Abstract
Anoctamin (ANO)2 (or TMEM16B) forms a cell membrane Ca2+-activated Cl− channel that is present in cilia of olfactory receptor neurons, vomeronasal microvilli, and photoreceptor synaptic terminals. Alternative splicing of Ano2 transcripts generates multiple variants with the olfactory variants skipping exon 14 and having alternative splicing of exon 4. In the present study, 5′ rapid amplification of cDNA ends analysis was conducted to characterize the 5′ end of olfactory Ano2 transcripts, which showed that the most abundant Ano2 transcripts in the olfactory epithelium contain a novel starting exon that encodes a translation initiation site, whereas transcripts of the publically available sequence variant, which has an alternative and longer 5′ end, were present in lower abundance. With two alternative starting exons and alternative splicing of exon 4, four olfactory ANO2 isoforms are thus possible. Patch-clamp experiments in transfected HEK293T cells expressing these isoforms showed that N-terminal sequences affect Ca2+ sensitivity and that the exon 4–encoded sequence is required to form functional channels. Coexpression of the two predominant isoforms, one with and one without the exon 4 sequence, as well as coexpression of the two rarer isoforms showed alterations in channel properties, indicating that different isoforms interact with each other. Furthermore, channel properties observed from the coexpression of the predominant isoforms better recapitulated the native channel properties, suggesting that the native channel may be composed of two or more splicing isoforms acting as subunits that together shape the channel properties.
INTRODUCTION
Anoctamin (ANO)1 and ANO2 (also known as TMEM16A and TMEM16B) belong to a recently described ANO family of 10 proteins that are implicated in a wide range of physiological functions (Duran and Hartzell, 2011; Kunzelmann et al., 2011; Pifferi et al., 2012). ANOs are predicted to have eight transmembrane domains with C and N termini located intracellularly and have been found to form cell membrane Ca2+-activated Cl− channels (Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008; Pifferi et al., 2009; Stephan et al., 2009; Stöhr et al., 2009; Hengl et al., 2010; Rasche et al., 2010), which may also be gated by temperature (Cho et al., 2012). Other members of the ANO family may only localize intracellularly (Duran et al., 2012, but see Martins et al., 2011, and Tian et al., 2012).
One of the founding members of the ANO family, ANO2, has been described to either be a subunit of or to form Ca2+-activated Cl− channels in photoreceptor synaptic terminals (Stöhr et al., 2009). ANO2, which shares a high level of amino acid homology to ANO1 (∼60%), also forms Ca2+-activated Cl− channels in the cilia of olfactory receptor neurons (ORNs) and in the microvilli of vomeronasal neurons (Stephan et al., 2009; Hengl et al., 2010; Rasche et al., 2010; Sagheddu et al., 2010; Dibattista et al., 2012). In ORN cilia, ANO2 amplifies the initial odorant-induced current from the Ca2+-permeable olfactory CNG channel (Kleene, 2008; Kaupp, 2010) and is responsible for up to 80–90% of the receptor current (Kurahashi and Yau, 1993; Lowe and Gold, 1993; Boccaccio and Menini, 2007; Billig et al., 2011), but its role in mouse behavior remains unclear (Billig et al., 2011). Ca2+-activated Cl− channels have been proposed to play a similar role in the amplification of vomeronasal neuron responses (Yang and Delay, 2010; Kim et al., 2011), and both ANO1 and ANO2 have been found in vomeronasal neurons, targeting to the apical microvillar layer of the vomeronasal epithelium (Rasche et al., 2010; Billig et al., 2011; Dauner et al., 2012). It has been demonstrated recently that ANO1 and ANO2 are coexpressed in the microvilli of individual vomeronasal neurons (Dibattista et al., 2012) and, considering that ANO1 channels form homo-multimers (Fallah et al., 2011; Sheridan et al., 2011), it raises the question whether ANO1 and ANO2 may form hetero-multimers.
Alternative splicing can greatly increase the differential functionality of proteins. Ano1 has at least six exons that are alternatively spliced, of which two encode for segments in the cytosolic N terminus (Caputo et al., 2008; Ferrera et al., 2009; O’Driscoll et al., 2011). ANO1 isoforms with different N termini display varied Ca2+ sensitivity (Ferrera et al., 2009). We previously found that the olfactory Ano2 transcripts lack exon 14 (formerly annotated as exon 13 in Stephan et al., 2009), which is present in retinal Ano2 transcripts. Also, in the olfactory epithelium, alternative splicing occurs at exon 4 (exon 3 in Stephan et al., 2009), generating Ano2 variants either with or without exon 4. The exon 4–containing transcripts are present in greater abundance than the exon 4–lacking transcripts (Stephan et al., 2009). Exon 4 encodes a 33–amino acid segment within the cytoplasmic N terminus. The functional significance of such alternative splicing is unknown.
Previously, comparison between biophysical properties of native olfactory Ca2+-activated Cl− channels and heterologously expressed ANO2 channels revealed similar channel properties with respect to the sensitivity of the channel to Ca2+, single-channel conductance, “rundown” of the current, halide permeability, and current inactivation at negative holding potentials (Stephan et al., 2009). One notable exception is that, at positive holding potentials, unlike the native channel, the heterologously expressed ANO2 channel does not inactivate (Reisert et al., 2003; Pifferi et al., 2006, 2009, 2012; Stephan et al., 2009). This difference may reflect the absence of modulatory factors within the heterologous system, or it may also reflect isoform-specific properties.
To better understand the molecular constituents and physiological roles of the olfactory Ca2+-activated Cl− channel, it is important to characterize Ano2 splicing variants and determine how the resulting ANO2 protein isoforms may contribute to channel function. In this study, we mapped the 5′ end of Ano2 transcripts from the mouse olfactory epithelium. We found that the most abundant Ano2 transcripts contain an alternative starting exon that encodes a translation initiation site upstream of the previously described transcription initiation site (Stephan et al., 2009) that yields an ANO2 variant with a 27–amino acid longer N terminus, which we termed “Isoform B.” Additionally, we detected a second, already described mRNA species in rare abundance that encodes an even longer N terminus (named “Isoform A”) that is similar to the retinal isoform (Stöhr et al., 2009). Thus, with two alternative starting exons and alternative splicing of exon 4 (Stephan et al., 2009), four Ano2 mRNA variants are possible in ORNs.
We analyzed the functional significance of the two alternative N-terminal start sequences and of the exon 4–encoded sequence. We expressed the four ANO2 isoforms in HEK293T cells and examined the current properties using excised patch-clamp and whole-cell electrophysiology. We found that the N-terminal sequences affect Ca2+ sensitivity and that the exon 4 sequence is required for channel function in this heterologous system. We also coexpressed the two variants of ANO2 (Isoform A or B), together with their respective isoforms lacking the exon 4 sequence (Isoforms AΔ4 or BΔ4), and found that channel properties resulting from the coexpression of B isoforms, but not A isoforms, recapitulated properties of the native channel better than that of the individual expression of ANO2 isoforms. In particular, the channel resulting from coexpression of Isoforms B and BΔ4, like the native channel, showed inactivation at positive holding potentials. These results suggest that the splicing isoforms form hetero-multimers and that the native channel may be composed of two or more splicing isoforms.
MATERIALS AND METHODS
5′ rapid amplification of cDNA ends (RACE)
Mice were handled and euthanized with methods approved by the Animal Care and Use Committees of The Johns Hopkins University. Total olfactory epithelial RNA was extracted from adult C57BL/6 mice using Trizol (Invitrogen). The RNA was reverse transcribed using a RETROscript kit (Invitrogen) and an Ano2-specific primer (AS15) 5′-TCTTTGCGATGCTGCCTCCTG-3′, which binds within Ano2 exon 5. The single-stranded cDNA was purified using a Qiaquick PCR Purification kit (QIAGEN). A poly-dT tail was added to the 3′ end of the single-stranded cDNA (corresponding to the 5′ end of the RNA) by a terminal transferase (New England Biolabs, Inc.) reaction for 5 min at 37°C. The enzyme was inactivated by incubation at 75°C for 10 min. Second-strand cDNA synthesis was performed by PCR with the AS15 primer and a primer that binds to the poly-dT tail and adds an “adapter sequence,” 5′-GACTCGAGTCGACATCGTTTTTTTTTTTTTTTTT-3′ (Scotto-Lavino et al., 2006). A second round of PCR amplification was performed using a primer that only includes the adapter sequence 5′-GACTCGAGTCGACATCG-3′ and an Ano2-specific primer nested upstream of AS15 primer, (AS134) 5′-CTCCAAGTCCTTCTCCAGCTC-3′. The resulting PCR product was divided in two. One half of the product was column-purified by a Qiaquick PCR Purification kit and sequenced using an Ano2-specific primer nested upstream of AS134 primer, (Primer R1) 5′-GCCCAGCAGCCATCAGGTTG-3′. The other half of the reaction was analyzed by agarose gel electrophoresis. The gel was sliced into six sections of varying molecular weights, and DNA was extracted from each of the bands using a Qiaquick Gel Purification kit (QIAGEN). The extracted DNA was TOPO-TA cloned into pCR2.1 (Invitrogen), and the individual clones were sequenced by a plasmid-specific primer (M13).
Differentiation of Ano2 mRNA variants by RT-PCR
Single-stranded cDNA from the 5′ RACE analysis was used as a template for PCR analysis. The reverse primer used in each of the reactions was Primer R1 (see 5′ RACE above), and the forward primers were: F1, 5′-ATGCACTTTCACGACAACCA-3′; F2, 5′-GCAGAAAGTCCAGTGAATTCC-3′; F3, 5′-GAGAGAGTTCCCGAGACCGT-3′; F4, 5′-CCCTCTGCTCCCTGGATCTC-3′; F5, 5′-GAGGAGCGCCAGCAGCATTT-3′. Each of the PCR products was excised from the gel, reamplified by PCR, and TOPO-TA cloned and sequenced to verify their identities. A clone containing DNA amplified by the F5/R1 primer pair was used as a positive control template to test the primer efficacy for primers F3, F4, and F5. Clone 2-4 (Table S1), which contains the novel exon 1b, was used as a positive control template to test the primer efficacy of primer F2. A mixture of both positive control plasmids was used as a template to test the efficacy of Primer F1. Band densitometry quantification was performed using the “Analyze Gels” tool in ImageJ/FIJI (Fig. S1 C).
ANO2 isoform expression constructs
Expression constructs were all derived from the pAdTrack-cloned ANO2 expression construct used previously (Stephan et al., 2009). This construct encodes EGFP from a separate CMV promoter for positive identification of transfected cells but does not contain the full-length N terminal–encoding sequence of either known ANO2 isoform. Therefore, the complete N terminal–encoding sequences beginning at the most 5′ in-frame ATG were individually cloned at the 5′ end of this parent clone. To clone an expression construct encoding Isoform B, exon 1b and part of exon 3 were PCR amplified from clone 2-4 (Table S1) using the forward primer 5′-AGTCGAGGTACCATGGATCCAGAACACCTGCC-3′, which contains a KpnI recognition sequence, and the reverse primer 5′-CCAGCAGCCATCAGGTTGTG-3′, which binds downstream of a SalI recognition sequence. The PCR product and parent plasmid were cut by KpnI and SalI, and the PCR insert was cloned into the parent plasmid. A similar approach was taken to clone the expression construct encoding Isoform A, but using the forward primer 5′-AGTCGAGGTACCATGGCGGCCCCTGGGCT-3′ and the clone containing DNA amplified by the F5/R1 primer pair as a PCR template.
To generate the clones encoding the two “Δ4” isoforms, exon 4 was deleted from the above two clones by inverse PCR using primers 5′-ATGTACGAGATCAAGCAGGAGG-3′ and 5′-CTCCAAGTCCTTCTCCAGCTC-3′, followed by 5′ phosphorylation and blunt-end self-ligation.
Immunohistochemistry
To generate C-terminal GFP fusion constructs with ANO2 Isoform B and Isoform BΔ4, the clones used for electrophysiological measurements were modified. Intervening plasmid sequence between the Ano2 open reading frame and the GFP open reading frame was deleted from each of the corresponding clones by inverse PCR with primers 5′-GTGAGCAAGGGCGAGGAGCTG-3′ and 5′-TACATTGGTGTGCTGGGACCCT-3′. The PCR products were 5′ phosphorylated and blunt-end self-ligated, and the sequences were confirmed by sequencing.
HEK293T cells were grown in a 6-well plate on coverslips that were coated with poly-d-lysine. HEK293T cells (at ∼50% confluency) were transfected with 0.5 µg of ANO2 Isoform B::GFP-encoding plasmid or ANO2 Isoform BΔ4::GFP-encoding plasmid using Lipofectamine 2000 (Invitrogen). After 36 h, cells were washed with 1× PBS, pH 7.4, and a biotin solution (1.5 mg/ml biotin in 1× PBS, 1 mM CaCl2, and 0.5 mM MgCl2; EZ-Link Sulfo-NHS-LC-Biotin; Thermo Fisher Scientific) was applied to the cells for 30 min at 4°C. The biotin solution was washed off thoroughly with 1× PBS and 50 mM glycine, and cells were fixed in 4% paraformaldehyde for 25 min. Cells were washed with 1× PBS and blocked at room temperature for 2 h in 1× PBS containing 0.1% (vol/vol) Triton X-100 and 5% (vol/vol) donkey serum (blocking buffer). Cells were incubated at 4°C overnight with primary antibody in blocking buffer. Primary antibody, anti-GFP, was used at a dilution of 1:500 (Invitrogen). After washing, the coverslips were incubated for 2 h at room temperature with a secondary antibody conjugated to Alexa Fluor 488 (Invitrogen) and a streptavidin Alexa Fluor 546 conjugate (Invitrogen). Coverslips were mounted in Vectashield (Vector Laboratories) containing DAPI stain and imaged with a confocal microscope (LSM 510 META; Carl Zeiss).
Electron microscopy
HEK293T cells were grown in a 6-well plate and transfected (at ∼30% confluency) with 0.5 µg of either ANO2 Isoform B::GFP-encoding plasmid, ANO2 Isoform BΔ4::GFP-encoding plasmid, TrkA::GFP (provided by the Kuruvilla laboratory, Johns Hopkins University, Baltimore, MD), or pEGFP-N1 plasmid using Lipofectamine 2000 (Invitrogen). After 48 h, cells fixed in a sodium cacodylate (100 mM), 0.1% (vol/vol) gluteraldehyde, and 4% (vol/vol) paraformaldehyde solution in 1× PBS and stored in 100 mM sodium cacodylate. Next, cells were washed briefly in 1× PBS and resuspended in 1% low gelling–temperature agarose. The agarose blocks were trimmed into 1-mm3 pieces, cryoprotected by infiltration with 2.3 M sucrose/30% polyvinyl pyrrolidone (10,000 mol wt)/PBS, pH 7.4, for 2 h, and mounted onto cryo-pins and rapidly frozen in liquid nitrogen. Ultrathin cryosections were cut on an ultramicrotome (UCT; Leica) equipped with an FCS cryo-attachment and collected onto formvar/carbon-coated copper grids. The grids were then washed through several drops of 1× PBS containing 2.5% FCS and 10 mM glycine, pH 7.4, and then blocked in 10% FCS for 30 min and incubated overnight in chicken anti-GFP diluted to 1:400 (Abcam). After washing, the grids were incubated for 2 h in donkey anti–chicken 12-nm Au (Jackson ImmunoResearch Laboratories, Inc.). The grids were washed through several drops of 1× PBS followed by several drops of ddH2O; floated on a 1-ml drop of neutral uranyl acetate, pH 7.4, for 10 min; quickly washed through five drops of ddH2O; and floated onto an aqueous solution containing 3.2% polyvinyl alcohol (10,000 mol wt), 0.2% methyl cellulose (400 centiposes), and 0.1% uranyl acetate. The grids were then embedded by removing excess solution using hardened filter paper (no. 50; Whatman) and examined in a transmission electron microscope (Tecnai 12 Twin; FEI) operating at 100 kV. Images were collected using a digital camera (Soft Imaging System Megaview III; Olympus), and figures were assembled in Photoshop (Adobe) using only linear adjustments in brightness and contrast.
To quantify distances of each gold particle from the cell membrane, XY coordinates were obtained in ImageJ/FIJI of each particle and of a freehand line tracing the cell membrane. The distances between each particle and the closest point to the cell membrane line were calculated.
Patch-clamp electrophysiology
HEK293T cells were transfected with individual ANO2 expression constructs along with an expression plasmid for the principal subunit of the olfactory CNG channel CNGA2, which serves as a control for patch integrity (Stephan et al., 2009). The transfections were performed using the Lipofectamine 2000 reagent (Invitrogen). For coexpression experiments, the different ANO2 splice variants were expressed from individual plasmids transfected at equal amounts. Inside-out patches were excised using borosilicate glass pipettes fabricated using a puller (P97; Sutter Instrument). The pipettes had an open tip resistance of ∼3 MΩ. Transfected cells were identified by their GFP fluorescence. The patches were pulled after a gigaohm seal was established. If, after excision, the membrane patch formed a vesicle at the pipette tip, it was opened by contact with debris at the bottom of the recording chamber. The patches were exposed to Ca2+ and cAMP immediately (15 s) after patch excision, and the protocol was continued for 20 min during which the chloride current stabilized after rundown. Peak ANO2 and CNGA2 currents were obtained from the first trace (15 s). Dose–response currents were recorded after the initial rundown of the chloride current subsided by exposing the patch to increasing concentrations of Ca2+. For quantifying the dose response, the currents from each patch were normalized to the maximal current and the normalized values were averaged across multiple patches and fit with Hill functions to yield the K1/2 (µM) and Hill coefficient (n) values. I-V relationships were obtained by preexposing the patch to 67 µM Ca2+ for 4 s to eliminate the possibility of rectification induced as a result of inactivation of the ANO2 channel. The preexposure was followed by two voltage ramps (at 100 mV/s) from −50 to +50 mV and back to −50 mV, from which the traces were averaged to yield the I-V relationship. The currents were recorded using a patch-clamp amplifier (PC501A; Warner) and digitized using a data acquisition unit (Micro1401) and acquisition software (Signal; Cambridge Electronic Design). The currents were low-pass filtered (eight-pole Bessel; Krohn-Hite Corp.) at DC-100 Hz and DC-2 kHz, and digitized and sampled at 5 kHz. Two-way ANOVA was performed for comparing the inactivation of the splice variants (factor A, two levels) using Imax/I10s values at the four concentrations (Factor B, 4 levels). The post-hoc test used was Bonferroni (P < 0.05). The analysis was performed using Origin (V.8.5) software. Shifts in dose–response relations were statistically compared using the K1/2 values and errors obtained from the relevant dose–response relations.
For whole-cell recordings, the pipettes had an open tip resistance of 3–5 MΩ. The recordings were made by varying the holding potential between −100 and +100 mV in steps of 20 mV, followed by a final step to −100 mV. Signals were filtered at 5 kHz and sampled at 10 kHz.
Solutions
For excised patch-clamp experiments, the bath solution and pipette solution contained (mM): 140 NaCl, 10 HEDTA, and 10 HEPES. For dose–response experiments, the different solutions contained 0.25, 0.75, 2.4, 11, and 67 µM of free Ca2+ (Reisert et al., 2003), and patches were exposed from 0 to 10 s. To estimate the patch current magnitudes for the ANO2 and the CNGA2 currents, patches were exposed to 67 µM Ca2+ from 0 to 3 s and to 1 mM cAMP from 6 to 9 s. The bath solution was used for “zero” Ca2+ exposure. The pH was adjusted to 7.2 with NMDG. A fast-step perfusion system (Warner) was used to rapidly expose the patch and to switch solutions.
For whole-cell experiments, the extracellular solution contained (mM): 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, and 10 HEPES, with pH adjusted to 7.4 using NaOH. The pipette solution contained (mM): 140 choline chloride, 10 HEDTA, 10 HEPES, 3.209 CaCl2 (1.5 µM of free Ca2+) (Patton et al., 2004), with pH adjusted to 7.2 using NMDG. The holding potentials were corrected offline for liquid junction potential (−2.5 mV) calculated using Clampex software.
Online supplemental material
Fig. S1 A shows an agarose gel of the 5′ RACE PCR products for Ano2, whereas B and C show the reverse complement sequencing chromatogram of the predominant RACE PCR products including the new exon, named exon 1b (red), and densitometry quantification of bands from Fig. 1 (B and C). Fig. S2 shows current–voltage relationships and their rectification for Isoforms A and B at three different Ca2+ concentrations. Fig. S3 quantifies the distance of immunogold particles from the cell membrane for Isofrom B, BΔ4, the TrkA receptor, and GFP when expressed in HEK cells. Figs. S1–S3 and Table S1 are available at http://www.jgp.org/cgi/content/full/jgp.201210937/DC1.
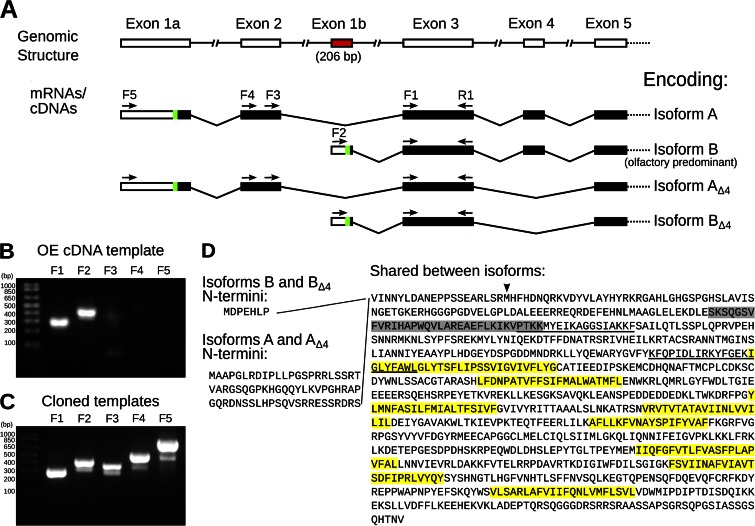
Figure 1.
Characterization of Ano2 mRNA variants present in mouse olfactory epithelium. (A) Diagram summarizing the 5′ Ano2 exon splicing structure. The green sections indicate the most 5′ AUG translation start codons, and the subsequent black bars indicate predicted protein-coding sequence. The variants containing exons 1a and 2 are less abundant in the olfactory epithelium than the variants containing the newly discovered exon 1b (red), as determined in B and C. The five forward (F1–F5) and one reverse (R1) PCR primer–binding sites are indicated as arrows. Given two alternative 5′ ends and alternative splicing of exon 4, these mRNA variants may encode up to four ANO2 isoforms, named on the right. (B and C) Ethidium bromide–stained agarose gels showing RT-PCR products with primers specific for each 5′ variant of Ano2. Primer F1 is universal for both variants, F2 is specific for exon 1b, F3 and F4 are specific for exon 2, and F5 is specific for exon 1a. The reverse primer (R1) was the same for all PCR reactions. The expected PCR product sizes are 267, 409, 350, 511, and 830 bp, respectively. Quantification of the bands by densitometry is shown in Fig. S1 C. (B) PCR template was olfactory epithelium cDNA. (C) Positive control PCR using cloned templates of equal amounts. (D) Corresponding amino acid sequence of the olfactory ANO2 isoforms. The amino acids encoded by exon 4 are highlighted in gray. Predicted transmembrane domains are highlighted in yellow. Putative CaM-binding sites are underlined (Yap et al., 2000; Tian et al., 2011). The start site used in Stephan et al. (2009) is indicated by an arrowhead.
RESULTS
Olfactory Ano2 transcripts are expressed predominantly from a novel transcription initiation site
Subsequent to our publication in 2009 (Stephan et al., 2009), the mouse Ano2 sequence entry in GenBank was updated from NM_153589.1 to NM_153589.2, which indicated a change in the N-terminal amino acid sequence. To determine the 5′ end of Ano2 transcripts in the olfactory epithelium, we conducted 5′ RACE analysis (Scotto-Lavino et al., 2006). The 5′ RACE PCR products were analyzed by agarose gel electrophoresis, which showed two predominant bands at 532 and 436 bp, as well as a background smearing of lower molecular weights (Fig. S1 A). These PCR products were directly sequenced (Fig. S1 B) and were also cloned and then sequenced (Table S1). Contrary to the publicly available mouse Ano2 sequences (e.g., GenBank accession no. NM_153589.2), we found that the predominant olfactory Ano2 transcripts did not contain sequences from the first two exons. Instead, all the olfactory Ano2 transcripts detected by 5′ RACE contained a novel starting exon, 206 bp in the longest RACE products, which we named exon 1b (Figs. 1 A and S1, A and B, and Table S1). Exon 1b is then joined to exon 3 and subsequent exons (Fig. 1 A). Based on the location of the most 5′ in-frame ATG codon, we determined that Ano2 transcripts containing exon 1b encode a shorter N terminus, containing a 7–amino acid sequence unique to this novel ANO2 isoform (Fig. 1 D).
Because this novel Ano2 exon 1b–containing variant has never been described, we asked whether the canonical Ano2 variant containing exon 1a and exon 2 was detectable in the olfactory epithelium, and if so, what its abundance is relative to the novel Ano2 variant containing exon 1b. We performed RT-PCR on olfactory epithelium cDNA with various primers that differentiate between the two variants. PCR using primers that bind to exon 3, common to all mRNA variants, readily amplified the target sequence, as did the primers specific to the novel exon 1b (Fig. 1 B; primer binding sites indicated in Fig. 1 A). Conversely, PCR using primers specific to exon 1a or exon 2 only weakly amplified the target sequences (Figs. 1 B and S1 C). To control for the possibility that this weak amplification reflected poor priming efficiency, we performed PCR under the same conditions but this time using cloned templates in equal amounts as positive controls. In this case, PCRs using each of the primer pairs readily amplified the target sequences (Figs. 1 C and S1 C). Thus, mRNA species representing both Ano2 5′ variants are present within the olfactory epithelium, but the novel exon 1b–containing Ano2 variant predominates over the canonical Ano2 isoform containing exon 1a and exon 2. For the sake of brevity, the two mRNA variants—the canonical exon 1a– and exon 2–containing variant and the novel exon 1b–containing variant—are referred to as encoding ANO2 Isoforms A and B, respectively (Fig. 1 A).
Given the two N termini of ANO2, along with the two additional possible splicing variants either containing or lacking exon 4 (Stephan et al., 2009), up to four different ANO2 isoforms may exist within the olfactory epithelium: Isoforms A, B, AΔ4, and BΔ4 (Fig. 1 A). The mRNA sequences encoding these four isoforms have been made available publically in GenBank under accession numbers KC164759, KC164761, KC164760, and KC164762, respectively.
The N terminus of ANO2 affects the sensitivity of channel activation by Ca2+
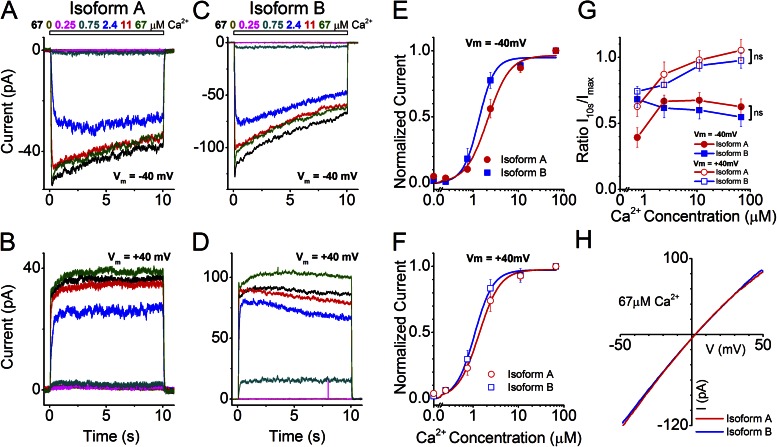
To determine whether the alternative N termini differing between ANO2 Isoforms A and B confer any functional differences to heterologously expressed ANO2 channels, we individually expressed both isoforms in HEK293T cells and performed excised inside-out patch-clamp analysis (Fig. 2). The patches were exposed to a series of Ca2+ concentrations ranging from 0 to 67 µM for 10 s each. The recordings were made after the “rundown” phase of the current was over (typically ∼20 min after excision; see also Fig. 6) to ensure current stability (Stephan et al., 2009). We observed that expression of either Isoforms A or B showed robust Ca2+ dose-dependent channel activity: currents rapidly peaked (Imax) and then inactivated at negative but less so at positive holding potentials (see below) over the course of the 10-s stimulation period to a lower current (I10s) (Fig. 2, A–D). The normalized Imax values for the two individual ANO2 isoforms were fitted with Hill functions to obtain K1/2 values, which reflect the sensitivity of the channel activation by Ca2+ (Fig. 2, E and F). Comparison of K1/2 values between Isoforms A and B shows that Isoform B displayed an increased sensitivity to Ca2+ at a holding potential of −40 mV as compared with Isoform A (Table 1; P = 0.04). This sensitivity increase is also apparent as a leftward shift of the dose–response relationship shown in Fig. 2 E. No significant sensitivity difference between the two isoforms was seen at the +40-mV holding potential (Table 1; P > 0.05; Fig. 2 F).
Figure 2.
Effects of alternative N termini on ANO2 channel properties. (A–D) Inside-out excised patch-clamp traces of ANO2 isoforms expressed individually in HEK293T cells. Dose–response traces were recorded ∼20 min after patch excision when the rundown phase was over. Patches were exposed to various concentrations of Ca2+ for 10 s as indicated in the figures. The current traces are color-coded to match their respective stimulus concentrations. (A and B) Typical currents from a patch expressing Isoform A at −40- and +40-mV holding potentials. (C and D) Currents from the expression of Isoform B. (E and F) Hill fits of the maximal currents from Isoform A (11 patches) and Isoform B (7 patches) at negative and positive holding potentials, respectively. (G) Inactivation properties of currents from Isoforms A and B are quantified using the ratio of current amplitudes at 10 s (I10s) after Ca2+ exposure to the peak I value (Imax). ns, no significant difference in inactivation between the isoforms at positive and negative holding potentials across all concentrations of Ca2+ (two-way ANOVA). Data are means of I10s/Imax values (±SEM) from 7–11 patches for the four Ca2+concentrations (0.75, 2.4, 11, and 67 µM Ca2+). (H) I-V relationship for the two isoforms in response to saturating Ca2+ (67 µM).
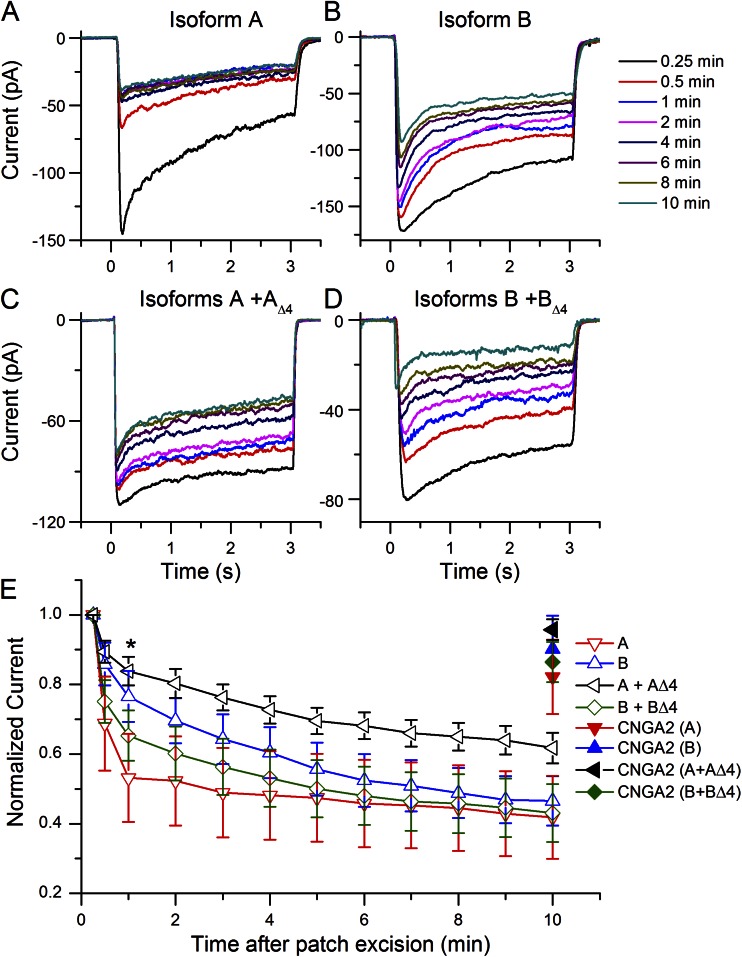
Figure 6.
Rundown of the Cl− current upon coexpression of ANO2 isoforms. Expression of Isoforms A and B alone and with their respective isoforms lacking exon 4 yielded channels with different rundown properties. (A) Isoform A. (B) Isoform B. (C) Isoform A + AΔ4. (D) Isoform B + BΔ4. (E) Rundown as a function of time after patch excision. All currents were normalized to the first current recorded 15 s after excision. *, P < 0.05 for Isoform A compared with A + AΔ4 at t = 1 min. Number of patches recorded from for Isoform A, B, A + AΔ4, and B + BΔ4 were 7, 5, 12, and 7, respectively. The holding potential was −40 mV.
Table 1.
Summary of Ca2+ sensitivities of the ANO2 isoforms
| −40 mV | +40 mV | |||||||||||
| Values derived from Imax | Isoform A | Isoforms A + AΔ4 | Isoform B | Isoforms B + BΔ4 | Native channel (mouse) | ANO2 variant Stephan et al., 2009 | Isoform A | Isoforms A + AΔ4 | Isoform B | Isoforms B + BΔ4 | Native channel (mouse) | ANO2 variant Stephan et al., 2009 |
| K1/2 (µM) | 2.1 ± 0.26 | 1.65 ± 0.13 | 1.33 ± 0.13 | 2.04 ± 0.20 | 3.46 ± 0.21 | 1.83 ± 0.03 | 1.35 ± 0.11 | 0.96 ± 0.09 | 1.08 ± 0.07 | 1.45 ± 0.19 | 1.48 ± 0.08 | 1.18 ± 0.11 |
| n Hill Coeff. | 1.79 ± 0.38 | 1.91 ± 0.23 | 2.49 ± 0.39 | 2.61 ± 0.81 | 1.41 ± 0.1 | 2.3 ± 0.07 | 1.87 ± 0.23 | 2.32 ± 0.47 | 2.13 ± 0.25 | 2.99 ± 0.65 | 3.12 ± 0.27 | 1.92 ± 0.27 |
| I10s/Imax at 67 µM Ca2+ | 0.62 ± 0.06 | 0.4 ± 0.05 | 0.55 ± 0.07 | 0.43 ± 0.07 | 0.59 ± 0.05 | 0.41 ± 0.06 | 1.05 ± 0.08 | 1 ± 0.07 | 0.97 ± 0.06 | 0.79 ± 0.07 | 0.50 ± 0.06 | 0.98 ± 0.04 |
All values are mean ± SEM (6–11 patches). Mouse-native channel values are from Reisert et al., 2005, and Table S2 in Stephan et al., 2009.
Although currents from Isoforms A and B showed inactivation at −40 mV (Fig. 2, A and C), inactivation was not apparent at +40 mV, particularly at high Ca2+ concentrations (Fig. 2, B and D). To quantify inactivation, the ratios of Imax /I10s were calculated (Fig. 2 G). The Imax /I10s ratios approach unity at +40 mV for both Isoforms A and B at high Ca2+ concentrations, indicating a lack of inactivation under these conditions (Fig. 2 G). It has been shown that native olfactory Ca2+-activated Cl− channels exhibit inactivation even at +40 mV and at high Ca2+ concentrations (Reisert et al., 2003); thus, neither Isoform A nor Isoform B in this heterologous system fully recapitulates the native olfactory channel properties.
The I-V relationship for Isoforms A and B were similar, with the current showing a slight inward rectification and a reversal potential close to 0 mV (Fig. 2 H) at saturating (67 µM) Ca2+ concentrations. At intermediate Ca2+ concentration, both Isoform A and B channels displayed linear I-V relationships, and at low Ca2+ concentrations, both channels became outwardly rectifying (Fig. S2).
Exon 4 is required to encode functional ANO2 channels
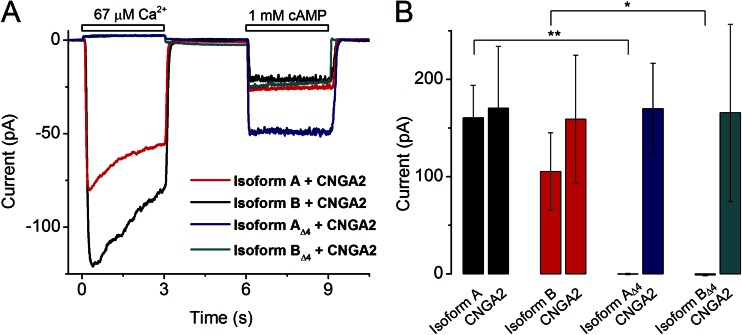
To determine if the exon 4–encoded protein sequence confers any functional differences to the heterologously expressed ANO2 channels, we expressed Isoforms A, B, AΔ4, and BΔ4 individually in HEK293T cells and performed excised inside-out patch-clamp analysis. Whereas Isoforms A and B produced robust chloride currents in response to a saturating Ca2+concentration (67 µM), Isoforms AΔ4 and BΔ4 failed to produce any measurable currents (Fig. 3 A). The traces in Fig. 3 A represent results from an experimental protocol where membrane patches from cells cotransfected with plasmids encoding each of the four isoforms of ANO2 along with a plasmid encoding the principal subunit of the olfactory CNG channel CNGA2 were exposed to 67 µM Ca2+ for 3 s followed by 1 mM cAMP for 3 s. The traces were quickly recorded 15 s after patch excision to minimize the effect of Cl− current rundown. The cAMP-induced currents through CNGA2 served as controls for patch integrity. Although cells expressing the “Δ4” isoforms did not show any chloride currents, cAMP-generated currents were still observed. A quantification of the amplitude of the ANO2 and CNGA2 currents is shown in Fig. 3 B. Although Isoforms A and B exhibited considerable chloride current amplitudes (160 ± 34 and 105 ± 40 pA, respectively), Isoforms AΔ4 and BΔ4 yielded negligible current amplitudes (−1.2 ± 0.7 and −0.3 ± 0.7 pA, respectively). The cAMP currents for all four isoforms remained, on average, the same (159 ± 66, 171 ± 63, 166 ± 91, and 170 ± 47 pA).
Figure 3.
Patch-clamp analysis of ANO2 isoforms containing or lacking exon 4–encoded sequence. (A) Ca2+ and cAMP-gated currents from HEK293T patches expressing the individual isoforms of ANO2 along with the CNGA2 channel. Patches were exposed immediately (15 s) after excision, and the holding potential was −40 mV. (B) Quantification of the peak ANO2 and CNGA2 currents for each expression condition derived from experiments as shown in A. Current (pA) values are means from 9–11 patches ± SEM. *, P < 0.01; **, P < 0.001; Student’s t test.
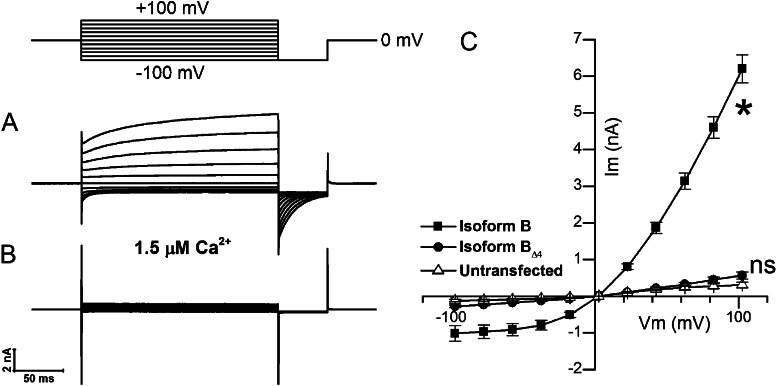
To determine whether the absence of currents from ANO2 “Δ4” isoforms is caused by “instantaneous rundown” after patch excision, we performed experiments in the whole-cell configuration where rundown is not observed (Pifferi et al., 2009). We transfected HEK293T cells with Isoform B, which we determined to be the predominant isoform in the olfactory epithelium, and Isoform BΔ4 individually and used a pipette solution containing 1.5 µM Ca2+ to activate whole-cell currents (Fig. 4, A and B). The cells expressing Isoform B showed robust outwardly rectifying currents (Fig. 4 C), which is a known property of ANO2 at subsaturating Ca2+ levels (Pifferi et al., 2009). In cells expressing Isoform BΔ4, the currents were similar in magnitude to those in untransfected cells (Fig. 4 C). Collectively, the results from excised patch recordings and whole-cell recordings indicate that the protein sequence encoded by Ano2 exon 4 is necessary for channel function or membrane targeting in this heterologous system.
Figure 4.
Whole-cell voltage-clamp analysis of predominant ANO2 isoforms containing or lacking exon 4–encoded sequence. (A) Whole-cell currents from a HEK293T cell expressing the olfactory ANO2 Isoform B obtained with a pipette solution containing 1.5 µM Ca2+. (B) Negligible currents from a cell expressing the BΔ4 ANO2 isoform. (C) Mean I-V relation from cells expressing either Isoform B or Isoform BΔ4 and untransfected cells (n = 10 for each condition; error bars are ±SEM). Current amplitudes were measured at the end of the voltage-step protocol shown above A. *, currents from Isoform B were significantly different from untransfected control (P < 0.0001; two-way ANOVA). ns, nonsignificant difference between Isoform BΔ4 and untransfected control (P > 0.5; two-way ANOVA).
Cellular localization of alternatively spliced isoforms
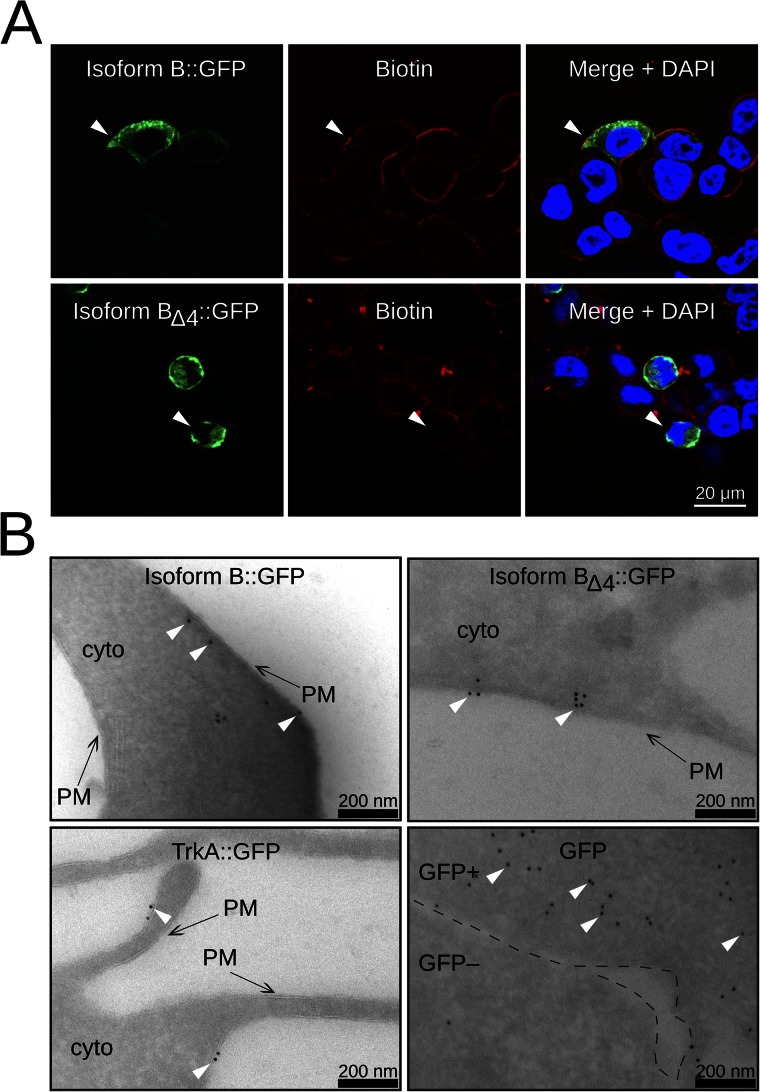
To address if exon 4–lacking isoforms traffic to the cell membrane and thus if exon 4 is required for cell membrane targeting, we investigated the protein localization of GFP-tagged ANO2 isoforms expressed in HEK cells. First, we expressed either Isoform B::GFP or BΔ4::GFP and visualized the fusion proteins with anti-GFP immunofluorescence staining. The cell membrane of HEK293T cells was labeled with biotin and stained with a streptavidin Alexa Fluor 546 conjugate (Fig. 5 A). Biotin labeling (red) was clearly limited to the cell membrane where it colocalized with Isoform B::GFP staining (green), which was more broadly distributed throughout the cell (Fig. 5 A, top row, arrowhead). A similar membrane colocalization of the GFP and biotin staining was also observed for the B isoform lacking exon 4 (Fig. 5 A, bottom row, arrowhead).
Figure 5.
Localization of ANO2 Isoform B and Isoform BΔ4 in HEK cells. (A) Immunofluorescence images of HEK293T cells transfected with either ANO2 Isoform B::GFP (top row) or Isoform BΔ4::GFP (bottom row). The left panels show Isoform B::GFP or BΔ4::GFP staining. Biotin labeling, shown in the middle panels, is used as a membrane marker. The right panels show merged images with DAPI as a nuclear marker. White arrowheads point to areas of Isoform B::GFP or BΔ4::GFP colocalization with biotin. (B) Immunogold labeling against GFP in HEK293T cells. TrkA::GFP is used as a membrane localization control (bottom left panel), and GFP is used as a cytoplasmic control (bottom right panel). Black arrows point to the cell membrane. White arrowheads point to several immunogold particles. PM, cell membrane; cyto, cytoplasm. The GFP panel shows two adjacent cells: one expressing a high level of GFP (GFP+) and the other expressing little or no GFP (GFP−). The black dashed lines demarcate the GFP+ and the GFP− cell. For quantification of the spatial distribution of gold particles, see Fig. S3.
In a second approach, we investigated protein localization at a higher resolution using electron microscopy and immunogold labeling. We expressed Isoform B::GFP and BΔ4::GFP, as well as TrkA::GFP and GFP as controls. TrkA, the nerve growth factor receptor, is a well-characterized membrane protein. As shown in Fig. 5 B (bottom left), TrkA::GFP immunogold particles are clearly localized to the cell membrane, whereas GFP particles are distributed throughout the cytoplasm (bottom right). Immunogold particles for Isoform B::GFP (Fig. 5 B, top left) and Isoform B lacking exon 4 (Fig. 5 B, top right) are, similar to TrkA::GFP particles, found predominately close to the cell membrane. We measured the distances of gold particles to the cell membrane for all four proteins (Fig. S3). The distance distributions for B::GFP and BΔ4::GFP, which are similar to TrkA:GFP distribution, suggest a membrane localization. These data show that BΔ4::GFP is indeed capable of targeting to the cell membrane and that the exon 4 sequence is necessary for channel function.
Rundown properties of individual and coexpressed splice variants
Given that ANO2 isoforms lacking exon 4 cannot confer ANO2 channel activity to HEK293T cells when expressed individually, we considered the possibility that the “Δ4” isoforms could constitute modulatory subunits of a hetero-multimeric ANO2 channel. To test this possibility, we cotransformed HEK293T cells with expression constructs encoding both Isoforms A and AΔ4 and alternatively also B and BΔ4, performed excised inside-out patch-clamp analysis, and asked whether coexpression affected channel properties. Some of the analyses were performed as described above for individual expression of isoforms; thus, comparisons were made directly to the channel properties of Isoforms A or B, respectively, when expressed alone.
We first investigated the rundown properties of channels arising from expression of a single isoform or when coexpressed with the corresponding isoform lacking exon 4. Patches were repeatedly exposed to 67 µM Ca2+ and 1 mM cAMP using the same protocol as shown in Fig. 3 A. The CNG current was also recorded to monitor patch integrity over time. When Isoform A was expressed alone, the peak Cl− current declined quickly with each repeated Ca2+ exposure (Fig. 6 A) and lost nearly half its current within 1 min (Fig. 6 E). In contrast, coexpression of A with AΔ4 greatly slowed the rundown of the Cl− current (Fig. 6, C and E). Expression of Isoform B alone or together with BΔ4 yielded channels that had similar rundown rates falling in between those of A and A + AΔ4 (Fig. 6, B, D, and E). We tested for statistical difference between the different expression conditions at 1 min after patch excision, where the difference in rundown was largest. The level of rundown was different between A and A + AΔ4 (P = 0.024) but not between other combinations. After 10 min after excision, all four channels decreased to similar current levels. The CNG current remained quite stable over time, as indicated by the closed symbols at t = 10 min in Fig. 6 E.
Coexpression of Isoforms A and AΔ4 results in altered channel sensitivity
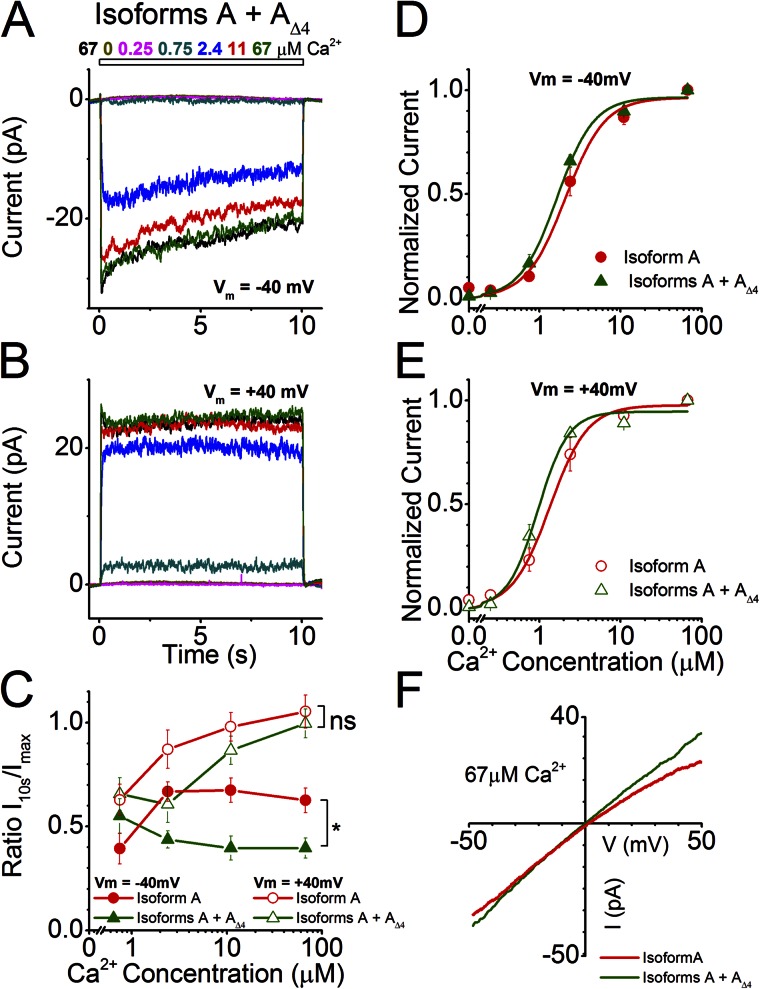
We exposed patches that coexpressed Isoforms A and AΔ4 to the same set of Ca2+ concentrations used in Fig. 2 for 10 s and determined the Ca2+ sensitivity and inactivation properties of the resulting channel at a holding potential of −40 and +40 mV (Fig. 7, A and B). At −40 mV, the channel resulting from coexpression of Isoforms A and AΔ4 had similar sensitivities (Table 1; K1/2 = 1.65 ± 0.13 µM) compared with Isoform A alone (Fig. 7 D), whereas at +40 mV, coexpression yielded a more sensitive channel (Fig. 7 E; K1/2 = 0.96 ± 0.09 µM; P = 0.017). However, the coexpression increased the inactivation of the current during the 10-s Ca2+ exposure at −40 mV (P < 0.01), a property that remained unaltered at +40 mV (Fig. 7 C). The inactivation was again quantified by dividing the current at 10 s (end of stimulation, I10s) by the maximal current (Imax) and was plotted as a function of Ca2+ concentration. Little difference was seen between the I-V relationship of Isoform A and Isoform A coexpressed with Isoform AΔ4 (Fig. 7 F).
Figure 7.
Channel properties of Isoform A coexpressed with Isoform AΔ4. (A and B) Currents from the coexpression of ANO2 Isoforms A and AΔ4 at holding potentials of −40 and +40 mV in response to varying concentrations of Ca2+. (C) Comparison of current inactivation of the coexpression with that of Isoform A alone across all Ca2+ concentrations (data for A replotted from Fig. 2 G for comparison). *, P < 0.01; two-way ANOVA. (D and E) Hill fits of normalized current values from the coexpression of Isoforms A and AΔ4 at holding potentials of −40 and +40 mV (eight and six patches, respectively). Hill fits of currents from Isoform A (from Fig. 2, E and F) are also drawn for comparison. (F) I-V relationship at saturating Ca2+ for the coexpression and its comparison with Isoform A.
Coexpression of Isoforms B and BΔ4 results in altered channel inactivation properties
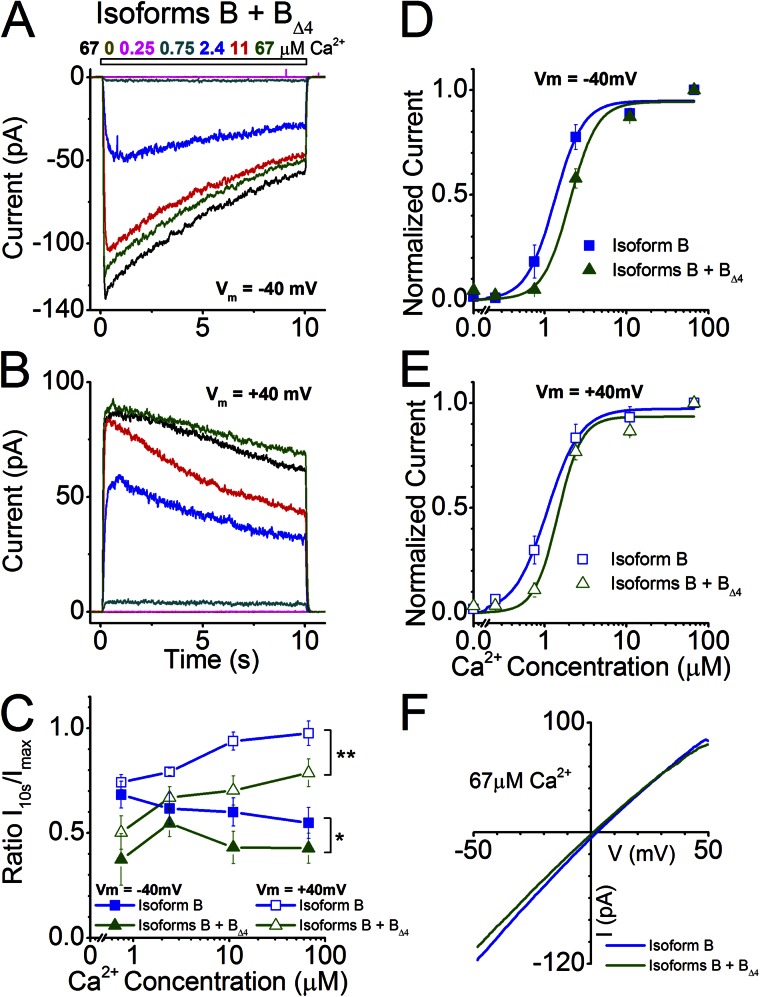
We found that when Isoforms B and BΔ4 were coexpressed (Fig. 8, A and B), ANO2 channel activity was less sensitive to Ca2+ than when Isoform B was expressed alone, which is opposite to what was observed for Isoforms A and AΔ4 at positive holding potentials. This decreased sensitivity was apparent as a rightward shift of the Ca2+ dose–response curve (Fig. 8, D and E) and as an increase in K1/2 values (Table 1; P = 0.01). The K1/2 value for the coexpression condition (2.04 ± 0.2 µM) is closer to that of the native channel (3.46 ± 0.21 µM) at a holding potential of −40 mV. At a holding potential of +40 mV, the K1/2 value for the coexpression (1.45 ± 0.19 µM) is comparable to that of the native channel (1.48 ± 0.08 µM; Table 1) (Stephan et al., 2009).
Figure 8.
Channel properties of Isoform B coexpressed with Isoform BΔ4. (A and B) Currents from the coexpression of ANO2 Isoforms B and BΔ4 at holding potentials of −40 and +40 mV in response to varying concentrations of Ca2+. (C) Comparison of current inactivation of the coexpression (seven patches) with that of Isoform B alone across all Ca2+ concentrations (data for Isoform B replotted from Fig. 2 G for comparison). *, P < 0.01; **, P < 0.0001; two-way ANOVA. (D and E) Hill fits of normalized current values from the coexpression of Isoforms B and BΔ4 at holding potentials of −40 and +40 mV (seven patches). Hill fits of currents from Isoform B (from Fig. 2, E and F) are also drawn for comparison. (F) I-V relationship at saturating Ca2+ for the coexpression and its comparison with Isoform B.
We also found that coexpression changed channel inactivation properties to more closely resemble the native olfactory channel. At a holding potential of +40 mV, whereas the individually expressed Isoforms A or B, or the coexpressed Isoforms A and AΔ4, did not show inactivation at higher concentrations of Ca2+ (Figs. 2, B, D, and G, and 7, B and C), coexpression of Isoforms B and BΔ4 showed remarkable inactivation (Fig. 8, B and C). A significant difference between Isoform B and the coexpression is observed in the inactivation properties at both positive and negative holding potentials (P < 0.05). This display of inactivation even at positive holding potentials is reminiscent of what is observed for the native olfactory channel (Stephan et al., 2009) (Table 1). However, a complete recapitulation in the degree of inactivation at high Ca2+ levels seen in the native channel is not observed in the coexpression (Table 1). The I-V relationship of Isoform B is not different from the coexpression; both Isoform B alone and Isoform B coexpressed with Isoform BΔ4 showed a slight inward rectification and a reversal potential close to 0 mV (Fig. 8 F).
DISCUSSION
In this study, we describe the identification of a novel exon encoding a novel N terminus of the olfactory Ca2+-activated chloride channel ANO2. The ANO2 isoform containing this novel N terminus along with the exon 4 sequence (Isoform B) is the predominant olfactory isoform and has current properties very similar to the previously reported isoform with the truncated N terminus starting at exon 3 (Stephan et al., 2009). Probably most importantly for this study, the inactivation properties at positive and negative holding potentials were similar between these two isoforms. We also found the presence of a rarer isoform (Isoform A) that conforms to the longer N terminus encoded by the publically available sequence (GenBank accession no. NM_153589.2). When activated by Ca2+, Isoform A produced similarly sized chloride currents and had similar current inactivation kinetics at +40 and −40 mV compared with Isoform B. However, Isoform A was less sensitive to Ca2+ at −40 mV, indicating that the N terminus is involved in Ca2+ sensitivity at negative potentials. A similar scenario has been reported for ANO1, where an alternatively spliced protein segment (22 amino acids) in the cytosolic N terminal results in a fourfold difference in Ca2+ sensitivity among the isoforms, with the shorter isoform being more sensitive than the longer isoform, especially at negative holding potentials (Ferrera et al., 2009). Although the difference in K1/2 values in the ANO2 counterparts are not as drastically different as in the ANO1 isoforms, the results nonetheless point to the importance of the N terminus in determining the Ca2+ sensitivity of these channels (Table 1).
We also exposed patches expressing two additional isoforms that splice alternatively at exon 4 and contain the alternative N termini (exon 1b or exon 1a) to Ca2+ (Isoform AΔ4 and Isoform BΔ4). Regardless of the starting exon, the isoforms lacking the exon 4 sequence failed to generate any current when exposed to Ca2+, indicating that without the exon 4 sequence, these isoforms either do not target to the cell membrane or the channels are nonfunctional. Immunohistochemical and immunogold data (Fig. 5) show that isoforms lacking exon 4 can traffic to the cell membrane, suggesting that exon 4 is integral to channel function and not required for membrane trafficking. The ANO channel pore is predicted to be between transmembrane domains 5 and 6 (Caputo et al., 2008; Das et al., 2008; Yang et al., 2008), thus excluding the exon 4 sequence, which is predicted to be within the N-terminal cytoplasmic domain. Therefore, it is possible that exon 4 encodes a regulatory domain that is crucial for channel function. A web-based calmodulin (CaM)-binding prediction application shows Ano2 exon 4 partially coding for a putative CaM-binding domain, which may act as a regulator of channel activity (Calmodulin Target Database; Ikura laboratory, University of Toronto, Toronto, Canada) (Fig. 1 D). In the case of ANO1, it has been demonstrated that CaM physically interacts with homologous N-terminal putative CaM-binding sites and that CaM is necessary for channel activation (Tian et al., 2011). However, the splicing variants of Ano1 not encoding the CaM-binding domain were still able to elicit currents and did not depend on CaM for activation. In the absence of any evidence to the contrary, CaM-dependent activation of ANO2 cannot be completely ruled out, although the native channel does not seem to be affected by exogenously applied CaM (Reisert et al., 2003).
As exon 4–lacking isoforms do not form functional channels on their own, we investigated if these isoforms could modulate channel properties of the exon 4–containing isoforms when expressed together. Indeed, coexpression of Isoform A with AΔ4 yielded a channel that displayed significantly slowed rundown properties compared with those when A was expressed alone. The latter displayed very fast rundown kinetics, losing nearly half its current within 1 min after patch excision (Fig. 6). Such a difference was not observed when Isoform B and BΔ4 were coexpressed, indicating that the speed of rundown might be determined by the interplay of the exon 4 sequence of Isoform A with its longer N terminus.
A second biophysical property altered by coexpression of isoforms containing and lacking exon 4 is the inactivation property of the channel during longer Ca2+ exposures. Interestingly, when Isoform A was coexpressed with AΔ4, the channel showed increased inactivation at −40 mV but did not alter its inactivation properties at +40 mV. This is in contrast to the channel resulting from coexpression of Isoform B and BΔ4, where the channel inactivated more at both holding voltages and became less sensitive to Ca2+ (Figs. 7 and 8). Current properties of the coexpression resemble the native olfactory Ca2+-activated Cl− channel more closely (see Table 1). In particular, the currents of the Isoform B coexpression show a slow inactivation even at positive holding potentials, similar to what is observed for the native channel. The peak current value (198 ± 50 pA) of the coexpressed B isoforms is not significantly different from the expression of Isoform B alone (105 ± 40 pA; P > 0.05), indicating that coexpression does not alter membrane trafficking of the predominant isoform. It is tempting to speculate that the native olfactory Ca2+-activated Cl− channel might be a heteromer composed of (at least) two ANO2 isoforms, predominantly the B + BΔ4 isoforms, as coexpression of A isoforms or expression of Isoform B alone does not recapitulate the native channel. It has been shown that ANO1 strictly forms homodimers by associating before reaching the cell membrane (Fallah et al., 2011; Sheridan et al., 2011) and that ANO1 and ANO2 are both present in microvilli of vomeronasal neurons (Dibattista et al., 2012). It has also been shown that different ANOs can interact and alter the activities of each other. For example, ANO9 can negatively affect the activity of ANO1 when expressed in FRT cells (Schreiber et al., 2010). Therefore, it is also possible that ANO2 B isoforms, instead of forming heteromers, may be interacting in other ways to modify their current properties.
To summarize, we showed that the major olfactory isoform of ANO2 is composed of a novel N terminus that contributes toward the Ca2+ sensitivity of the channel. The previously reported splice site exon 4 sequence is indispensable for channel function through a yet unidentified mechanism. Coexpression of the predominant B isoforms with and without exon 4 sequence yields currents that better recapitulate the native channel current properties, indicating that the native channel may be a heteromer composed of two or more ANO2 splicing isoforms.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Frank Margolis for pointing out discrepancies in Ano2 mRNA sequences within the early database entries and for annotation of predicted CaM-binding sites. The authors would like to thank Dr. Michael Tordoff for providing access to cell culture equipment and Dr. Rejji Kuruvilla for the TrkA::GFP plasmid. The authors would also like to thank Sonia Caraballo for help with Ano2 molecular characterization as a JHU SURE student, and Eric Yang for scripting the immunogold distance calculations. Special thanks to Dr. J. Michael McCaffery of the JHU Integrated Imaging Center for his input on the electron microscopy.
This work was supported by National Institutes of Health (grant DC009613 to J. Reisert and grant DC007395 to H. Zhao) and the Human Frontiers Science Program (to J. Reisert). A.B. Stephan is currently a Department of Energy Biosciences Fellow of the Life Sciences Research Foundation.
Author contributions: Ano2 sequencing, 5′ RACE, and cloning were performed by A.B. Stephan and H. Zhao; electrophysiology was performed by S. Ponissery Saidu and J. Reisert; A.K. Talaga assayed subcellular localization of ANO2 isoforms; and data were interpreted and paper was written by all the authors.
Edward N. Pugh Jr. served as editor.
Note added in proof. In a recent paper by Tien et al. (2013. Proc. Natl. Acad. Sci. USA. http://dx.doi.org/10.1073/pnas.1303672110), the authors describe a 19–amino acid domain in the N terminus of Anoctamin 1 that can support dimerization. Interestingly, the region homologous to this domain in Anoctamin 2 is entirely contained within exon 4, the exon that can be spliced out in olfactory receptor neurons.
Footnotes
Abbreviations used in this paper:
- ANO
- Anoctamin
- CaM
- calmodulin
- ORN
- olfactory receptor neuron
- RACE
- rapid amplification of cDNA ends
References
- Billig G.M., Pál B., Fidzinski P., Jentsch T.J. 2011. Ca2+-activated Cl− currents are dispensable for olfaction. Nat. Neurosci. 14:763–769 10.1038/nn.2821 [DOI] [PubMed] [Google Scholar]
- Boccaccio A., Menini A. 2007. Temporal development of cyclic nucleotide-gated and Ca2+-activated Cl− currents in isolated mouse olfactory sensory neurons. J. Neurophysiol. 98:153–160 10.1152/jn.00270.2007 [DOI] [PubMed] [Google Scholar]
- Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., Galietta L.J. 2008. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 322:590–594 10.1126/science.1163518 [DOI] [PubMed] [Google Scholar]
- Cho H., Yang Y.D., Lee J., Lee B., Kim T., Jang Y., Back S.K., Na H.S., Harfe B.D., Wang F., et al. 2012. The calcium-activated chloride channel anoctamin 1 acts as a heat sensor in nociceptive neurons. Nat. Neurosci. 15:1015–1021 10.1038/nn.3111 [DOI] [PubMed] [Google Scholar]
- Das S., Hahn Y., Walker D.A., Nagata S., Willingham M.C., Peehl D.M., Bera T.K., Lee B., Pastan I. 2008. Topology of NGEP, a prostate-specific cell:cell junction protein widely expressed in many cancers of different grade level. Cancer Res. 68:6306–6312 10.1158/0008-5472.CAN-08-0870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauner K., Lissmann J., Jeridi S., Frings S., Möhrlen F. 2012. Expression patterns of anoctamin 1 and anoctamin 2 chloride channels in the mammalian nose. Cell Tissue Res. 347:327–341 10.1007/s00441-012-1324-9 [DOI] [PubMed] [Google Scholar]
- Dibattista M., Amjad A., Maurya D.K., Sagheddu C., Montani G., Tirindelli R., Menini A. 2012. Calcium-activated chloride channels in the apical region of mouse vomeronasal sensory neurons. J. Gen. Physiol. 140:3–15 10.1085/jgp.201210780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran C., Hartzell H.C. 2011. Physiological roles and diseases of Tmem16/Anoctamin proteins: are they all chloride channels? Acta Pharmacol. Sin. 32:685–692 10.1038/aps.2011.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran C., Qu Z., Osunkoya A.O., Cui Y., Hartzell H.C. 2012. ANOs 3-7 in the anoctamin/Tmem16 Cl− channel family are intracellular proteins. Am. J. Physiol. Cell Physiol. 302:C482–C493 10.1152/ajpcell.00140.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallah G., Romer T., Detro-Dassen S., Braam U., Markwardt F., Schmalzing G. 2011. TMEM16A(a)/anoctamin-1 shares a homodimeric architecture with CLC chloride channels. Mol. Cell. Proteomics. 10:M110.004697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrera L., Caputo A., Ubby I., Bussani E., Zegarra-Moran O., Ravazzolo R., Pagani F., Galietta L.J. 2009. Regulation of TMEM16A chloride channel properties by alternative splicing. J. Biol. Chem. 284:33360–33368 10.1074/jbc.M109.046607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengl T., Kaneko H., Dauner K., Vocke K., Frings S., Möhrlen F. 2010. Molecular components of signal amplification in olfactory sensory cilia. Proc. Natl. Acad. Sci. USA. 107:6052–6057 10.1073/pnas.0909032107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp U.B. 2010. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat. Rev. Neurosci. 11:188–200 [DOI] [PubMed] [Google Scholar]
- Kim S., Ma L., Yu C.R. 2011. Requirement of calcium-activated chloride channels in the activation of mouse vomeronasal neurons. Nat Commun. 2:365 10.1038/ncomms1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene S.J. 2008. The electrochemical basis of odor transduction in vertebrate olfactory cilia. Chem. Senses. 33:839–859 10.1093/chemse/bjn048 [DOI] [PubMed] [Google Scholar]
- Kunzelmann K., Tian Y., Martins J.R., Faria D., Kongsuphol P., Ousingsawat J., Thevenod F., Roussa E., Rock J., Schreiber R. 2011. Anoctamins. Pflugers Arch. 462:195–208 10.1007/s00424-011-0975-9 [DOI] [PubMed] [Google Scholar]
- Kurahashi T., Yau K.W. 1993. Co-existence of cationic and chloride components in odorant-induced current of vertebrate olfactory receptor cells. Nature. 363:71–74 10.1038/363071a0 [DOI] [PubMed] [Google Scholar]
- Lowe G., Gold G.H. 1993. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature. 366:283–286 10.1038/366283a0 [DOI] [PubMed] [Google Scholar]
- Martins J.R., Faria D., Kongsuphol P., Reisch B., Schreiber R., Kunzelmann K. 2011. Anoctamin 6 is an essential component of the outwardly rectifying chloride channel. Proc. Natl. Acad. Sci. USA. 108:18168–18172 10.1073/pnas.1108094108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll K.E., Pipe R.A., Britton F.C. 2011. Increased complexity of Tmem16a/Anoctamin 1 transcript alternative splicing. BMC Mol. Biol. 12:35 10.1186/1471-2199-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton C., Thompson S., Epel D. 2004. Some precautions in using chelators to buffer metals in biological solutions. Cell Calcium. 35:427–431 10.1016/j.ceca.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Pifferi S., Pascarella G., Boccaccio A., Mazzatenta A., Gustincich S., Menini A., Zucchelli S. 2006. Bestrophin-2 is a candidate calcium-activated chloride channel involved in olfactory transduction. Proc. Natl. Acad. Sci. USA. 103:12929–12934 10.1073/pnas.0604505103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifferi S., Dibattista M., Menini A. 2009. TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflugers Arch. 458:1023–1038 10.1007/s00424-009-0684-9 [DOI] [PubMed] [Google Scholar]
- Pifferi S., Cenedese V., Menini A. 2012. Anoctamin 2/TMEM16B: a calcium-activated chloride channel in olfactory transduction. Exp. Physiol. 97:193–199 [DOI] [PubMed] [Google Scholar]
- Rasche S., Toetter B., Adler J., Tschapek A., Doerner J.F., Kurtenbach S., Hatt H., Meyer H., Warscheid B., Neuhaus E.M. 2010. Tmem16b is specifically expressed in the cilia of olfactory sensory neurons. Chem. Senses. 35:239–245 10.1093/chemse/bjq007 [DOI] [PubMed] [Google Scholar]
- Reisert J., Bauer P.J., Yau K.W., Frings S. 2003. The Ca-activated Cl channel and its control in rat olfactory receptor neurons. J. Gen. Physiol. 122:349–363 10.1085/jgp.200308888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert J., Lai J., Yau K.W., Bradley J. 2005. Mechanism of the excitatory Cl− response in mouse olfactory receptor neurons. Neuron. 45:553–561 10.1016/j.neuron.2005.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagheddu C., Boccaccio A., Dibattista M., Montani G., Tirindelli R., Menini A. 2010. Calcium concentration jumps reveal dynamic ion selectivity of calcium-activated chloride currents in mouse olfactory sensory neurons and TMEM16b-transfected HEK 293T cells. J. Physiol. 588:4189–4204 10.1113/jphysiol.2010.194407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber R., Uliyakina I., Kongsuphol P., Warth R., Mirza M., Martins J.R., Kunzelmann K. 2010. Expression and function of epithelial anoctamins. J. Biol. Chem. 285:7838–7845 10.1074/jbc.M109.065367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B.C., Cheng T., Jan Y.N., Jan L.Y. 2008. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 134:1019–1029 10.1016/j.cell.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G., Frohman M.A. 2006. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 1:2555–2562 10.1038/nprot.2006.480 [DOI] [PubMed] [Google Scholar]
- Sheridan J.T., Worthington E.N., Yu K., Gabriel S.E., Hartzell H.C., Tarran R. 2011. Characterization of the oligomeric structure of the Ca2+-activated Cl− channel Ano1/TMEM16A. J. Biol. Chem. 286:1381–1388 10.1074/jbc.M110.174847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A.B., Shum E.Y., Hirsh S., Cygnar K.D., Reisert J., Zhao H. 2009. ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc. Natl. Acad. Sci. USA. 106:11776–11781 10.1073/pnas.0903304106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr H., Heisig J.B., Benz P.M., Schöberl S., Milenkovic V.M., Strauss O., Aartsen W.M., Wijnholds J., Weber B.H., Schulz H.L. 2009. TMEM16B, a novel protein with calcium-dependent chloride channel activity, associates with a presynaptic protein complex in photoreceptor terminals. J. Neurosci. 29:6809–6818 10.1523/JNEUROSCI.5546-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Kongsuphol P., Hug M., Ousingsawat J., Witzgall R., Schreiber R., Kunzelmann K. 2011. Calmodulin-dependent activation of the epithelial calcium-dependent chloride channel TMEM16A. FASEB J. 25:1058–1068 10.1096/fj.10-166884 [DOI] [PubMed] [Google Scholar]
- Tian Y., Schreiber R., Kunzelmann K. 2012. Anoctamins are a family of Ca2+-activated Cl− channels. J. Cell Sci. 125:4991–4998 10.1242/jcs.109553 [DOI] [PubMed] [Google Scholar]
- Yang C., Delay R.J. 2010. Calcium-activated chloride current amplifies the response to urine in mouse vomeronasal sensory neurons. J. Gen. Physiol. 135:3–13 10.1085/jgp.200910265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.D., Cho H., Koo J.Y., Tak M.H., Cho Y., Shim W.S., Park S.P., Lee J., Lee B., Kim B.M., et al. 2008. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 455:1210–1215 10.1038/nature07313 [DOI] [PubMed] [Google Scholar]
- Yap K.L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. 2000. Calmodulin target database. J. Struct. Funct. Genomics. 1:8–14 10.1023/A:1011320027914 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.