Cytosolic pH is a cellular signal involved both in the glucose sensing that mediates proteasome storage granule formation and in a more general mechanism for signaling carbon source exhaustion.
Abstract
The 26S proteasome is the major protein degradation machinery of the cell and is regulated at many levels. One mode of regulation involves accumulation of proteasomes in proteasome storage granules (PSGs) upon glucose depletion. Using a systematic robotic screening approach in yeast, we identify trans-acting proteins that regulate the accumulation of proteasomes in PSGs. Our dataset was enriched for subunits of the vacuolar adenosine triphosphatase (V-ATPase) complex, a proton pump required for vacuole acidification. We show that the impaired ability of V-ATPase mutants to properly govern intracellular pH affects the kinetics of PSG formation. We further show that formation of other protein aggregates upon carbon depletion also is triggered in mutants with impaired activity of the plasma membrane proton pump and the V-ATPase complex. We thus identify cytosolic pH as a specific cellular signal involved both in the glucose sensing that mediates PSG formation and in a more general mechanism for signaling carbon source exhaustion.
Introduction
The 26S proteasome is a 2.5-MD protease that is composed of the 19S regulatory particle (RP) and the 20S core particle (CP), which possesses the protease activity. In eukaryotes, the major mechanism for targeting proteins to the proteasome is through the process of ubiquitination. Through this pathway the ubiquitin–proteasome system exerts its influence by selectively degrading misfolded, damaged, or regulatory proteins (Hershko and Ciechanover, 1998).
Proteolytic function must be tightly regulated in a cellular environment, and therefore, mechanisms exist to turn the proteasome off. This massive complex is highly abundant (∼1% of the yeast proteome; Russell et al., 1999) and therefore costly to synthesize and assemble. Therefore, degradation can be considered to be the least favored regulation in this case. The alternate option, that is widely used, is to assemble it into proteasome storage granules (PSGs) during cellular stress conditions such as glucose starvation or quiescence (Laporte et al., 2008). In these conditions, nonselective autophagy is activated in yeast cells, and it is thought that the PSGs represent a protective mechanism that shelters the proteasome from autophagic degradation. The utilization of PSGs enables cells to avoid the replenishment of large amounts of active proteasome subunits and the possible delay in reentry into the proliferative state. In fact, many other proteins that are localized diffusely in the cytoplasm of dividing yeast cells relocalize into cytoplasmic foci upon glucose starvation or entry into quiescence. The widespread reorganization of nuclear and cytoplasmic proteins into reversible assemblies suggests that it is a general mechanism for coping with starvation (Narayanaswamy et al., 2009).
Attempts to comprehend which nutrients or chemicals trigger the formation of cytoplasmic granules showed that depletion or addition of glucose is sufficient and that glucose has to be catabolized through the glycolytic pathway to exert this effect (Daignan-Fornier and Sagot, 2011; Laporte et al., 2011). Still, at the present time, it is not clear which metabolite resulting from glucose catabolism is required for glucose induced cytoplasmic granule mobilization.
In this paper, we describe a whole-genome systematic screening approach that allowed us to discover mutants that affect the shuttling of the proteasome into PSGs (Fig. 1 A). The functional distribution of the identified mutants reveals enrichment for subunits of the vacuolar ATPase (V-ATPase; Table 1). We show that the kinetics of PSG formation in the mutants upon carbon depletion is different from that seen in the wild type (wt), probably because of the impaired ability of the V-ATPase mutants to properly adjust their intracellular pH (pHi), in response to carbon starvation. Further characterization identified cytosolic pH as a novel regulator that mediates the formation of PSGs and other protein aggregates that form upon sugar depletion. We therefore propose that cytosolic pH acts as the cellular signal for glucose sensing and mediates the formation of PSGs in response to carbon depletion.
Figure 1.
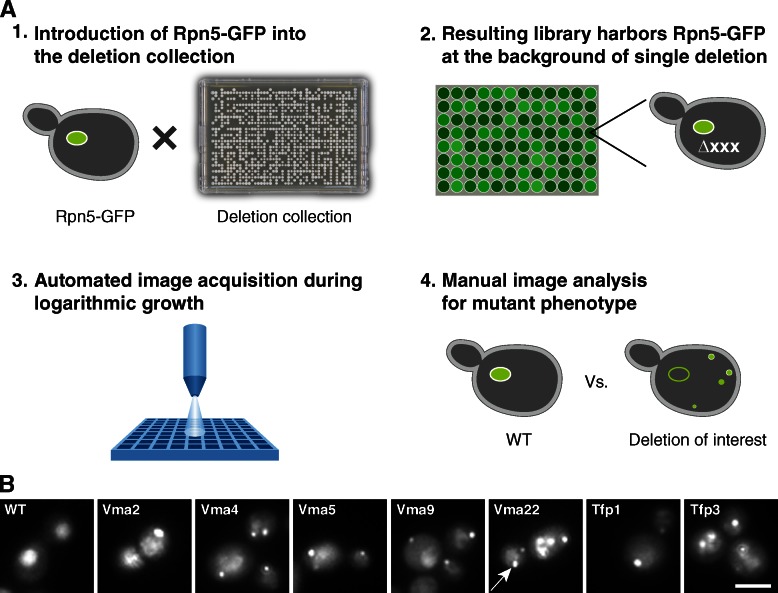
A systematic approach for the identification of trans-acting proteins regulating proteasome sequestering into PSGs. (A) To uncover novel regulators of proteasome localization into PSGs, we integrated Rpn5-GFP into every strain of the yeast deletion collection (1), resulting in a systematic library harboring the tagged proteasome on the background of single gene deletions (2). After high content screening (3), we manually categorized the localization of Rpn5-GFP for all strains tested (4) and recognized hits. (B) Deletion of V-ATPase subunits results in proteasome failure to leave PSGs during logarithmic growth. Representative images of data acquired during the systematic screening approach described in A. Although Rpn5-GFP accumulates in the nucleus in the control cells (wt), it is still found in PSGs in cells deleted for V-ATPase subunits (pointed to by a white arrow in vma22 mutant) despite the abundance of glucose. Bar, 5 µm.
Table 1.
List of genes identified through deletion mutants affecting proteasome sequestering into PSGs
| Description | Gene name | ORF name |
| Subunit d of the five-subunit V0 integral membrane domain of V-ATPase | VMA6 | YLR447C |
| Peripheral membrane protein that is required for V-ATPase function | VMA22 | YHR060W |
| V-ATPase V0 domain subunit c′, involved in proton transport activity; hydrophobic integral membrane protein | TFP3 | YPL234C |
| Subunit C of the eight-subunit V1 peripheral membrane domain of V-ATPase | VMA5 | YKL080W |
| Subunit B of the eight-subunit V1 peripheral membrane domain of the V-ATPase | VMA2 | YBR127C |
| Dubious ORF; ORF overlaps the verified gene VMA4/YOR332W: subunit E of the eight-subunit V1 peripheral membrane domain of the V-ATPase | YOR331C | YOR331C |
| Subunit A of the eight-subunit V1 peripheral membrane domain of the V-ATPase | TFP1 | YDL185W |
| Dubious ORF; overlaps verified ORF YCL005W-A\VMA9: V-ATPase subunit e of the V-ATPase V0 subcomplex | YCL007C | YCL007C |
| DNA damage-inducible V-SNARE binding protein contains a ubiquitin-associated domain, may act as a negative regulator of constitutive exocytosis, and may play a role in S-phase checkpoint control | DDI1 | YER143W |
| Component of the GTPase-activating Bfa1p–Bub2p complex involved in multiple cell cycle checkpoint pathways that control exit from mitosis | BFA1 | YJR053W |
| Protein that stimulates the activity of serine palmitoyltransferase (Lcb1p and Lcb2p) severalfold; involved in sphingolipid biosynthesis | TSC3 | YBR058C-A |
Results and discussion
A systematic approach for the identification of trans-acting proteins regulating proteasome sequestering into PSGs
The regulation of proper partitioning of the proteasome into PSGs is essential for maintaining the correct level of the proteasome in the cytosol and nucleus. Therefore, we set out to uncover additional factors that regulate this process. To visually screen for such factors, we first created a yeast strain (based on the yeast S288C background; Brachmann et al., 1998) expressing a fluorescently tagged RP proteasome subunit (Rpn5-GFP). Indeed, as expected, this fusion protein localized correctly to both the cytosol and the nucleus (Fig. 1 B, wt). Using synthetic genetic array (SGA) methodology (Tong and Boone, 2006; Cohen and Schuldiner, 2011), we crossed this strain against the yeast deletion library (Giaever et al., 2002), which consists of ∼5,000 strains, each harboring a deletion in one nonessential gene. This procedure allowed us to create a library of nearly 5,000 haploid cells, each expressing the Rpn5-GFP fusion protein on the background of a mutation in a single gene (Fig. 1 A). The freshly grown colonies were copied from agar plates into liquid cultures in synthetic medium containing glucose (synthetic defined [SD]) for overnight growth. After back dilution into fresh medium for 3.5 h, cells were transferred into glass-bottom 384-well microscope plates. We then visualized the cellular localization of Rpn5-GFP in each of these mutated strains during logarithmic growth in SD using a high throughput microscopy setup (for details see Materials and methods; Fig. 1 A; Cohen and Schuldiner, 2011). The pictures were then manually visualized to uncover strains in which the normal localization was disturbed.
In the majority of the cases, cells responded as expected to the increase in glucose levels (caused during the back dilution into fresh medium) and relocalized their proteasomes from PSGs back into the cytosol and the nucleus. However, the time of recovery in 11 mutants was significantly delayed, enabling us to identify genes that abrogate the ability of cells to sense the changes in glucose levels and respond properly by releasing proteasomes from PSGs (Table 1). Interestingly, the functional distribution of the identified mutants reveals that our data were highly enriched for genes that govern pHi. These genes include seven subunits of the V-ATPase complex and one subunit required for its assembly (Table 1).
Impaired ability of V-ATPase mutants to control pHi significantly affects the redistribution of the proteasome from PSGs
V-ATPase, a proton pump required for the acidification of vacuoles, plays a central role in vacuolar and cytosolic pH homeostasis in all eukaryotic cells. V-ATPases are activated by glucose and function together with Pma1 to pump protons out of the cytosol to help regulate cytosolic pH. Reduction in cytosolic pH as a result of glucose deprivation is necessary for the inactivation of V-ATPase by their reversible disassembly (Dechant et al., 2010). In V-ATPase mutants, even at optimal extracellular pH, cytosolic pH was previously shown to be much lower, and cells failed to respond properly when glucose was added (Dechant et al., 2010; Young et al., 2010). Given that it is known that PSGs are regulated by glucose availability, we were intrigued by the idea that V-ATPase function could be the missing link between glucose levels and signaling of PSG formation.
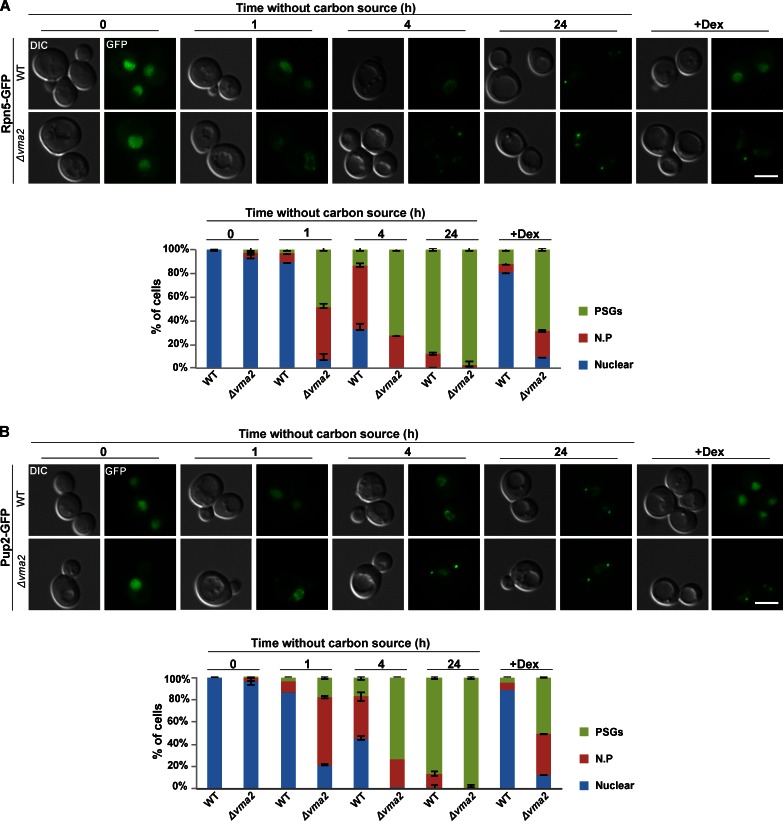
Because the screen was performed at a single time point and measured mostly the dissociation of PSGs, we decided to look at the effect of V-ATPase mutants on the kinetics of PSGs formation. We focused on a representative mutant, Δvma2 and followed the localization of Rpn5-GFP after depletion of glucose relative to control (wt) cells. At t = 0, Rpn5-GFP localized to the nucleus both in wt and Δvma2 mutants (Fig. 2 A, t = 0). Resuspension of wt cells in glucose-free synthetic media (synthetic complete [SC]) led to the expected relocalization of Rpn5-GFP to PSGs in 87% of the cells within 24 h. Strikingly, in Δvma2 cells, the kinetics of PSGs formation was significantly faster with 44% of the cells displaying Rpn5-GFP localization to the nuclear periphery after 1 h and 72% of the cells displaying PSGs after <4 h. Furthermore, as expected from the phenotypes observed in our screen, the kinetics of Rpn5-GFP disassembly and relocalization back into the nucleus in response to glucose replenishment was significantly slower in Δvma2 mutants (Fig. 2 A, +Dex; the full kinetics of disassembly and relocalization is shown in Fig. S1 C). Similar kinetics were obtained with cells mutated in another V-ATPase subunit (Δvma5; Fig. S1 D), with the CP protein Pup2 fused to GFP (Fig. 2 B), and with cells treated with the V-ATPase–specific inhibitor concanamycin A (Fig. S1 E; Krogan et al., 2004; Dechant et al., 2010). We also verified that Pup2 (tagged with mCherry) and Rpn5 (tagged with GFP) colocalize in the PSGs formed in Δvma2 to ensure that this mutation did not create a change in PSG physiology (Fig. S1 F). Collectively, these results suggest that the impaired ability to control pHi in V-ATPase mutants increased the sensitivity of cells to lower levels of the carbon source and induced the simultaneous relocalization of both core and regulatory proteasomal subunits into PSGs (Fig. 2).
Figure 2.
The kinetics of PSG formation is affected in cells deleted in V-ATPase subunit VMA2 (Δvma2). (A and B) Logarithmic wt and Δvma2 cells expressing the lid subunit Rpn5-GFP (A) and the core subunit Pup2-GFP (B) were washed and resuspended in glucose-free medium (t = 0) for 24 h and then transferred back into a glucose-rich medium (+dextrose [+DEX]). Cells were visualized by differential interference contrast (DIC) and GFP at the indicated time points. The localization of Rpn5-GFP (A) and Pup2-GFP (B) was scored as nuclear, nuclear periphery (N.P), or PSGs. Unless otherwise stated, the representative images in all figures show the dominant localization pattern for each of the indicated time points. For each time point, a minimum of 200 cells was counted (n > 200); error bars show the standard deviation between two independent experiments. Bars, 5 µm. Images represent high resolution (63×) images of a single plane chosen from z series images extending above and below the entire cell.
Rapid drop of internal pH is sufficient for formation of PSGs at constant glucose levels
Our screen identified that mutated subunits of V-ATPase, key regulators of pHi homeostasis, affect the kinetics of PSGs formation. Previous studies have shown that glucose-starved yeast exhibit a rapid drop in pHi (Martínez-Muñoz and Kane, 2008; Dechant et al., 2010; Young et al., 2010). Because both glucose-starved yeast and mutants that control pHi (Fig. 2) affect the formation of PSGs, we hypothesized that the internal drop in cytosolic pH might be the physiological signal for PSGs formation. To test this hypothesis, we assayed whether changes in pHi are sufficient to mediate the formation of PSGs, even when glucose levels are kept high (2%).
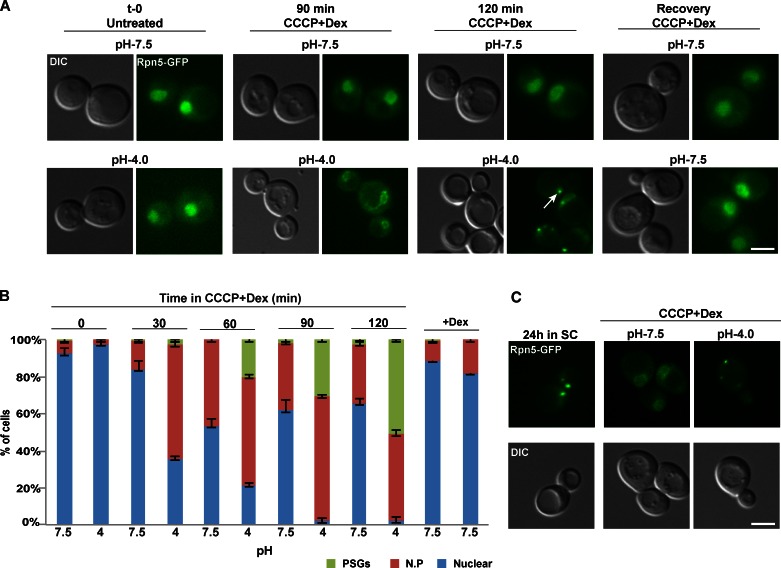
First, we analyzed the localization of Rpn5-GFP at timely intervals, after cells were shifted to dextrose-containing buffers, buffered to normal and low pH (pH 7.5 and 4.0, respectively). The buffers also contained the proton ionophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which affects the plasma membrane proton gradient and therefore prevents cells from controlling their internal pH (Orij et al., 2009). The results clearly show that after 120 min, under normal pH (7.5), the GFP signal was mainly localized to the nucleus (∼63% of the cells) and, in ∼30% of the cells, to the nuclear periphery (Fig. 3, A [top] and B). In contrast, at a lower pH, the proteasome formed PSGs in 55% of the cells (Fig. 3 A, bottom, white arrow) and relocalized to the nuclear periphery in the others. When the cells were shifted back to physiological pH and allowed to recover in the presence of dextrose + CCCP, the GFP signal in most cells relocated back into the nucleus within 30 min (Fig. 3 A, right). Furthermore, the PSGs formed after 24 h in minimal medium (Fig. 3 C, left) relocalized back into nucleus (in 95% of the cells) when cells were resuspended in a medium adjusted to pH 7.5, supplemented with CCCP and glucose (Fig. 3 C, middle). In contrast, relocalization was impaired when cells were resuspended in a similar medium that was buffered to a low pH (4.0; Fig. 3 C, right).
Figure 3.
Failure to control internal pH affects PSG formation. (A) Logarithmically growing culture (untreated), expressing the lid subunit Rpn5-GFP, was washed and resuspended in glucose (dextrose [Dex])-containing buffers, buffered to physiological pHi, pH 7.5 (control), and to pH 4.0, supplemented with CCCP. Both samples were then washed and allowed to recover for 30 min in pH 7.5 +CCCP (Recovery). Cells were visualized by DIC and GFP at the indicated time points. The localization of Rpn5-GFP was scored as described in Fig. 2. PSGs are pointed out by a white arrow. (B) Representative images in all figures show the dominant localization pattern for each of the indicated time points. Error bars show the standard deviation between two independent experiments. (C) Similar experimental setup as was described in A, this time, PSG relocalization in cells grown in a low glucose media for 24 h (24 h in SC) was examined after the resuspension in the indicated medium (CCCP + dextrose). N.P, nuclear periphery. Bars, 5 µm.
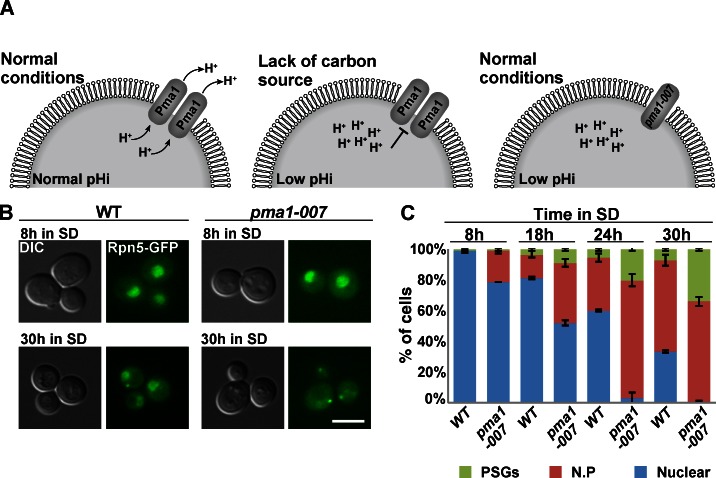
Next, we decided to test the effect of physiological changes in pHi by analyzing the localization of Rpn5-GFP on cells expressing reduced levels of Pma1. It has been previously shown that in wt cells, in response to glucose starvation, Pma1 is repressed, resulting in a rapid drop in pHi (Fig. 4 A, left and middle; Young et al., 2010). pma1-007 is a hypomorphic allele of PMA1 that was previously shown to reduce the activity of the Pma1 protein by 50% (Porat et al., 2005; Young et al., 2010). This results in a decreased capacity to pump protons out of the cell and a concomitant reduction in cytosolic pHi, similar to that which occurs when CCCP is added externally (Fig. 4 A, right). The results clearly show that although both strains were grown in glucose-rich medium for 30 h, the rate of PSG formation was significantly higher in pma1-007 mutants, with 31% of the cells forming PSGs in the mutants, (n = 210) versus only 8% in the control (n = 204; Fig. 4, B and C). Under these conditions, the proteasome CP subunit Pre6-mCherry was similarly affected and clearly colocalized with the Rpn5-GFP (Fig. S2 A). To further support these results, we transferred the same strains to a medium that lacks any carbon source. The lack of carbon should result in a faster reduction in pHi in control cells, and to a greater extent in pma1-007 mutants, when compared with SD. As expected, under such conditions, the kinetics of PSGs formation were dramatically faster and were detected after 4 h, with 62% of the cells exhibiting PSGs in the mutants, (n = 200) versus 18% in the control (n = 200; Fig. S2 B). Furthermore, we have repeated the experiment described in Fig. 4 A, this time in the presence of 100 µM ebselen, both in wt cells and pma1-007 mutants. A previous study has shown that the drug ebselen inhibits Pma1 in vitro and causes an immediate drop in pHi of both wt and pma1-007 cells to ∼6.3. Whereas wt cells stabilized at pHi ∼6.4, pHi of pma1-007 cells continued to decrease (Chan et al., 2007). As expected from the immediate drop in internal pHi, we have observed that the addition of ebselen resulted in faster kinetics of nuclear periphery and PSG formation in pma1-007 cells, indicating that the mutant is more sensitive, likely as a result of reduced gene dosage (Fig. S2 C).
Figure 4.
Impaired capacity to pump protons out of cells affects the kinetics of PSG formation. (A) Schematic illustration of PMA1 regulation. (B) pma1-007 and wt cells were grown in SD for 30 h. Cells were visualized by DIC and GFP at the indicated time points. The representative images shown at t = 30 h show the PSG localization pattern that was significantly enriched in pma1-007 mutants, comparing with the nuclear localization pattern that still appeared in wt cells. Bar, 5 µm. (C) Cells were visualized by DIC and GFP at the indicated time points. The localization of Rpn5-GFP was scored as described in Fig. 2. Error bars show the standard deviation between two independent experiments. N.P, nuclear periphery.
In total, these results strongly suggest that the rapid drop in pHi and the acidification of the cytosol (either by external treatment with CCCP or genetically by reducing Pma1 levels) are sufficient to trigger PSG formation. Overall, our findings demonstrate for the first time that the change in cytosolic pH in response to glucose levels serves as a messenger and a major cellular signal to mediate the activation of PSGs reorganization. pH homeostasis is critical for the survival of yeast cells, as it is for all eukaryotic cells. Indeed, as expected, previous studies have shown a link between cytosolic pH and various cellular activities. For example, Young et al., 2010 have demonstrated a link between nutrient availability, pH biosensing, and membrane biogenesis. A very recent study identified that an early age increase in vacuolar pH limits mitochondrial function and life span in yeast (Hughes and Gottschling, 2012). Other work has shown that the increase in cytosolic pH as a result of addition of glucose in starved cells promotes V-ATPase assembly and that this activation positively regulates the cAMP-dependent PKA pathway in yeast (Dechant et al., 2010). Similarly, our study suggests that the reformation of PSGs is also mediated by V-ATPase reassembly and that the V-ATPase complex functions as an intracellular glucose sentinel. Failure of the VMA mutants to sense the changes in glucose levels misregulates cytosolic pH levels and leads to reduced efficiency of PSGs dynamics.
Drop in pHi affects additional starvation markers
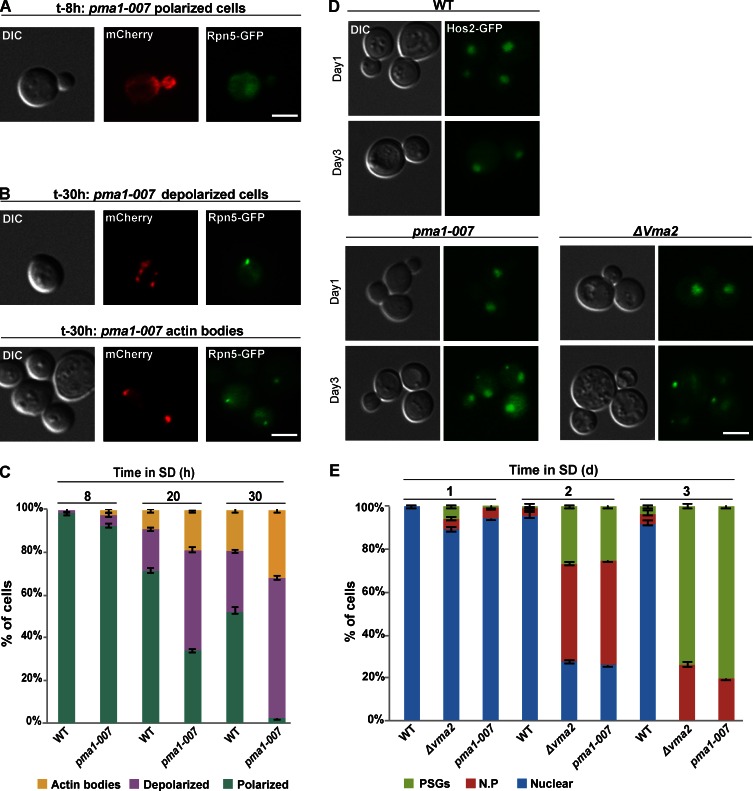
Previous studies have shown that many diffusely localized cytosolic proteins relocalize into cytoplasmic foci, such as PSGs or actin bodies upon carbon exhaustion (Sagot et al., 2006; Laporte et al., 2008). Because we observed that drop in pHi as a result of carbon source exhaustion triggers the formation of PSGs, we wanted to test whether pHi mediates the signal for a larger spectrum of cellular reprogramming that takes place upon glucose deprivation. To test this idea, we asked whether pma1-007 mutants also affected changes in actin structure. Indeed, after 8 h in SD, when the proteasomes are still localized into the nucleus, actin cables and patches were polarized in structures similar to those previously described in proliferating cells in 92% of the cells (Fig. 5, A and C; Pruyne et al., 2004). In contrast, after 30 h, when PSGs appeared in 40% of the cells as a result of drop in pHi, the actin cytoskeleton reorganized into structures typical of cells exposed to low carbon source (Sagot et al., 2006), including 65% showing depolarization of actin patches and 31% that formed actin bodies (Fig. 5, B and C). The kinetics of actin cytoskeleton reorganization is shown in Fig. 5 C. Similarly, using a stationary-phase granule (SPG) marker, Hos2-GFP (Liu et al., 2012), we found increased formation of SPGs both in pma1-007 and in Δvma2 mutants (Fig. 5, D and E). Similar results were obtained when we analyzed the localization of Hos2-GFP 120 min after cells were shifted to CCCP-containing buffers (similar to the experiment described in Fig. 3 A). The results clearly show that after 120 min, under normal pH (7.5), the GFP signal was mainly localized to nucleus (∼75% of the cells). In contrast, at a lower pH, Hos2-GFP localized to the nuclear periphery or formed SPGs in 76% of the cells. These results suggest that, similar to PSGs, the effect on Hos2-GFP in pma1-007 and Δvma2 mutants is the result of low cytosolic pH (Fig. S2 D).
Figure 5.
Impaired capacity to control pHi in pma1-007 mutants results in actin cytoskeleton reorganization. pma1-007 mutants were grown in SD for 30 h. At the indicated time points, samples were taken, stained with Alexa Fluor–phalloidin, and visualized by DIC, GFP (Rpn5-GFP), and mCherry (actin). (A) Examples of polarized actin when most cells (95%) show proteasome nuclear localization (time point 8 h, n > 100). (B) Examples of actin reorganization (time point 30 h) in cells with PSGs (40% of the cells, n > 100). (top) Examples for depolarized cells (65% of the PSG-containing cells). (bottom) Examples for cells with actin bodies (31% of the PSG-containing cells). (C) Quantitation of the localization patterns for the experiment described in A and B. The localization of actin at the indicated time points was scored as actin bodies, depolarized actin, or polarized actin. Impaired capacity to control pHi results in SPG formation. (D) Logarithmic wt, Δvma2, and pma1-007 cells expressing the SPG marker Hos2-GFP were washed and resuspended in glucose-containing medium (t = 0) for 3 d. Cells were visualized by DIC and GFP. The images represent the dominant pattern at the first and third day. (E) The Hos2-GFP localization at the indicated time points was scored as nuclear, nuclear periphery (N.P), or SPGs. Error bars show the standard deviation between two independent experiments. Bars, 5 µm.
Collectively, the relocalization of actin cytoskeleton and Hos2 suggests that cytosolic acidification represents a general signal that affects not only PSG formation but more broadly may be the signal for cytosolic protein relocalization in response to carbon source exhaustion. Cytosolic pH is regulated not only by glucose but also by other fermentable carbon sources (Dechant et al., 2010). This suggests that the yeast cell may use cytosolic pH as a downstream reporter on the availability of various carbon sources and enables cells to avoid monitoring the abundance of a specific carbon source or metabolite.
Materials and methods
Yeast strains and growth conditions
All the strains used in this study are isogenic to BY4741, BY4742, or BY4743 (Brachmann et al., 1998). The relevant genotypes are presented in Table S1. Gene deletions, GFP, and mCherry fusions were performed using one-step PCR-mediated homologous recombination (Longtine et al., 1998; Goldstein and McCusker, 1999). For all deletions, the selection markers replaced the coding region of the targeted genes. GFP and mCherry were fused at the 3′ of the coding region of the targeted genes, by replacing their stop codons. pma1-007 strain was a gift from C. Lowen’s laboratory (University of British Columbia, Vancouver, British Columbia, Canada). In this strain, insertion of the KanMX selection marker upstream of the coding region of PMA1 resulted in reduced activity of the Pma1 protein by 50%. We have verified that the proteasome is expressed at the same level in wt and in the mutants used in this study (Δvma2, pma1-007, and Δvma5; Fig. S1 A). We also show that the expression of the fusion proteins Rpn5-GFP, Pup2-GFP, and Pre6-mCherry have no effect on proteasome activity (Fig. S1 B).
Yeast cells were grown in synthetic medium (SD; 0.17% yeast nitrogen base, 0.5% (NH4)2SO4, 2% glucose, and amino acids). Glucose starvation was performed in SC (SD without glucose). Unless stated otherwise, the pH of the synthetic media was buffered to pH 4.6. Yeast cells were grown in liquid medium at 30°C in glass tubes. For logarithmic culture, yeast cells were grown for 16–18 h and then back diluted 10× with fresh media and allowed to grow for 2 h. For glucose starvation experiments, strains were grown in medium containing 2% glucose to logarithmic phase, collected by centrifugation, washed once, and resuspended in the same medium lacking glucose.
CCCP treatment
CCCP treatment was performed as previously described (Orij et al., 2009). In brief, cells were reinoculated at an OD600nm of 0.6–1 to Hepes buffer (25 mM Hepes, pH 7.4, 200 mM KCl, 1 mM CaCl2, and 2% dextrose, pH 4.0 and 7.5) in the presence of 100 µM CCCP (Sigma-Aldrich).
Actin staining
Actin staining with phalloidin was performed as previously described (Sagot et al., 2006). In brief, cells were fixed with formaldehyde (3.7% final), washed, and stained overnight with Alexa Fluor 568–phalloidin (Invitrogen). Cells were then washed twice, resuspended in a mounting solution containing 70% glycerol and 5 mg/liter paraphenylenediamine, and imaged at room temperature.
Microscopy
Cells were observed in a fully automated inverted microscope (Axio Observer.Z1; Carl Zeiss) equipped with an automated stage (MS-2000; Applied Scientific Instrumentation), a 300-W xenon light source (Lambda DG-4 LS; Sutter Instrument), a 63× oil, 1.4 NA Plan Apochromat objective lens, and a six-position filter cube turret with a GFP filter (excitation, bp 470/40 nm; emission, bp 525/50 nm; beamsplitter, FT 495 nm) and an HcRed filter (excitation, bp 592/24 nm; emission, bp 675/100 nm; beamsplitter, FT 615 nm) obtained from Chroma Technology Corp. Images were acquired using a camera (CoolSnap HQ2; Roper Scientific). The microscope, camera, and shutters (Uniblitz) were controlled by AxioVision Rel. 4.8.2 (Carl Zeiss). Images are a single plane of z stacks performed using a 0.5-µm step.
Insertion of Rpn5-GFP to the deletion library
The SGA technique was performed as described in the following paragraph, to allow efficient introduction of Rpn5-GFP into the yeast deletion collection (for more details see Tong and Boone, 2006). To manipulate the collection in high-density format (384-well plates), we used a bench top colony arrayer (RoToR; Singer Instruments). Specifically, a MATα haploid strain harboring Rpn5-GFP::URA3 integrated into the endogenous locus (Ben-Aroya et al., 2010) and the required markers for SGA (can1Δ::STE2pr-his5 and lyp1Δ) were mated on rich media plates against the deletion collection. Diploid cells were selected on plates lacking uracil and provided with G418. Sporulation was then induced by transferring cells to nitrogen starvation plates for 5 d. Haploid cells containing all desired mutations were selected by transferring cells to plates containing all selection markers alongside the toxic amino acid derivatives Canavanine and Thialysine (Sigma-Aldrich) to select against remaining diploids and, lacking histidine, to select for MATa haploid spores. SGA procedure was validated by inspecting representative strains for the presence of the GFP-tagged Rpn5 and deletion of the chosen genes.
High-throughput fluorescence microscopy
Microscopic screening was performed using an automated microscopy setup as previously described (Herzig et al., 2012). Cells were moved from agar plates into liquid 384-well polystyrene growth plates using the RoToR arrayer. Liquid cultures were grown overnight in SD medium in a shaking incubator (LiCONiC Instruments) at 30°C. A liquid handler (JANUS; PerkinElmer), which is connected to the incubator, was used to back dilute the strains to ∼0.25 OD into plates containing the same medium. Plates were then transferred back to the incubator and were allowed to grow for 3.5 h at 30°C to reach logarithmic growth phase as was validated in preliminary calibration. The liquid handler was then used to transfer strains into glass-bottom 384-well microscope plates (MatriCal Bioscience) coated with Concanavalin A (Sigma-Aldrich) to allow cell adhesion. Wells were washed twice in medium to remove floating cells and reach cell monolayer. Plates were then transferred into an automated inverted fluorescent microscopic scan^R system (Olympus) using a swap robotic arm (Hamilton Robotics). The scan^R system is designed to allow autofocus and imaging of plates in 384-well format using a 60× air lens and is equipped with a cooled charge-coupled device camera (ORCA-ER; Hamamatsu Photonics). Images were acquired at GFP (excitation at 490/20 nm and emission at 535/50 nm).
Online supplemental material
Fig. S1 shows controls for the experiments described in Fig. 2. Fig. S2 shows controls for the experiments described in Fig. 4 (A–C) and Fig. 5 (D). Table S1 shows the relevant genotype of the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211146/DC1.
Supplementary Material
Acknowledgments
We thank Martin Kupiec and the Ben-Aroya laboratory members for helpful comments on an earlier version of the manuscript.
The work was supported in part by Human Frontier Science Program career development award grant (career development award 00005/2010) and Israeli Cancer Research Fund career development grant (research career development award 2011-713) to S. Ben-Aroya. M. Schuldiner was funded by a European Research Council grant (starting grant 26039).
Footnotes
Abbreviations used in this paper:
- CCCP
- carbonyl cyanide 3-chlorophenylhydrazone
- CP
- core particle
- DIC
- differential interference contrast
- pHi
- intracellular pH
- PSG
- proteasome storage granule
- RP
- regulatory particle
- SC
- synthetic complete
- SD
- synthetic defined
- SGA
- synthetic genetic array
- SPG
- stationary-phase granule
- V-ATPase
- vacuolar ATPase
- wt
- wild type
References
- Ben-Aroya S., Agmon N., Yuen K., Kwok T., McManus K., Kupiec M., Hieter P. 2010. Proteasome nuclear activity affects chromosome stability by controlling the turnover of Mms22, a protein important for DNA repair. PLoS Genet. 6:e1000852 10.1371/journal.pgen.1000852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132 [DOI] [PubMed] [Google Scholar]
- Chan G., Hardej D., Santoro M., Lau-Cam C., Billack B. 2007. Evaluation of the antimicrobial activity of ebselen: role of the yeast plasma membrane H+-ATPase. J. Biochem. Mol. Toxicol. 21:252–264 10.1002/jbt.20189 [DOI] [PubMed] [Google Scholar]
- Cohen Y., Schuldiner M. 2011. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 781:127–159 10.1007/978-1-61779-276-2_8 [DOI] [PubMed] [Google Scholar]
- Daignan-Fornier B., Sagot I. 2011. Proliferation/Quiescence: When to start? Where to stop? What to stock? Cell Div. 6:20 10.1186/1747-1028-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R., Binda M., Lee S.S., Pelet S., Winderickx J., Peter M. 2010. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29:2515–2526 10.1038/emboj.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Herzig Y., Sharpe H.J., Elbaz Y., Munro S., Schuldiner M. 2012. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 10:e1001329 10.1371/journal.pbio.1001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L., Gottschling D.E. 2012. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 492:261–265 10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Lam M.H., Fillingham J., Keogh M.C., Gebbia M., Li J., Datta N., Cagney G., Buratowski S., Emili A., Greenblatt J.F. 2004. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell. 16:1027–1034 10.1016/j.molcel.2004.11.033 [DOI] [PubMed] [Google Scholar]
- Laporte D., Salin B., Daignan-Fornier B., Sagot I. 2008. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181:737–745 10.1083/jcb.200711154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Lebaudy A., Sahin A., Pinson B., Ceschin J., Daignan-Fornier B., Sagot I. 2011. Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 192:949–957 10.1083/jcb.201009028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I.C., Chiu S.W., Lee H.Y., Leu J.Y. 2012. The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol. Biol. Cell. 23:1231–1242 10.1091/mbc.E11-09-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Martínez-Muñoz G.A., Kane P. 2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283:20309–20319 10.1074/jbc.M710470200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G.M., O’Connell J.D., Mirrielees J., Ellington A.D., Marcotte E.M. 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA. 106:10147–10152 10.1073/pnas.0812771106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orij R., Postmus J., Ter Beek A., Brul S., Smits G.J. 2009. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology. 155:268–278 10.1099/mic.0.022038-0 [DOI] [PubMed] [Google Scholar]
- Porat Z., Wender N., Erez O., Kahana C. 2005. Mechanism of polyamine tolerance in yeast: novel regulators and insights. Cell. Mol. Life Sci. 62:3106–3116 10.1007/s00018-005-5341-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559–591 10.1146/annurev.cellbio.20.010403.103108 [DOI] [PubMed] [Google Scholar]
- Russell S.J., Steger K.A., Johnston S.A. 1999. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 274:21943–21952 10.1074/jbc.274.31.21943 [DOI] [PubMed] [Google Scholar]
- Sagot I., Pinson B., Salin B., Daignan-Fornier B. 2006. Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol. Biol. Cell. 17:4645–4655 10.1091/mbc.E06-04-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Boone C. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313:171–192 [DOI] [PubMed] [Google Scholar]
- Young B.P., Shin J.J., Orij R., Chao J.T., Li S.C., Guan X.L., Khong A., Jan E., Wenk M.R., Prinz W.A., et al. 2010. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 329:1085–1088 10.1126/science.1191026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.