In mammalian cells, the phospho-dependent interaction between BRCA1 and CtIP is not required for homology-directed DNA repair or tumor suppression.
Abstract
The CtIP protein facilitates homology-directed repair (HDR) of double-strand DNA breaks (DSBs) by initiating DNA resection, a process in which DSB ends are converted into 3′-ssDNA overhangs. The BRCA1 tumor suppressor, which interacts with CtIP in a phospho-dependent manner, has also been implicated in DSB repair through the HDR pathway. It was recently reported that the BRCA1–CtIP interaction is essential for HDR in chicken DT40 cells. To examine the role of this interaction in mammalian cells, we generated cells and mice that express Ctip polypeptides (Ctip-S326A) that fail to bind BRCA1. Surprisingly, isogenic lines of Ctip-S326A mutant and wild-type cells displayed comparable levels of HDR function and chromosomal stability. Although Ctip-S326A mutant cells were modestly sensitive to topoisomerase inhibitors, mice expressing Ctip-S326A polypeptides developed normally and did not exhibit a predisposition to cancer. Thus, in mammals, the phospho-dependent BRCA1–CtIP interaction is not essential for HDR-mediated DSB repair or for tumor suppression.
Introduction
At least three distinct pathways for repair of DNA double-strand breaks (DSBs) have been identified in eukaryotic cells: homology-directed repair (HDR), Ku-dependent nonhomologous end joining (NHEJ), and Ku-independent microhomology-mediated end joining (MMEJ; Symington and Gautier, 2011). In vivo, the pathway used for repair of a given DSB is governed in part by DNA resection. This nucleolytic process converts DSB ends into 3′-ssDNA overhangs that inhibit NHEJ repair, but act as essential intermediates for both HDR and MMEJ (Symington and Gautier, 2011). In addition, the 3′-ssDNA tails generated by resection are bound initially by RPA protein complexes to form ssDNA–RPA nucleoprotein filaments that trigger ATR-dependent checkpoint signaling and subsequently by Rad51 polypeptides to form the ssDNA–Rad51 filaments that mediate HDR.
As shown in yeast, DNA end resection involves at least two mechanistically distinct stages (Mimitou and Symington, 2008; Zhu et al., 2008; Nicolette et al., 2010; Niu et al., 2010; Symington and Gautier, 2011). During an initiation stage, the yeast MRX (Mre11–Rad50–Xrs1) complex, together with the Sae2 protein, mediates a limited degree of resection to yield short ssDNA tails of roughly 100–400 nucleotides. In a subsequent extension stage, ssDNA tails greater than a kilobase in length can be generated by the Exo1 exonuclease or through the coordinated action of the DNA2 endonuclease and a RecQ-family helicase. As the human orthologue of yeast Sae2, the CtIP protein collaborates with MRN (Mre11–Rad50–Nbs1) to promote DNA resection, ATR signaling, and HDR repair in mammalian cells (Sartori et al., 2007; Bennardo et al., 2008; Chen et al., 2008). Indeed, CtIP/Sae2 and their orthologues have now been implicated in DNA resection across a vast phylogenetic spectrum that encompasses fungi, plants, insects, and vertebrates (Limbo et al., 2007; Penkner et al., 2007; Uanschou et al., 2007; You et al., 2009; You and Bailis, 2010; Peterson et al., 2011). Recent studies show that CtIP/Sae2-mediated resection is also required to expose as ssDNA the microhomologies necessary for MMEJ repair of DSBs (Lee and Lee, 2007; Bennardo et al., 2008). In addition, CtIP can facilitate the conversion of chromosomal DSBs into aberrant chromosome translocations in mouse embryonic stem (ES) cells, suggesting a potential pathological role for this protein (Zhang and Jasin, 2011). In any event, as a key effector for the initiation step of DNA resection, CtIP generates essential intermediates for checkpoint signaling (ssDNA–RPA filament), HDR (ssDNA–Rad51 filament), and MMEJ (ssDNA). Apart from its well-defined role in DNA resection, CtIP has also been implicated in other cellular processes, including transcriptional regulation and cell cycle progression (Chinnadurai, 2006).
In early studies, CtIP was identified as a major in vivo partner of the BRCA1 tumor suppressor (Wong et al., 1998; Yu et al., 1998; Yu and Baer, 2000). Although germline mutations of the BRCA1 gene are a major cause of the familial breast and ovarian cancer syndrome, the mechanisms by which BRCA1 suppresses tumor formation are still unclear (Huen et al., 2010; Moynahan and Jasin, 2010; Li and Greenberg, 2012; Roy et al., 2012). BRCA1 has been implicated in multiple aspects of the DNA damage response and it plays an essential, but undefined, role in the HDR pathway of DSB repair. At its C terminus, BRCA1 harbors two tandem BRCT repeats that form a single phospho-recognition surface. Of note, the BRCT surface of BRCA1 can bind the phosphorylated isoforms of several important DNA repair proteins, including Abraxas/CCDC98, BACH1/FancJ/BRIP1, and CtIP. Because BRCA1 interacts with each of these BRCT phospho-ligands in a mutually exclusive manner, it has the potential to form at least three distinct protein complexes (BRCA1 complexes A, B, and C, respectively) that appear to influence different aspects of the DNA damage response (Yu and Chen, 2004; Greenberg et al., 2006; Kim et al., 2007; Liu et al., 2007; Wang et al., 2007).
Because the BRCA1 lesions associated with familial breast cancer are usually frameshift or nonsense mutations, most tumorigenic BRCA1 alleles encode truncated polypeptides that have lost one or both BRCT motifs (Huen et al., 2010; Moynahan and Jasin, 2010; Li and Greenberg, 2012; Roy et al., 2012). Moreover, in some breast cancer families, tumor susceptibility can be ascribed to single amino acid substitutions (e.g., the S1655F mutation) that disrupt the interaction between the BRCT domain and its cognate phospho-ligands (Botuyan et al., 2004; Clapperton et al., 2004; Shiozaki et al., 2004; Williams et al., 2004; Varma et al., 2005). Indeed, using a mouse model of hereditary breast cancer, we recently showed that BRCT phospho-recognition is essential for both the HDR and tumor suppression activities of BRCA1 (Shakya et al., 2011). Thus, these two critical functions of BRCA1 are dependent on its ability to interact with one or more of its BRCT phospho-ligands (Shakya et al., 2011).
Of the known BRCA1 phospho-ligands, CtIP is especially intriguing given its central role in DNA resection and DSB repair (Sartori et al., 2007; You and Bailis, 2010). BRCA1 specifically binds human CtIP isoforms that are phosphorylated at serine residue S327, primarily during the G2 phase of the cell cycle (Yu and Chen, 2004). At present, however, the function of the BRCA1–CtIP interaction is poorly understood. In particular, it remains unclear whether this interaction is required for the tumor suppression activity of BRCA1 and/or the DNA resection activity of CtIP. Although the former possibility has not been tested experimentally, the latter has been addressed in studies of chicken DT40 cells that express nonphosphorylatable forms of CtIP. For example, Yun and Hiom (2009) reported that CtIP-null chicken DT40 cells reconstituted with an exogenous expression vector encoding human S327A mutant CtIP (CtIP−/−/− + hCtIP-S327A) are defective for HDR, but not NHEJ or MMEJ. On this basis, they concluded that the BRCA1–CtIP interaction is required for CtIP-mediated resection and, in turn, for DSB repair through pathways, such as HDR, that entail extensive DNA resection. In contrast, Nakamura et al. (2010) observed normal levels of HDR in DT40 cells (CtIPS332A/−/−) that express endogenous CtIP bearing the corresponding mutation (S332A) of chicken CtIP, suggesting that the BRCA1–CtIP interaction is dispensable for resection-dependent repair.
To examine this issue in mammalian cells, and to ascertain whether the BRCA1–CtIP interaction is required for BRCA1-mediated tumor suppression, we have introduced the corresponding mutation (S326A) into mouse embryonic stem (ES) cells to generate cells and mice expressing Ctip–S326A polypeptides that fail to interact with mouse Brca1. Here we show that the S326A mutation does not impair resection-dependent pathways of DSB repair (e.g., HDR and MMEJ) and does not abrogate Brca1-mediated tumor suppression. These findings indicate that, at least in mammalian cells, the BRCA1–CtIP interaction is dispensable for these aspects of BRCA1 function. Moreover, because the HDR and tumor suppression activities of BRCA1 are dependent on the phospho-recognition property of its BRCT sequences (Shakya et al., 2011), these results suggest that the interactions of BRCA1 with one or more of its other BRCT phospho-ligands are critical for these functions.
Results
The CtipS326A mutation ablates the Brca1–Ctip interaction in mouse cells
In human cells, the phospho-dependent interaction between BRCA1 and CtIP can be disrupted by an alanine substitution of the relevant CtIP phosphorylation site (S327A; Yu and Chen, 2004). Therefore, we introduced the corresponding mutation (S326A) into the Ctip gene of mouse embryonic stem (ES) cells using either of two targeting constructs. The CtipS326A-neo construct contains the S326A mutation in exon 11 and a loxP-flanked PGK promoter-driven neomycin gene cassette in the adjacent upstream intron (Fig. 1 B). An analogous CtipS326A-hyg construct was generated by replacing the neomycin cassette in intron 10 with a loxP-flanked hygromycin gene cassette. Ctip+/− ES cells, which are heterozygous for a CtipneoR-null allele (Ctip−; unpublished data; see Materials and methods), were then electroporated with the CtipS326A-hyg targeting construct, and Southern analysis was used to identify hygromycin-resistant colonies that had undergone homologous recombination at the Ctip+ allele to yield CtipS326A-hyg/− ES subclones (Fig. 1 E). To excise the hygromycin gene cassette from the knock-in allele, CtipS326A-hyg/− cells were infected with an adenovirus expressing Cre recombinase and properly recombined CtipS326A/− ES clones (Fig. 1 D) were identified by Southern analysis and confirmed by nucleotide sequencing. The Ctip+/− and CtipS326A/− clones, which represent isogenic ES cell lines expressing either wild-type or S326A mutant Ctip, were then used to study the DSB repair functions of Ctip (e.g., see next section).
Figure 1.
Design of the mutant CtipS326A allele and identification of heterozygous ES cells with the CtipS326A-hyg or CtipS326A-neo knock-in alleles. The wild-type Ctip locus encompassing exons 9–14 is shown (A), along with the CtipS326A-neo targeting vector (B), and maps of the Ctip locus after homologous recombination (C) and cre-mediated recombination (D). For the targeting vector, a neomycin expression cassette flanked by loxP signals (closed triangles) was inserted into the HpaI site of intron 10, whereas the CtipS326A mutation (asterisk) and an AgeI restriction site were introduced into exon 11. An HSV thymidine kinase (HSV-TK) gene cassette was included in the targeting vector for negative selection. The wavy line represents plasmid sequences of the targeting vector. Relevant restriction enzyme sites are: PvuII (P), EcoRI (E), HpaI (H), and AgeI (A). The Ctip probe used for Southern analysis and the sizes of the PvuII fragments recognized by the probe are shown. An analogous CtipS326A-hyg targeting construct was prepared by replacing the loxP-flanked neomycin resistance cassette in intron 10 with a loxP-flanked hygromycin selection marker (note: the hygromycin cassette lacks a PvuII restriction site). To identify heterozygous ES cells with the CtipS326A-hyg or CtipS326A-neo knock-in alleles, Southern analysis of PvuII-digested genomic DNA with a 5′ flanking probe (A) was used to screen (E) hygromycin-resistant Ctip+/− ES cell subclones targeted with the CtipS326A-hyg construct and (F) neomycin-resistant 129/Sv ES cell subclones targeted with the CtipS326A-neo construct. The 7.9-kb PvuII germline fragment is converted into a 10.4-kb fragment in properly targeted CtipS326A-hyg/− ES subclones (lanes 2, 4, and 7; panel E) or a 5.8-kb fragment in properly targeted CtipS326A-neo/+ ES subclones (lanes 1, 2, and 11; panel F).
To produce isogenic mouse embryonic fibroblast (MEF) lines expressing either wild-type or S326A mutant Ctip, 129/Sv ES cells were electroporated with the CtipS326A-neo targeting construct and selected for neomycin resistance. Independent neomycin-resistant CtipS326A-neo/+ 129/Sv ES clones were then identified (Fig. 1 F) and injected into C57BL/6J blastocysts to establish the mutant allele in the mouse germline. To excise the loxP-flanked neomycin expression cassette from the targeted allele (Fig. 1 C) and produce offspring expressing the desired CtipS326A allele (Fig. 1 D), chimeric male CtipS326A-neo/+ mice were mated with females carrying a ubiquitously expressed Cre transgene driven by the mouse Rosa26 gene promoter (RosaCre). Significantly, when heterozygous CtipS326A/+ mice were intercrossed, CtipS326A/S326A pups were born at the expected (25%) Mendelian ratio, indicating that the S326A mutation does not affect embryonic development. Thus, unlike animals homozygous for either a Brca1- or Ctip-null allele, which undergo embryonic lethality before gastrulation (precluding the generation of MEFs; Liu et al., 1996; Hakem et al., 1997; Ludwig et al., 1997; Chen et al., 2005), CtipS326A/S326A mice are viable. Isogenic primary Ctip+/+, CtipS326A/+, and CtipS326A/S326A MEFs were prepared from day E13.5 embryos, and immortalized MEF lines were established by transfection with simian virus 40 large T antigen.
The mutant Ctip polypeptide of CtipS326A/S326A MEFs is readily detected by immunoblot analysis (see Figs. 2 A and 6 C), although its steady-state levels appear to be slightly but consistently reduced relative to wild-type Ctip. To ascertain whether S326A mutant Ctip interacts with mouse Brca1, nuclear extracts of Ctip+/+, CtipS326A/+, and CtipS326A/S326A MEFs were immunoprecipitated with a mouse Brca1-specific antiserum (B1) or the corresponding preimmune serum (Pre) and immunoblotted with a CtIP-specific monoclonal antibody. As shown in Fig. 2 B, Ctip was efficiently coimmunoprecipitated with Brca1 from lysates of wild-type Ctip+/+ and heterozygous CtipS326A/+ MEFs. In contrast, Ctip failed to coimmunoprecipitate with Brca1 from homozygous CtipS326A/S326A MEFs (Fig. 2 B), indicating that Ctip-S326A polypeptides do not interact with Brca1 in vivo. In accord with this result, the in situ association of Brca1 and Ctip is markedly reduced in cells (CtipS326A/S326A) that express the mutant Ctip-S326A polypeptide (Fig. S1), as measured by the proximity ligation assay (PLA; Söderberg et al., 2006). In contrast, mutant Ctip-S326A retained the ability to interact with Mre11 and to support IR-induced phosphorylation of the Chk1 kinase (Fig. S2). Moreover, Ctip-staining nuclear foci are assembled with similar kinetics in wild-type (Ctip+/+) and mutant (CtipS326A/S326A) MEFs, indicating that the S326A mutation does not impair the recruitment of Ctip to sites of DNA damage (Fig. S3).
Figure 2.
The Ctip–S326A (SA) polypeptide does not interact with Brca1 in vivo. (A) Nuclear extracts of Ctip+/+, CtipS326A/+, and CtipS326A/S326A MEFs were fractionated by SDS-PAGE and immunoblotted with antibodies specific for α-tubulin, Brca1, and Ctip. A nonspecific band in the Brca1 immunoblot is designated as “ns”. (B) To evaluate the Brca1–Ctip interaction, the extracts were immunoprecipitated with mouse Brca1-specific antiserum (B1) or the corresponding preimmune serum (Pre) and immunoblotted with Ctip-specific monoclonal antibodies. As shown, Ctip was coimmunoprecipitated with Brca1 from Ctip+/+ and CtipS326A/+ cells, but not CtipS326A/S326A cells. Note: the amount of nuclear extract used for immunoblotting (A) represents 6.25% of the total extract used for coimmunoprecipitation analysis (B).
Figure 6.
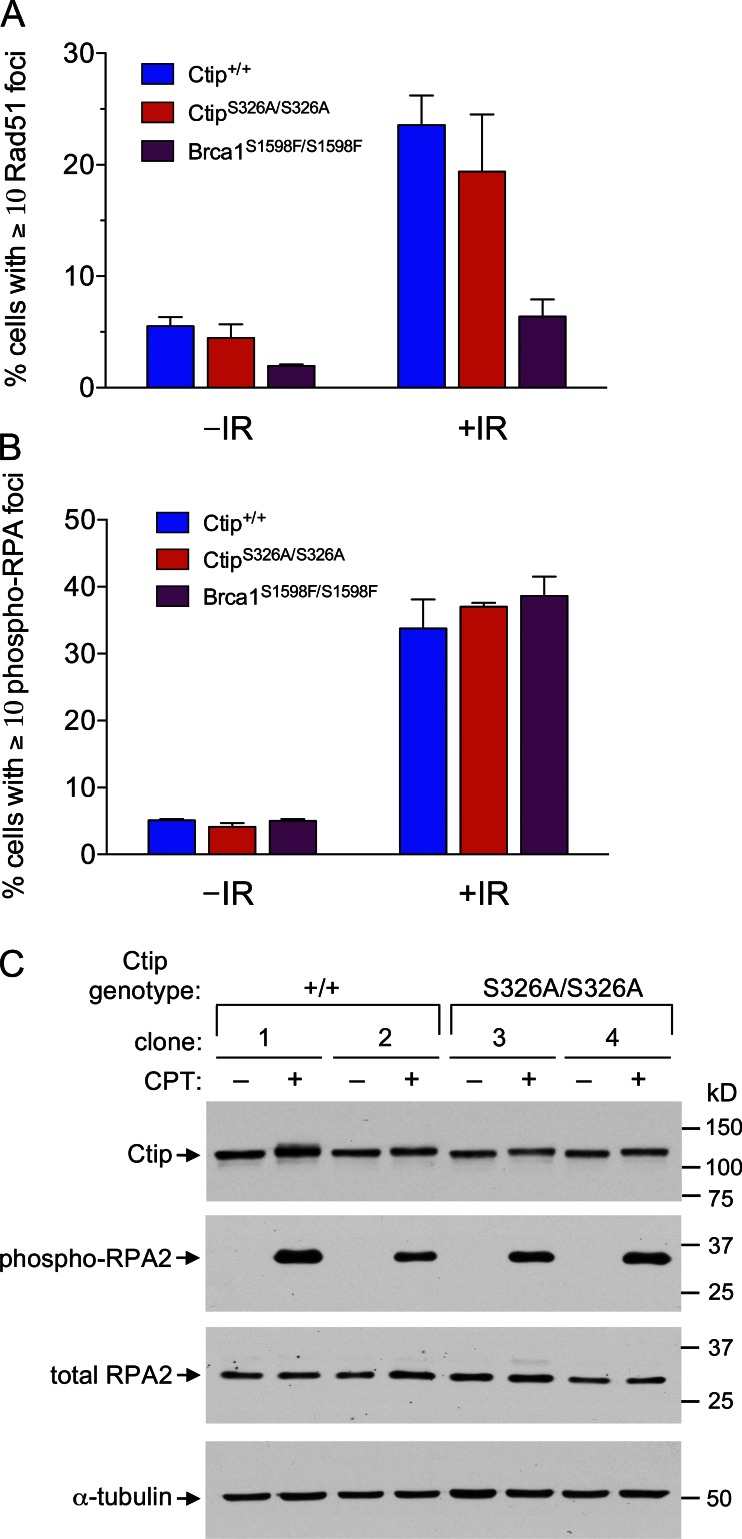
CtipS326A/S326A MEFs are proficient for assembly of Rad51 and RPA nuclear foci in response to DNA damage. (A and B) Ctip+/+ and CtipS326A/S326A MEFs were exposed to IR (10 Gy) and IRIF formation was assessed 1 h later by immunostaining with rabbit antisera specific for Rad51 (A) or Thr21-phosphorylated RPA2 (B). Cells containing 10 or more distinct Rad51- or phosphorylated RPA2-staining nuclear foci were counted in at least 500 nuclei of two independent MEF lines for each genotype, and the error bars represent SEM. IR treatment strongly induced the number of Rad51 foci in Ctip+/+ and CtipS326A/S326A MEFs, but not in Brca1S1598F/S1598F MEFs, which are known to have reduced IRIF assembly of Rad51 (Shakya et al., 2011). IR treatment also strongly induced the number of Thr21-phosphorylated RPA2 IRIFs in Ctip+/+ and CtipS326A/S326A MEFs, as well as in Brca1S1598F/S1598F MEFs. (C) Independent clones of Ctip+/+ and CtipS326A/S326A MEFs were cultured in the presence or absence of 1.0 µM camptothecin (CPT) and harvested 1 h later. Total cell extracts of each culture were then fractionated by SDS-PAGE and immunoblotted with antibodies specific for Ctip, Ser4/Ser8-phosphorylated RPA2, total RPA2, or α-tubulin.
Cells expressing the Ctip–S326A mutant show limited sensitivity to genotoxic agents
Brca1S1598F/S1598F cells, which harbor a missense mutation (S1598F) that ablates the BRCT phospho-recognition activity of Brca1, are hypersensitive to the DNA cross-linking agent mitomycin C (MMC; Shakya et al., 2011). This suggests that cellular resistance to MMC is dependent on the interaction of BRCA1 with one or more of its BRCT phospho-ligands. To determine whether the BRCA1–CtIP interaction is required for MMC resistance, isogenic Ctip+/− and CtipS326A/− ES subclones were evaluated in a clonogenicity assay in parallel with Brca1Δ223–763/Δ223–763 ES cells, which express an internally deleted Brca1 polypeptide that renders cells hypersensitive to MMC (Moynahan et al., 2001a). As shown in Fig. 3, the MMC survival curves of CtipS326A/− cells, unlike Brca1Δ223–763/Δ223–763 cells, overlap with those of the Ctip+/− and Brca1+/− control cells. Thus, MMC resistance is dependent on the interaction of BRCA1 with one or more of its BRCT phospho-ligands (Shakya et al., 2011), but not solely on its interaction with CtIP.
Figure 3.
CtipS326A/− ES cells are resistant to MMC-induced genotoxic stress. Isogenic Ctip+/− and CtipS326A/− ES cells were examined for mitomycin C (MMC) sensitivity in clonogenic survival assays, together with ES cells homozygous for the hypomorphic Brca1Δ223–763 mutation (Brca1Δ223–763/Δ223–763) and control ES cells (Brca1+/−). Cells were treated with various concentrations of MMC for 4 h, allowed to recover for 7–9 d, and surviving colonies were stained with Crystal violet. Survival is calculated as a percentage of colonies in the mock-treated plates. Each subclone was tested in triplicate, and the error bars represent the SEM of survival for each subclone.
CtIP-depleted cells are hypersensitive to both the topoisomerase I inhibitor camptothecin (CPT) and the topoisomerase II inhibitor etoposide (ETO; Sartori et al., 2007). By stabilizing their respective Topo cleavage complexes, these agents can block DNA replication and elicit DSB formation. To study the function of the BRCA1–CtIP interaction, Nakamura et al. (2010) generated chicken DT40 cells (CtIPS332A/−/−) that express a CtIP protein lacking the phosphorylation site (S332) required for its interaction with BRCA1. Interestingly, although these cells were proficient for DSB repair by HDR, they displayed hypersensitivity to both CPT and ETO, suggesting a specific role for the BRCA1–CtIP interaction in processing DSB ends that possess covalently bound polypeptides (Nakamura et al., 2010). To assess the requirement for this interaction in mammalian cells, clonogenicity assays were conducted with Ctip+/− and CtipS326A/− ES clones. As shown in Fig. 4, A and B, CtipS326A/− cells displayed modest but reproducible sensitivity to both CPT and ETO relative to the isogenic Ctip+/− control cells. In contrast, Brca1Δ223–763/Δ223–763 cells showed significant hypersensitivity to CPT (Fig. 4 A) and modest sensitivity to ETO (Fig. 4 B). These results suggest that the Brca1–Ctip interaction is required for some, but not all, of the cellular resistance mediated by Brca1 in response to the topoisomerase inhibitors CPT and ETO.
Figure 4.
CtipS326A/− ES cells are moderately sensitive to CPT- and ETO-induced genotoxic stress. Ctip+/− and CtipS326A/− ES cells were examined for (A) camptothecin (CPT) and (B) etoposide (ETO) sensitivity in clonogenic survival assays, together with ES cells homozygous for the hypomorphic Brca1Δ223–763 mutation (Brca1Δ223–763/Δ223–763). Cells were exposed for 24 h to varying doses of CPT and ETO, allowed to recover for 7–9 d, and surviving colonies were stained with Crystal violet. Survival is calculated as a percentage of colonies in the mock-treated plates. Each subclone was tested in triplicate, and the error bars represent the SEM of survival for each subclone.
CtipS326A cells maintain chromosomal stability at normal rates
Because chromosomal rearrangements arise frequently in Brca1S1598F/S1598F cells, chromosomal stability is likely to require the interaction of BRCA1 with one or more of its BRCT phospho-ligands (Shakya et al., 2011). To ascertain the role of the BRCA1–CtIP interaction, we examined metaphase spreads of early passage Ctip+/+ and CtipS326A/S326A primary MEF subclones. As shown in Fig. 5 A and Table 1, CtipS326A/S326A MEFs displayed low levels of spontaneous chromosomal rearrangements, comparable to those of wild-type Ctip+/+ cells. Moreover, aneuploidy was not observed in CtipS326A/S326A cells, which contained on average the expected number of 40 mouse chromosomes. In addition, when subjected to DNA damage by MMC treatment, CtipS326A/S326A cells acquired cytogenetic defects to the same extent as control Ctip+/+ MEFs (Fig. 5 B; Table 1). Thus, the Brca1–Ctip interaction is dispensable for suppression of spontaneous and genotoxic-induced chromosomal instability.
Figure 5.
Low levels of spontaneous and MMC-induced chromosomal aberrations in CtipS326A/S326A primary MEFs. Primary Ctip+/+ and CtipS326A/S326A MEFs were cultured in the (A) absence or (B) presence of mitomycin C (MMC; 40 ng/ml) for 16 h and subjected to karyotype analysis by Giemsa staining. 25 metaphase spreads per cell line were examined for each treatment condition for numerical and structural chromosomal aberrations.
Table 1.
Spontaneous and induced chromosomal aberrations in primary MEFs with different Ctip genotypes
| Aberrations | |||||
| Genotype | Metaphases analyzed | MMC treatment | Metaphase aberrations | Chr/Cht breaks and gaps | Exchange/other |
| % | |||||
| Ctip+/+ | 25 | − | 16 | 4 | 0 |
| 25 | + | 44 | 14 | 1 | |
| CtipS326A/S326A clone 1 | 25 | − | 32 | 9 | 0 |
| 25 | + | 52 | 15 | 2 | |
| CtipS326A/S326A clone 2 | 25 | − | 20 | 7 | 0 |
| 25 | + | 52 | 14 | 3 | |
The percentage of metaphases containing one or more aberrations and a breakdown of aberration type is shown for each primary MEF cell line in both the absence (−) and presence (+) of MMC. Cht, chromatid; Chr, chromosome; MMC, mitomycin C.
The Brca1–Ctip interaction is not required for assembly of Rad51 or RPA nuclear foci
In cells treated with ionizing radiation (IR), Rad51 polypeptides accumulate at sites of DNA damage to form IR-induced foci (IRIFs) that can be visualized by immunofluorescent microscopy. These structures are thought to represent the recruitment of Rad51 polypeptides to sites of DSBs and formation of the ssDNA–Rad51 nucleofilaments necessary for HDR. Because the assembly of Rad51 IRIFs is dependent on the BRCT phospho-recognition property of BRCA1 (Shakya et al., 2011), we examined whether the BRCA1–CtIP interaction is also required for this process. As expected, formation of Rad51-staining IRIFs was markedly reduced in Brca1S1598F/S1598F MEFs (Fig. 6 A and Fig. S4). In contrast, assembly of Rad51 IRIFs occurred at normal levels in MEFs expressing either wild-type or S326A mutant Ctip polypeptides. These data indicate that the Brca1–Ctip interaction is not required for Rad51 recruitment to sites of DNA damage, a key step in DSB repair by homologous recombination.
Upon DNA resection of a DSB, the nascent ssDNA tail is initially coated with the RPA heterotrimer to form an ssDNA–RPA filament which can be observed cytologically by the appearance of nuclear foci that stain with RPA-specific antibodies or biochemically by hyperphosphorylation of the RPA2 subunit. Therefore, to examine whether the BRCA1–CtIP interaction is required for DNA resection, we compared the assembly of damage-induced ssDNA–RPA filaments in isogenic cells that express either wild-type or S326A mutant Ctip. Of note, similar levels of IR-induced RPA foci (Fig. 6 B and Fig. S5) and camptothecin-induced RPA2 hyperphosphorylation (Fig. 6 C) were observed in Ctip+/+ and CtipS326A/S326A MEFs. Interestingly, normal assembly of IR-induced RPA foci was also observed in Brca1S1598F/S1598F MEFs (Fig. 6 B and Fig. S5). Thus, the BRCT phospho-recognition property of BRCA1 and, more particularly, the BRCA1–CtIP interaction are dispensable for resection of damage-induced DSB ends in mammalian cells.
The Brca1–Ctip interaction is not essential for the HDR pathway of DSB repair in mammalian cells
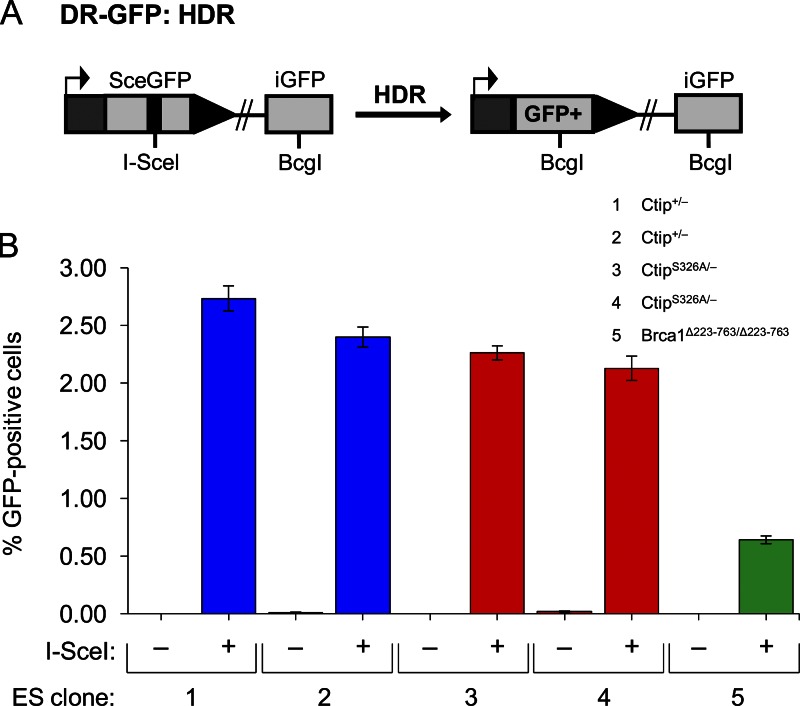
Previous studies have established that DSB repair by the HDR pathway is dependent on both BRCA1 and CtIP (Moynahan et al., 1999, 2001a; Sartori et al., 2007; Bennardo et al., 2008; Chen et al., 2008). Therefore, to determine whether the BRCA1–CtIP interaction is also required we measured HDR at a defined chromosomal break using an integrated DR-GFP recombination substrate (Pierce et al., 2001). The DR-GFP substrate consists of two defective GFP genes: SceGFP, which contains the cleavage site for the I-SceI endonuclease; and iGFP, which lacks the N- and C-terminal coding sequences of GFP (Fig. 7 A). I-SceI expression triggers cleavage of SceGFP, resulting in a chromosomal DSB at the I-SceI site. Repair of this DSB by HDR using iGFP as a template generates a functional GFP gene, such that the frequency of HDR can be quantified as the percentage of GFP-positive cells using flow cytometry (Pierce et al., 2001).
Figure 7.
Cells lacking the Brca1–Ctip interaction are proficient for DSB repair by HDR. ES cells harboring the DR-GFP substrate (A) integrated into the Pim1 locus were transfected with either an I-SceI expression vector or the empty vector. (B) I-SceI expression induced the appearance of GFP-positive cells to comparable levels in cultures of isogenic Ctip+/− (subclones 1 and 2) and CtipS326A/− (subclones 3 and 4) ES cells, indicating that the Ctip-S326A mutation does not appreciably affect HDR. In contrast, a Brca1 mutant ES cell line 236.44 (Brca1Δ223–763/Δ223–763) with the DR-GFP substrate integrated at the same position of the Pim1 locus (subclone 5) was deficient for HDR, as previously reported (Moynahan et al., 1999). Each ES subclone was assayed in triplicate with three independent transfections, and the error bars represent SEM. Similar results were also observed in separate experiments using additional independently derived Ctip+/− and CtipS326A/− ES subclones.
To measure HDR of DSBs at a defined chromosomal site, Ctip+/− and CtipS326A/− ES cells were electroporated with p59xDR-GFP6, a DNA construct that contains the DR-GFP recombination substrate and a promoterless hygromycin-resistance marker flanked by targeting arms comprised of mouse Pim1 genomic DNA (Moynahan et al., 2001b). After hygromycin selection, drug-resistant colonies were examined by Southern analysis to identify ES subclones that possess the DR-GFP substrate at an identical position within the Pim1 locus. To measure HDR of an induced chromosomal DSB, the Ctip+/−DR-GFP and CtipS326A/− DR-GFP subclones were evaluated for the appearance of GFP-positive cells after transient transfection with an I-SceI expression vector. In the absence of I-SceI, GFP-positive cells were seldom detected (<0.02%) in cells of either Ctip genotype (Fig. 7 B), indicating that spontaneous intrachromosomal gene conversion is rare. As expected, I-SceI expression induced HDR of the DR-GFP substrate in control Ctip+/− ES cells, as reflected by the increased percentage of GFP-positive cells (2.4–2.74%; Fig. 7 B). Surprisingly, I-SceI expression induced similar proportions of CtipS326A/− GFP-positive cells (2.13–2.26%; Fig. 7 B), indicating that the Brca1–Ctip interaction is not essential for proficient HDR. Consistent with previous reports (Moynahan et al., 1999, 2001a; Snouwaert et al., 1999), HDR efficiency was significantly reduced (0.64%) in Brca1 mutant ES cells (Brca1Δ223–763/Δ223–763) with the DR-GFP substrate integrated at the same position of the Pim1 locus (Fig. 7 B). Thus, although the BRCT phospho-recognition activity of Brca1 is required for HDR (Shakya et al., 2011), the Brca1–Ctip interaction appears to be dispensable for DSB repair by this pathway.
The Brca1–Ctip interaction is also dispensable for DSB repair by MMEJ and single-strand annealing
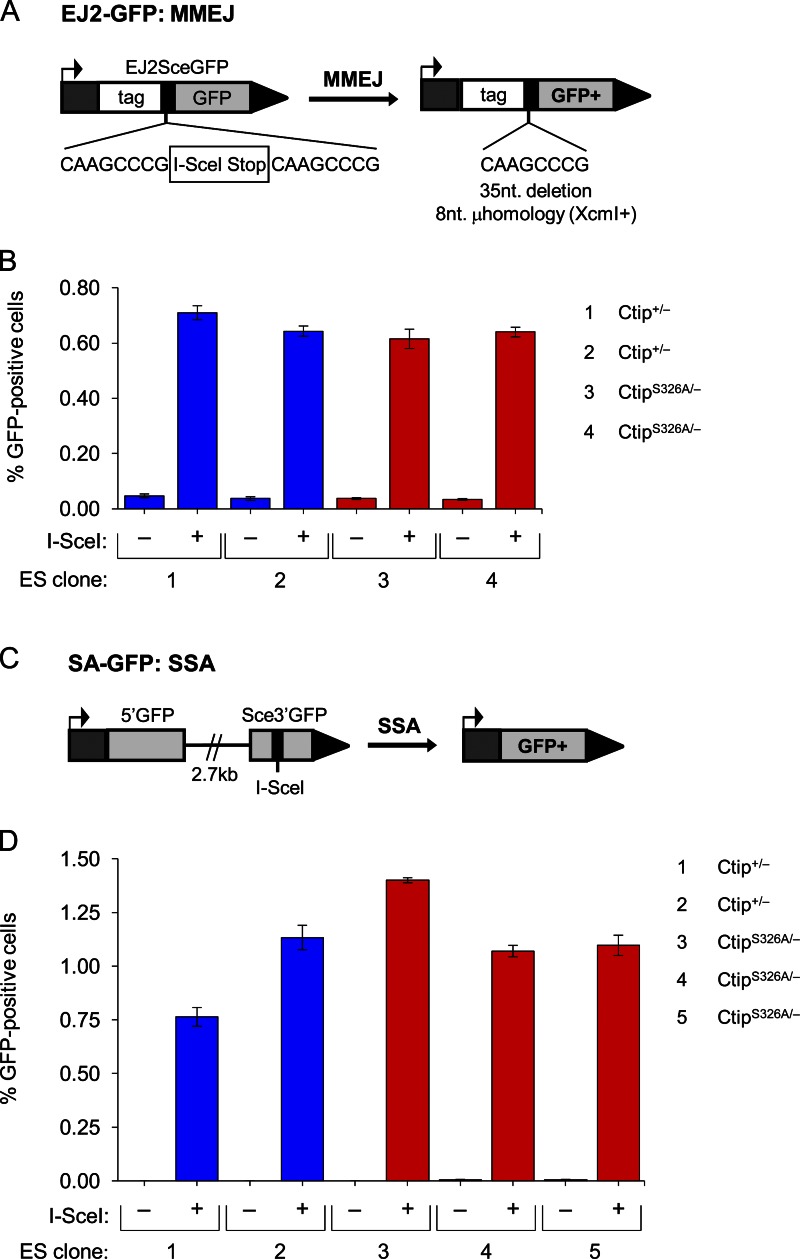
Unlike HDR, Ku-dependent NHEJ ligates DSB ends without a requirement for extensive sequence homology (Symington and Gautier, 2011). In contrast to this classical pathway of NHEJ, microhomology-mediated end joining (MMEJ) employs short sequence homologies to align broken DNA ends before ligation and, as such, is dependent on DNA resection to expose microhomologies within the ssDNA overhangs (Nussenzweig and Nussenzweig, 2007; McVey and Lee, 2008). To examine whether the BRCA1–CtIP interaction is required for MMEJ, we electroporated isogenic Ctip+/− and CtipS326A/− ES cells with pim-EJ2-GFP-hyg, a targeting vector that contains the EJ2-GFP recombination reporter, a hygromycin resistance cassette, and genomic sequences for targeting the Pim1 locus (Bennardo et al., 2008). The EJ2-GFP reporter consists of an N-terminal tag fused to GFP, which is disrupted by an 8-nucleotide microhomology repeat that flanks an I-SceI site and stop codons in all three reading frames (Fig. 8 A). If MMEJ occurs by annealing of the microhomology repeats, the intervening 35-nucleotide sequence is deleted, the coding frame between the N-terminal tag and GFP is restored, and a functional GFP gene is reconstructed (Bennardo et al., 2008). After electroporation with the pim-EJ2-GFP-hyg targeting vector, Ctip+/− and CtipS326A/− ES cells were selected with hygromycin and DNA was prepared from the surviving colonies. Southern analysis revealed proper homologous integration of the EJ2-GFP reporter into the mouse Pim1 locus of several Ctip+/− and CtipS326A/− ES subclones. To measure MMEJ repair of an I-SceI–induced chromosomal break, Ctip+/− EJ2-GFP and CtipS326A/− EJ2-GFP subclones were transiently transfected with an I-SceI expression vector and evaluated for the appearance of GFP-positive cells by flow cytometry. As expected, very few GFP-positive cells were detected (<0.05%) in ES subclones transfected with an empty expression vector (Fig. 8 B). However, after I-SceI expression, the proportions of GFP-positive cells were increased to a comparable extent in both the Ctip+/− EJ2-GFP (0.64–0.71%) and CtipS326A/− EJ2-GFP (0.62–0.64%) subclones (Fig. 8 B). Thus, as with the resection-dependent HDR pathway, the Brca1–Ctip interaction appears to be dispensable for MMEJ repair of chromosomal DSBs.
Figure 8.
Cells lacking the Brca1–Ctip interaction are proficient for the MMEJ and SSA pathways of DSB repair. ES cells harboring either the EJ2-GFP (A and B; to measure MMEJ repair) or SA-GFP (C and D; to measure SSA repair) substrate integrated into the Pim1 or Hprt locus, respectively, were transfected with either an I-SceI expression vector or the empty vector. (B) I-SceI expression induced similar levels of GFP-positive cells in cultures of isogenic Ctip+/− (subclones 1 and 2) and CtipS326A/− (subclones 3 and 4) cells with the EJ2-GFP substrate, indicating that the Ctip–S326A mutation does not affect MMEJ. (D) Likewise, I-SceI expression induced comparable levels of GFP-positive cells in cultures of isogenic Ctip+/− (subclones 1 and 2) and CtipS326A/− (subclones 3, 4, and 5) cells with the SA-GFP substrate, indicating that the Ctip–S326A mutation does not affect SSA.
Single-strand annealing (SSA) is a mutagenic mode of DSB repair that shares some features with MMEJ (Bennardo et al., 2008; McVey and Lee, 2008). In SSA, annealing occurs between long (>30 nucleotides) direct repeats that flank the DSB, allowing for ligation of the ends and deletion of the intervening sequences. Although the late stages of repair by SSA and MMEJ are distinct, both pathways require CtIP-mediated DNA resection to expose complementary sequences within the ssDNA overhangs (Bennardo et al., 2008; McVey and Lee, 2008). Previous studies have established that efficient DSB repair by SSA is dependent on both BRCA1 and CtIP (Stark et al., 2004; Bennardo et al., 2008). Therefore, to ascertain whether the BRCA1–CtIP interaction is also required for SSA, isogenic Ctip+/− and CtipS326A/− ES cells were electroporated with the hprtSAGFP targeting construct (Stark et al., 2004), and subclones that carry the SA-GFP recombination reporter integrated into the Hprt locus were derived. The SA-GFP reporter, which contains a restriction site for I-SceI cleavage, generates a functional GFP gene when repaired by SSA (Fig. 8 C; Stark et al., 2004). Therefore, to measure the efficiency of SSA in Ctip+/− and CtipS326A/− ES subclones with an integrated SA-GFP substrate, these cells were transiently transfected with an I-SceI expression vector. As shown in Fig. 8 D, I-SceI expression elicited comparable levels of GFP-positive cells in both the Ctip+/− SA-GFP (0.76–1.13%) and CtipS326A/− SA-GFP (1.07–1.40%) subclones. Thus, although Brca1 and Ctip are each essential for SSA (Stark et al., 2004; Bennardo et al., 2008), the Brca1–Ctip interaction is not required for this mode of DSB repair.
Tumor suppression is not dependent on the Brca1–Ctip interaction
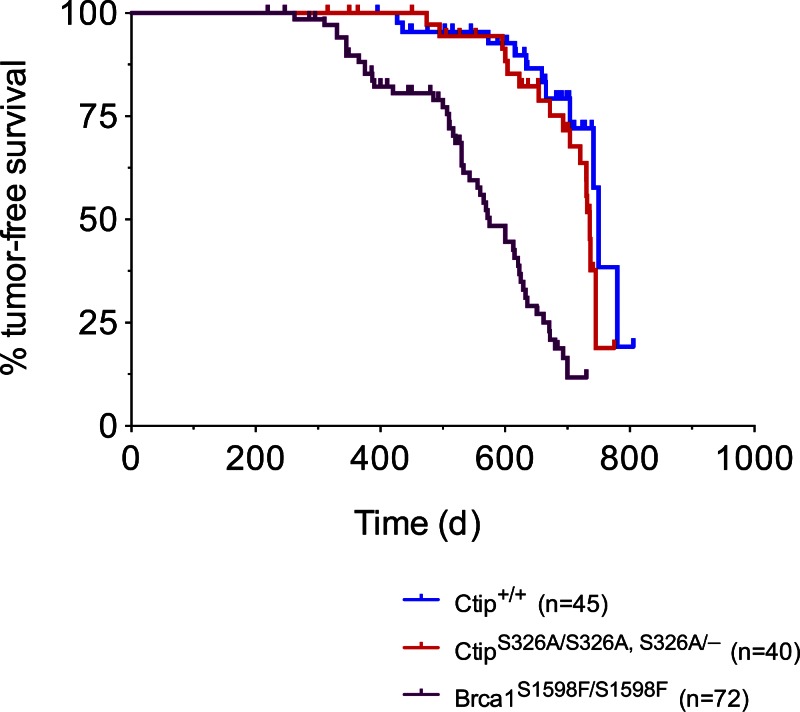
Homozygous Brca1S1598F/S1598F mice, which express a mutant Brca1 protein defective for BRCT phospho-recognition, are prone to tumor development (Shakya et al., 2011). This observation implies that the interaction of BRCA1 with one or more of its BRCT phospho-ligands is required for tumor suppression. To assess the role of the Brca1–Ctip interaction in tumor suppression, we monitored cohorts of 45 control (Ctip+/+) and 40 mutant (CtipS326A/S326A, CtipS326A-neo/S326A-neo, or CtipS326A-neo/−) mice for tumor development over a 24-month observation period. As shown in Fig. 9, a few mutant mice developed tumors at a very advanced age, with kinetics (frequency and latency) statistically indistinguishable from that of the control cohort (750 days; P = 0.2099), but significantly delayed relative to the tumor-prone Brca1S1598F/S1598F mice (P < 0.0001). Thus, although tumor suppression by BRCA1 is dependent on the phospho-recognition potential of its BRCT repeats (Shakya et al., 2011), solely disrupting the Brca1–Ctip interaction does not predispose mice to tumor formation.
Figure 9.
The Brca1–Ctip interaction is not required for tumor suppression. The Kaplan-Meier tumor-free survival curves of the control (Ctip+/+; blue curve; n = 45; T50 = 750 d) and experimental (CtipS326A/S326A and CtipS326A/− mice; red curve; n = 40; T50 = 736 d) cohorts are shown. Statistical significance (P = 0.2099) was estimated with the log-rank test using Prism software (version 4; GraphPad Software); values were considered significant at P < 0.05. For comparison, the Kaplan-Meier curve of tumor-prone Brca1S1598F/S1598F mice (purple curve; n = 72; T50 = 575 d; P < 0.0001) is also displayed (Shakya et al., 2011).
Discussion
The BRCA1 tumor suppressor has emerged as a central player in the cellular response to DNA damage (Huen et al., 2010; Moynahan and Jasin, 2010; Li and Greenberg, 2012; Roy et al., 2012). Of particular interest, BRCA1 is required for homology-directed repair (HDR), a relatively error-free pathway for repair of DSBs. Because HDR defects can lead to both chromosome rearrangements and aneuploidy, loss of HDR function may be a primary source of the genomic instability that is characteristic of BRCA1 mutant cells (Huen et al., 2010; Moynahan and Jasin, 2010; Li and Greenberg, 2012; Roy et al., 2012). Interestingly, the BRCA2 tumor suppressor has also been implicated in HDR (Moynahan et al., 2001b) and ascribed a specific biochemical function in formation of the ssDNA–Rad51 nucleoprotein filament, an essential HDR intermediate (Yang et al., 2005). Thus, HDR deficiency may be a common determinant of breast cancer susceptibility in both BRCA1 and BRCA2 mutation carriers (Huen et al., 2010; Moynahan and Jasin, 2010; Li and Greenberg, 2012; Roy et al., 2012). Although the precise biochemical role of BRCA1 in HDR remains unclear, the phospho-recognition property of its BRCT repeats is required for both HDR and tumor suppression (Shakya et al., 2011). In this regard, it is noteworthy that each of the three phospho-ligands known to form in vivo protein complexes with BRCA1 (i.e., Abraxas/CCDC98, BACH1/FancJ/BRIP1, and CtIP) has also been implicated in DSB repair by the HDR pathway (Huen et al., 2010; Moynahan and Jasin, 2010; Li and Greenberg, 2012; Roy et al., 2012).
CtIP collaborates with the MRN (Mre11–Rad50–Nbs1) complex to initiate resection of DSB ends and formation of the 3′-ssDNA overhangs required for HDR and MMEJ (Sartori et al., 2007; Bennardo et al., 2008; Chen et al., 2008). Because BRCA1 has been implicated in these same repair pathways, it is conceivable that BRCA1, and more particularly the BRCA1–CtIP interaction, is involved in the DNA resection functions of CtIP. This notion is very attractive, as it could provide a biochemical mechanism to explain how BRCA1 promotes the HDR pathway of DSB repair. A requirement for BRCA1 in CtIP-mediated resection would also be consistent with genetic data that place BRCA1 upstream of BRCA2 in the HDR pathway (Stark et al., 2004). Moreover, BRCA1, CtIP, and MRN are known to form a discrete protein complex in mammalian cells that could potentially mediate the resection activities ascribed to CtIP and MRN (Greenberg et al., 2006; Chen et al., 2008). At present, however, published data regarding the role of BRCA1 in DNA resection are contradictory. On the one hand, Schlegel et al. (2006) assessed ssDNA formation in BrdU-labeled cells cytologically by the appearance of nuclear foci that stain with BrdU-specific antibodies. Using this approach, they showed that ssDNA focus formation in response to ionizing radiation (IR) is abolished by siRNA-mediated depletion of BRCA1 (Schlegel et al., 2006). Moreover, on the basis of BRCA1 reconstitution experiments in HCC1937 cells, a human breast tumor line that expresses a truncated BRCA1 polypeptide lacking its C-terminal BRCT motif, they concluded that the ability of BRCA1 to promote end resection is independent of its BRCT sequences. On the other hand, Chen et al. (2008) found that IR-induced assembly of nuclear ssDNA/RPA foci, detectable by staining with RPA-specific antibodies, is impaired in HCC1937 cells, but not in HCC1937 cells reconstituted with wild-type BRCA1. Thus, they also concluded that BRCA1 promotes end resection, but that it does so in a manner dependent on its BRCT sequences (Chen et al., 2008). Finally, Escribano-Díaz et al. (2013) described a modest reduction in IR-induced RPA focus formation upon siRNA depletion of BRCA1 in U2OS cells, but Zhao et al. (2007) observed no effect in HeLa cells.
The role of the BRCA1–CtIP interaction itself in DNA resection and DSB repair is also controversial. To address this issue, Yun and Hiom (2009) generated CtIP-null cells (CtIP−/−/−) from the chicken B cell tumor line DT40. The efficiency of the major DSB repair pathways was then measured in CtIP-null cells reconstituted with either a wild-type (CtIP−/−/− + hCtIP) or S327A mutant (CtIP−/−/− + hCtIP-S327A) form of human CtIP. Notably, cells expressing mutant CtIP were competent for DSB repair by NHEJ and MMEJ, but displayed marked defects in DSB repair by pathways that entail extensive DNA resection (i.e., HDR and SSA). These striking results implicated BRCA1 in CtIP-mediated DNA resection and suggested a key role for the BRCA1–CtIP interaction in directing the pathway choice for DSB repair (Yun and Hiom, 2009). Subsequently, however, Nakamura et al. (2010) reported that HDR function is proficient in DT40 cells (CtIPS332A/−/−) that express endogenous CtIP with the corresponding mutation (S332A in chickens). Moreover, Peterson et al. (2011) showed that mutation of the corresponding phosphorylation site in frog xCtIP (S328A) does not affect its ability to stimulate DNA resection in Xenopus cell-free extracts. Therefore, to determine whether the BRCA1–CtIP interaction is required for DNA resection and DSB repair in mammals, we generated isogenic subclones of mouse ES cells that express mouse Ctip with and without the S326A mutation. Our results indicate that the BRCA1–CtIP interaction in mammalian cells is dispensable for the major resection-dependent pathways of DSB repair, including HDR, MMEJ, and SSA. In accord with these results, CtipS326A/S326A mouse embryonic fibroblasts displayed normal suppression of spontaneous and genotoxic-induced chromosomal instability as well as normal assembly of IR-induced Rad51 and RPA foci.
Although two independent studies using chicken DT40 cells have generated contradictory conclusions about the function of the BRCA1–CtIP interaction (Yun and Hiom, 2009; Nakamura et al., 2010), our results indicate that, in mammalian cells, this interaction is not required for DNA resection or the resection-dependent modes of DSB repair. As such, our findings concur with those of Nakamura et al. (2010), who observed normal HDR levels in DT40 cells (CtIPS332A/−/−) expressing CtIP polypeptides that fail to bind BRCA1. Interestingly, these cells also displayed marked hypersensitivity to camptothecin (CPT), a topoisomerase 1 (Topo1) inhibitor that ultimately generates 3′-DSB ends covalently linked to Topo1, and to a lesser degree etoposide (ETO or VP16), a topoisomerase 2 (Topo2) inhibitor that yields Topo2-linked 5′-DSB ends. On this basis, Nakamura et al. (2010) proposed that the BRCA1–CtIP interaction promotes the endonucleolytic cleavage of oligonucleotide-bearing covalently bound polypeptides from DSB ends. Although yeast do not possess an obvious orthologue of BRCA1, this idea is appealing given that Sae2 is required for removal of Spo11-bound oligonucleotides during meiotic recombination in Saccharomyces cerevisiae (McKee and Kleckner, 1997; Prinz et al., 1997) and that the CtIP orthologue Ctp1 has been implicated in excision of covalently bound Topo2 protein from DSB ends in Schizosaccharomyces pombe (Hartsuiker et al., 2009). We observed a very modest, but reproducible, sensitivity of CtipS326A/− ES cells to camptothecin and, to a lesser extent, etoposide. Although the pattern of drug sensitivity in CtipS326A/− ES cells mirrors that seen in CtIPS332A/−/− chicken DT40 cells, the magnitude of drug sensitivity is much more pronounced in the mutant chicken cells. On one hand, this quantitative difference may simply represent species-specific variation in cellular resistance to the topoisomerase inhibitors. On the other hand, it may reflect the fact that chicken DT40 cells, unlike mouse embryonic stem cells, are fully transformed tumor cells that harbor a variety of tumor-associated genetic lesions, one or more of which might augment the drug sensitivity of its CtIP mutant subclones.
Our results indicate that the BRCA1–CtIP interaction is dispensable for CtIP-mediated resection and for the resection-dependent modes of DSB repair, such as HDR, MMEJ, and SSA. These conclusions are supported by biochemical data in cell-free extracts (Peterson et al., 2011) and genetic data in chicken DT40 cells (Nakamura et al., 2010). What, then, are the cellular functions of the evolutionarily conserved BRCA1–CtIP interaction? Previous studies have implicated this interaction in activation of the G2/M cell cycle checkpoint (Yu and Chen, 2004) and, more recently, in preventing aberrant recruitment of the RIF1 repair protein to damaged DNA in S/G2 cells (Escribano-Díaz et al., 2013). Although both of these activities likely facilitate the cellular response to DNA damage, the absence of heightened tumor susceptibility in CtipS326A/S326A mice suggests that they are not required for tumor suppression.
Although not yet formally proven, the HDR function of BRCA1 is thought to be a critical, if not essential, aspect of its tumor suppression activity. Indeed, a homozygous BRCA1 mutation that ablates BRCT phospho-recognition (Brca1S1598F/S1598F) also disrupts HDR and renders mice prone to tumor development (Shakya et al., 2011). This implies that the ability of BRCA1 to bind one or more of its BRCT phospho-ligands is critical for both HDR and tumor suppression. Here we show that tumor susceptibility is not enhanced by a mutation that specifically ablates the Brca1–Ctip interaction in mice (CtipS326A/S326A and CtipS326A/−). These results suggest that the BRCA1–CtIP interaction is dispensable for BRCA1-mediated tumor suppression altogether or that its role in tumor suppression can be compensated by one or more of the other BRCT phospho-ligands. In this regard, it will be interesting to test whether BRCA1-mediated tumor suppression is affected by specific disruption of the BRCA1–Abraxas and/or BRCA1–BACH1 interactions.
Materials and methods
The CtipS326A targeting constructs
The homology arms of the CtipS326A-neo targeting construct (Fig. 1 B) were derived from a 6.0-kb EcoRI fragment of genomic DNA, encompassing Ctip exons 10–12, from strain 129-derived E14 TG2a ES cells (Warren et al., 1994). Site-directed mutagenesis of exon 11 was used to (1) replace the natural serine codon (TCT) for residue 326 with an alanine codon (GCA), and (2) convert the sequence TCCGGT into an AgeI restriction site (ACCGGT). A loxP-flanked PGK promoter-driven neomycin selection cassette was inserted into a unique HpaI site in intron 10 (Fig. 1 B). A gene cassette encoding herpes simplex virus type 1 thymidine kinase (HSV-TK) was also included in the construct as a negative selection marker. The CtipS326A-hyg targeting construct was prepared by replacing the loxP-flanked neomycin cassette of the CtipS326A-neo construct with a loxP-flanked hygromycin cassette (lacking a PvuII site).
Analysis of isogenic ES cell subclones
Embryonic stem (ES) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Cellgro) supplemented with 15% heat-inactivated FBS (HyClone ES Cell Screened; Thermo Fisher Scientific), 2 mM l-glutamine, 1% nonessential amino acids, 100 µg/ml penicillin/streptomycin, 0.1 mM 2-mercaptoethanol, 1.25 µg/ml Plasmocin (InvivoGen), and 1,000 units/ml LIF (Esgro; EMD Millipore) at 37°C in 5% CO2/95% humidity. Unless otherwise noted, ES cells were grown on a layer of mitotically inactive primary embryonic fibroblasts (feeders) in the presence of LIF to maintain their pluripotency. NotI-linearized DNA of the CtipS326A-hyg targeting construct was introduced by electroporation (30 µg of DNA at 0.8 kV/3 µF) into Ctip+/− ES cells; the functionally null Ctip− allele of these cells was generated previously by replacing 1.2 kilobases of Ctip genomic DNA, including part of exon 11 and all of exon 12, with an MC1 promoter-driven neomycin expression cassette. After hygromycin selection, drug-resistant ES cell subclones were examined for gene targeting by Southern analysis of PvuII-digested genomic DNA with a 5′ flanking probe that spans Ctip exon 9 (Fig. 1 A). Correctly targeted CtipS326A-hyg/− clones were then infected with Adeno-Cre virus, and DNA from individually picked ES cell colonies were screened by Southern analyses for removal of the loxP-flanked hygromycin cassette and generation of the desired CtipS326A/− subclones (Fig. 1 D). To conduct clonogenic survival assays, ES cells were exposed for 4 h (mitomycin C) or 24 h (camptothecin and etoposide) to various concentrations of the drug, washed twice with 1× PBS, provided fresh media, and allowed to grow at 37°C for 7–9 d. Colonies of surviving cells were then fixed in 10% buffered formalin for 30 min, stained with 0.5% Crystal violet (Sigma-Aldrich) for 15 min, washed in water three times, and then counted. Survival experiments were performed in triplicates.
Recombination reporter assays
To assess homology-directed repair (HDR), ES subclones containing the DR-GFP recombination reporter integrated into the Pim1 locus were generated by electroporating (30 µg of DNA at 0.8 kV/3 μF) Ctip+/− and CtipS326A/− ES cells with the XhoI-linearized p59xDR-GFP6 targeting vector (Moynahan et al., 2001b). Hygromycin-resistant colonies were then evaluated by Southern analysis for proper integration of the DR-GFP reporter. To assess microhomology-mediated end joining (MMEJ), ES subclones containing the EJ2-GFP recombination reporter integrated into the Pim1 locus were generated by electroporating Ctip+/− and CtipS326A/− ES cells with the XhoI-linearized pim-EJ2-GFP-hyg targeting vector (Bennardo et al., 2008). Hygromycin-resistant colonies were then evaluated by Southern analysis for proper integration of the EJ2-GFP reporter. To assess single-strand annealing (SSA), ES subclones containing the SA-GFP recombination reporter integrated into the Hprt locus were generated by electroporating Ctip+/− and CtipS326A/− ES cells with the SacI–KpnI-linearized hprtSAGFP targeting vector (Stark et al., 2004). Colonies that were resistant to both puromycin and 6-thioguanine were then evaluated by Southern analysis for proper integration of the SA-GFP reporter.
To measure repair of I-SceI-induced chromosomal DNA breaks, each ES clone carrying an integrated GFP recombination reporter was trypsinized and seeded in 6 wells of a 12-well gelatinized plate in the absence of a feeder layer (∼106 cells/well). The next day, ES cells were provided fresh media (ES media minus penicillin/streptomycin, Plasmocin, and LIF) and transfected with 2.5 µg of the empty vector (pCAGGS) or the I-SceI expression vector (pCβASce) using Lipofectamine 2000 (Invitrogen). After a 24-h incubation, each well of transfected cells was washed twice with 1× PBS, trypsinized, and replated in a well of a 6-well gelatinized plate. 48 h after replating and 72 h after transfection, the cells were harvested and analyzed on a FACSCalibur using CellQuest software (BD). The proportion of GFP-positive events (at least 70,000 events were scored per sample) provided a measure of DSB repair.
Generation of mice and tumor monitoring
129/Sv ES cells were electroporated with the CtipS326A-neo targeting construct and independent neomycin-resistant clones of CtipS326A-neo/+ cells were injected into C57BL/6J blastocysts to establish the mutant allele in the mouse germline. The resulting CtipS326A-neo/+ mice were then intercrossed to generate homozygous CtipS326A-neo/S326A-neo mutants or mated with Ctip+/− animals to obtain CtipS326A-neo/− mice. In addition, heterozygous CtipS326A-neo/+ animals were crossed with RosaCre mice, which ubiquitously express Cre recombinase, to produce CtipS326A/+ mice with the loxP-flanked neomycin cassette removed (Fig. 1 D). Subsequent intercrossing of these animals yielded homozygous CtipS326A/S326A mice. For tumor monitoring (Fig. 9), the experimental cohort was comprised of 26 CtipS326A-neo/S326A-neo mice, 3 CtipS326A-neo/− mice, and 11 CtipS326A/S326A mice (total n = 40), while the control cohort included 45 Ctip+/+ mice. Mice were sacrificed when they were moribund or showed overt pathological signs. Upon autopsy, all major organs and any identified tumors were dissected and fixed overnight in 10% buffered formalin. Fixed specimens were dehydrated and embedded in paraffin. Paraffin blocks were sectioned at 4 µm and stained with hematoxylin and eosin for histopathology analyses.
Analysis of isogenic mouse embryonic fibroblasts
Primary Ctip+/+, CtipS326A/+, and CtipS326A/S326A mouse embryonic fibroblasts (MEFs) were derived from E13.5 embryos and cultured in DMEM (Cellgro) supplemented with 10% heat-inactivated FBS (Tissue Culture Biologicals), 100 µg/ml penicillin/streptomycin, 2 mM l-glutamine, and 1.25 µg/ml Plasmocin (InvivoGen) at 37°C in 5% CO2/95% humidity. Immortalized MEFs were then generated by transfecting primary MEFs at early passage with the pMSSVLT expression plasmid (10 µg/10-cm plate), which encodes SV40 large T antigen (Schuermann, 1990), using Lipofectamine 2000 (Invitrogen). Cells were passaged until only immortalized MEFs remained in culture (∼12 passages/4 wk). Established lines of immortalized MEFs were then frozen down and re-genotyped.
For analyses of Ctip protein expression, Brca1–Ctip coimmunoprecipitation, and Ctip–Mre11 coimmunoprecipitation nuclear extract lysates were prepared from immortalized MEFs. The cells were harvested in 1× PBS and lysed for 15 min in buffer (10 mM Hepes, pH 7.9, 10 mM KCl, and 0.1 mM EGTA) supplemented with 1 mM dithiothreitol (DTT), 5 mM sodium fluoride, 0.1 mM sodium orthovanadate, and complete protease inhibitor (Roche). A 1:16 volume of 10% Nonidet P-40 (NP40) was added to each sample before centrifugation to isolate the nuclear pellet. The pellet was resuspended in 1.5 times its volume with buffer (20 mM Hepes, pH 7.9, 10% glycerol, 0.4 M NaCl, 0.5 mM EDTA, 0.5 mM EGTA, and 0.1% NP-40) supplemented with 1 mM DTT, 5 mM sodium fluoride, 0.1 mM sodium orthovanadate, and complete protease inhibitor (Roche) and put at 4°C to rotate for 15 min. Nuclear extract lysates were then clarified by high-speed centrifugation. For coimmunoprecipitation analyses, the nuclear extract lysates (800–1,000 µg) were co-incubated at 4°C overnight with either mouse Brca1-specific antiserum (Shakya et al., 2011), antibodies specific for Mre11 (ab397; Abcam), or preimmune serum. The next day, the samples were incubated with protein A–Sepharose beads (20% slurry; GE Healthcare) at 4°C for 1 h, washed three times in buffer (20 mM Hepes, pH 7.9, 10% glycerol, 0.4 M NaCl, 0.5 mM EDTA, 0.5 mM EGTA, and 0.1% NP-40) supplemented with 1 mM DTT, 5 mM sodium fluoride, 0.1 mM sodium orthovanadate, and complete protease inhibitor (Roche), boiled for 4 min in 40 µl of 2× SDS loading buffer (0.1 M Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 0.1% 2-mercaptoethanol, and 0.004% bromophenol blue), and the supernatant was fractionated by electrophoresis. Immunoblotting was then performed with a mouse monoclonal antibody (14-1) raised against human CtIP (Yu and Baer, 2000). For straight Western analyses, the same nuclear extract lysates were fractionated by SDS-PAGE and immunoblotted with a rabbit antiserum raised against mouse Brca1 (Shakya et al., 2011), a mouse monoclonal antibody (14-1) specific for CtIP (Yu and Baer, 2000), a rabbit polyclonal antibody raised against human Mre11 (ab397; Abcam), a goat polyclonal antibody raised against mouse Lamin B (M-20, Santa Cruz Biotechnology, Inc.), and an α-tubulin–specific mouse monoclonal antibody (DM1A; EMD Millipore). To prepare total cell extracts for Western analyses, immortalized MEFs were harvested in 1× PBS and lysed in low salt Nonidet P-40 (NP40) buffer (10 mM Hepes, pH 7.6, 250 mM NaCl, 0.1% NP-40, 5 mM EDTA, and 10% glycerol) supplemented with 1 mM DTT, 50 mM sodium fluoride, 0.5 mM phenylmethanesulfonyl fluoride, and complete protease inhibitor (Roche). Analysis of RPA expression was conducted using a monoclonal antibody that recognizes total RPA2 (Ab-3; EMD Millipore) and a rabbit polyclonal antiserum raised against Ser4/Ser8-phosphorylated RPA2 (A300-245A; Bethyl Laboratories, Inc.). Analyses of Chk1 expression were conducted using a monoclonal antibody that recognizes total Chk1 (G-4; Santa Cruz Biotechnology, Inc.), a rabbit polyclonal antiserum raised against S345-phosphorylated Chk1 (#2341; Cell Signaling Technology), and an α-Na,K-ATPase–specific rabbit polyclonal antibody (RDI-ATPASEabr, Research Diagnostics Inc.).
For karyotyping, metaphase spreads were prepared from primary MEFs treated with 0.05 µg/ml of KaryoMAX Colcemid solution (Gibco/Invitrogen) for 2 h and with or without mitomycin C (40 ng/ml; Sigma-Aldrich) for 16 h before harvest (Reid et al., 2008). For Rad51 and phospho-RPA immunostaining, cells were grown on poly-l-lysine (Sigma-Aldrich)–coated coverslips and fixed at 1 h after IR treatment (10 Gy) in 1:10 diluted 37% formaldehyde/PBS, permeabilized in 1% Triton X-100/PBS, and either extracted (phospho-RPA) in 100 mM Pipes, pH 6.8, 2 mM EGTA, 1 mM MgCl2, and 0.5% Triton X-100 for 5 min and stripped for 5 min in 10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.5% Triton X-100 before blocking, or blocked directly (Rad51) in 5% BSA/PBS. The cells were then stained with rabbit monoclonal antibodies for Thr21-phosphorylated RPA2 (EPR2846(2); Abcam) or rabbit polyclonal antibodies for Rad51 (Ab-1; EMD Millipore). Next, the cells were washed several times in 1× PBS, incubated with Alexa Fluor 488 goat anti–rabbit (Invitrogen) secondary antibody, stained with DAPI (Sigma-Aldrich), and mounted onto a glass slide with Aqua-Poly/Mount medium (Polysciences Inc.). Ctip immunostaining was performed at various time points after IR treatment (10 Gy) by fixing cells grown on poly-l-lysine (Sigma-Aldrich)–coated coverslips in 3.7% paraformaldehyde/PBS, pH 7.4, for 15 min and permeabilizing in 0.5% Triton X-100/net gel (150 mM NaCl, 5 mM EDTA, 50 mM Tris-Cl, 0.05% NP-40, 0.25% Gelatin IV bloom 75, Type B [Sigma-Aldrich], and 0.02% NaN3, pH 7.4) for 10 min. The cells were then stained with a mouse monoclonal antibody (14-1) raised against human CtIP (Yu and Baer, 2000) and further incubated with Alexa Fluor 488 goat anti–mouse (Invitrogen) secondary antibody. The cells were stained with DAPI (Sigma-Aldrich) for 5 min and mounted onto a glass slide with Aqua-Poly/Mount medium (Polysciences Inc.).
To assess the in situ association of Brca1 and Ctip using the Duolink proximity ligation assay (PLA; Olink Bioscience), cells grown on poly-l-lysine (Sigma-Aldrich)–coated coverslips were fixed in 3.7% paraformaldehyde/PBS, pH 7.4, permeabilized in 0.5% Triton X-100/net gel, and coimmunostained with a mouse monoclonal antibody (14-1) specific for Ctip (Yu and Baer, 2000) and a rabbit polyclonal antibody that detects mouse Brca1 (Shakya et al., 2011). Next, the cells were incubated with secondary antibodies conjugated with oligonucleotides (PLA probe anti–mouse MINUS and PLA probe anti–rabbit PLUS), ligation solution consisting of two oligonucleotides and ligase, and amplification solution consisting of nucleotides, fluorescently labeled oligonucleotides (Detection Reagent Red), and Polymerase (Duolink). The cells were then mounted onto a glass slide using Duolink in situ mounting medium with DAPI. Immunostaining analysis and images were acquired at room temperature using a microscope (Axio Imager.Z2; EC Plan-Neofluar 40×/0.75 NA objective lens; Carl Zeiss) equipped with a CoolCube 1 camera (MetaSystems), a motor-controlled stage (MetaSystems), and the Isis and Metafer 4 software packages (MetaSystems). Green (Alexa Fluor 488) and blue (DAPI) images of the same cells were merged using ImageJ software (National Institutes of Health) and the final images were prepared in Adobe Photoshop CS5 extended version 12.0.
Online supplemental material
Fig. S1 shows that the in situ association of Brca1 and Ctip is markedly reduced in CtipS326A/S326A MEFs. Fig. S2 shows that the Ctip–S326A polypeptide interacts with Mre11 and supports Chk1 phosphorylation. Fig. S3 shows that CtipS326A/S326A MEFs are proficient for assembly of Ctip nuclear foci in response to DNA damage. Fig. S4 shows that CtipS326A/S326A MEFs are proficient for assembly of Rad51 nuclear foci in response to DNA damage. Fig. S5 shows that CtipS326A/S326A MEFs are proficient for assembly of RPA nuclear foci in response to DNA damage. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201302145/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201302145.dv.
Supplementary Material
Acknowledgments
We are especially grateful to Jean Gautier, Max Gottesman, Kenta Yamamoto, and Shan Zha for advice and critical comments.
This work was supported by the Mary Kay Foundation (008-10) and Public Health Service grants from the National Cancer Institute (P01-CA097403, R01-CA137023, and R01-CA120954). C.R. Reczek was supported by fellowships from the National Cancer Institute (T32-CA09503) and the DoD Breast Cancer Research Program (BC083089). These studies were facilitated by the Transgenic Mouse, Confocal & Specialized Microscopy, Molecular Cytogenetics and Epigenetics, Molecular Pathology, and Radiation Research Shared Resources of the Herbert Irving Comprehensive Cancer Center at Columbia University Medical Center.
Footnotes
Abbreviations used in this paper:
- CPT
- camptothecin
- DSB
- double-strand DNA break
- ES
- embryonic stem
- ETO
- etoposide
- HDR
- homology-directed repair
- HSV-TK
- herpes simplex virus type 1 thymidine kinase
- IR
- ionizing radiation
- IRIF
- ionizing radiation-induced foci
- MEF
- mouse embryonic fibroblast
- MMC
- mitomycin C
- MMEJ
- microhomology-mediated end joining
- NHEJ
- nonhomologous end joining
- PLA
- proximity ligation assay
- SSA
- single-strand annealing
- Topo
- topoisomerase
References
- Bennardo N., Cheng A., Huang N., Stark J.M. 2008. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4:e1000110 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan M.V., Nominé Y., Yu X., Juranic N., Macura S., Chen J., Mer G. 2004. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 12:1137–1146 10.1016/j.str.2004.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Nievera C.J., Lee A.Y., Wu X. 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283:7713–7720 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- Chen P.L., Liu F., Cai S., Lin X., Li A., Chen Y., Gu B., Lee E.Y., Lee W.H. 2005. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25:3535–3542 10.1128/MCB.25.9.3535-3542.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. 2006. CtIP, a candidate tumor susceptibility gene is a team player with luminaries. Biochim. Biophys. Acta. 1765:67–73 [DOI] [PubMed] [Google Scholar]
- Clapperton J.A., Manke I.A., Lowery D.M., Ho T., Haire L.F., Yaffe M.B., Smerdon S.J. 2004. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 11:512–518 10.1038/nsmb775 [DOI] [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 49:872–883 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M. 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20:34–46 10.1101/gad.1381306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R., de la Pompa J.L., Elia A., Potter J., Mak T.W. 1997. Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nat. Genet. 16:298–302 10.1038/ng0797-298 [DOI] [PubMed] [Google Scholar]
- Hartsuiker E., Neale M.J., Carr A.M. 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell. 33:117–123 10.1016/j.molcel.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M.S., Sy S.M., Chen J. 2010. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11:138–148 10.1038/nrm2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Huang J., Chen J. 2007. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat. Struct. Mol. Biol. 14:710–715 10.1038/nsmb1277 [DOI] [PubMed] [Google Scholar]
- Lee K., Lee S.E. 2007. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 176:2003–2014 10.1534/genetics.107.076539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Greenberg R.A. 2012. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem. Sci. 37:418–424 10.1016/j.tibs.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O., Chahwan C., Yamada Y., de Bruin R.A., Wittenberg C., Russell P. 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 28:134–146 10.1016/j.molcel.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.-Y., Flesken-Nikitin A., Li S., Zeng Y., Lee W.-H. 1996. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 10:1835–1843 10.1101/gad.10.14.1835 [DOI] [PubMed] [Google Scholar]
- Liu Z., Wu J., Yu X. 2007. CCDC98 targets BRCA1 to DNA damage sites. Nat. Struct. Mol. Biol. 14:716–720 10.1038/nsmb1279 [DOI] [PubMed] [Google Scholar]
- Ludwig T., Chapman D.L., Papaioannou V.E., Efstratiadis A. 1997. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11:1226–1241 10.1101/gad.11.10.1226 [DOI] [PubMed] [Google Scholar]
- McKee A.H., Kleckner N. 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 146:797–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Lee S.E. 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24:529–538 10.1016/j.tig.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 455:770–774 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11:196–207 10.1038/nrm2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell. 4:511–518 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Cui T.Y., Jasin M. 2001a. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61:4842–4850 [PubMed] [Google Scholar]
- Moynahan M.E., Pierce A.J., Jasin M. 2001b. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 7:263–272 10.1016/S1097-2765(01)00174-5 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., et al. 2010. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6:e1000828 10.1371/journal.pgen.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette M.L., Lee K., Guo Z., Rani M., Chow J.M., Lee S.E., Paull T.T. 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17:1478–1485 10.1038/nsmb.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 467:108–111 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A., Nussenzweig M.C. 2007. A backup DNA repair pathway moves to the forefront. Cell. 131:223–225 10.1016/j.cell.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Penkner A., Portik-Dobos Z., Tang L., Schnabel R., Novatchkova M., Jantsch V., Loidl J. 2007. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 26:5071–5082 10.1038/sj.emboj.7601916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.E., Li Y., Chait B.T., Gottesman M.E., Baer R., Gautier J. 2011. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J. Cell Biol. 194:705–720 10.1083/jcb.201103103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.J., Hu P., Han M., Ellis N., Jasin M. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15:3237–3242 10.1101/gad.946401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Amon A., Klein F. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 146:781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid L.J., Shakya R., Modi A.P., Lokshin M., Cheng J.-T., Jasin M., Baer R., Ludwig T. 2008. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. USA. 105:20876–20881 10.1073/pnas.0811203106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Chun J., Powell S.N. 2012. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 12:68–78 10.1038/nrc3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel B.P., Jodelka F.M., Nunez R. 2006. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 66:5181–5189 10.1158/0008-5472.CAN-05-3209 [DOI] [PubMed] [Google Scholar]
- Schuermann M. 1990. An expression vector system for stable expression of oncogenes. Nucleic Acids Res. 18:4945–4946 10.1093/nar/18.16.4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R., Reid L.J., Reczek C.R., Cole F., Egli D., Lin C.-S., deRooij D.G., Hirsch S., Kandasamy R., Hicks J.B., et al. 2011. BRCT phosphoprotein recognition, but not E3 ligase activity, is essential for BRCA1 tumor suppression. Science. 334:525–528 10.1126/science.1209909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki E.N., Gu L., Yan N., Shi Y. 2004. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol. Cell. 14:405–412 10.1016/S1097-2765(04)00238-2 [DOI] [PubMed] [Google Scholar]
- Snouwaert J.N., Gowen L.C., Latour A.M., Mohn A.R., Xiao A., DiBiase L., Koller B.H. 1999. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 18:7900–7907 10.1038/sj.onc.1203334 [DOI] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M. 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 24:9305–9316 10.1128/MCB.24.21.9305-9316.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S., Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- Uanschou C., Siwiec T., Pedrosa-Harand A., Kerzendorfer C., Sanchez-Moran E., Novatchkova M., Akimcheva S., Woglar A., Klein F., Schlögelhofer P. 2007. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 26:5061–5070 10.1038/sj.emboj.7601913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A.K., Brown R.S., Birrane G., Ladias J.A. 2005. Structural basis for cell cycle checkpoint control by the BRCA1-CtIP complex. Biochemistry. 44:10941–10946 10.1021/bi0509651 [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 316:1194–1198 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren A.J., Colledge W.H., Carlton M.B.L., Evans M.J., Smith A.J.H., Rabbitts T.H. 1994. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 78:45–57 10.1016/0092-8674(94)90571-1 [DOI] [PubMed] [Google Scholar]
- Williams R.S., Lee M.S., Hau D.D., Glover J.N. 2004. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat. Struct. Mol. Biol. 11:519–525 10.1038/nsmb776 [DOI] [PubMed] [Google Scholar]
- Wong A.K., Ormonde P.A., Pero R., Chen Y., Lian L., Salada G., Berry S., Lawrence Q., Dayananth P., Ha P., et al. 1998. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene. 17:2279–2285 10.1038/sj.onc.1202150 [DOI] [PubMed] [Google Scholar]
- Yang H., Li Q., Fan J., Holloman W.K., Pavletich N.P. 2005. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 433:653–657 10.1038/nature03234 [DOI] [PubMed] [Google Scholar]
- You Z., Bailis J.M. 2010. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 20:402–409 10.1016/j.tcb.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T. 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 36:954–969 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Baer R. 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275:18541–18549 10.1074/jbc.M909494199 [DOI] [PubMed] [Google Scholar]
- Yu X., Chen J. 2004. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24:9478–9486 10.1128/MCB.24.21.9478-9486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273:25388–25392 10.1074/jbc.273.39.25388 [DOI] [PubMed] [Google Scholar]
- Yun M.H., Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jasin M. 2011. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 18:80–84 10.1038/nsmb.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.Y., Sonoda E., Barber L.J., Oka H., Murakawa Y., Yamada K., Ikura T., Wang X., Kobayashi M., Yamamoto K., et al. 2007. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell. 25:663–675 10.1016/j.molcel.2007.01.029 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 134:981–994 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.