Abstract
Plant seeds naturally accumulate storage reserves (proteins, carbohydrates, lipids) that are mobilized during germination to provide energy and raw materials to support early seedling growth. Seeds have been exploited as bioreactors for the production to foreign materials, but stable, high level expression has been elusive, in part due to the intrinsic bias for producing the natural reserves in their typical proportions. To identify mutants governing seed filling, we screened a population of mutagenized Arabidopsis plants for a mutant that failed to fill its seeds. Here we report the identification of ssp1, a recessive, viable mutant that accumulates approximately 15% less protein than wildtype seeds. Molecular analyses revealed that ssp1 is due to the introduction of a premature stop codon in CRU3, one of the major cruciferin genes. Unlike many other reserve mutants or transgenic lines in which seed storage protein levels are reduced by antisense/RNAi technologies, ssp1 exhibits low level compensation by other reserves, and represents a mutant background that might prove useful for high level expression of foreign proteins. To test this hypothesis, we used a bean phytohemagglutinin (PHA) gene as a reporter and compared PHA expression levels in single copy insertion lines in ssp1 vs. wildtype. These near isogenic lines allow reporter protein levels to be compared without the confounding and sometimes unknown influences of transgene copy number and position effects on gene expression. The ssp1 lines consistently accumulated more PHA than the backcrossed counterparts, with increases ranging from 12% to 126%. This proof of principle study suggests that similar strategies in crop plants may improve the yield of foreign proteins of agronomic and economic interest.
Introduction
Historically, bacterial, fungal and animal cell cultures have been exploited to produce chemicals and macromolecules of importance to human health, agriculture, and a variety of industrial processes. The advent of recombinant DNA and transgenic technologies has revolutionized biomolecular engineering to facilitate large-scale production of such molecules. In general, these systems employ bioreactors that necessitate sterility and require considerable capital investment and maintenance expenses. Plants have long been viewed as attractive alternatives to bioreactors, as existing mechanical harvesting, cropping systems, and processing technologies can be employed to reduce infrastructure costs. Additionally, research on the biology of naturally occurring metabolites (e.g. seed storage proteins, lipids, starch) and knowledge of the regulatory control of the expression, modification, maturation, targeting, and catabolism of these macromolecules have informed strategies for plant improvement. Translational research successes have been reported in many areas of agriculture and include yield enhancement [1], food quality [2], pathogen resistance [3], and the targeted expression of specific biomolecules of interest to the fields of medicine, pharmacology and industrial processes. In addition, value-added products such as vitamins [4], vaccines [5], biodegradable plastics [6], and biofuels [7], extend the potential economic and agronomic benefits of transgenic plants.
Numerous approaches for various vector delivery systems, promoter types, and targeting sequences have been explored [8]–[9], and focusing expression of desirable proteins in naturally occurring storage organs such as seeds and tubers has met with some success [10]–[13]. Nevertheless, high-level expression of foreign proteins has proven to be elusive in many cases. Seeds, in particular, may contain processing proteases that may cleave the foreign proteins, reducing the stability/activity of the intended product. The lack of robust expression of transgenes also is due to the apparent bias of the seed towards filling with endogenous reserves. Several lines of evidence suggest a seed filling sensor exists to monitor reserve deposition and composition. For example, transgenic rice harboring a sulfur rich sunflower protein gene express the transgene at relatively high levels, but the total protein and amino acid composition is relatively unchanged due to the downregulation of endogenous reserve proteins [14]. Similarly, Scossa and coworkers [15], found that overexpression of a glutenin gene in wheat resulted in a compensatory reduction in the endogenous prolamins. These and other such studies have revealed that grain quality can be manipulated either by induced mutations or by the expression of specific transgenes. For example, high lysine corn varieties have been developed by exploiting mutations (e.g. opaque 2) that reduce the level of lysine-poor zein proteins, which may also be reduced by employing RNAi technology [16]. Reduction in α zein levels is often associated with rebalancing of the amino acid content to increase the relative proportions of essential amino acids [17]. Likewise, in Phaseolus vulgaris, removal of the 7S globulin phaseolin and the lectin phytohemagglutinin results in the up-regulation of several sulphur-rich proteins, and increased levels of cysteine and methionine, at the expense of the non-protein amino acid, S-methylcysteine [18]. In the vast majority of studies, reserve compensation occurs in one of the following forms: enhanced expression of the target protein results in reduced levels of other proteins to maintain either a constant protein level and/or a rebalanced proteome that exhibits a similar amino acid composition profile as wildtype. Alternatively or in addition, manipulation of the levels of one type of reserve material (e.g. lipids) can influence the level of other reserve macromolecules (e.g. starch). These experiments reveal that metabolite/volume sensor(s) exist to monitor and adjust seed reserve contents. Despite evidence for a filling sensor(s), no such molecule has been identified by mutant screens or genomics approaches. An alternative strategy has been employed to reduce the levels of endogenous reserves by employing antisense/RNAi technologies directed against one or more seed storage protein genes/classes. This approach, often coupled with cotransformation of transgenes that express a desired protein driven by a seed specific promoter, has met with success in several different systems. For example, in rice, suppression of both the prolamins and glutelins by RNAi was associated with relatively high level expression of a human growth hormone polypeptide [19]. Similarly, in soybeans, suppression of conglycinin expression was correlated with enhanced expression of a GFP reporter driven by a glycinin promoter [20], yet this same effect was not observed in plants in which both glycinin and conglycinin levels were reduced [21]. In many such studies, detailed analyses of seed reserves are not reported such as to evaluate compensation, and/or the copy number and position effect of transgenes is not presented to unequivocally show that confusing the filling sensor can be exploited to enhance the production of a desirable product.
We screened mutagenized Arabidopsis lines for seeds that are defective in seed filling in an attempt to discover a sensor. To this end, we have identified a viable mutant, ssp1, which exhibits low-level compensation, and we describe the testing and validation of ssp1 for enhancing the expression of a transgene, based on comparisons between single locus reporter isogenic lines in which the expression of the reporter is evaluated in both wildtype and ssp1 genetic backgrounds.
Results
Screen for mutants defective in storage protein accumulation
To identify mutants defective in seed filling and which might represent lesions in sensor/regulatory genes that could be exploited for overexpression of foreign genes, we generated a population of Arabidopsis EMS-mutagenized lines and screened seed protein extracts by gel electrophoresis. The polypeptide pattern of Laemmli buffer extracted seeds is simple due to the abundance of two classes of seed storage proteins, the 2S albumins and the 12S cruciferins. Cruciferins are the major type of seed protein in Arabidopsis, and are encoded by three genes, CRU1, CRU2 and CRU3 [22]. The cruciferins are 12S globulins that are synthesized as preproproteins, and are ultimately cleaved into α (30–35 kD) and β (21–25 kD) polypeptides and assembled into hexamers [23]. CRU1 and CRU2 exhibit 70% identity and are of similar size, while CRU3 has an extended glutamine rich region in the center of a larger polypeptide that contributes to its divergence from the other family members (CRU3 exhibits approximately 50% identity to CRU1/CRU2). In general, the posttranslational cleavage events that generate the α and β polypeptides give rise to two distinct clusters of bands when visualized by Coomassie staining of SDS gels. Of over 3000 seed extracts examined, one exhibited a dramatic deviation from the characteristic polypeptide pattern of wildtype seeds (Figure 1). This mutant was designated ssp1 (seed storage protein mutant 1). In the ssp1 mutant, the largest α and βpolypeptides are apparently missing or reduced in size, while the pattern of the other polypeptides is generally unchanged.
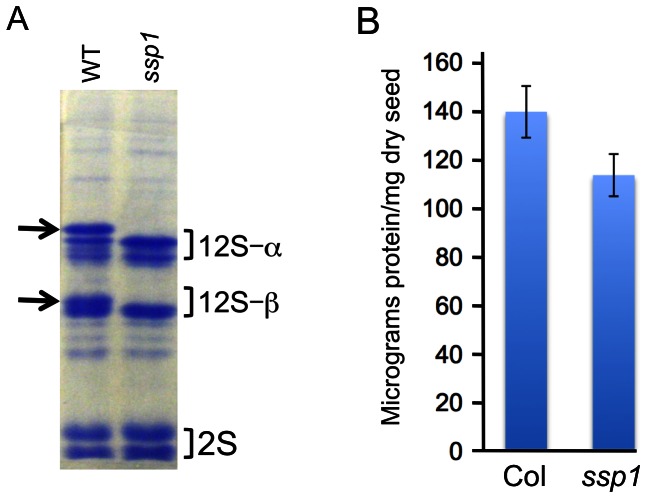
Figure 1. ssp1 gel profile and seed protein content.
A. Mature seeds from Columbia (WT) and ssp1 were extracted in Laemmli buffer and subjected to SDS-PAGE and staining with Coomassie Brilliant Blue. The 12S cruciferins are post-translationally cleaved into stable a (30–35 kD) and b (21–25 kD) polypeptides, and are predominant components of the gel profile, as are the smaller 2S albumins (2S). The arrows point to two cruciferin polypeptides that are missing from the ssp1 profile. B. Relative protein levels in mature seed extracts from WT and ssp1. Lowry assays were performed on triplicate aliquots of seed extracts and the optical density values were mathematically transformed to yield micrograms of protein per milligram of dry seed, based on the use of BSA as a protein standard. The data represent the average of values from three independent experiments. T-test statistics for ssp1 vs. wildtype reveal a p-value of less than 0.001.
To be potentially useful as a vehicle for overproduction of foreign proteins, ssp1 or other mutants in which the sensor/filling roles are compromised should fail to fill their seeds properly and as such, would possess increased storage capacity for foreign proteins. To evaluate the potential of ssp1 as an ‘empty container’, we conducted protein assays on seed extracts from wildtype and ssp1 seeds, and found that ssp1 seeds contain approximately 15% less protein than wildtype (Figure 1b). The Students T-test was employed to compare the triplicate values and support the significance of this difference with a p value of less than 0.01.
Biochemical analyses of ssp1 and wildtype seeds
To more carefully assess the levels of seed reserves in ssp1, protein, lipid, amino acid, and starch analyses were undertaken. Quantitative two dimensional gel analysis of phenol extracted seeds and comparison of the staining patterns to the 2-D gel map of Higashi et al. [24], confirmed that the CRU3 bands are missing from ssp1, and mass spectroscopy of excised trypsin digested proteins revealed that there is some compensation in ssp1 by elevated levels of CRU1 (Figure 2 and data not shown). In wildtype plants, CRU3 consists of two prominent bands, though molecular and biochemical analyses (2-D gels followed by immunoblotting) revealed that a variety of processed/phosphorylated forms of CRU3 contribute to polypeptide diversity [25]. In general, the wildtype and ssp-1 profiles are remarkably similar, with minor differences in protein abundance as gauged by stained gels. Amino acid analyses of total cellular proteins in the two backgrounds was undertaken to determine if the amino acid composition differs (Figure 3). Although it is clear that CRU1 is more abundant in ssp1 than wildtype (Figure 2), there appear to be only minor changes to the 2D protein profile that lead to a rebalancing of the amino acid composition to be reflective of the wildtype profile. There are minor differences in the levels of many amino acids, but statistically, these differences are not significant. Nevertheless, despite this rebalancing, the level of total seed protein in ssp1 is significantly lower than wildtype.
Figure 2. Two dimensional protein profiles of wildtype vs. ssp1.
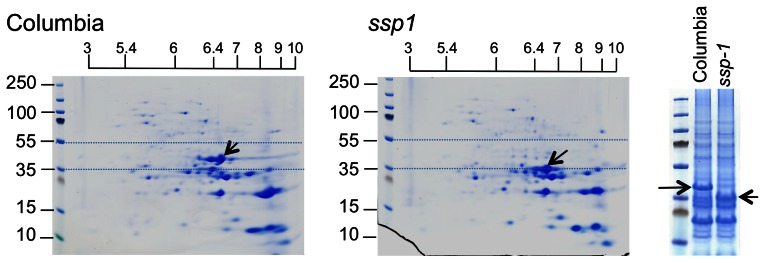
Total seed proteins were extracted and equivalent amounts of protein subjected to 2D gel analysis. Note the general concordance of spots in each profile. The major differences (for CRU3, ssp1 panel; and CRU1, Columbia panel) are highlighted by the arrows.
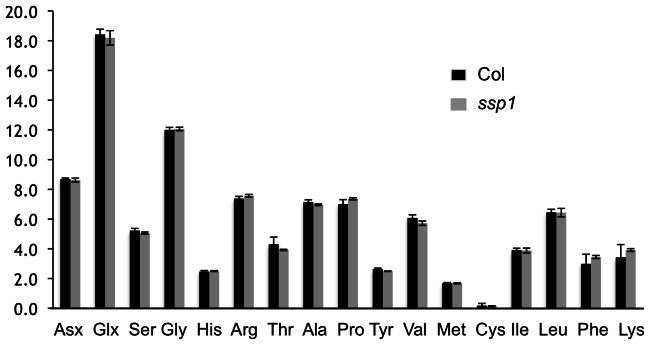
Figure 3. The amino acid composition of ssp1 seeds is largely unchanged.
Total seed protein was extracted from wildtype and ssp1 in triplicate and analyzed by reverse phase HPLC. The three letter amino acid code applies except to Asx (Asn+Asp) and Glx (Gln+Glu). Statistical analyses revealed no significant changes for any amino acid, suggesting that the loss of the CRU3 component is adjusted by a rebalancing of the proteome.
Lipid profiling was conducted to ascertain if the ssp1 mutation alters oil content or composition. Triplicate samples of both Columbia and ssp1 were analyzed by NMR for oil content and by gas chromatography for fatty acid composition (Figure 4). Interestingly, ssp1 contains less oil than wildtype (26.6% vs. 31.5%), yet the percentage of most major classes of fatty acids are slightly elevated in ssp1 relative to wildtype. Two exceptions are palmitic acid (16∶0) and linolenic acid (18∶3), which exhibit slightly higher levels in wildtype. With respect to the linolenic acid levels, a reasonable conclusion is that the delta 15 desaturase level/activity is reduced in ssp1. It is unclear how oil content and composition are altered by ssp1, but this observation may be related to a filling sensor effect whereby the protein-to-oil ratio is maintained in the ssp1 background by a concomitant reduction in lipids.
Figure 4. Lipid profiles of wildtype and ssp1 seeds.
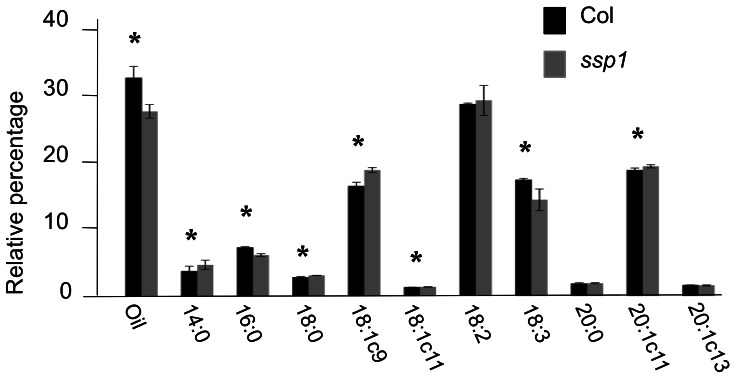
Aliquots of wildtype and ssp1 were analyzed in triplicate by NMR for oil content, and by gas chromatography for the fatty acid species indicated. The oil content of ssp1 is lower than wildtype, but several fatty acid species are slightly enriched. The asterisks indicate pairwise measurements where T-test statistics revealed significant (p<0.05) differences.
Starch measurements revealed no significant differences between ssp1 and wildtype (data not shown). In summary, ssp1 seeds contain about 15% less protein than wildtype, and exhibit proteome rebalancing to maintain a wildtype amino acid composition. Unlike other naturally occurring or induced seed protein mutants (see the Discussion) which compensate by increasing the levels of proteins or other seed reserves, ssp1 does not adjust the level of protein reserves to compensate for the loss of CRU3, and thus meets the criterion for a genetic background that might be exploited to overexpress foreign proteins.
Molecular analyses of ssp1
The absence of one α and one β cruciferin polypeptide in ssp1 could be due to a lesion in a regulatory gene that governs one of the three CRU genes or alternatively, a mutation in one of the structural genes. RNA gel blotting was employed to determine the relative expression of CRU1, CRU2 and CRU3 in both wildtype and ssp1 (Figure 5). CRU1 and CRU2 are expressed at similar levels in ssp1 and wildtype seeds, but the ssp1 mutant contains no detectable mRNA for CRU3. This analysis assigns the largest α andβ polypeptides as products of CRU3 mRNA translation and indicates that the ssp1 defect is CRU3 specific.
Figure 5. The ssp1 phenotype is due to a premature stop codon in CRU3.
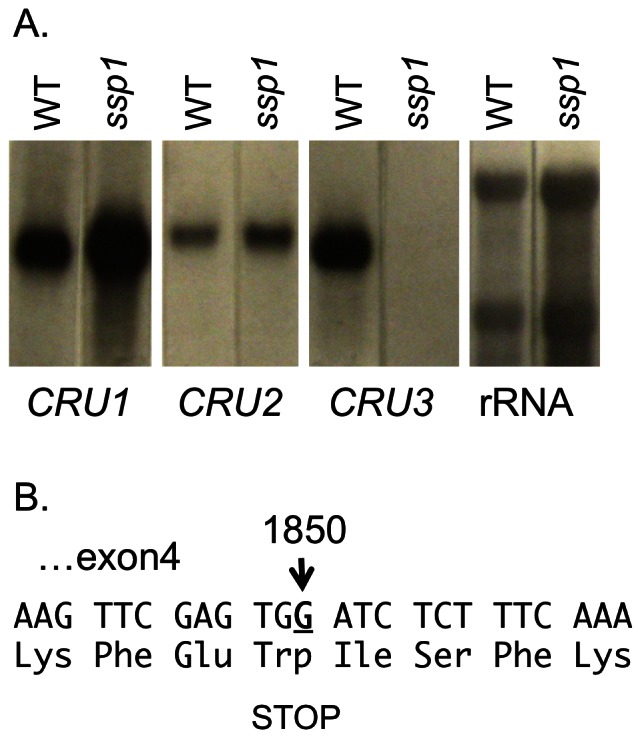
Aliquots of total RNA from mature seeds were purified and subjected to RNA gel blot analysis with probes prepared from cloned CRU1, CRU2, CRU3, and rRNA genes. CRU1 and CRU2 are expressed at approximately equal levels in wildtype and ssp1, while there is no CRU3 signal from ssp1 mRNA. B. Sequence analysis of the CRU3 gene in ssp1 shows that EMS induced a G to A transition mutation in exon 4, converting a tryptophan codon to a premature stop codon.
In parallel, we conducted map-based cloning of ssp1, and observed enhanced segregation of F2 mutants with the MI422 and MI232 RFLP markers and the nga1139 SSLP marker, located on the distal end of chromosome 4 (data not shown). Thus, marker segregation analysis positions the ssp1 mutation near CRU3, which resides at 14.2 Mb. Direct sequencing of the CRU3 gene from ssp1 revealed a G to A transition mutation at position 1850 of the genomic sequence, resulting in the conversion of a TRP codon into a premature stop codon. Thus, it is likely that the molecular basis of the ssp1 mutant is a defective CRU3 structural gene, and the lack of detectable mRNA implies that nonsense mediated mRNA destruction operates to rid the cell of compromised CRU3 mRNAs.
To confirm that ssp1 represents a defect in CRU3, we introduced a wildtype CRU3 gene into ssp1 and found that the seed protein profile reverted to the wildtype polypeptide pattern (data not shown).
Strategy for testing the utility of ssp1 for overproducing foreign proteins
A tenet of our ‘empty container’ hypothesis is that there exists a seed filling sensor mechanism governing reserve accumulation, and that if compromised by a mutation, this genetic background might be exploited to overexpress proteins of economic/agronomic interest. A proof-of-principle strategy was developed to examine the level of a foreign protein in both ssp1 and wildtype backgrounds in which the transgene copy number and chromosomal context were fixed (Figure 6). The reporter gene employed was the Phaseolus vulgaris phytohemagglutinin (PHA) gene, selected for use because PHA is a seed specific gene, and the polypeptide is processed and stable within protein bodies of transgenic seeds; in addition, a strong specific antiserum exists that would facilitate expression analyses [26], [27]. Our strategy was to generate primary transgenic lines having a single T-DNA insertion. We employed DNA gel blotting to screen for transgenics that give rise to single hybridizing bands with a PHA probe (see Figures S1, S2, S3). Of 53 independent transformants assayed, 49 possessed multiple insertions, and the four single copy insertion lines were selected and confirmed as single locus sites by observing a 3∶1 ratio of hygromycin resistance∶sensitivity when outcrossed. The primary transformants (cru3/cru3; PHA/-; note that with the identification of a premature stop codon in the CRU3 gene in ssp1, the nomenclature now changes to represent ssp1 as cru3/cru3) were permitted to self, and T2 seeds germinated on hygromycin. For transformants bearing a single copy of the PHA gene, the T3 homozygotes would be expected to produce T4 seeds all of which are hygromycin resistant. We identified the T3 lines that were gauged to be homozygous based on hygromycin resistance and used immunoblotting to confirm that these express PHA (data not shown). These lines are genetically cru3/cru3; PHA/PHA, and were termed the tester lines.
Figure 6. Strategy for testing the empty container hypothesis.
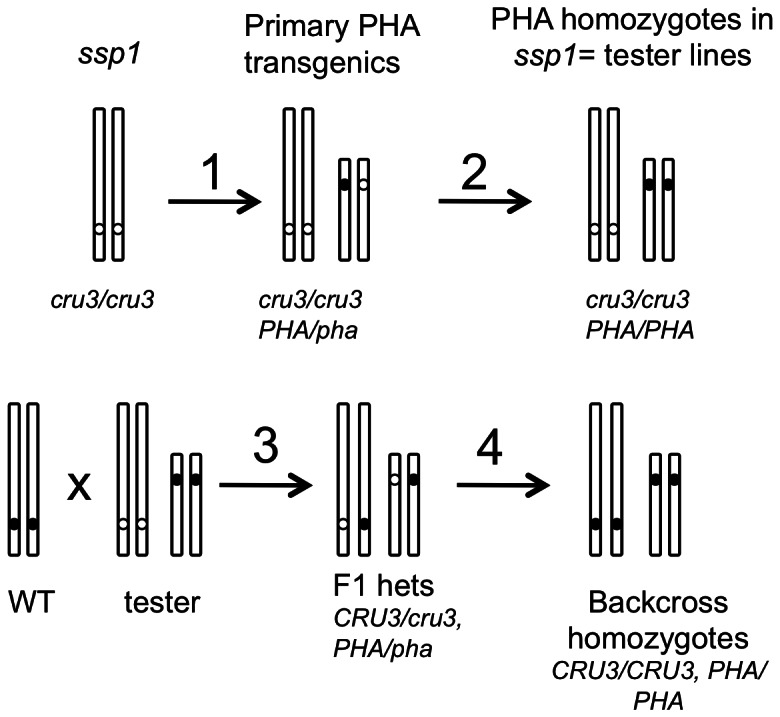
Homozygous ssp1 mutants were transformed with a PHA transgene and single copy insertion lines were selected by Southern blotting (step 1). The primary transgenic lines were selfed and homozygous tester lines were selected by examination of SDS-PAGE profiles of seed extracts (ssp1 pattern) and 100% hygromycin segregation in the T3 lines (step 2). The tester lines were crossed to wildtype with the resulting F1 plants being both heterozygous for CRU3 (ssp1) and for the PHA transgene (step 3). Selfing was conducted and F3 lines screened by SDS-PAGE for normal seed protein profiles, by sequencing of CRU3 to determine homozygosity, and by hygromycin segregation to determine PHA homozygosity (step 4). The resulting tester and backcross lines are homozygous for a single PHA insertion such that reporter gene copy number and context are fixed in both the tester (cru3 background) and the wildtype (CRU3) background.
To unequivocally test the empty container hypothesis, it is necessary to backcross the tester lines with wildtype and to select lines that are homozygous for both CRU3 and the PHA transgene. A three step screening protocol was used to identify such plants. SDS-PAGE was initially used to screen seed extracts for a wildtype protein profile, which would imply either a CRU3/cru3 or CRU3/CRU3 genotype (see Figures S4, S5, S6, S7). Homozygotes were distinguished from heterozygotes by direct DNA sequencing of PCR products, such that the single base change in ssp1 could be discerned. Plants having only a wildtype G signal at the TGG codon were identified and simultaneously examined for hygromycin segregation to determine the zygosity of the PHA transgene. This strategy enabled us to select CRU3/CRU3; PHA/PHA plants, which we refer to as the backcross homozygotes. Each reporter line therefore is represented in either an ssp1 background or a wildtype background in which copy number and position effect variation is fixed by a single PHA reporter transgene. If the ssp1 mutant could be used to overexpress a foreign (PHA) gene, it is expected that the tester homozygotes would have higher levels of the protein than the backcross homozygotes. Immunoblotting of mature fifth generation seed extracts from four independent lines (1, 3, 5, 7) was undertaken to facilitate this test (Figure 7).
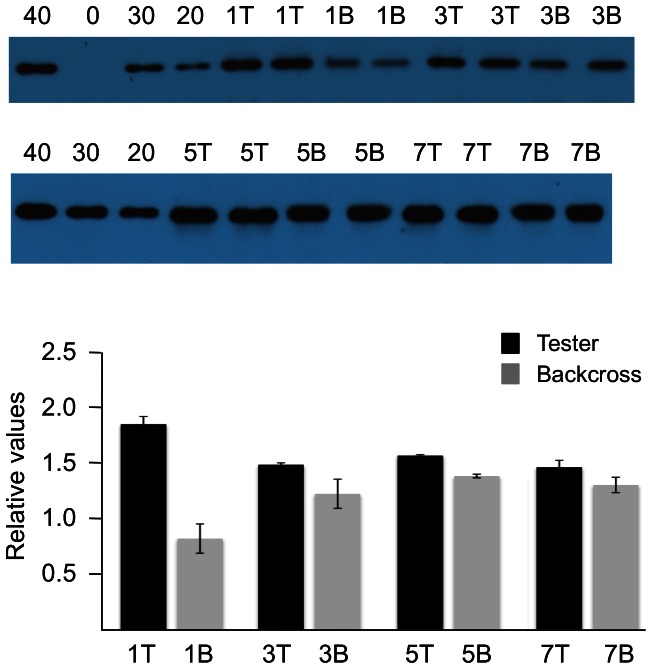
Figure 7. Immunoblotting validates ssp1 as an empty container.
Duplicate one microgram aliquots of total seed protein from tester and backcross homozygotes were subjected to SDS-PAGE and immunoblotting with an anti-PHA antibody. Reporter lines 1, 3, 5, and 7 are shown in duplicate as testers (T) and the corresponding backcrosses (B). Aliquots of purified PHA were included as standards. The autoradiographs were scanned by densitometry and quantified by employing Image J software. The bar graphs represent the average PHA values relative to 30 ng of purified PHA. T-test statistical analyses revealed that lines 1, 3, and 5 have p-values less than 0.05, while the line 7 p-value was greater than 0.05. An independent experiment on the previous generation of these tester/backcross lines revealed similar enrichment trends and p-values.
Aliquots of purified PHA were used to construct standard curves and to compare the relative signals in one microgram aliquots of seed protein extracts. This strategy also allowed us to estimate the amount of PHA produced as a percentage of total seed protein. In lines 1 and 3, PHA accounted for 5.54% and 4.44%, respectively, of the total soluble protein, while their backcross derivatives gave levels of 2.45% and 3.66%, respectively. The ratio of these terms indicates that PHA expression in line 1 is 125.9% greater than its backcross counterpart, while line 3 is 21.4% higher. Lines 5 and 7 exhibited less dramatic results with the ratio of differential expression being 13.3% and 12.25%, respectively. Statistical analysis validated lines 1,3, and 5 as significant (p<0.05), while the difference for line 7 was not (p>0.05). Collectively, three of the four lines exhibited enhanced PHA reporter expression in the ssp1 background, validating the empty container hypothesis.
Discussion
The use of plants as bioreactors for the economical production of proteins, oils, and specialty chemicals continues to be an area of intense interest (reviewed in [28]). A variety of genetic elements (e.g. promoters, enhancers) have been used to drive the expression of target genes, often in specific organs or tissues [8]. While many successes have been reported, there remain difficulties in obtaining and sustaining high level expression, particularly in storage organs such as seeds. The existence of a seed filling sensor and a natural bias towards the accumulation of endogenous reserves has led to attempts to find mutants that do not fill their seeds, and to antisense/RNAi approaches to genetically reduce the endogenous reserves.
Genetic manipulation of endogenous seed reserves by employing antisense technology has been reported in numerous species, ranging from cereals such as wheat [29], rice [19], and maize [16], [17], to dicotyledonous crops such as soybean [20], [21], and to the model plant Arabidopsis [30]. In some instances, gene expression and proteome changes occur in response to reduced levels of the targeted gene to alter the nutritional quality of the seed [16], [17]. For example, antisense suppression of the C-hordeins of barley resulted in lower levels of these nutritionally poor seed proteins, and the concomitant compensation by enhanced expression of other seed proteins produced seeds with elevated levels of several essential amino acids [31]. In other situations, ‘neutral’ compensation occurs, whereby the enhanced expression of other seed proteins facilitate proteome rebalancing to normalize the amino acid composition of the seed [21], [32].
In antisense lines, the fidelity of the RNAi machinery may be variable in different lines [33], and/or over generations [34], whereas stable mutants with defined roles are expected to yield more consistent results. In addition, some antisense strategies have had unanticipated consequences. For example, targeting of the soybean glycinin mRNAs had an ancillary effect of also reducing the level of conglycinins [21], despite very limited sequence identity amongst the two classes of soybean mRNAs. Nevertheless, this SP- line has been stably maintained for many generations, but the proteome rebalancing that occurs in this line is restricted to endogenous genes and an introgressed glycinin-GFP mimic construct was expressed at levels comparable to the parent lines [21]. This contrasts with a previous report in which the suppression of glycinin created a background in which the glycinin-GFP mimic could be overexpressed [20]. From all of these studies, it is clear that there are differences in the capacity to overexpress transgenes that are dependent on the species, the targets of antisense suppression and the degree to which it manifests its effect, and the localization and amino acid composition of the target protein. Therefore, success in any system will depend upon application of several factors to enhance the expression and stability of the desired trait.
Unlike many genetic backgrounds in which seed storage protein accumulation has been modified, either as a result of a mutation or due to RNAi strategies, ssp1 is a non-compensating mutant, in the sense that reduced CRU3 expression/protein levels are not normalized by enhanced expression of other proteins. Although it is unclear why ssp1 has 15% lower protein levels than wildtype, it is possible that the implied nonsense mediated mRNA decay of mutant CRU3 transcripts in some way bypasses the filling sensor that activates compensation, resulting in a seed that does not completely fill.
While other reports of reserve mutants or antisense suppression of endogenous reserves have been coupled with analysis of the expression of a transgene [19], [20], [29], [35], our strategy of selecting single copy lines and examining expression of the reporter in near isogenic lines in which the copy number and position of the transgene are fixed, has, to our knowledge, not been reported. In most cases, the transgene copy number has not been investigated, and hence neither its correlation with transgene expression nor the effect of the integration sites on transgene expression can be known. In cases where transgene copy number has been determined, there are many instances in which copy number cannot be correlated with expression levels [27], [30], [36], suggesting that the chromosomal context of the insertion site is a primary determinant of the expressivity of a transgene. In this regard, it has been established that position effect variation may partly be compensated for by employing a matrix attachment region in the transforming construct [37].
Analysis of the four tester lines in our studies clearly show variability in PHA expression, and in three of four lines, reporter expression was significantly higher in the ssp1 background than the corresponding backcross line. Increases in yield of 10% or greater are agriculturally significant. Our proof-of-principle studies with PHA indicate that it is possible to generate a genetic background that can be useful for overexpression of foreign proteins. It is conceivable that this strategy could be applied to agronomically important members of the Brassicaceae, for example, canola. Based on the available data, the targeted reduction of specific seed proteins is likely the most promising strategy and appropriate backgrounds could be identified by screening TILLING lines [38] for specific mutants. Coupling an empty container background that overexpresses a protein of interest with other transgenic approaches, such as the overexpression of the BBX32 gene, proven to increase yield [39], could be a powerful combination in future strategies to combat global food security problems.
Materials and Methods
Plant materials and growth conditions
Wildtype and ssp1 seeds were grown either in a greenhouse under ambient conditions, or in growth chambers at 22°C with a 16 hr/8 hr day/night cycle under fluorescent lighting (100 mE/m2)
Mutagenesis and screening for seed storage protein mutants
Approximately 1 µg of Columbia seeds were suspended in 50 ml of 50 mM phosphate buffer, pH 7.2, containing 0.2% ethyl methanesulfonate and left at room temperature for 16 hours. Seeds were then washed extensively with water, planted in 8 inch pots, and allowed to grow and self in a greenhouse. The M2 seeds were collected en masse and planted at low density in 4 inch pots. Seeds from individual plants were then collected and cataloged, and approximately 100 seeds were ground in hot (>90°C) denaturation buffer (2% SDS, 1% β-mercaptoethanol, 20 mM Tris pH 8.6, 10% glycerol). Following a 5 minute incubation at 95°C, the extract was clarified by centrifugation and the supernatant stored at −20°C until needed. Protein extracts were subjected to electrophoresis on 12% SDS-PAGE gels by standard techniques and protein profiles revealed by staining with Coomassie Brilliant Blue.
Biochemical techniques
Seed protein content
To evaluate seed protein content, 10 mg aliquots of mature seeds were homogenized in 200 µl of hot extraction buffer (1% SDS, 0.5% β-mercaptoethanol, 20 mMTris, pH 8.6, 10% glycerol) and kept at 95°C for 10 minutes, followed by clarification in a microfuge for 5 minutes. One hundred microliters of the supernatant was collected and diluted with three volumes of 20 mM Tris, pH 8.2. Triplicate 25 µl aliquots of the diluted extracts were assayed for protein content by employing the Pierce BCA kit as described by the manufacturer. The raw absorbance values for three independent experiments (each measuring triplicates of both wildtype and ssp1) were converted to micrograms of protein/milligram of seed extract using absorbance values of known amounts of bovine serum albumin as protein standards. The converted values were statistically evaluated by using Microsoft Excel, and TTEST p-values were determined.
Isolation of Arabidopsis protein for two-dimensional electrophoresis
Total protein from Arabidopsis seed was isolated by a procedure from Mooney and Thelen [40] with the following modifications. Dry seed (150 mg) was pulverized to a fine powder in a mortar and pestle using liquid nitrogen. Powder was resuspended in the mortar with 8 mL of homogenization buffer (50% phenol, 0.45 M sucrose, 5 mM EDTA, 0.2% (v/v) β-mercaptoethanol, 50 mM Tris–HCl pH 8.8) with continued homogenization until the homogenate reached room temperature. The homogenate was transferred to a phenol-resistant screw cap tube, mixed for 30 min at 4°C, then centrifuged at 5000 g for 15 min at 4°C. The top phenol layer was transferred to a fresh tube, mixed with five volumes of ice cold 0.1 M ammonium acetate in 100% methanol and placed at −20°C for 2 hrs. Precipitated protein was collected by centrifugation at 5000 g for 10 min at 4°C. The precipitate was washed twice with 20 mL of 0.1 M ammonium acetate in 100% methanol followed by two washes with ice-cold 80% acetone. The sample was then resuspended in ice-cold 70% ethanol, divided into 3 tubes, centrifuged and either stored as a precipitate at −20°C or dried and resuspended for immediate isoelectric focusing.
Isoelectric focusing using immobilized pH gradient strips
Protein pellet (from 50 mg of dry seed material) was resuspended in 200 µL of isoelectric focusing buffer (8 M urea/2 M thiourea (deionized), 2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 2% Triton X-100, 50 mM DTT, 2 mM Tris (2-carboxyethyl)phosphine hydrochloride (TCEP), 0.8% carrier ampholytes) and incubated for 1 h at room temperature. The sample was centrifuged for 20 min at 14,000 g and the supernatant was transferred into a separate tube. Protein concentration was determined by Bradford assay using BioRad dye reagent (Bio-Rad, Hercules, CA). Protein quantitation was performed in triplicate, and quantitated against a standard curve of bovine serum albumin standard. Exactly 68 µg of protein was added to a separate tube and volume was brought up to 155 µL with IEF buffer/0.001% Bromophenol blue and mixed before pipetting into a loading well of a ZOOM IPGRunner cassette (Life Technologies Inc., Carlsbad, CA). Immobilized ZOOM pH 3–10 NL gradient 7 cm strips (Life Technologies Inc.) were carefully placed into enclosed channel and rehydrated for 18 hrs at room temperature. IPGRunner cell was assembled and isoelectric focusing was performed as follows: 200 V/20 min, 450 V/15 min, 750 V/15 min, 2,000 V/55 min.
SDS–PAGE for 2-D electrophoresis
Following isoelectric focusing, IPG strips were incubated in 5 mL 1× NuPAGE LDS sample buffer (Life Technologies Inc.) supplemented with 1% (w/v) DTT for 20 min with gentle agitation, followed by incubation in buffer supplemented with 2.5% (w/v) iodoacetamide for 20 min with gentle agitation. IPG strips were then rinsed with NuPAGE MES SDS running buffer (Life Technologies Inc.) and placed onto 4–12% Bis-Tris ZOOM gels (Life Technologies Inc.). Strips were then overlayed with agarose solution (1× MES SDS running buffer, 0.5% (w/v) agarose). Second dimension SDS–PAGE was conducted in XCell SureLock unit (Life Technologies Inc.) for 43 min at 200 V. Following SDS–PAGE gels were washed with deionized water three times 15 min each and stained with SimplyBlue stain (Life Technologies Inc.). Each Arabidopsis seed preparation was resolved by 2-D electrophoresis in at least three independent experiments.
Amino acid composition
For amino acid composition analysis, total seed proteins were extracted as described above and precipitated with four volumes of cold acetone. Following centrifugation for 10 minutes, the pellets were dried, and hydrolyzed by the addition of 100 µl of 6N HCl. After 24 hours at 110°C, the acid was removed by vacuum and the hydrolyzates derivatized using PITC. The derivatized amino acids were redissolved in 100 ul of phosphate buffer and analyzed by reverse phase HPLC using a Waters Pico-Tag system at the Advanced Protein Technology Centre, Hospital for Sick Children, Toronto. Triplicate samples of wildtype and ssp1 were quantified and statistical analyses were carried out using the TTEST function of Microsoft Excel, which revealed no significant differences (p>0.5).
Lipid analysis
Triplicate aliquots of mature seeds of both ssp1 and wildtype were analyzed for oil content by NMR (MARAN Ultra, Oxford Instruments) as described by O'Neill et al. [41] Gas chromatography analysis of seed fatty acid composition was performed as described previously [42]. The triplicate values were statistically evaluated by employing the TTEST function of Microsoft Excel.
Molecular techniques
Genomic DNA was purified by a miniprep procedure [43], or by using Sigma GeneElute columns as described by the manufacturer. Total RNA was prepared by employing a Qiagen RNeasy kit. Five microgram aliquots of DNA or RNA were used for DNA/RNA gel blotting. Probes were prepared from DNA inserts of the plasmids harboring the CRA, CRB, CRC, and 18S rRNA genes using α 32P dCTP (Amersham) and the Rediprime random primer labeling kit (GE Healthcare).
Empty container hypothesis testing strategy
ssp1 homozygotes were transformed with a reporter construct consisting of a 2.8 kbp fragment harboring the Phaseolus vulgaris phytohemagglutinin (PHA) gene (pDR214, [27]), which was mobilized into the binary vector pGVPT-HPT [44], and used to transform plants via the method of Martinez-Trujillo et al. [45]. Transgenic plants were selected on 0.5× MS media containing 20 µg/ml hygromycin B (Invitrogen). DNA gel blotting was employed to determine gene copy number by employing a restriction enzyme that cuts at the end of the cloned sequence and therefore single hybridizing bands are indicative of single insertion events (see Figures S1, S2, S3). Single copy lines were selfed and hygromycin segregation examined in both self crosses (100% hygromycin resistant) and backcrosses (75% hygromycin resistance) to confirm the homozygosity of the reporter constructs. These lines are referred to as the tester lines.
The tester lines were backcrossed to wildtype and taken through 3 generations to ensure that both the reporter gene and the wildtype CRU3 gene were in a homozygous state. The CRU3 gene region that encompasses the region of the ssp1 mutation was amplified from these plants by employing the primers: FOR; 5′ATGAGGTCCCACGAGAACATTG3′ and BACK: 5′CGTTAAAGGTAAAGGGACGTGA3′, and the status of CRU3 assessed by sequencing with the BACK primer (The Centre for Applied Genomics, Toronto). These lines are referred to as the backcross homozygotes.
Seed protein extracts from tester homozygotes and backcross homozygotes were prepared as described above and used for immunoblotting by employing standard techniques. Duplicate one microgram aliquots of seed extracts along with 20,30 and 40 ng of purified PHA (Boehringer-Mannheim) were separated by SDS-PAGE and blotted to nitrocellulose. An anti-PHA antibody [46] was employed, followed by a secondary goat anti-rabbit IgG conjugated to horseradish peroxidase. Detection was facilitated by chemiluminescence, utilizing the Pierce ECL substrate. Multiple exposures on BioFlex x-ray films were taken to insure the linearity of the signals. Autoradiographs were scanned by densitometry and Image J (NIH) was employed to quantify pixel density. Values are expressed as relative to the signals for 30 ng of purified PHA.
Supporting Information
Schematic representation of the 2.8 kb PHA EcoRI/HindIII fragment together with the HYG selectable marker.
(TIFF)
Analysis of T2 transgenic lines showing that most transformants harbor multiple inserts.
(TIFF)
PHA lines 1, 3, 5 and 7 are single locus insertions.
(TIFF)
Validation of PHA1 backcross lines.
(TIFF)
Validation of PHA3 backcross lines.
(TIFF)
Validation of PHA5 backcross lines.
(TIFF)
Validation of PHA7 backcross lines.
(TIFF)
Acknowledgments
The authors are grateful to Jitao Zou for conducting the lipid analysis, Rey Interior for the amino acid composition analyses, and Annette Rzepczyk and Lakshmi Pelecanda for expert technical assistance. We acknowledge Jane Lee and Gabriele Ferraioli for early stage screening for mutants and preliminary characterization of ssp1.
Funding Statement
This research was supported by: Ontario Research Fund, Research Excellence Programme of the Ontario Ministry of Economic Development and Innovation (Grant # ORF-RE04-043) to FM; Natural Sciences and Engineering Research Council of Canada (NSERC) (Discovery Grant # 90057) to CDR; and NSERC Strategic Grant (Grant # STR0167294) to PM and CDR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Edgerton MD (2009) Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiology 149: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Newell-McGloughlin M (2008) Nutritionally Improved Agricultural Crops. Plant Physiology 147: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465. [DOI] [PubMed] [Google Scholar]
- 4. Fitzpatrick TB, Basset GJC, Borel P, Carrari F, DellaPenna D, et al. (2012) Vitamin deficiencies in humans: Can plant science help? Plant Cell 24: 395–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lau OS, Ng DWK, Chan WWL, Chang SP, Sun SSM (2010) Production of the 42-kDa fragment of Plasmodium falciparum merozoite surface protein 1, a leading malaria vaccine antigen, in Arabidopsis thaliana seeds. Plant Biotechnol J 8: 994–1004. [DOI] [PubMed] [Google Scholar]
- 6. Somleva MN, Snell KD, Beaulieu JJ, Peoples OP, Garrison BR, et al. (2008) Production of polyhydroxybutyrate in switchgrass, a value-added co-product in an important lignocellulosic biomass crop. Plant Biotechnol J 6: 663–678. [DOI] [PubMed] [Google Scholar]
- 7. Vega-Sanchez ME, Ronald PC (2010) Genetic and biotechnological approaches for biofuel crop improvement. Current Opinion in Biotechnology 21: 218–224. [DOI] [PubMed] [Google Scholar]
- 8. Egelkrout E, Rajan V, Howard JA (2012) Overproduction of recombinant proteins in plants. Plant Sci 184: 83–101. [DOI] [PubMed] [Google Scholar]
- 9. Werner S, Breus O, Symonenko Y, Marillonnet S, Gleba Y (2011) High-level recombinant protein expression in transgenic plants by using a double-inducible viral vector. Proc Natl Acad Sci USA 108: 14061–14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boothe J, Nykiforuk C, Shen Y, Zaplachinski S, Szarka S, et al. (2010) Seed-based expression systems for plant molecular farming. Plant Biotechnol J 8: 588–606. [DOI] [PubMed] [Google Scholar]
- 11. Naqvi S, Ramessar K, Farre G, Sabalza M, Miralpeix B, et al. (2011) High-value products from transgenic maize. Biotech Adv 29: 40–53. [DOI] [PubMed] [Google Scholar]
- 12. Zhang JA, Nallamilli BR, Mujahid H, Peng ZH (2010) OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J 64: 604–617. [DOI] [PubMed] [Google Scholar]
- 13. Peng FY, Weselake RJ (2011) Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genomics 12: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hagan ND, Upadhyaya N, Tabe LM, Higgins TJV (2003) The redistribution of protein sulfur in transgenic rice expressing a gene for a foreign, sulfur-rich protein. Plant J 34: 1–11. [DOI] [PubMed] [Google Scholar]
- 15. Scossa F, Laudencia-Chingcuanco D, Anderson OD, Vensel WH, Lafiandra D, et al. (2008) Comparative proteomic and transcriptional profiling of a bread wheat cultivar and its derived transgenic line overexpressing a low molecular weight glutenin subunit gene in the endosperm. Proteomics 8: 2948–2966. [DOI] [PubMed] [Google Scholar]
- 16. Frizzi A, Caldo RA, Morrell JA, Wang M, Lutfiyya LL, et al. (2010) Compositional and transcriptional analyses of reduced zein kernels derived from the opaque2 mutation and RNAi suppression. Plant Mol Biol 73: 569–585. [DOI] [PubMed] [Google Scholar]
- 17. Segal G, Song RT, Messing J (2003) A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics 165: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marsolais F, Pajak A, Yin F, Taylor M, Gabriel M, et al. (2010) Proteomic analysis of common bean seed with storage protein deficiency reveals up-regulation of sulfur-rich proteins and starch and raffinose metabolic enzymes, and down-regulation of the secretory pathway. J Proteomics 73: 1587–1600. [DOI] [PubMed] [Google Scholar]
- 19. Shigemitsu T, Ozaki S, Saito Y, Kuroda M, Morita S, et al. (2012) Production of human growth hormone in transgenic rice seeds: co-introduction of RNA interference cassette for suppressing the gene expression of endogenous storage proteins. Plant Cell Reports 31: 539–549. [DOI] [PubMed] [Google Scholar]
- 20. Schmidt MA, Herman EM (2008) Proteome rebalancing in soybean seeds can be exploited to enhance foreign protein accumulation. Plant Biotechnol J 6: 832–842. [DOI] [PubMed] [Google Scholar]
- 21. Schmidt MA, Barbazuk WB, Sandford M, May G, Song Z, et al. (2011) Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiology 156: 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pang PP, Pruitt RE, Meyerowitz EM (1988) Molecular cloning, genomic organization expression and evolution of 12S –seed storage protein genes of Arabidopsis thaliana. Plant Mol Biol 11: 805–820. [DOI] [PubMed] [Google Scholar]
- 23. Dickinson CD, Hussein EHA, Nielsen NC (1989) Role of postranslational cleavage in glycinin assembly. Plant Cell 1: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Higashi Y, Hirai MY, Fujiwara T, Naito S, Noji M, et al. (2006) Proteomic and transcriptomic analysis of Arabidopsis seeds: molecular evidence for successive processing of seed proteins and its implication in the stress response to sulfur nutrition. Plant J 48: 557–571. [DOI] [PubMed] [Google Scholar]
- 25. Wan LL, Ross ARS, Yang JY, Hegedus DD, Kermode AR (2007) Phosphorylation of the 12 S globulin cruciferin in wild-type and abi1-1 mutant Arabidopsis thaliana (thale cress) seeds. Biochem J 404: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sturm A, Voelker TA, Herman EM, Chrispeels MJ (1988) Correct glycosylation, golgi-processing and targeting to protein bodies of the vacuolar protein phytohemagglutinin in transgenic tobacco. Planta 175: 170–183. [DOI] [PubMed] [Google Scholar]
- 27. Riggs CD, Voelker TA, Chrispeels MJ (1989) Cotyledon nuclear proteins bind to DNA fragments harboring regulatory elements of phytohemagglutinin genes. Plant Cell 1: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahmad P, Ashraf M, Younis M, Hu XY, Kumar A, et al. (2012) Role of transgenic plants in agriculture and biopharming. Biotechnol Adv 30: 524–540. [DOI] [PubMed] [Google Scholar]
- 29. Piston F, Gil-Humanes J, Rodriguez-Quijano M, Barro F (2011) Down-Regulating gamma-Gliadins in Bread Wheat Leads to Non-Specific Increases in Other Gluten Proteins and Has No Major Effect on Dough Gluten Strength. PLoS One 6 10.1371/journal.pone.0024754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goossens A, Van Montagu M, Angenon G (1999) Co-introduction of an antisense gene for an endogenous seed storage protein can increase expression of a transgene in Arabidopsis thaliana seeds. FEBS letters 456: 160–164. [DOI] [PubMed] [Google Scholar]
- 31. Lange M, Vincze E, Wieser H, Schjørring JK, Holm PB (2007) Effect of an antisense C-hordein gene on the storage protein composition in the barley seed. J Agri Food Chem 55: 6074–6081. [DOI] [PubMed] [Google Scholar]
- 32. Kawakatsu T, Hirose S, Yasuda H, Takaiwa F (2010) Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol 154: 1842–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang SS, Kruger DE, Frizzi A, D'Ordine RL, Florida CA, et al. (2005) High-lysine corn produced by the combination of enhanced lysine biosynthesis and reduced zein accumulation. Plant Biotechnol J 3: 555–569. [DOI] [PubMed] [Google Scholar]
- 34. Dietz-Pfeilstetter A (2010) Stability of transgene expression as a challenge for genetic engineering. Plant Sci 179: 164–167. [Google Scholar]
- 35. Tada Y, Utsumi S, Takaiwa F (2003) Foreign gene products can be enhanced by introduction into low storage protein mutants. Plant Biotechnol J 1: 411–422. [DOI] [PubMed] [Google Scholar]
- 36. Rai M, Datta K, Parkhi V, Tan J, Oliva N, et al. (2007) Variable T-DNA linkage configuration affects inheritance of carotenogenic transgenes and carotenoid accumulation in transgenic indica rice. Plant Cell Rep 26: 1221–1231. [DOI] [PubMed] [Google Scholar]
- 37. Peterson K, Leah R, Knudsen S, Cameron-Mills V (2002) Matrix attachment regions (MARs) enhance transformation frequencies and reduce variance of transgene expression in barley. Plant Mol Biol 49: 45–58. [DOI] [PubMed] [Google Scholar]
- 38. Stephenson P, Baker D, Girin T, Perez A, Amoah S, et al. (2010) A rich TILLING resource for studying gene function in Brassica rapa . BMC Plant Biol 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Preuss SB, Meister R, Xu QZ, Urwin CP, Tripodi FA, et al. (2012) Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS One 7 10.1371/journal.pone.0030717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mooney BP, Thelen JJ (2004) High-throughput peptide mass fingerprinting of soybean seed proteins: automated workflow and utility of UniGene expressed sequence tag databases for protein identification. Phytochemistry 65: 1733–1744. [DOI] [PubMed] [Google Scholar]
- 41. O'Neill CM, Gill S, Hobbs D, Morgan C, Bancroft I (2003) Natural variation for seed oil composition in Arabidopsis thaliana. Phytochemistry 64: 1077–1090. [DOI] [PubMed] [Google Scholar]
- 42. Li YH, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915. [DOI] [PubMed] [Google Scholar]
- 43. McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, et al. (1995) Sequence based identification of T-DNA insertion mutants in Arabidopsis- Actin mutants act2-1 and act4-1 . Plant J 8: 613–622. [DOI] [PubMed] [Google Scholar]
- 44. Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197. [DOI] [PubMed] [Google Scholar]
- 45. Martinez-Trujillo M, Limones-Briones V, Cabrera-Ponce JL, Herrera-Estrella L (2004) Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol Biol Rep 22: 63–70. [Google Scholar]
- 46. Voelker TA, Sturm A, Chrispeels MJ (1987) Differences in expression between two seed lectin alleles obtained from normal and lectin-deficient beans are maintained in transgenic tobacco. EMBO Journal 6: 3571–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the 2.8 kb PHA EcoRI/HindIII fragment together with the HYG selectable marker.
(TIFF)
Analysis of T2 transgenic lines showing that most transformants harbor multiple inserts.
(TIFF)
PHA lines 1, 3, 5 and 7 are single locus insertions.
(TIFF)
Validation of PHA1 backcross lines.
(TIFF)
Validation of PHA3 backcross lines.
(TIFF)
Validation of PHA5 backcross lines.
(TIFF)
Validation of PHA7 backcross lines.
(TIFF)