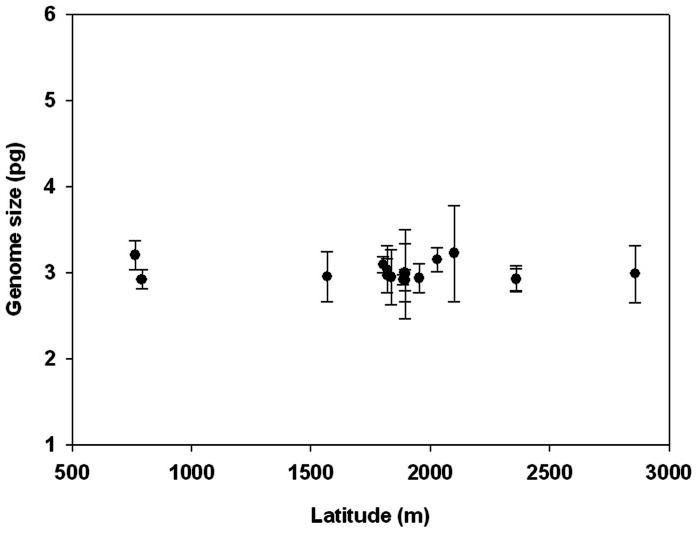
To determine the extent and patterns of intraspecific nuclear DNA content variation, we sampled a total of 17 cultivars of C. sinensis var. assamica, which extensively represent different geographic and ecological origins of the species in Yunnan Province, China (Table 2). The 2C DNA content varied only 1.1-fold among different cultivars from 5.82±0.119 pg in C. sinensis var. assamica cv. zijuan to 6.45±0.559 pg in C. sinensis var. assamica cv. manghui, with a standard deviation of 0.20. Based on the mean DNA content of all the measured cultivars (1C = 3.01 pg), the genome size of C. sinensis var. assamica was estimated to be 2940 Mb by using 1 pg DNA = 978 Mb [42]. To determine the relationship between latitudes and DNA contents of those measured C. sinensis var. assamica cultivars, we further performed the regression analysis of them. The results exhibited an R2 value of 0.033 and a low slope value of -7.418e-5, which was not statistically different from zero (Fig. 2).
Table 2. Nuclear DNA amount of C. sinensis var. assamica cultivars estimated with flow cytometry.
| Species | Chromosome Number (2n) | Estimation of Ploidy Levels | 2C-value (pg) | SD | Latitude/Longitude |
| C. sinensis var. assamica cv. bijiang | 30 | 2n = 2x | 5.97 | 0.333 | 26°55′N/98°51′E |
| C. sinensis var. assamica cv. bingdaohei | 30 | 2n = 2x | 5.91 | 0.291 | 23°38′N/99°53′E |
| C. sinensis var. assamica cv. changning | 30 | 2n = 2x | 5.93 | 0.196 | 24°50′N/99°36′E |
| C. sinensis var. assamica cv. dasiyuantou | 30 | 2n = 2x | 5.83 | 0.132 | 24°32′N/99°55′E |
| C. sinensis var. assamica cv. datuan | 30 | 2n = 2x | 5.84 | 0.06 | 21°54′N/100°26′E |
| C. sinensis var. assamica cv. fengqing | 30 | 2n = 2x | 5.86 | 0.156 | 24°32′N/99°55′E |
| C. sinensis var. assamica cv. manghui | 30 | 2n = 2x | 6.45 | 0.559 | 24°25′N/100°07′E |
| C. sinensis var. assamica cv. manluo | 30 | 2n = 2x | 5.87 | 0.174 | 22°59′N/102°24′E |
| C. sinensis var. assamica cv. mengtong | 30 | 2n = 2x | 6.07 | 0.274 | 24°50′N/99°36′E |
| C. sinensis var. assamica cv. mengyang | 30 | 2n = 2x | 6.40 | 0.168 | 22°05′N/100°53′E |
| C. sinensis var. assamica cv. naka | 30 | 2n = 2x | 6.18 | 0.093 | 23°29′N/100°42′E |
| C. sinensis var. assamica cv. nongdaoqin | 30 | 2n = 2x | 5.84 | 0.108 | 24°00′N/97°51′E |
| C. sinensis var. assamica cv. tuantian | 30 | 2n = 2x | 5.89 | 0.321 | 25°02′N/98°29′E |
| C. sinensis var. assamica cv. xiaogude | 30 | 2n = 2x | 6.30 | 0.139 | 25°03′N/100°30′E |
| C. sinensis var. assamica cv. xishelu | 30 | 2n = 2x | 5.96 | 0.520 | 25°01′N/101°32′E |
| C. sinensis var. assamica cv. yunkangshihao | 30 | 2n = 2x | 6.00 | 0.333 | 25°02′N/102°43′E |
| C. sinensis var. assamica cv. zijuan | 30 | 2n = 2x | 5.82 | 0.119 | 25°02′N/102°43′E |
Z. mays L. cv. B73 was employed as a standard. Chromosome number was taken from Min et al. (2010) [28]. All materials were collected from Tea Research Institute, Yunnan Academy of Agricultural Sciences (TRIYAAS), China. The information of latitude, longitude and altitude of germplasm origins was kindly provide by TRIYAAS.
Figure 2. The relationship between genome size (pg) and latitudinal origins of 17 cultivars of C. sinensis var. assamica.
Interspecific genome size variation of sections Thea and Camellia
The 2C-values of the 31 diploid species were measured in the section Thea [43] (Table 3). The 2C DNA contents varied 1.5-fold among these species, ranging from 4.45±0.293 pg in C. gymnogyna to 6.51±0.085 pg in C. ptilophylla. The overall mean nuclear 2C DNA content of all studied species was 5.60 pg with a 0.63 standard deviation. The DNA contents of interspecific variation (1.5-fold) in the section Thea, as expected, was somewhat larger than intraspecific variation (1.1-fold) among the representative cultivars of C. sinensis var. assamica. Apparently, our estimates of DNA ploidy (2n = 2x) based on DNA contents of these measured species were confirmed by conventional chromosome counting (2n = 30) (Table 3). The estimated 2C-values of the 22 species from the section Thea were then marked along the phylogenetic tree to show genome size variation and evolutionary relationships among species (Fig. 3). The phylogenetic tree was constructed by using UPGMA and Nei and Li's similarity coefficient from pairwise comparisons between the species based on RAPD markers [44]. In spite of slight variations, nuclear DNA contents were not randomly distributed and appeared largely conserved across the majority of the species under investigation. However, C. fengchengensis (4.64±0.341 pg) and C. pubescens (4.74±0.223 pg) were apparently found to exhibit lower DNA content than other species. Such decreased estimates of DNA content seemingly led to counterpart differences between two pairs of closely related species, C. parvisepaloides (5.94±0.243 pg) and C. fengchengensis (4.64±0.341 pg), C. pubicosta (6.24±0.196 pg) and C. pubescens (4.74±0.223 pg), with Δ 2C DNA contents of 1.3 and 1.5 pg, respectively.
Table 3. Nuclear DNA amount of the section Thea species estimated with flow cytometry.
| Species (Min et al. 2010) [28] | Species (Chang and Ren, 1998) [31] | Chromosome Number (2n) | Estimation of Ploidy Levels | 2C-value (pg) | SD | Origins |
| C. costata | C. kwangtungensis | 30 | 2n = 2x | 4.59 | 0.402 | ICSG |
| C. costata | C. danzaiensis | 30 | 2n = 2x | 4.97 | 0.540 | ICSG |
| C. crassicolumna | C. crassicolumna | 30 | 2n = 2x | 6.01 | 0.134 | TRIYAAS |
| C. crassicolumna var. crassicolumna | C. atrothea | 30 | 2n = 2x | 6.14 | 0.188 | TRIYAAS |
| C. crassicolumna var. crassicolumna | C. makuanica | 30 | 2n = 2x | 6.27 | 0.267 | TRIYAAS |
| C. crassicolumna var. crassicolumna | C. rotundata | 30 | 2n = 2x | 6.04 | 0.218 | TRIYAAS |
| C. fangchengensis | C. fengchengensis | 30 | 2n = 2x | 4.64 | 0.341 | ICSG |
| C. grandibracteata | C. grandibracteata | 30 | 2n = 2x | 5.98 | 0.233 | TRIYAAS |
| C. gymnogyna | C. gymnogyna | 30 | 2n = 2x | 4.45 | 0.293 | ICSG |
| C. kwangsiensis | C. kwangsiensis | 30 | 2n = 2x | 5.86 | 0.420 | ICSG |
| C. kwangsiensis var. kwangnanica | C. kwangnanica | 30 | 2n = 2x | 5.94 | 0.471 | TRIYAAS |
| C. leptophylla | C. leptophylla | 30 | 2n = 2x | 4.49 | 0.236 | ICSG |
| C. ptilophylla | C. ptilophylla | 30 | 2n = 2x | 6.51 | 0.085 | ICSG |
| C. ptilophylla | C. pubescens | 30 | 2n = 2x | 4.74 | 0.223 | ICSG |
| C. pubicosta | C. pubicosta | 30 | 2n = 2x | 6.24 | 0.196 | TRIYAAS |
| C. sinensis | C. sinensis | 30 | 2n = 2x | 5.81 | 0.171 | TRIYAAS |
| C. sinensis var. assamica | C. assamica | 30 | 2n = 2x | 6.00 | 0.333 | TRIYAAS |
| C. sinensis var. assamica | C. manglaensis | 30 | 2n = 2x | 6.00 | 0.182 | TRIYAAS |
| C. sinensis var. assamica | C. polyneura | 30 | 2n = 2x | 5.86 | 0.214 | TRIYAAS |
| C. sinensis var. assamica | C. sinensis var. kucha | 30 | 2n = 2x | 5.92 | 0.185 | TRIYAAS |
| C. sinensis var. assamica | C. yunkiangica | 30 | 2n = 2x | 6.02 | 0.233 | TRIYAAS |
| C. sinensis var. dehungensis | C. dehungensis | 30 | 2n = 2x | 5.48 | 0.060 | TRIYAAS |
| C. sinensis var. dehungensis | C. parvisepaloides | 30 | 2n = 2x | 5.94 | 0.243 | TRIYAAS |
| C. sinensis var. pubilimba | C. angustifolia | 30 | 2n = 2x | 4.75 | 0.237 | ICSG |
| C. sinensis var. pubilimba | C. parvisepala | 30 | 2n = 2x | 4.59 | 0.249 | ICSG |
| C. sinensis var. sinensis | C. arborescens | 30 | 2n = 2x | 5.67 | 0.343 | TRIYAAS |
| C. tachangensis | C. tachangensis | 30 | 2n = 2x | 5.97 | 0.009 | TRIYAAS |
| C. tachangensis var. remotiserrata | C. gymnogynoides | 30 | 2n = 2x | 5.96 | 0.167 | TRIYAAS |
| C. tachangensis var. remotiserrata | C. jinyunshanica | 30 | 2n = 2x | 4.77 | 0.345 | ICSG |
| C. taliensis | C. irrawadiensis | 30 | 2n = 2x | 5.91 | 0.213 | TRIYAAS |
| C. taliensis | C. taliensis | 30 | 2n = 2x | 6.11 | 0.108 | TRIYAAS |
Z. mays L. cv. B73 was employed as a standard. Chromosome numbers were adopted from previous studies and the index to Plant Chromosome Numbers (http://mobot.mobot.org/W2T/Search/ipch.html). ICSG: International Camellia Species Garden; TRIYAAS: Tea Research Institute, Yunnan Academy of Agricultural Sciences.
Figure 3. Nuclear DNA contents and evolutionary relationships among members of the section Thea [31].
The phylogenetic tree of the section Thea was constructed by using UPGMA and Nei and Li's similarity coefficient from pairwise comparisons between the 22 species and varieties based on RAPD markers [44]. The estimated 2C-values for each species are shown on the right of species, while the 1C DNA amount (pg) which also equals the genome size is shown by •.
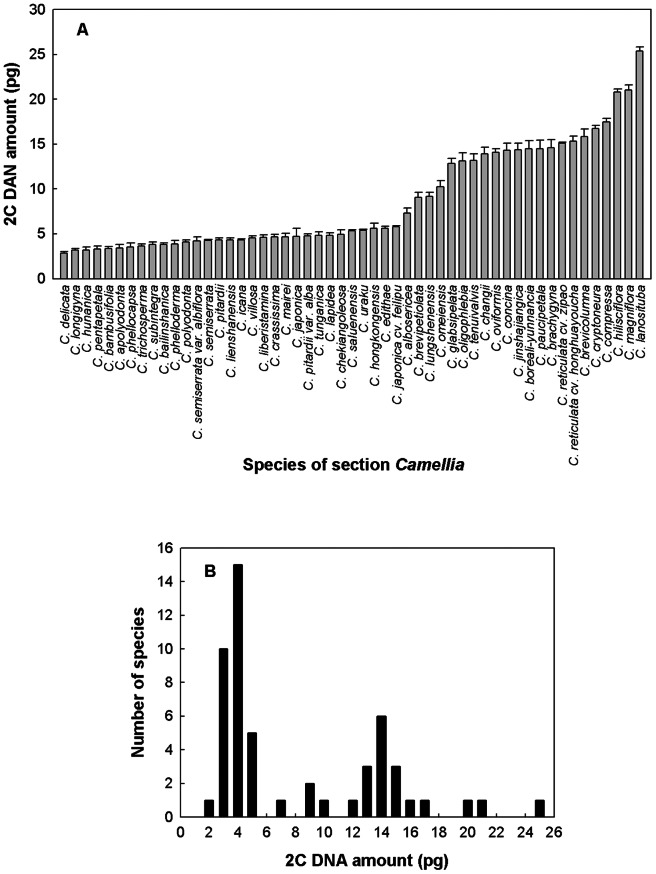
To investigate variations of DNA contents and polyploidy levels in the section Camellia, we measured 2C-values for a total of 53 species (CRTS) which were commonly recognized by the two taxonomical treatments [28], [31] (Table 4). All studied species mentioned below were followed by Chang and Ren's taxonomic system (CRTS). The 2C -values varied 8.9-fold from 2.86±0.171 pg in C. delicata to 25.35±0.484 pg in C. lanosituba (Table 4). The mean 2C-value of the section Camellia species was 8.61 pg, with a 5.78 standard deviation, larger than that of the section Thea (5.60 pg) with a 0.63 standard deviation. Figure 4a showed that the changes in DNA 2C-values of the 53 examined species arranged by increasing DNA amount in the section Camellia. Their 2C-values were greatly lower than 6 pg, and a small part of them were larger than 20 pg. Based on our results, these 2C-values were classified into the four groups (Group 1: <6 pg, Group 2: 6–10 pg, Group 3: 10–20 pg, and Group 4: >20 pg) (Fig. 4a, b). The 2C DNA contents of 31, 4, 15 and 3 species were found to fall into groups 1, 2, 3 and 4 with the percentages of 58.5%, 7.5%, 28.3% and 5.7%, respectively (Fig. 4b).
Table 4. Nuclear DNA amount of the section Camellia species estimated with flow cytometry.
| Species (Min et al. 2010) [28] | Species (Chang and Ren, 1998) [31] | Chromosome Number (2n) | 2C-value (pg) | SD | Estimation of Ploidy Levels |
| C. azalea | C. changii | NA* | 13.92 | 0.718 | 2n = 6x |
| C. chekiangoleosa | C. chekiangoleosa | 30 | 4.94 | 0.502 | 2n = 2x |
| C. chekiangoleosa | C. crassissima | 30,90 | 4.63 | 0.289 | 2n = 2x |
| C. chekiangoleosa | C. liberistamina | 30 | 4.59 | 0.304 | 2n = 2x |
| C. concina | C. concina | NA* | 14.30 | 0.819 | 2n = 6x |
| C. edithae | C. edithae | 30 | 5.61 | 0.240 | 2n = 2x |
| C. glabsipelata | C. glabsipelata | NA* | 12.83 | 0.563 | 2n = 6x |
| C. hongkongensis | C. hongkongensis | 30 | 5.60 | 0.569 | 2n = 2x |
| C. icana | C. icana | NA* | 4.31 | 0.115 | 2n = 2x |
| C. japonica | C. japonica | 30, 45 | 4.69 | 0.940 | 2n = 2x |
| C. japonica cv. feilipu | C. japonica cv. feilipu | 30 | 5.76 | 0.123 | 2n = 2x |
| C. mairei | C. mairei | 90 | 4.64 | 0.385 | 2n = 2x |
| C. mairei var. lapidea | C. delicata | 60, 90 | 2.86 | 0.171 | 2n = 2x |
| C. mairei var. lapidea | C. lanosituba | 60, 90 | 25.35 | 0.484 | 2n = 10x |
| C. mairei var. lapidea | C. lapidea | 60 | 4.85 | 0.271 | 2n = 2x |
| C. mairei var. lapidea | C. longigyna | 60, 90 | 3.19 | 0.171 | 2n = 2x |
| C. mairei var. mairei | C. omeiensis | NA* | 10.23 | 0.664 | 2n = 4x |
| C. mairei var. lapidea | C. phelloderma | 60 | 3.89 | 0.349 | 2n = 2x |
| C. pitardii | C. pitardii | 30 | 4.30 | 0.230 | 2n = 2x |
| C. pitardii var. compressa | C. compressa | 120 | 17.46 | 0.419 | 2n = 8x |
| C. pitardii var. compressa | C. magniflora | 45, 90 | 21.04 | 0.561 | 2n = 10x |
| C. pitardii var. cryptoneura | C. cryptoneura | 90 | 16.71 | 0.384 | 2n = 8x |
| C. pitardii var. cryptoneura | C. lungshenensis | 90 | 9.18 | 0.470 | 2n = 4x |
| C. pitardii var. pitardii | C. hunanica | 30 | 3.19 | 0.335 | 2n = 2x |
| C. pitardii var. pitardii | C. pitardii var. alba | 30 | 4.76 | 0.240 | 2n = 2x |
| C. pitardii var. pitardii | C. tunganica | 30 | 4.81 | 0.436 | 2n = 2x |
| C. polyodonta | C. polyodonta | 30 | 4.09 | 0.224 | 2n = 2x |
| C. polyodonta var. longicaudata | C. apolyodonta | 30 | 3.40 | 0.379 | 2n = 2x |
| C. polyodonta var. polyodonta | C. oviformis | 30 | 14.08 | 0.375 | 2n = 6x |
| C. polyodonta var. polyodonta | C. villosa | 30 | 4.57 | 0.210 | 2n = 2x |
| C. reticulata | C. albosericea | 30,60,90 | 7.30 | 0.571 | 2n = 4x |
| C. reticulata | C. bailinshanica | 60 | 3.82 | 0.170 | 2n = 2x |
| C. reticulata | C. bambusifolia | 30 | 3.33 | 0.250 | 2n = 2x |
| C. reticulata | C. boreali-yunnancia | 90 | 14.48 | 0.905 | 2n = 8x |
| C. reticulata | C. brachygyna | 60 | 14.61 | 0.875 | 2n = 8x |
| C. reticulata | C. brevicolumna | 90 | 15.85 | 0.824 | 2n = 8x |
| C. reticulata | C. brevipetiolata | 60 | 9.03 | 0.581 | 2n = 4x |
| C. reticulata | C. jinshajiangica | 90 | 14.38 | 0.725 | 2n = 8x |
| C. reticulata | C. hilisciflora | 90 | 20.80 | 0.325 | 2n = 12x |
| C. reticulata | C. oligophlebia | 60 | 13.12 | 0.902 | 2n = 6x |
| C. reticulata | C. paucipetala | 90 | 14.49 | 0.939 | 2n = 8x |
| C. reticulata | C. pentapetala | 30,60,90 | 3.32 | 0.311 | 2n = 2x |
| C. reticulata cv. honghuayoucha | C. reticulata cv. honghuayoucha | 90 | 15.34 | 0.550 | 2n = 8x |
| C. reticulata cv. zipao | C. reticulata cv. zipao | 90 | 15.10 | 0.094 | 2n = 8x |
| C. saluenensis | C. saluenensis | 30 | 5.33 | 0.125 | 2n = 2x |
| C. saluenensis | C. tenuivalvis | 30 | 13.19 | 0.712 | 2n = 6x |
| C. semiserrata | C. semiserrata | 30 | 4.27 | 0.114 | 2n = 2x |
| C. semiserrata var. semiserrata | C. phellocapsa | 30 | 3.51 | 0.441 | 2n = 2x |
| C. semiserrata var. semiserrata | C. semiserrata var. albiflora | 30 | 4.22 | 0.424 | 2n = 2x |
| C. semiserrata var. semiserrata | C. trichosperma | 30 | 3.62 | 0.235 | 2n = 2x |
| C. subintegra | C. lienshanensis | 30 | 4.30 | 0.221 | 2n = 2x |
| C. subintegra | C. subintegra | 30 | 3.80 | 0.269 | 2n = 2x |
| C. uraku | C. uraku | 30 | 5.37 | 0.150 | 2n = 2x |
Z. mays L. cv. B73 was employed as a standard. Chromosome numbers were adopted from previous studies and the index to Plant Chromosome Numbers (http://mobot.mobot.org/W2T/Search/ipch.html). All materials were collected from International Camellia Species Garden (ICSG).
NA indicates that the information of chromosome number is not available.
Figure 4. Histograms of the distribution of DNA 2C-values for the 53 species of the section Camellia [31].
The DNA 2C-values arranged by increasing DNA content (a) and the distribution of DNA 2C-values (b) for the 53 species of the section Camellia.
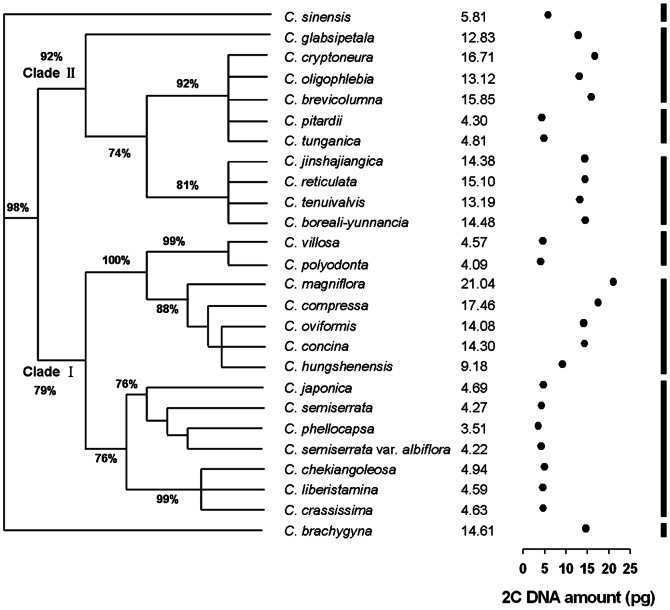
The estimated 2C-values were then marked to the phylogenetic tree of the section Camellia constructed based on ITS sequences [45] (Fig. 5). The results revealed that DNA contents were mainly conserved among closely related species. Within Clade I (79%), for example, C. japonica, C. semiserrata, C. phellocapsa, C. semiserrata var. albiflora, C. chekiangoleosa, C. liberistanmina and C. crassissima closely clustered together (76%) and displayed a fairly conservation of DNA contents of approximately 3.51±0.441 pg (C. phellocapsa) - 4.94±0.502 pg (C.chekiangoleosa). Nevertheless, C. magniflora, C. compressa, C. oviformis, C. concina and C. lungshenensis clustered together (88%), but their DNA contents increased from C. lungshenensis (2C = 9.18±0.470 pg) to C. magniflora (2C = 21.04±0.561 pg). In addition, C. polyodonta appeared closely related with C. villoda (99%) and exhibited a conserved DNA content which was much smaller than the above-mentioned species within Clade I. Those species included within Clade II (92%) showed a conserved DNA content of up to 10 pg except for C. pitardii (2C = 4.30±0.230 pg) and C. tunganica (2C = 4.81±0.436 pg), which were much lower than that of other species from the same lineage.
Figure 5. Nuclear DNA contents and evolutionary relationships among species of the section Camellia [31].
The phylogenetic tree was constructed based on ITS sequences [45]. The estimated 2C-values are shown on the right of each species, while the 2C DNA amount (pg) is given by • for each species.
Genome size variation among the Camellia species from representative sections of the genus
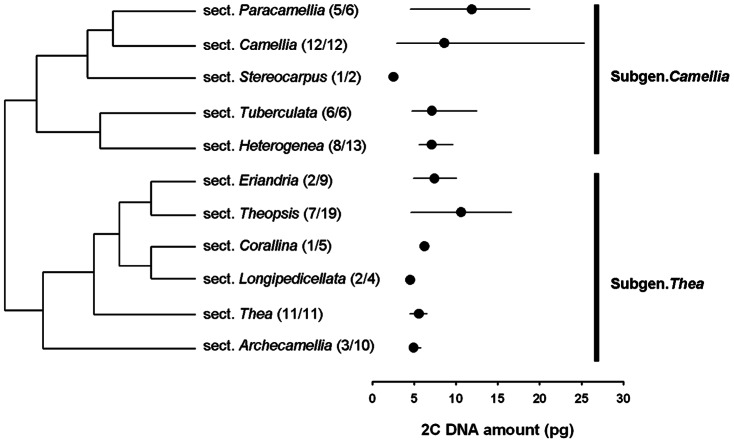
Nuclear DNA contents were more extensively sampled and examined, in addition to the above-described sections of Thea and Camellia, for a total of 38 representative species from the 10 sections [28] or 13 sections [31] in the genus Camellia (Table 5). The chromosome numbers of those measured species which were adopted from previous studies and ploidy levels which were estimated based on DNA contents were showed in Table 5. The genus Camellia was phylogenetically split into the two subgenera, Camellia and Thea [28]. Superimposing 2C-values onto a phylogenetic tree provides an interpretation of the evolutionary direction(s) of genome size evolution in the genus Camellia (Fig. 6). Increases in DNA content have apparently occurred not only in the subgenus Thea but also in the subgenus Camellia. The subgenus Camellia apparently exhibited a larger DNA content variation (10.0-fold, 2C = 2.54–25.35 pg) probably due to the polyploidization than the subgenus Thea.
Table 5. Nuclear DNA amount of representing species in the genus Camellia estimated with flow cytometry.
| Species (Min et al. 2010) [28] | Species (Chang and Ren, 1998) [31] | Chromosome Number (2n) | 2C-value (pg) | SD | Estimation of Ploidy Levels |
| sect. Paracamellia | sect. Oleifera | ||||
| C. fluviatilis var. megalantha | C. lanceoleosa | NA* | 4.53 | 0.469 | 2n = 2x |
| C. gauchowensis | C. gauchowensis | 75 | 17.98 | 0.992 | 2n = 8x |
| C. oleifera | C. oleifera | 30,45,90 | 17.47 | 0.970 | 2n = 8x |
| C. sasanqua | C. sasanqua | 45–120 | 18.79 | 0.868 | 2n = 8x |
| sect. Paracamellia | sect. Paracamellia | ||||
| C. brevistyla var. microphylla | C. microphylla | 30 | 5.48 | 0.325 | 2n = 2x |
| C. grijsii | C. grijsii | 60 | 4.93 | 0.446 | 2n = 2x |
| C. grijsii var. grijsii | C. yuhsienensis | NA* | 15.22 | 0.262 | 2n = 6x |
| C. kissii var. confusa | C. confusa | NA* | 10.86 | 1.207 | 2n = 4x |
| sect. Tuberculata | sect. Tuberculata | ||||
| C. anlungensis var. anlungensis | C. obovatifolia | 30 | 9.38 | 0.619 | 2n = 4x |
| C. ilicifolia var. ilicifolia | C. rubimuricata | 30 | 5.36 | 0.439 | 2n = 2x |
| C. parvimuricata var. hupehensis | C. hupehensis | NA* | 12.48 | 0.425 | 2n = 6x |
| C. pyxidiacea var. rubituberculata | C. rubituberculata | 30 | 4.59 | 0.272 | 2n = 2x |
| C. rhytidocarpa | C. rhytidocarpa | NA* | 4.75 | 0.299 | 2n = 2x |
| C. tuberculata | C. tuberculata | NA* | 8.52 | 1.130 | 2n = 4x |
| sect. Archecamellia | sect. Chrysantha | ||||
| C. huana | C. liberofilamenta | 30 | 5.79 | 0.233 | 2n = 2x |
| C. impressinervis | C. impressinervis | 30 | 4.59 | 0.167 | 2n = 2x |
| C. petelotii | C. nitidissima | 30 | 4.48 | 0.194 | 2n = 2x |
| sect. Theopsis | sect. Theopsis | ||||
| C. costei | C. dubia | 30 | 12.25 | 0.772 | 2n = 6x |
| C. crassipes | C. crassipes | 30 | 16.64 | 1.713 | 2n = 8x |
| C. fraterna | C. fraterna | 90 | 12.71 | 0.439 | 2n = 6x |
| C. rosthorniana | C. rotsthorniana | NA* | 4.61 | 0.237 | 2n = 2x |
| C. synaptica var. synaptica | C. tsaii | 30 | 12.72 | 0.710 | 2n = 6x |
| C. transarisanensis | C. handelii | NA* | 4.83 | 0.217 | 2n = 2x |
| sect. Longipedicellatae | sect. Longissima | ||||
| C. longissima | C. longissima | NA* | 4.67 | 0.185 | 2n = 2x |
| sect. Longipedicellatae | sect. Longipedicellatae | ||||
| C. longipedicellata | C. longipedicellata | NA* | 4.38 | 0.206 | 2n = 2x |
| sect. Stereocarpus | sect. Luteoflora | ||||
| C. luteoflora | C. luteoflora | 30 | 2.54 | 0.209 | 2n = 2x |
| sect. Eriandria | sect. Eriandria | ||||
| C. lawii | C. lawii | 30 | 4.86 | 0.275 | 2n = 2x |
| C. salicifolia | C. salicifolia | NA* | 10.02 | 0.523 | 2n = 4x |
| sect. Tuberculata | sect. Pseudocamellia | ||||
| C. tuberculata var. tuberculata | C. chungkingensis | NA* | 4.83 | 0.772 | 2n = 2x |
| sect. Heterogenea | sect. Furfuracea | ||||
| C. crapnelliana | C. crapnelliana | 30 | 5.58 | 0.263 | 2n = 2x |
| C. crapnelliana | C. gigantocarpa | 30 | 5.61 | 0.386 | 2n = 2x |
| C. pubifurfuracea | C. pubifurfuracea | NA* | 6.77 | 0.274 | 2n = 2x |
| sect. Heterogenea | sect. stereocarpus | ||||
| C. yunnanensis var. camellioides | C. liberistyloides | NA* | 5.85 | 0.197 | 2n = 2x |
| C. yunnanensis | C. yunnanensis | 30 | 5.91 | 0.007 | 2n = 2x |
| sect. Heterogenea | sect. Pseudocamellia | ||||
| C. yunnanensis var. camellioides | C. trichocarpa | NA* | 8.46 | 0.356 | 2n = 4x |
| sect. Heterogenea | sect. Archecamellia | ||||
| C. granthamiana | C. albogigas | 60 | 8.98 | 0.202 | 2n = 4x |
| C. granthamiana | C. granthamiana | 60 | 9.66 | 0.705 | 2n = 4x |
| sect. Corallinae | sect. Thea | ||||
| C. pubicosta | C. pubicosta | 30 | 6.24 | 0.323 | 2n = 2x |
Z. mays L. cv. B73 was employed as a standard. Chromosome numbers were adopted from previous studies and the index to Plant Chromosome Numbers (http://mobot.mobot.org/W2T/Search/ipch.html). All germplasms were collected from International Camellia Species Garden (ICSG).
NA indicates that the information of chromosome number is not available.
Figure 6. Nuclear DNA contents and evolutionary relationships among members of the genus Camellia.
The indicated phylogenetic relationships of the genus were constructed by using morphological data and adopted from Min et al. [28]. The numbers in brackets for each section represent the number of species with the measured nuclear DNA content followed by the total number of species comprising the section. The mean 2C DNA amount is indicated by • for each section, while the range is shown as a line from the minimum to maximum 2C DNA amounts. The two subgenera recognized in Camellia are given on the right side of the figure.
Discussion
Performance of flow cytometry for the Camellia species
High content of cytosolic compounds in the tissues of plants like the Camellia species often attracts the attention to facilitate the selection of the most appropriate buffer [46]. In addition to releasing nuclei from intact cells, lysis buffers must ensure the stability of nuclei throughout the experiment, protect DNA from degradation and ease stoichiometric staining. We finally selected and employed an improved WPB isolation buffer in the flow cytometry, which was able to counteract the negative effects of tannic acid (TA) [41] and reliably provided excellent results with lower CV<5%. In the improved WPB isolation buffer, PVP was added to bind the phenolics kept in a reduced state [34] and thus suppressed the TA effect [41]. The antioxidant dithiothreitol, a substance that preserves chromatin integrity and minimizes stoichiometric errors in the DNA staining was also added in the experiments. Loureiro et al. [34] also confirmed that WPB is suitable for the analysis of problematic tissue or species. The explanation for our excellent results of this WPB buffer may be able to improve chromatin accessibility and ‘homogenizes’ chromatin structure, eliminating differences in staining intensity among nuclei with the same DNA content. The suitable plant tissues for flow cytometry should ideally contain rapidly dividing cell without substances that interfere with the experiment. In the eight investigated species of Camellia, comparisons of flow cytometry data obtained from the flowers, leaves and buds showed little discrepancy of DNA contents among different tissues. Accordingly, leaves were selected for the evaluation of DNA contents in the next experiments in the present study. In the leaves of Camellia, specialized cells often accumulate different phenolic compounds, such as tannins in particular, which may interfere with the flow cytometry [47], [48]. Because phenolic compounds and other oxypurines are known to bind with DNA, modify DNA-supercoiling, and form a complex with intercalating dye [49]. The experimental artifacts were observed in Pinaceae species [50], which was called as ‘tannic acid effect’ [40]. However, the opposite results were obtained in the nuclei of sunflower leaves isolated in Galbraith's buffer, despite increasing the variance of the peaks [14]. Other oxypurines and alkaloids could interfere with the phenolic effect [51]. For the tea tree, dye accessibility variations are likely to be the result of caffeine-chlorogenic acids (CGA) interactions, which is often rich in secondary metabolites [15]. In our experiment, we found that C. sinensis var. assamica brought impurity into the solution showing with low intensity peaks, and thus led to the slight variation of PI fluorescence of maize when they were treated simultaneously (Fig. 1). The competition between PI and phenolic compound is thus expected, resulting in a drop in PI accessibility to DNA. Nevertheless, the impact of secondary metabolite on the fluorescence of Camellia nuclei is slight with a 0.1 pg/2C discrepancy so that it is enough to gain credible estimates of Camellia DNA content by flow cytometry.
In this study, maize (Z. mays L. cv. B73) with a DNA content of 1C = 2.35 pg was used as the standard to estimate nuclear DNA contents of the Camellia representative sections and species. An ideal scenario is to use the plant species whose genome has been completely sequenced as a reference standard and thus the genome size may accurately be determined. However, up to date, there are not any genomes have been fully sequenced, given the assembly difficulties of repeat sequences and particularly heterochromatin regions in telomeres and centromere that cannot be easily sequenced. While it is certainly true that the C-values assumed for standards can vary depending on a number of factors [52], [53], [54], this study selected maize as a reference since genome size of the species has been roughly determined comparing with numerous plants without genome sequences available [35]. Among the other sequenced plants, the estimated genome size of maize (∼2300 Mb) is comparatively close to the tea tree, and thus may be suitable to serve as a standard and obtain a relatively reliable estimation of the Camellia species.
Genome size estimation of C. sinensis var. assamica and its intraspecific variation
As C. sinensis var. assamica was reported as a diploid (2n = 30) [55], karyological uniformity and the characteristic of all cultivars of the species make it a suitable example to study intraspecific genome size variation. The 2C DNA content varied 1.1-fold among 17 cultivars of C. sinensis var. assamica, indicated that there was a low level of intraspecific variation of the genome size among the measured cultivars of C. sinensis var. assamica. Despite the fact that genome size is more likely constant at species level, intraspecific variation was indeed observed and characterized in various plant species [19]. Genome size variation is common among congeneric species [56], subspecies [57] and populations [58], [59]. This is particularly noticeable in the species with extensive geographic distribution that shows high morphological differentiation and includes several subspecific categories. In the absence of polyploidy and changes in chromosome number [60], significant variations in genome size could be due either to fluctuations within highly repetitive DNA such as retrotransposons [27], [61] or to structural rearrangements such as small amplifications and deletions at the individual chromosomal level [62]. In addition, the simultaneous presence of ‘phenolics-alkaloids’ could lead to interactions and slight intraspecific variations in nuclear DNA content of C. sinensis var. assamica [15]. In this study, our results showed that there was a lack of latitudinal effect on intraspecific variation in genome size of the examined cultivars of C. sinensis var. assamica.
Based on the mean DNA content of all the measured cultivars (1C = 3.01 pg), the genome size of C. sinensis var. assamica was estimated to be 2940 Mb by using 1 pg DNA = 978 Mb [42]. Our result apparently conflicted with a previous estimation that genome size of C. sinensis was estimated to be 4000 Mb [63]. The discrepancy might originate from RNA digestion by RNase and fluorescent-dye which were simultaneously performed [63], resulting in an overestimation due to the interference of RNA binding with PI. Note that this is the first effort to estimate genome size of the Camellia species by using a standard with which genome size is better known from the sequenced genome. Thus, another likely explanation is that the internal standards formerly employed were based on uninsurable estimates of genome size from organisms (e.g. soybean and wheat) yet to be sequenced.
Interspecific genome size variation in the genus Camellia
The DNA contents of interspecific variation (1.5-fold) in the section Thea, as expected, was somewhat larger than intraspecific variation (1.1-fold) among the representative cultivars of C. sinensis var. assamica. Apparently, our estimates of DNA ploidy (2n = 2x) based on DNA contents of these measured species were confirmed by conventional chromosome counting (2n = 30). Given the absence of polyploidization and changes in chromosome number in the section Thea [43], [55], it is likely that the variations in genome size among different species might be caused by fluctuations within highly repetitive DNA such as retrotransposons [27], [61] and structural rearrangements [62]. The present study revealed that, in spite of slight variations, nuclear DNA contents were not randomly distributed and appeared largely conserved across the majority of the species under investigation. There were different opinions with regard to taxonomic treatment on C. pubicosta, which was classified into the section Thea by Chang et al. [31] but was recently treated as a member of the section Corallinae by Min et al. [28]. Considering that differences within related species were much fewer than those irrelevant species [11], the finding suggests that C. pubicosta and C. pubescens might have a distant relationship at least in term of genome size evolution and thus require to further study the taxonomic treatment on C. pubicosta.
The section Camellia is a taxonomically complicated group of plants that is substantially influenced by frequent interspecific hybridization and polyploidization [28]. The mean 2C-value of the section Camellia species was 8.61 pg, with a 5.78 standard deviation, larger than that of the section Thea (5.60 pg) with a 0.63 standard deviation. While levels of polyploidy used in this study were based on previous chromosome counts, the results should always be designated as “DNA ploidy” or “DNA aneuploidy” as some chromosome counts are lacking [64]. Only with the aid of FCM, has it been possible to reliably assess ploidy distribution at various spatial scales, interactions among cytotypes, and evolutionary processes in diploid-polyploid sympatric populations [65], [66]. Based on the estimation of DNA contents, DNA ploidy levels for the 53 studied species were approximately determined (Table 4; Fig. 4). We inferred that DNA ploidy levels of the studied species ranged largely including 2n = 2x, 4x, 6x, 8x, 10x and 12x when an average estimation of ∼4.91 pg was applied at the diploid level. Although ploidy estimation by cytometric techniques is generally considered to be a trivial task, some precautions should be taken during data interpretation [64]. For example, there is a possibility that changes in genome size independent of polyploidy could be taking place within the genus Camellia. Our estimates of different DNA ploidy levels of these measured species should be further confirmed by conventional chromosome counting. Chromosome counts (2n = 30, 45, 60, 90, 120) [55], [67], [68] and our estimates of different DNA ploidy levels (2n = 2x–12x) of these measured species (Table 4) together indicate that the polyploidization and interspecific hybridization may mainly account for the patterns of large DNA content variation in this section. It is the polyploidization that has made the evolution of DNA content within the section appears phasic variation rather than gradual. In addition, our results showed that DNA content varied among different diploid species, suggesting that there may be the other factors causing the difference of genome size in this section. The most likely explanation is the varied extent of amplification of repeat sequences [4], [60] occurred in different species and possible hybridization between closely related taxa [58]. We further showed that DNA contents were mainly conserved among closely related species and its variation is nearly consistent to evolutionary relationships of the section Camellia species, as indicated by molecular phylogenetic evidence [45]. Accordingly, our results further support that nuclear DNA content has a predictive value for inferring evolutionary relationships [32]. While genome size data can help to understand evolutionary relationships, there are many cases where the variation between species is not at all helpful as one can get big differences in genome size between closely related species.
Genome size evolution of the genus Camellia
Many studies on a currently unresolved question on the variation of DNA contents from a phylogenetic perspective suggested that the evolutionary direction(s) of DNA content in plants could increase [27], decrease [22], [57], or exhibit a bio-directional dynamic [1]. The genus Camellia was phylogenetically split into the two subgenera, Camellia and Thea [28]. Increases in DNA content have apparently occurred not only in the subgenus Thea but also in the subgenus Camellia. Our results suggested that the ‘increase’ hypothesis for genome size evolution may hold true in the genus Camellia. There are a small number of reductions of DNA content in certain lineages might due to an incomplete sampling. We found that the diploid species account for a large percentage of those measured species, representing in all those sampled sections. It seems likely that the speciation occurred among different sections of the genus earlier than polyploidization events, leading to that all sections contained diploids in addition to polyploidy species. It is clear that polyploidization occurred more frequently in the recently diverged sections (e.g. sections Paracamellia and Camellia, MTS) than other sections (e.g. section Stereocarpus, MTS) in the two subgenera. In addition, the majority of the 26 studied Camellia species are hexaploid. It may be inferred that the polyploidization may main lead evolutionary direction of the genus Camellia, which is consistent to the previous study [69]. Moreover, artificial selection might have played an ineligible role in genome size evolution of the genus Camellia on account of the advantages and ornamental value of polyploidy with large flowers. With the hope of outlining a full picture of genome size variation and evolution of the genus Camellia, the future work is needed to investigate phylogenetic relationships, karyotypes and genome sizes of other undetermined species.
Funding Statement
This work was supported by National Science Foundation of China (U0936603), Top Talents Program of Yunnan Province (20080A 009), and Hundreds of Oversea Talents Program of Yunnan Province to L.Z. Gao, and grants from National Science Foundation of China (31200515) and Natural Science Foundation of Yunnan Province to H. Huang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Soltis DE, Soltis PS, Bennett MD, Leitch IJ (2003) Evolution of genome size in the angiosperms. Am J Bot 90: 1596–1603. [DOI] [PubMed] [Google Scholar]
- 2. Chase MW, Hanson L, Albert VA, Whitten WM, Williams NH (2005) Life history evolution and genome size in subtribe Oncidiinae (Orchidaceae). Ann Bot 95: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beaulieu JM, Moles AT, Leitch IJ, Bennett MD, Dickie JB, et al. (2007) Correlated evolution of genome size and seed mass. New Phytol 173: 422–437. [DOI] [PubMed] [Google Scholar]
- 4. Wakamiya I, Newton R, Johnston JS, Price HJ (1993) Genome size and environmental factors in Pinus . Am J Bot 80: 1235–1241. [Google Scholar]
- 5. Lee CE (2002) Evolutionary genetics of invasive species. Trend Ecol Evol 17: 386–391. [Google Scholar]
- 6. Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot 107: 467–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grover CE, Yu Y, Wing RA, Paterson AH, Wendel JF (2008) A phylogenetic analysis of indel dynamic in the cotton genus. Mol Biol Evol 7: 1415–1418. [DOI] [PubMed] [Google Scholar]
- 8. Price HJ (1988) Nuclear DNA content variation within angiosperm species. Evol Trend Plant 2: 53–60. [Google Scholar]
- 9. Leong-Skornickova J, Sida O, Jarolimova V, Sabu M, Fer T, et al. (2007) Chromosome numbers and genome size variation in Indian species of Curcuma (Zingiberaceae). Ann Bot 100: 505–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laurie DA, Bennett MD (1985) Nuclear DNA content in the genera Zea and Sorghum. Intergeneric, interspecific and intraspecific variation. Heredity 55: 307–313. [Google Scholar]
- 11. Cullis CA (2005) Mechanisms and control of rapid genomic changes in flax. Ann Bot 95: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greilhuber J (2005) Intraspecific variation in genome size in angiosperms: identifying its existence. Ann Bot 95: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greilhuber J (1998) Intraspecific variation in genome size: a critical reassessment. Ann Bot 82: 27–35. [Google Scholar]
- 14. Price HJ, Hodnett G, Johnston JS (2000) Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propidium iodide fluorescence. Ann Bot 86: 929–934. [Google Scholar]
- 15. Noirot M, Barre P, Duperrayy C, Louran J, Hamon S (2003) a. Effects of caffeine and chlorogenic acid on propidium iodide accessibility to DNA: consequences on genome size evaluation in coffee tree. Ann Bot 92: 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lysák MA, Rostková A, Dixon JM, Rossi G, Doležel J (2000) Limited genome size variation in Sesleria albicans . Ann Bot 86: 399–403. [Google Scholar]
- 17. Le Thierrry d'Ennequin M, Panaud O, Brown S, Siljak-Yakovlev A, Sarr A (1998) First evaluation of DNA content in Settaria genus by flow cytometry. J Hered 89: 556–559. [Google Scholar]
- 18. Ellul P, Boscaiu M, Vicente O, Moreno V, Roselló JA (2002) Intra- and interspecific variation in DNA content in Cistus (Cistaceae). Ann Bot 90: 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moscone EA, Baranyi M, Ebert I, Greilhuber J, Ehrendorfer F, et al. (2003) Analysis of nuclear DNA content in Capsicum (Solanaceae) by flow cytometry and Feulgen densitometry. Ann Bot 92: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baranyi M, Greilhuber J (1995) Flow cytometric analysis of genome size variation in cultivated and wild Pisum sativum (Fabaceae). Plant Syst Evol 194: 231–239. [Google Scholar]
- 21. Bennett MD, Johnston S, Hodnett GL, Price HJ (2000) Allium cepa L. cultivars from four continents compared by flow cytometry show nuclear DNA constancy. Ann Bot 85: 351–357. [Google Scholar]
- 22. Wendel JF, Cronn RC, Johnston JS, Price HJ (2002) Feast and famine in plant genomes. Genetica 115: 37–47. [DOI] [PubMed] [Google Scholar]
- 23. Barakat A, Carels N, Bernardi G (1997) The distribution of genes in the genomes of Gramineae . P Natl Acad Sci USA 94: 6857–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grover CE, Wendel JF (2010) Recent insights into mechanisms of genome size change in plants. J Bot doi:10.1155/2010/382732. [Google Scholar]
- 25. Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, et al. (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16: 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17: 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennetzen J, Ma J, Devos K (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min TL, Wu ZY, Li DZ, Hong DY, Zhang XC, et al.. (2010) Flora of China. Science Press. pp. 366–478.
- 29. Krahulcová A, Krahulec F (2000) Offspring diversity in Hieracium subgen. Pilosella (Asteraceae): new cytotypes from hybridization experiments and from open pollination. Fragm Flor Geobot 45: 239–255. [Google Scholar]
- 30. Kamemoto H (1987) Genome breeding in Dendrobium orchids. In: The breeding of horticultural crops Chang WN, Opena RT, editors. Taipei: FFTC; 35: 182–188. [Google Scholar]
- 31. Chang HD, Ren SX (1998) Flora of China. Science Press. Tomus 49 (3) 1–251. [Google Scholar]
- 32. Suda J, Krahulcová A, Trávnícek P, Rosenbaumová R, Peckert T, et al. (2007) Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Ann Bot 100: 1323–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, et al. (1983) Rapid flow cytometric analysis of the cell-cycle in intact plant-tissues. Science 220: 1049–1051. [DOI] [PubMed] [Google Scholar]
- 34. Loureiro J, Rodriguez E, Doležel J, Santos C (2007) Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot 100: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, et al. (2009) The B73 maize genome: complexity, diversity and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- 36. Doležel J, Binarová P, Lucretti S (1989) Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plantarum 31: 113–120. [Google Scholar]
- 37.Otto F (1992) Preparation and staining of cells for high-resolution DNA analysis. In: Radbruch A, editor. Flow cytometry and cell sorting. Berlin: Springer-Verlag, pp. 101–104.
- 38. Doležel J, Göhde W (1995) Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry 19: 103–106. [DOI] [PubMed] [Google Scholar]
- 39. Pfosser M, Amon A, Lelley T, Heberle-Bors E (1995) Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat-rye addition lines. Cytometry 21: 387–393. [DOI] [PubMed] [Google Scholar]
- 40. Loureiro J, Rodriguez E, Doležel J, Santos C (2006) a. Flow cytometric and microscopic analysis of the effect of tannic acid on plant nuclei and estimation of DNA content. Ann Bot 98: 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loureiro J, Rodriguez E, Doležel J, Santos C (2006) b. Comparison of four nuclear isolation buffers for plant DNA flow cytometry. Ann Bot 98: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doležel J, Bartoš J, Voglmayr H, Greilhuber J (2003) Nuclear DNA content and genome size of trout and human. Cytometry A 5: 127–128. [DOI] [PubMed] [Google Scholar]
- 43. Li GT, Liang T, Zhang RQ, Zhang ML (2007) Study of the karyotypes of some species of section Thea . J Tea Bus 1: 25–28. [Google Scholar]
- 44. Chen L, Yamaguchi S, Wang PS, Xu M, Song WX, et al. (2002) Genetic polymorphism and molecular phylogeny analysis of section Thea based on RAPD markers. J Tea Sci 22: 19–24. [Google Scholar]
- 45. Tian M, Li JY, Ni S, Fan ZQ, Li XL (2008) Phylogenetic study on section Camellia based on ITS sequences data. Acta Hort Sin 35: 1685–1688. [Google Scholar]
- 46.Kuo J, McComb AJ (1989) Seagrass taxonomy, structure and development. In: Larkum AWD, McComb A, Shepherd SA, editors. Biology of Seagrasses. Elsevier, Amsterdam. pp. 112–156.
- 47. Greilhuber J (1988) ‘Self-tanning’-a new and important source of stoichiometric error in cytophotometric determination of nuclear DNA content in plants. Plant Syst Evol 158: 87–96. [Google Scholar]
- 48. Greilhuber J (1986) Severly distorted Feulgen DNA amounts in Pinus (Coniferophytina) after nonadditive fixations as a result of meristematic self-tanning with vacuole contents. Can J Genet Cytol 28: 409–415. [Google Scholar]
- 49. Traganos F, Kapuscinski J, Darzynkiewicz Z (1991) Caffeine modulates the effects of DNA-intercalating drugs in vitro a flow cytometric and spectrophotometric analysis of caffeine interaction with novantrone, doxorubicine, ellipticine and the doxorubicne analogue AD198. Cancer Res 51: 3682–3689. [PubMed] [Google Scholar]
- 50. Greilhuber J (1986) Severly distorted Feulgen DNA amounts in Pinus (Coniferophytina) after nonadditive fixations as a result of meristematic self-tanning with vacuole contents. Can J Genet Cytol 28: 409–415. [Google Scholar]
- 51. Noirot M, Poncet V, Barre P, Hamon P, Hamon S, et al. (2003) b. Genome size variations in diploid African coffea species. Ann Bot 92: 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dolezel J, Greilhuber J (2010) Nuclear genome size: are we getting closer? Cytometry 77A: 635–642. [DOI] [PubMed] [Google Scholar]
- 53. Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot 107: 467–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Suda J, Leitch IJ (2010) The quest for suitable reference standards in genome size research. Cytometry 77A: 717–720. [DOI] [PubMed] [Google Scholar]
- 55. Gu ZJ, Sun XF (1997) A karyomorphological study of seventeen species of Chinese Camellia . Acta Bot Yunn 19: 159–170. [Google Scholar]
- 56. Price HJ (1976) Evolution of DNA content in higher plants. Bot Review 42: 27–52. [Google Scholar]
- 57. Price HJ, Dillon SL, Hodnett G, Rooney WL, Ross L, et al. (2005) Genome evolution in the genus Sorghum (Poaceae). Ann Bot 95: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hall SE, Dvorak WS, Johnston JS, Price HJ, Willians CG (2000) Flow cytometric analysis of DNA content for tropical and temperate new world pines. Ann Bot 86: 1081–1086. [Google Scholar]
- 59. Šmarda P, Bureš P (2006) Intraspecific DNA content variability in Festuca pallens on different geographical scales and ploidy levels. Ann Bot 98: 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ohri D, Khoshoo TN (1986) Genome size in gymnosperms. Plant Syst Evol 153: 119–132. [Google Scholar]
- 61. Šmarda P, Bureš P (2010) Understanding intraspecific variation in genome size in plants. Preslia 82: 41–61. [Google Scholar]
- 62. Williams RR, Broad S, Sheer D, Ragoussis J (2002) Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res 272: 163–175. [DOI] [PubMed] [Google Scholar]
- 63. Tanaka J, Taniguchi F (2006) Estimation of the genome size of Tea (Camellia sinensis), Camellia (C. japonica), and their interspecific hybrids by flow cytometry. J Tea Res 101: 1–7. [Google Scholar]
- 64. Soda J, Krahulcová A, Trávnícek P, Krahulec F (2006) Ploidy level versus DNA ploidy level: an appeal for consistent terminology. Taxon 55: 447–450. [Google Scholar]
- 65. Baack EJ (2004) Cytotype segregation on regional and microgeographic scales in snow buttercups (Ranunculus adoneus: Ranunculaceae). Amer J Bot 91: 1783–1788. [DOI] [PubMed] [Google Scholar]
- 66. Husband BC, Sabara HA (2004) Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytol 161: 701–711. [DOI] [PubMed] [Google Scholar]
- 67. Li GT, Liang T (1990) The chromosome counts and karyomorphological study of Camellia . Guihaia 10: 127–138. [Google Scholar]
- 68. Li GT (2001) New advance in karyotype studies of genus Camellia . Chinese Wild Plant Res 20: 9–14. [Google Scholar]
- 69.Chang HD (1981) Systemic research on Camellia. Zhongshan University Press. pp. 52–84.