Hif-1α is a novel and important regulator of EMT and chemotaxis during migration of neural crest cells.
Abstract
One of the most important mechanisms that promotes metastasis is the stabilization of Hif-1 (hypoxia-inducible transcription factor 1). We decided to test whether Hif-1α also was required for early embryonic development. We focused our attention on the development of the neural crest, a highly migratory embryonic cell population whose behavior has been likened to cancer metastasis. Inhibition of Hif-1α by antisense morpholinos in Xenopus laevis or zebrafish embryos led to complete inhibition of neural crest migration. We show that Hif-1α controls the expression of Twist, which in turn represses E-cadherin during epithelial to mesenchymal transition (EMT) of neural crest cells. Thus, Hif-1α allows cells to initiate migration by promoting the release of cell–cell adhesions. Additionally, Hif-1α controls chemotaxis toward the chemokine SDF-1 by regulating expression of its receptor Cxcr4. Our results point to Hif-1α as a novel and key regulator that integrates EMT and chemotaxis during migration of neural crest cells.
Introduction
Hypoxia, a reduction in the normal level of tissue oxygen tension, occurs in several pathologies, such as vascular and pulmonary diseases, as well as in cancer. Although hypoxia is a challenge to both cancer and normal cells, cancer cells undergo genetic and adaptive changes that allow them to thrive in a hypoxic environment. Some of these adaptive changes are likely related to genetic programs that are used by normal cells, usually during embryo development. Hypoxia influences many aspects of the biology of tumors, including cell survival (Graeber et al., 1996), suppression of apoptosis (Erler et al., 2004), autophagy (Rouschop et al., 2010), and the anabolic switch in central metabolism (Cairns et al., 2011). Importantly, hypoxia also promotes migration of tumor cells by enhancing receptor tyrosine kinase–mediated signaling (Wang and Ohh, 2010), tumor angiogenesis (Semenza, 2000), vasculogenesis (Kioi et al., 2010), epithelial to mesenchymal transition (EMT; Hill et al., 2009), invasiveness (Pennacchietti et al., 2003), and metastasis (Chang et al., 2011).
A key factor involved in the cell response to reduced oxygen levels is hypoxia-inducible transcription factor (HIF; Semenza et al., 1991). HIF is a heterodimer that consists of a hypoxic response α subunit and the constitutively expressed subunit HIF-1β. In the presence of oxygen, HIF-α is bound to the von Hippel–Lindau tumor suppressor protein. This interaction causes HIF-α to become ubiquitylated and targeted to the proteasome, where it is degraded (Cockman et al., 2000; Kamura et al., 2000; Ohh, et al., 2000; Tanimoto et al., 2000; Semenza, 2010). Under hypoxic conditions, HIF-α becomes stable and binds HIF-1β to form the active HIF transcription factor, which binds to hypoxia-response elements activating the expression of numerous hypoxia-responsive genes. Stabilization of HIF-α in cancer cells is believed to be one of the most important mechanisms that promotes metastasis and, thereby, increases tumor aggressiveness (Harris 2002; Semenza, 2002; Gupta and Massagué, 2006).
Programmed cellular migration is a major feature of embryonic development. We hypothesized that the role of HIF in promoting metastasis in tumors might be related to a role for this pathway in developmental migration programs. The neural crest is a highly migratory and multipotent embryonic cell population, whose behavior has been likened to malignant invasion (Kulesa et al., 2006; Hendrix et al., 2007; Kuriyama and Mayor, 2008). Neural crest cells are formed at the border of the neural plate and subsequently leave the neuroepithelium during a delamination phase to migrate all over the embryo, where they differentiate in many cell types, including some of the cartilages and bones of the head and neck, neurons, and glial cells of the peripheral nervous system, endocrine cells, smooth muscle cells, tendons, and pigment cells (Steventon et al., 2005; Theveneau and Mayor, 2011). Cancer metastasis and neural crest cell migration share many cellular and molecular features. Neural crest and malignant cancer cells undergo an EMT controlled by genes of the Snail, Sox, Twist, and Ets families (Theveneau and Mayor, 2011). In addition, guidance molecules controlling neural crest cell migration, such as SDF-1, are also involved in the homing of cancer cells into specific organs (Kucia et al., 2005). These similarities suggest that cancer cells may have hijacked the neural crest development program, which would make neural crest cells a good model to understand the dynamics of cancer progression.
Here, we show that Hif-1α is essential for neural crest migration in zebrafish and Xenopus laevis embryos. We use the advantages of these animal models to characterize the cell behavior that depends on Hif-1α and on hypoxic conditions and identify some of the downstream targets that are regulated by Hif-1α during neural crest migration. Our analysis of neural crest migration in vivo and in vitro reveals Hif-1α as a crucial regulator of EMT and chemotaxis in the neural crest cells during embryo development.
Results
Hif-1α is required for neural crest migration
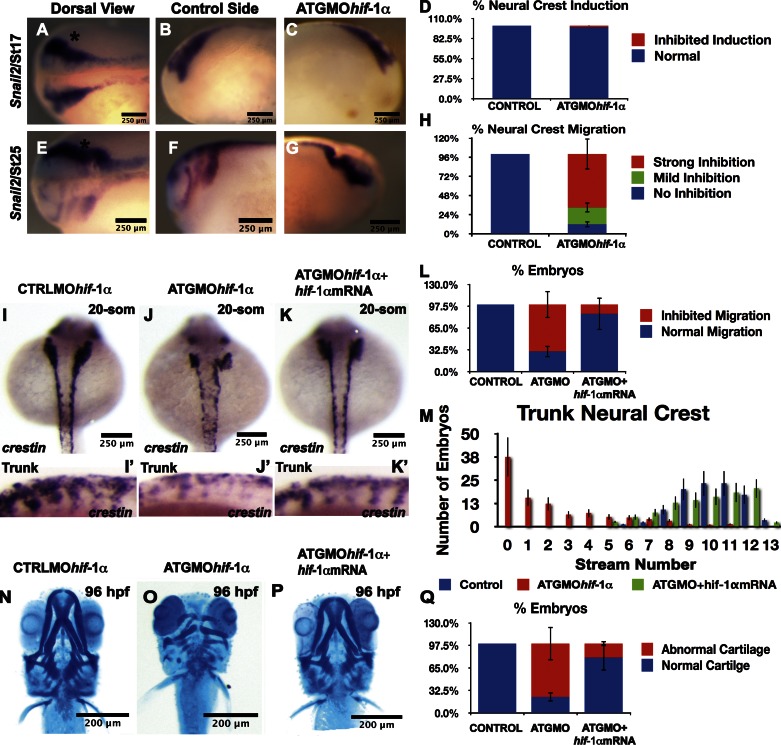
To test the role of Hif-1α on neural crest development, we proceeded to inhibit its activity in Xenopus and zebrafish embryos, as these two animal models offer complementary advantages to study neural crest cells. Injection of a previously characterized morpholino (MO) against Xenopus Hif-1α (Nagao et al., 2008) did not affect neural crest induction, as shown by the normal expression of the neural crest marker snail2 (Fig. 1, A–D). However, a strong inhibition in the formation of the cephalic neural crest streams was observed in embryos injected with Hif-1α MO (Fig. 1, E–H), which is consistent with a requirement of Hif-1α on neural crest migration.
Figure 1.
Hif-1α is required for neural crest migration. (A–Q) Xenopus (A–H) and zebrafish (I–Q) embryos were injected with antisense MOs as indicated in the figure, and the expression of the neural crest markers Snail2 (A–H) or crestin (I–M) was analyzed. Asterisks show the injected side. (A–D) No effect of ATGMOhif-1α on neural crest induction. (E–H) Strong effect of ATGMOhif-1α on neural crest migration. (I–J′) Dorsal (I and J) or lateral (I′ and J′) view of zebrafish embryos. (J) Note that in ATGMOhif-1α–injected embryos, neural crests are present around to the dorsal midline. (L) Percentage of embryos with the described phenotype. (M) The number of neural crest streams was counted by analyzing the expression of crestin for each treatment. (K, K′, and M) Note that the number of streams is reduced with the ATGMOhif-1α but rescued by the coinjection of Hif-1α mRNA. (N–Q) Analysis of cartilage development in zebrafish embryos. The bars in D, H, L, M, and Q histograms represent the standard deviations of three independent experiments, with ∼40 embryos used in each case. som, somite.
We inhibited Hif-1α in zebrafish using two different MOs: an upstream MO that overlaps the ATG region (ATGMOhif-1α) and an MO against the splicing acceptor between exons 2 and 3, splice site (SS) MOhif-1α. To test the efficiency of the ATGMOhif-1α, the 5′ region of hif-1α was fused to GFP, and the mRNA of the fused protein was injected into zebrafish embryos; this showed the expected fluorescence (Fig. S1 A). This fluorescence was dramatically reduced when ATGMOhif-1α, but not when SSMOhif-1α, was coinjected with the fusion protein (Fig. S1, B and C), indicating the ability of the ATG MO to block translation of Hif-1α–GFP protein. To test the efficiency of SSMOhif-1α to block splicing, we used RT-PCR to amplify the intron between exons 2 and 3, which should be absent after normal splicing (Fig. S1 D, lane 1); however, a clear amplification product was observed when SSMOhif-1α was injected (Fig. S1 D, lane 2). In addition, we amplified the Hif-1α mRNA, which has an expected size of 2.3 kb (Fig. S1 D, lane 3). When the splicing between exons 2 and 3 is disrupted by SSMOhif-1α, the expected fragment cannot be amplified by RT-PCR because of its large size, as shown by the absence of a band in lanes 4 and 5 of Fig. S1 D. Finally, we checked the ability of these two MOs to inhibit the expression of well-characterized Hif-1α targets, such as vegf and aldo (aldolase). These were present in zebrafish embryos injected with a control MO but absent after ATG or SSMOhif-1α injection (Fig. S1 E). Together, these experiments validate the use of the zebrafish MOs characterized here to study the effect of Hif-1α inhibition.
Different doses of the two MOs were injected into zebrafish embryos, and their development was analyzed. General morphology and development was normal, and no effect on neural crest induction, as analyzed by the expression of the neural crest markers foxd3 and ap2, was observed (Fig. S2, A–F). However, a clear effect on neural crest migration was observed by analyzing the migratory crest marker crestin (Fig. 1, I–K). The normal streams of trunk neural crest expressing crestin (Fig. 1, I and I′) were severely reduced after ATGMOhif-1α injection in embryos of the same age, as determined by the number of somites (Fig. 1, J and J′). It is important to note that a lateral view of Hif-1α morphant embryos can give a misleading impression: although these embryos appear to have a reduction in crestin expression (Fig. 1 J′), careful examination of a dorsal view revealed that, whereas similar in number, many crestin-positive cells were unable to emigrate from the dorsal midline (Fig. 1 J), consistent with a failure in migration. We counted the number of neural crest streams in control and ATGMOhif-1α embryos. Inhibition of Hif-1α caused a significant reduction in the number of neural crest streams (Fig. 1 M, compare blue and red bars). Importantly, both the number of embryos with reduced neural crest migration and the number of neural crest streams per embryo were rescued by coinjection of Hif-1α mRNA that does not bind to the ATGMOhif-1α (Fig. 1, K–M). As expected, neural crest derivatives, such as melanocytes (unpublished data), cranial ganglia (unpublished data), and cartilage, were affected by Hif-1α MO. Cartilage was severely distorted by ATGMOhif-1α injection (Fig. 1, N and O) and SSMOhif-1α injection (not depicted). This phenotype could be rescued efficiently by coinjection of a Hif-1α mRNA (Fig. 1, P and Q). These findings establish that Hif-1α is required for cephalic and trunk neural crest migration and development of neural crest derivatives.
Hif-1α is required within the neural crest to control their migration
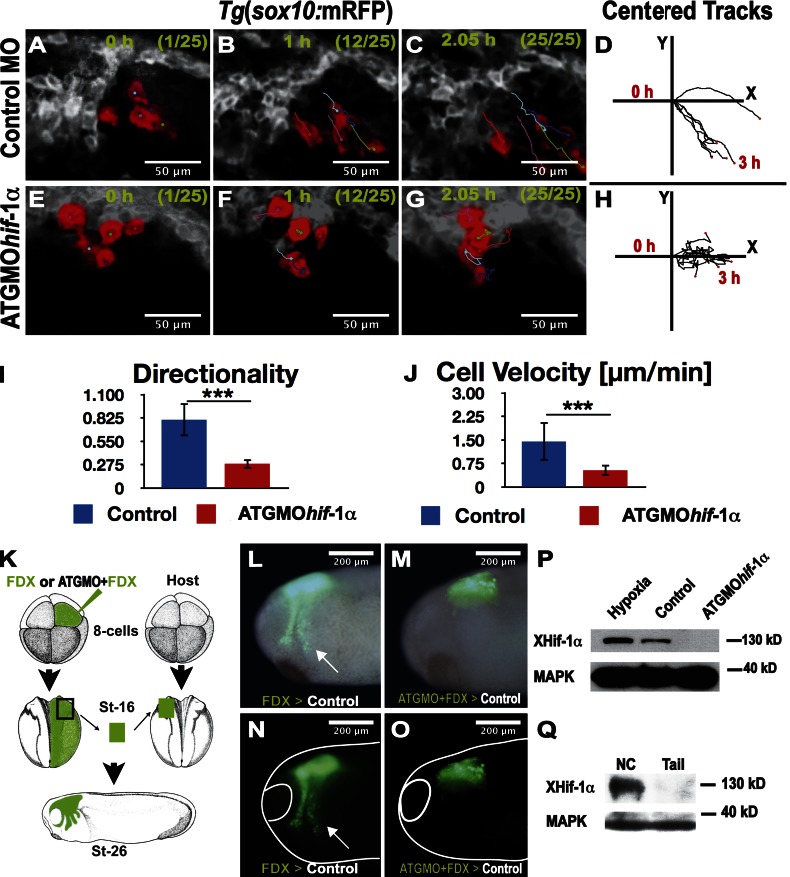
To observe the behavior of neural crest cells in the living embryo, we took advantage of the transgenic zebrafish line Tg(sox10:mRFP), which expresses membrane-RFP in the neural crest (Kirby et al., 2006). In contrast to control MO-injected embryos, embryos injected with ATGMOhif-1α had severe defects in neural crest migration (Video 1). Tracks of leader cells that can be easily individualized in the transgenic embryos (Fig. 2, A–H) show reduced directionality and velocity of migration (Fig. 2, I and J). This finding mirrors our observation of defective neural crest migration and supports the notion that Hif-1α is required for migration. However, as Hif-1α is ubiquitously expressed at these embryonic stages (de Beaucourt and Coumailleau, 2007; Rojas et al., 2007), it is possible that the effect on the aforementioned neural crest migration could be caused by an effect of Hif-1α MO on the development of another tissue required for normal crest migration. To test this possibility, we performed two experiments. First, we analyzed the expression in zebrafish embryos of several neural plate border and mesodermal markers after ATG or SS Hif-1α MO injection. No effect in the expression of the mesodermal markers gsc, ntl, or myod or the neural plate border marker dlx-3 was observed (Fig. S2, G–O). Second, Xenopus embryos were injected with fluorescein dextran (FDX) and MO at eight-cell stage; at neurula stage, the neural crest was transplanted from injected into noninjected embryos, and neural crest migration was analyzed 8–10 h later (Fig. 2 K). This generates embryos in which all the tissues are normal except the neural crest, which comes from the injected embryo. Controls injected with FDX show normal neural crest migration (Fig. 2, L and N, arrowheads), whereas coinjection with ATGMOhif-1α completely blocks migration (Fig. 2, M and O). In summary, inhibition of Hif-1α does not affect any neural or mesodermal tissue tested here, and it is not required for neural crest induction. However, Hif-1α is required for neural crest migration.
Figure 2.
Hif-1α is required cell autonomously for neural crest migration. (A–H) Frames of a time-lapse video (Video 1) of zebrafish embryos Tg(sox10:mRFP). Leader cells are shown pseudocolored in red for embryos injected with a control MO (A–C) or an ATGMOhif-1α (E–G). (D and H) Centered tracks of leader cells. (I and J) Analysis of cell directionality and velocity of leader cells under indicated conditions. Errors bars represent standard deviation of three independent experiments, with a minimal of 20 embryos in each case. ***, P < 0.005. (K–O) Xenopus graft experiments. (K) FDX/MOs were injected into the ectoderm of blastula embryos. At early neurula stages (St), the injected neural crest was grafted into noninjected embryos, and neural crest migration was analyzed in vivo. (L and N) Normal neural crest migration (n = 10, 80% migration). Arrows show migrating neural crest. (M and O) Inhibition of neural crest migration after ATGMOhif-1α injection (n = 10, 20% migration). (P) Western blot of whole neurula Xenopus embryos, under the indicated conditions. (Q) Western blot of cephalic Xenopus regions enriched in neural crest cells versus tail of young tadpoles that contain very few neural crest cells.
The expression of Hif-1α mRNA has been found to be low but ubiquitous in the embryo, including the neural crest territory (de Beaucourt and Coumailleau, 2007; Rojas et al., 2007). However, because Hif-1α is regulated posttranscriptionally at the level of protein stability, it is possible that Hif-1α protein may not be present in the neural crest cells at the time of their migration. To test this possibility, we performed Western analysis for Hif-1α. Western blot for Hif-1α using whole Xenopus embryos at neurula stage shows a clear band of the expected molecular mass (∼130 kD), which is absent after injecting the ATGMOhif-1α and which increases after incubating the embryos under hypoxic conditions (Fig. 2 P). Next, we used Western blot analysis to compare the presence of Hif-1α protein between two tissues that differ in their neural crest content: cephalic neural crest from Xenopus neurula and the tail, which lack neural crest at these stages of Xenopus development. A clear difference was observed (see Fig. 4 Q), consistent with Hif-1α protein being present in migrating neural crest.
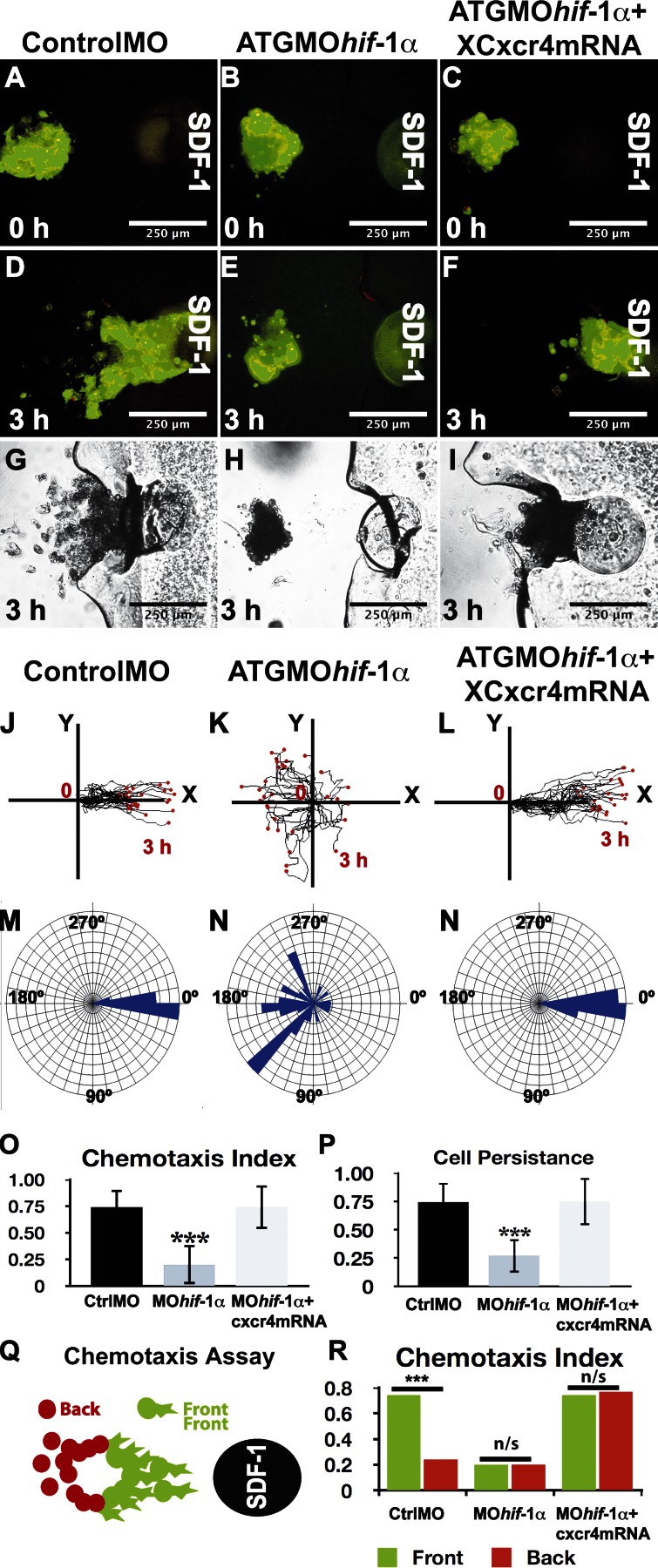
Figure 4.
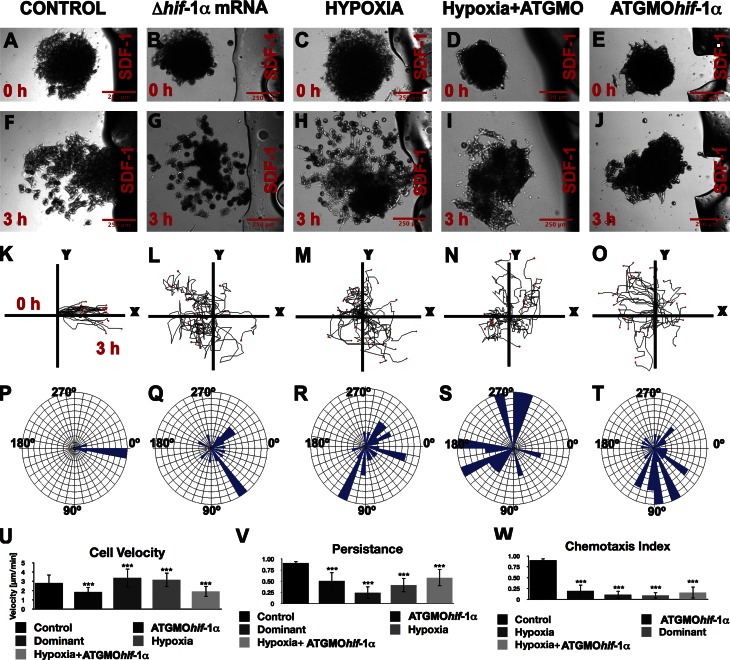
Hif-1α controls chemotaxis in a Cxcr4-dependent manner. Chemotaxis toward a localized source of SDF-1, which is located to the right of each panel, for each treatment as indicated at the top of the figure. (A–C) Time 0. (D–I) 3 h after culture. (A–I) Fluorescent (A–F) and bright-field (G–I) images. (J–L) Tracks of leader migrating cells. Chemotactic cells should move toward the right side of the panel, where SDF-1 is higher. (M and N) Angles of migration for individual cells. (O) Chemotaxis index for leader cells. (P) Persistence. (Q) Leader cells (green) are more efficiently attracted than trailing cells (red). (R) Chemotaxis of leader (green) and trailing (orange) cells under the indicated conditions. Bars represent the standard deviation from three independent experiments, with a minimal of 60 cells analyzed for each treatment. ***, P < 0.005. Ctrl, control.
Hif-1α controls the expression of Twist and Cxcr4 during neural crest migration
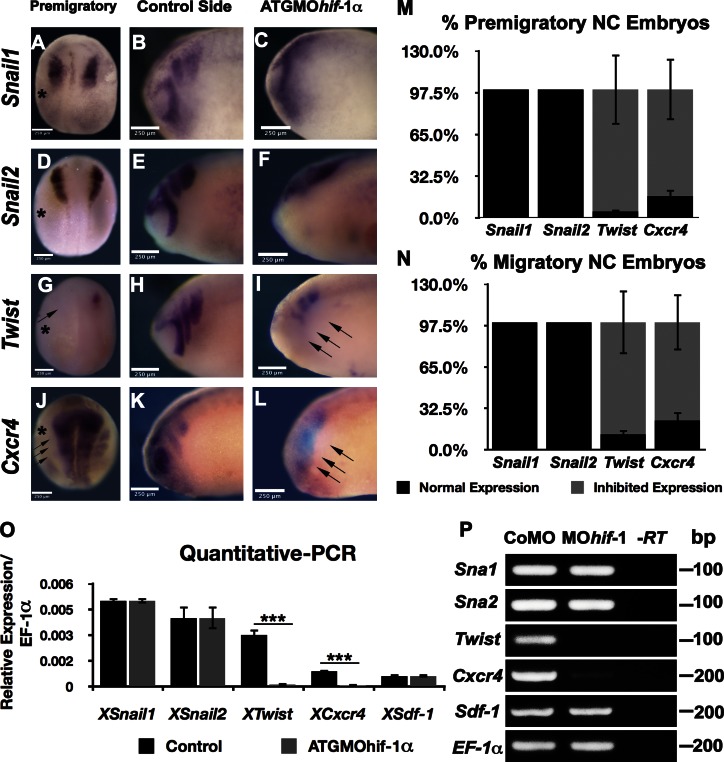
Studies in cancer have shown that Hif controls the expression of numerous genes, many of which are also important for driving neural crest development and migration. For example, it has been reported that HIF acts as a key regulator of EMT in cancer cells by up-regulating the expression of several genes, such as SNAIL, Zeb1, SIP1, E47, and TWIST (Cacheux et al., 2001; Gort et al., 2008; Yang et al., 2008) and that it plays a central role in chemotaxis by up-regulating the expression of the chemokine receptor Cxcr4 (Staller et al., 2003). Because several of these genes, including Snail1, Snail2, Twist, and Cxcr4, are indispensable for the proper development of the neural crest (Olesnicky Killian et al., 2009; Betancur et al., 2010; Theveneau et al., 2010), one might suppose that Hif-1α controls neural crest development by a similar mechanism to the one it uses to enable cancer cell metastasis. To investigate this possibility, we examined by RT-PCR, quantitative PCR, and in situ hybridization the expression of these genes in embryos lacking Hif-1α. Surprisingly, inhibition of Hif-1α by ATG MO has no effect on the expression of Snail1 or Snail2 in the neural crest, either at premigratory stages (Fig. 3, A, D, and M) or during neural crest migration (Fig. 3, B, C, E, F, and N–P). In contrast, blocking Hif-1α has a dramatic effect on Twist expression in the neural crest: expression of Twist is completely abolished before neural crest migration begins (Fig. 3, G and M) and is severely reduced at migratory stages (Fig. 3, H, I, and N–P). Similarly, injection of Hif-1α MO in Xenopus leads to complete inhibition of Cxcr4 expression in the premigratory neural crest (Fig. 3, J and M, arrows), and this inhibition persists in migrating neural crest (Fig. 3, K, L, and N–P). This finding is interesting in light of the fact that, in addition to its expression in the neural crest of Xenopus and zebrafish embryos (Olesnicky Killian et al., 2009; Theveneau et al., 2010), Cxcr4 expression is also found in the neural plate and hindbrain region (Fig. 3 J). Yet the MO had no effect on the neural plate expression of Cxcr4, demonstrating the specificity of the effect of Hif-1α on neural crest cells. Together, these observations show that Hif-1α up-regulates the expression of the EMT gene Twist and the SDF-1 receptor Cxcr4.
Figure 3.
Twist and Cxcr4, but not Snail1 or Snail2, are Hif-1α targets in the neural crest. (A–L) Xenopus embryos. In situ hybridization for Snail1, Snail2, Twist, and Cxcr4 after ATGMOhif-1α injection. Asterisks show the injected side. (A–F) Snail1 and Snail2 expression. Note that neither Snail1 nor Snail2 are affected by inhibition of Hif-1α, but a clear inhibition of neural crest migration is observed. (G–I) Twist expression is lost after ATGMOhif-1α injection (arrows). (J–L) Cxcr4 expression. (J) Note that strong expression in the neural plate is not affected, whereas the neural crest expression is visible only in the right (noninjected) side as three masses next to the neural plate. Arrows indicate absence of gene expression in the neural crest at the injected side. (M and N) Quantification of results shown in A–L. Bars represent the standard deviation from three independent experiments, a minimal of 40 embryos was analyzed for each case. (O and P) Quantitative PCR (O) and RT-PCR (P) of neural crest dissected from embryos injected with a control MO or with ATGMOhif-1α genes analyzed as indicated. EF-1α was used as a loading control. Bars represent the standard deviation from three independent experiments. A minimal of 30 embryos was analyzed for each case. ***, P < 0.005.
Hif-1α regulates neural crest chemotaxis by up-regulating the expression of Cxcr4
The inhibition of Cxcr4 expression by ATGMOhif-1α suggests that Hif-1α is required for neural crest chemotaxis. It has recently been shown that directional migration of neural crest is controlled, at least partially, by chemotaxis toward the chemokine SDF-1 (Theveneau et al., 2010). We have developed a chemotaxis assay in which neural crest cell explants are exposed to a localized source of SDF-1 and their migratory behavior monitored in vitro (Theveneau and Mayor, 2011). In this assay, normal control neural crest explants are attracted by SDF-1 (Fig. 4, A, D, G, J, and M; and Video 2) as previously described (Theveneau et al., 2010). Explants from ATGMOhif-1α–injected embryos, in contrast, failed to migrate toward the SDF-1 source (Fig. 4, B, E, H, K, and N; and Video 2), an effect consistent with the inhibition of the SDF-1 receptor Cxcr4 described in Fig. 3. To test whether the impaired chemotaxis induced by Hif-1α is caused by the absence of Cxcr4, we coinjected the ATGMOhif-1α together with Cxcr4 mRNA. A clear rescue of the chemotaxis was observed in the coinjected neural crest (Fig. 4, C, F, I, L, and N; and Video 2). Careful quantification of chemotaxis index and cell persistence confirmed that Hif-1α inhibition blocks chemotaxis and that this can be rescued by coinjection with Cxcr4 mRNA (Fig. 4, O and P). Thus, Hif-1α makes neural crest cells responsive to SDF-1 by up-regulating the expression of Cxcr4.
In our chemotaxis assays, we have observed that within an explant, cells at the leading front, the so-called “leader cells,” display different motile behavior than those at the back, presumably because of differences in local chemoattractant exposure between these two sites. Indeed, under control conditions, many trailing cells are left behind when the front cells move toward the chemoattractant (Fig. 4, G, Q, and R). Interestingly, we noticed a different behavior in explants from embryos coinjected with Hif-1α MO and Cxcr4 mRNA: unlike explants from control MO-injected embryos, in which only the leader cells exhibited efficient chemotaxis, the front and back of explants coinjected with Hif-1α MO and Cxcr4 mRNA moved equally efficiently toward the SDF-1 source (Fig. 4, I and R), suggesting that Hif-1 could be controlling cell clustering (see next section).
Next, we analyzed chemotaxis in Hif-1α gain-of-function experiments. Unexpectedly, activation of Hif-1α activity by overexpressing a constitutively active form of the protein (Nagao et al., 2008) or by hypoxia treatment impaired SDF-1 responsiveness (Fig. 5, A–C, F–H, K–M, P–R, and U–W; and Video 3). In addition, both treatments induce cell dispersion (Fig. 5, G and H). The unexpected loss of chemotaxis by activation of Hif-1α can be explained by the increased cell dissociation observed under this treatment. It is known that chemotaxis toward SDF-1 requires cell clustering, as it is severely decreased in single isolated cells (Theveneau and Mayor, 2010; Theveneau et al., 2010), which is likely to explain the failure to chemotax toward SDF-1 when cell dispersion is induced by Hif-1α activation. Alternatively, activation of Hif-1α is likely to induce an increase in Cxcr4 expression, which could lead to a hypermotility with no cell directionality. Interestingly, the dispersion phenotype induced by hypoxia is reversed by ATGMOhif-1α (Fig. 5, C–E, H–J, M–O, R–T, and U–W; and Video 3), suggesting that Hif-1α stability is oxygen dependent in the neural crest and that oxygen levels need to be tightly controlled, as an excess or deficit of Hif-1 can affect neural crest migration.
Figure 5.
Hif-1α gain of function induces cell dispersion and inhibits chemotaxis. Chemotaxis was analyzed as described in Fig. 4 for each treatment as indicated at the top of the figure. (A–E) Time 0. (F–J) 3 h after culture. (K–O) Tracks of migrating cells. (P–T) Angles of migration for individual cells. (U) Cell velocity. (V) Persistence. (W) Chemotaxis index. Each experiment was performed at least three times, and ∼60 cells were analyzed for each treatment. ***, P < 0.005. Errors bars represent standard deviation.
Hif-1α regulates neural crest dissociation by up-regulating Twist expression
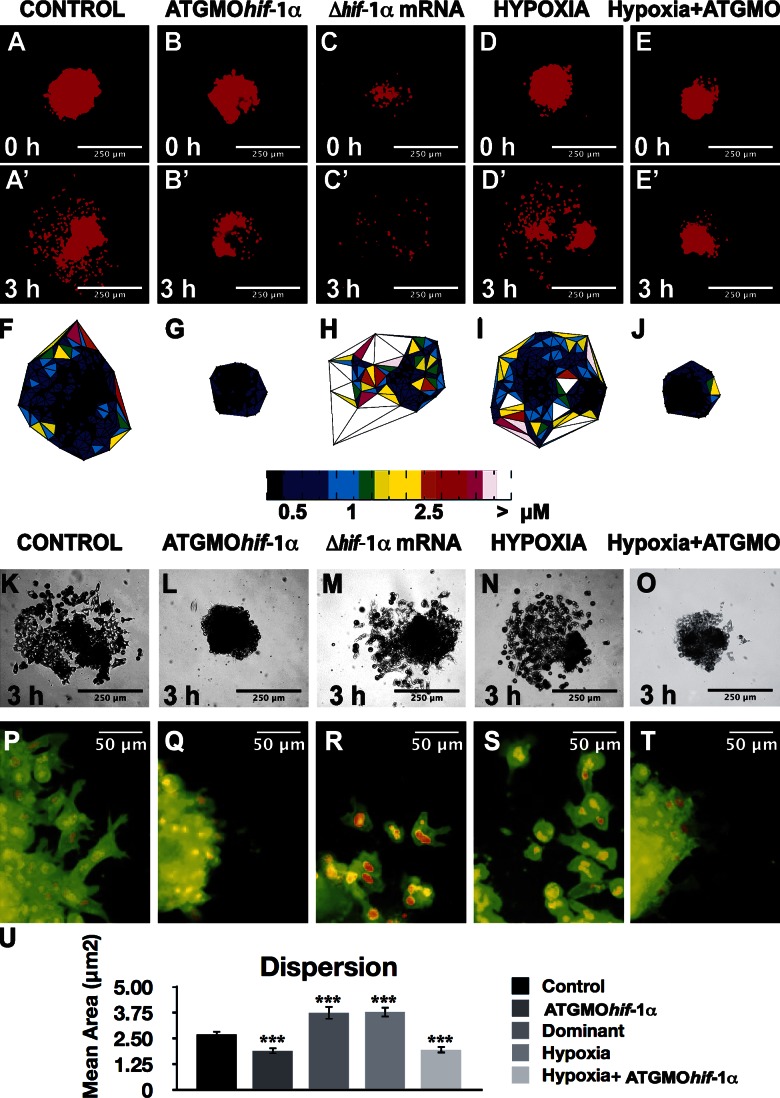
Loss and gain of Hif-1α function leads to cell clustering and dispersion, respectively, suggesting that in addition to regulating neural crest chemotaxis, Hif-1α could also regulate neural crest dispersion. Migration of neural crest requires an EMT, in which cell adhesion is decreased (Thiery et al., 2009). This EMT can be visualized in vitro, as cells disperse after a decrease in cell adhesion (Alfandari et al., 2003). When control neural crests taken from stage 16 neurula embryos are cultured in vitro, they start their dispersion after 3 h, and this continues for ≥8 h. To analyze this cell behavior, neural crest cells cultured in vitro were labeled with nuclear-RFP and membrane-GFP. To quantify the dispersion independent of cell number or density variation, we determined the closest neighbors of each cell using a Delaunay triangulation algorithm (Carmona-Fontaine et al., 2011). Control explants showed normal, moderate dispersion (Fig. 6, A and F; and Video 4), which was strongly reduced by the ATG MO (Fig. 6, B and G; and Video 4). Strikingly, expression of a constitutively active form of Hif-1α (Nagao et al., 2008) or exposure of the explants to hypoxia increased cell dispersion (Fig. 6, C, H, D, and I; and Video 4). This cell dispersion promoted by hypoxia requires Hif-1α, as it is completely blocked if the crest is obtained from embryos injected with the ATG MO (Fig. 6, E and J; and Video 4). The dispersion analyzed by looking at the nuclear-RFP fluorescence was consistent with the observation of the bright-field images (Fig. 6, K–O) and the morphology of single cells labeled with membrane-GFP (Fig. 6, P–T). Finally, quantification and statistical analysis of Delaunay triangulations show that inhibition of Hif-1α significantly inhibits cell dispersion, whereas activation of Hif-1α or hypoxia promotes it (Fig. 6 U).
Figure 6.
Hif-1α promotes neural crest dispersion. Neural crest cells labeled with nuclear-RFP and membrane-GFP were cultured in vitro for 3 h, and cells dispersion was analyzed. (A–E) Nuclear labeling is shown at time 0 and after 3 h. (F–J) Delaunay triangulation shown for a 3-h time to analyze the distance between neighbor cells. Color coded according to the area of the triangles. (K–O) Bright-field images after the indicate treatments. (P–T) Membrane-GFP merged with nuclear-RFP to observe cell morphology, after the indicated treatments. (P) Quantification of Delaunay triangulation for all the conditions analyzed. ***, P < 0.005. Errors bars represent standard deviation, calculated from three independent experiments, with a minimal of 60 cells for each condition.
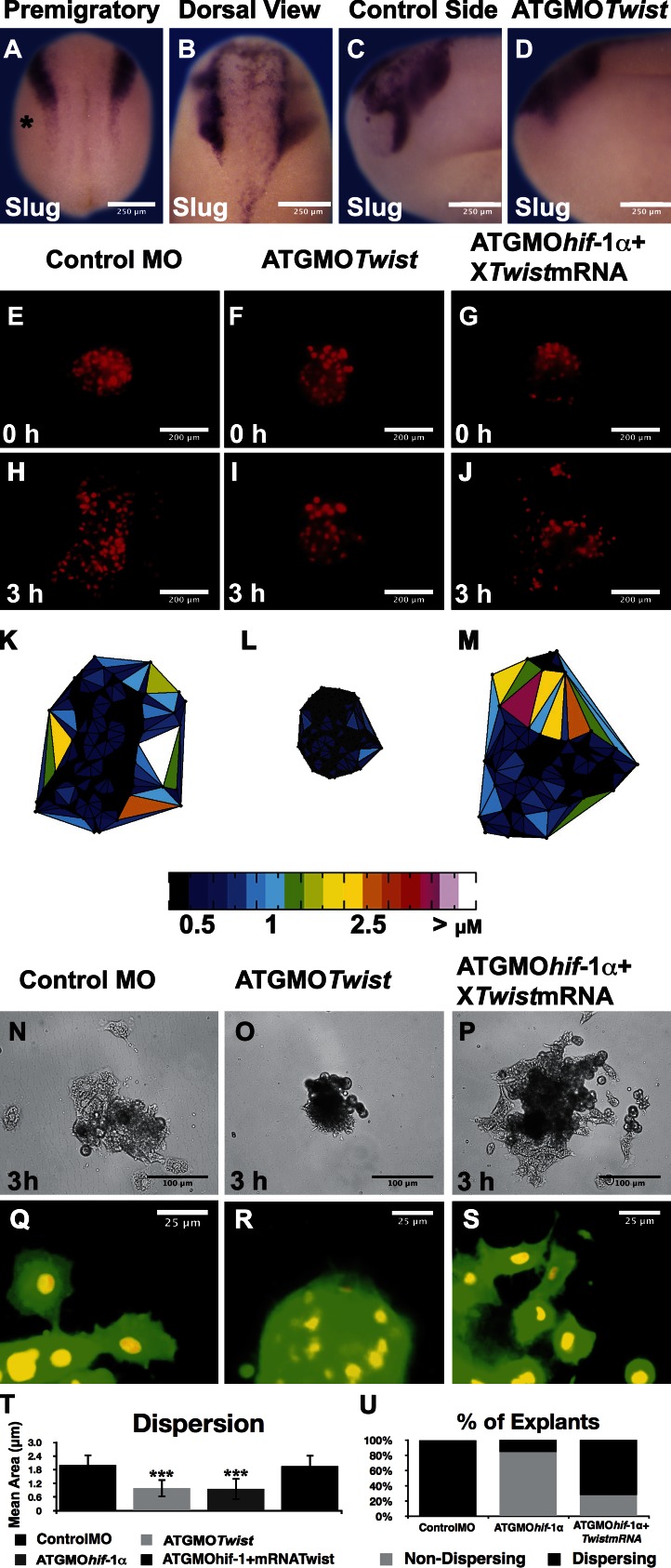
As we showed that Hif-1α controls Twist transcription and Twist has been implicated in EMT of cancer cells (Yang et al., 2008), we decided to study further the role of Twist on neural crest migration and its relationship with Hif-1α. Different doses of a previously characterized antisense MO against Twist (ATGMOtwist; Zhang and Klymkowsky, 2009) were injected into the 16- and 32-cell stage of Xenopus embryos, to avoid any effect on the mesoderm (Zhang and Klymkowsky, 2009). No effect on neural crest induction was observed under these conditions (Fig. 7 A); however, the same treatment lead to severe inhibition of neural crest migration (Fig. 7, B–D). Normal neural crest dissociation observed in vitro is strongly suppressed in explants injected with ATGMOtwist (Fig. 7, E, F, H, I, K, L, N, O, Q, and R). Importantly, inhibition of cell dispersion produced by ATGMOhif-1α is efficiently rescued by coinjection of Twist mRNA (Fig. 7, G, J, M, P, and S). Quantification and statistical analysis of Delaunay triangulations show that both loss of function of Hif-1α or Twist produce a similar inhibition in cell dispersion; however, the effect of loss of Hif-1α can be rescued by coinjection of Twist mRNA (Fig. 7, T and U). These results indicate that cell dispersion controlled by Hif-1α depends on the transcriptional up-regulation of Twist.
Figure 7.
Hif-1α controls cell dispersion in a Twist-dependent manner. (A–D) In situ hybridization against the neural crest marker Snail2. (A) Injection of ATGMOTwist into the prospective neural crest at the 16- and 32-cell stage embryo does not affect neural crest induction. Asterisk shows the injected side. (B–D) Injection of ATGMOTwist blocks neural crest migration. (E–S) Analysis of dispersion of cultured neural crest cells as described in Fig 6. (E–J) Nuclear labeling is shown at time 0 and after 3 h. (K–M) Delaunay triangulation shown for 3 h time to analyze the distance between neighbor cells. Color coded according to the area of the triangles after the indicate treatments. (N–P) Bright-field images after the indicated treatments. (Q–S) Membrane-GFP merged with nuclear-RFP to observe cell morphology. (T) Quantification of Delaunay triangulation for all the conditions analyzed. ***, P < 0.005. Errors bars represent standard deviation of three independent experiments, with a minimal of 60 cells analyzed for each condition. (U) Percentage of explants exhibiting cell dispersion.
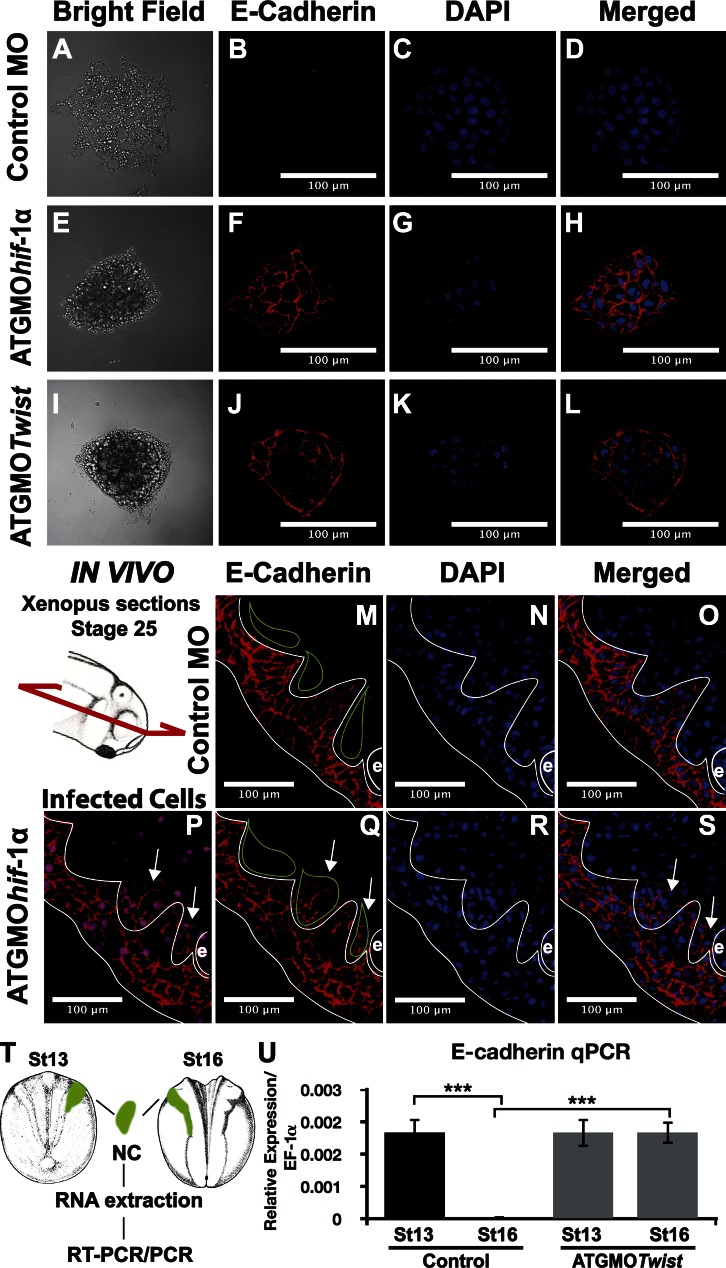
How is Twist controlling neural crest dispersion? It has been reported that Twist could be a repressor of E-cadherin in cancer cells (Yang et al., 2004); however, migrating neural crest do not express E-cadherin, instead they express N-cadherin, which is essential for its migration (Theveneau et al., 2010). Inhibition of Hif-1α or Twist by antisense MO injection does not affect N-cadherin expression (not depicted), but it has a strong effect on E-cadherin (Fig. 8, A–L). Control neural crest explants do not express E-cadherin (Fig. 8, A–D); however, ATGMOhif-1α and ATGMOTwist lead to an up-regulation of E-cadherin at the cell contact, as visualized by immunofluorescence (Fig. 8, E–L). A similar observation was made in vivo. Although control neural crest lack any E-cadherin expression (Fig. 8, M–O, bright green outlines), embryos injected with ATGMOhif-1α exhibit a clear up-regulation of E-cadherin in the neural crest (Fig. 8, P–S, arrows). Although neural crest does not express E-cadherin during development, our results suggest that prospective neural crest expresses it transiently and that the inhibition of E-cadherin in the neural crest required for proper EMT is Twist dependent. To test this idea, we performed a temporal analysis of E-cadherin in neural crest taken from embryos at different stages (Fig. 8, T and U). Prospective neural crest (stage 13) shows a clear expression of E-cadherin, which disappears at later stages (stage 16; Fig. 8 U). However, this disappearance of E-cadherin is suppressed in neural crest injected with ATGMOTwist (Fig. 8 U). Our results show that the repression of E-cadherin required for neural crest EMT is dependent on the Hif-1α/Twist axis.
Figure 8.
Hif-1α/Twist are repressors of E-cadherin in the neural crest. (A–L) Immunofluorescence against E-cadherin performed in neural crest cultured in vitro. (M–S) Histological sections of immunofluorescence against E-cadherin performed in whole embryos. The diagram shows the orientation of the sections. White outlines show epidermis and placodes; green outlines show three streams of cephalic neural crest; e, eye. Red staining shows E-cadherin; blue staining shows DAPI; purple staining in P and S shows cells injected with ATGMOhif-1α. Arrows show neural crest. Note that some of the neural crests injected with the MO show E-cadherin staining (arrows in Q). (T and U) Temporal analysis of E-cadherin expression. Prospective neural crest (stage 13) and premigratory neural crest (stage 16) were dissected, and quantitative PCR was performed for E-cadherin and EF-1α (loading control). Bars represent the standard deviation from three independent experiments. ***, P < 0.005. NC, neural crest; qPCR, quantitative PCR; St, stage.
Discussion
Here, we show that Hif-1α is essential for neural crest migration in zebrafish and Xenopus embryos. Inhibition of Hif-1α activity by antisense oligonucleotide MOs leads to a strong, and tissue-specific, effect on neural crest migration. Importantly, neural crest induction is not disrupted by any of these treatments, and Hif-1α seems specifically to control cell migration. Consistently, the Hif-1α protein is present in migrating neural crest, as determined by Western blot. We demonstrated that Hif-1α is needed for normal cell dissociation during EMT of neural crest cells as well as for the response to the external chemoattractant SDF-1. We identified Twist and Cxcr4 as Hif-1α targets, whose down-regulation by blocking Hif-1α activity explains the in vivo and in vitro neural crest phenotypes. Furthermore, we show that Twist represses E-cadherin in the premigratory neural crest cells, allowing them to undergo normal EMT.
Hif-1α is known to have numerous targets in different tissues, and it is likely that other genes, in addition to Twist and Cxcr4 reported here, are also regulated by Hif-1α activity. However, the down-regulation of these two is sufficient to explain the major neural crest phenotypes. It has been established that Twist is a major player in cancer metastasis by controlling expression of cell adhesion molecules during EMT (Yang et al., 2004). In addition, Twist is expressed in migrating neural crest and required for development of its derivatives (Hopwood et al., 1989; Chen and Behringer, 1995; Soo et al., 2002; Ishii et al., 2003; Ota et al., 2004; Bildsoe et al., 2009). Consequently, down-regulation of Twist should affect cell dissociation, as we observed here for Hif-1α MO-injected neural crest. We observed a clear down-regulation of Twist expression in Hif-1α morphant embryos. Furthermore, we found that Twist operates as a repressor of E-cadherin in the neural crest. It is tempting to speculate that in neural crest cells, this will operate similarly to a mechanism reported in cancer cells, whereby Hif-1α controls the expression of Twist by binding directly to the hypoxia-response element in the Twist proximal promoter (Yang et al., 2008) and that Twist represses the expression of E-cadherin by direct binding of Twist to the E boxes of the E-cadherin promoter in cancer cells (Yang et al., 2004). Other genes that are expressed in the neural crest and required for its migration are Snail1 and Snail2 (Barrallo-Gimeno and Nieto, 2005). The Snail genes have also been proposed as direct targets of Hif in cancer cells (Evans et al., 2007; Peinado et al., 2007); however, we found no evidence that Snail1 or Snail2 expression is regulated by Hif-1α in the neural crest cells, implying that this aspect of the gene regulatory network differs between different cells. It is important to mention that in Xenopus the earliest expression of Twist, but not of Snail1/2, correlates well with the beginning of neural crest migration (Linker et al., 2000), supporting our proposal that Twist could trigger EMT in Xenopus neural crest; however, a slightly different situation is observed in mouse embryos that show low levels of Twist expression during neural crest migration (Stoetzel et al., 1995). This low level of Twist could be sufficient to trigger EMT in mouse neural crest; alternatively, the specific details of the neural crest gene regulatory network could differ between species, as it has been shown, for example, with the differences between Snail1 and Snail2 across species (Sefton et al., 1998; Locascio et al., 2002). A second phenotype observed in our experiments is the loss of chemotaxis toward SDF-1 in Hif-1α morphant cells. The likely explanation of this phenotype is based on the down-regulation of the SDF-1 receptor Cxcr4 in neural crest cells that we observed in vivo. In fact, the chemotaxis phenotype can be rescued by coinjection of Cxcr4 mRNA. Surprisingly, activation of the Hif-1α pathway, by hypoxic treatment or by expression of a constitutively active form of Hif-1α, also inhibited chemotaxis. We have previously shown that chemotaxis toward SDF-1 requires cells to be previously polarized by cell contact, and consequently, cell dissociation is a strong inhibitor of chemotaxis (Theveneau et al., 2010). We propose here that hyperactivation of Hif-1α inhibits chemotaxis by promoting premature cell dissociation, which is incompatible with the cell contact–dependent polarity needed for chemotaxis. Indeed, we show that hypoxia and an activated form of Hif-1α produced a dramatic increase in cell dissociation, with cells hardly touching each other.
Our results show that Hif-1α is required for neural crest migration, and they suggest a role for hypoxia during embryo development. It is already known that hypoxia plays an important role in the development of blood vessels, by activating HIF signaling and a plethora of downstream targets (Semenza, 2007). However, a role of hypoxia in very early development, well before any blood or blood vessel is formed, is novel. Interestingly, a role for HIF in cardiogenesis has been shown in HIF-1α–deficient mice (Compernolle et al., 2003), and this phenotype has been attributed to a failure in the migration of neural crest cells that contribute to the heart. Although our results could potentially link hypoxia with neural crest migration, they do not prove that this is the case. It is possible that stability of Hif-1α is controlled in a hypoxia-independent manner in the neural crest as has been described in other systems (Zhong et al., 2000; Gray et al., 2005; Zhou and Brüne, 2006; Koh and Powis, 2009). However, our results imply that oxygen gradients may play a role in neural crest development. First, we demonstrated that cell dissociation can be promoted by hypoxia and that this phenotype is completely reversed by Hif-1α MO, indicating that in the neural crest cells, low oxygen levels promote dissociation via Hif-1α. Second, we showed that hypoxia leads to an increased level of Hif-1α protein, as analyzed by Western blot, indicating that in the neural crest Hif-1α is modulated by oxygen levels. Interestingly, we found that under standard conditions, Hif-1α protein was present in some migrating cells; however, we did not attempt to measure oxygen levels in situ, so it is not known what oxygen concentration these cells were experiencing. This opens the intriguing possibility that during neural crest migration, hypoxic conditions are present in the neural crest and drive migration. Interestingly, it has been shown that neural crest differentiation in vitro is favored by low oxygen levels (Morrison et al., 2000; Studer et al., 2000).
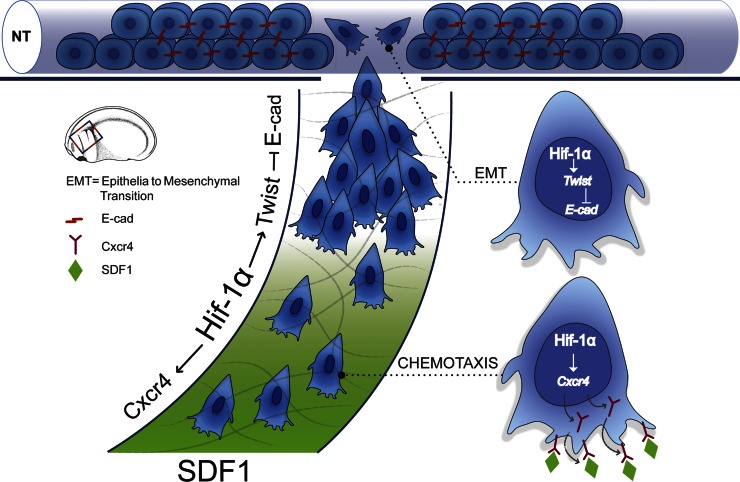
The interpretation of our observations is summarized in Fig. 9. Premigratory neural crest cells lie on the neuroepithelium where they express E-cadherin (Fig. 9, red lines). Stabilization of Hif-1α in a hypoxia-dependent or -independent manner leads to the up-regulation of Twist in the premigratory neural crest. Twist inhibits E-cadherin transcription, which allows neural crest cells to undergo EMT. In addition, Hif-1α also activates Cxcr4 transcription, which is required for chemotaxis toward Sdf-1. Thus, HIF activity is a key regulator of neural crest migration that integrates EMT and chemotaxis.
Figure 9.
Model of Hif-1 controlling neural crest migration. Neural crest, shown as blue cells, delaminates from the neural tube (NT). Prospective neural crest expresses E-cadherin (E-cad; red), just before EMT Hif-1α activates Twist, which in turn represses E-cadherin expression, allowing neural crest EMT. In addition, Hif-1α activates Cxcr4, which is required for chemotaxis toward SDF-1.
Our observations suggest that down-regulation of E-cadherin seems to be essential for neural crest EMT in Xenopus. Although it is widely accepted that EMT requires low levels of E-cadherin in different systems, it is not obvious that this down-regulation is triggering EMT in neural crest cells, as it is believed that E-cadherin is absent from the prospective neural crest much earlier than when its delamination starts. However, recent observations in chick and mouse embryos show important levels of E-cadherin expression just before neural crest delamination (Breau et al., 2008; Dady et al., 2012), consistent with the observations reported here in Xenopus embryos.
There is strong evidence that genes that are important in embryonic development are frequently involved in cancer. Conversely, genes identified on the basis of their oncogenic role are often found to be key players in embryogenesis. Our results add Hif-1α to the list of common regulators of cancer and embryo development. Specifically, we propose that Hif-1α regulates EMT and chemotaxis in the neural crest by a similar mechanism to that implicated in tumor progression and show that the neural crest provides a suitable model to understand cancer migration and metastasis. It has been recently shown that reversal of EMT is necessary for metastatic colonization (Ocaña et al., 2012) and that this EMT reversal can be regulated by Twist (Tsai et al., 2012). A similar reversal of EMT is observed in some neural crest at the end of their migration, which is accompanied by a down-regulation of Twist expression (Linker et al., 2000), supporting even further the parallel between neural crest development and metastasis.
Materials and methods
Zebrafish and Xenopus embryos, micromanipulation, and whole-mount in situ hybridization
Zebrafish strains were maintained and bred according to standard procedures (Westerfield, 2000). Transgenic Tg(sox10:mRFP) (Kirby et al., 2006) embryos were obtained by crossing heterozygous adults and staged according to Kimmel et al. (1995). Xenopus neural crest grafts were performed as previously described by De Calisto et al. (2005). Xenopus embryos were obtained as described previously (Gómez-Skarmeta et al., 1998). In brief, mature Xenopus females were injected with serum gonadotropin (Intervet) and chorionic gonadotropin (Intervet) to stimulate ovulation. Eggs were collected, and in vitro fertilization was performed by mixing the eggs with a sperm suspension. Embryos were staged according to Nieuwkoop and Faber (1967). Dissections and grafts were performed as previously described by Mancilla and Mayor (1996). Neural crest from injected embryos were dissected at stage 16 and transferred to wild-type host embryo, which had their neural crest removed. A fragment of glass coverslip was used to hold the grafted tissue in place until healing was complete. For in situ hybridization, antisense digoxigenin- or fluorescein-labeled RNA probes were used. Specimens were prepared, hybridized, and stained using the method of Harland (1991), and Nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate or 5-bromo-4-chloro-3-indolyl phosphate alone was used as a substrate for the alkaline phosphatase. The genes analyzed for zebrafish were ap2 (Knight et al., 2003), foxd3 (Kelsh et al., 2000), crestin (Luo et al., 2001), gsc (Stachel et al., 1993), dlx-3 (Akimenko et al., 1994), myod (Weinberg et al., 1996), ntl (Schulte-Merker et al., 1992), and cxcr4b (Chong et al., 2001), and the genes analyzed for Xenopus were Snail2 (Mayor et al., 1995), twist (Hopwood et al., 1989), and Cxcr4 (Moepps et al., 2000). For cartilage staining, 96-h postfertilization zebrafish were stained according to Barrallo-Gimeno et al. (2004). Embryos were photographed in a camera (DFL420; Leica) attached to a dissecting microscope (MZ FL III; Leica) using the IM50 software (Leica). The magnification was 3.2× for Xenopus or 8× for zebrafish embryos. All pictures were captured at room temperature in an agarose dish filled with PBS.
MO and RNA injections
For zebrafish microinjection, 4 nl was injected at the one- or two-cell stage. For injection and lineage tracing in Xenopus, nuclear-RFP or membrane-GFP RNA (500 pg per embryo each) was coinjected with fluoresceinated lysine-fixable dextran (Molecular Probes) using 8–12-nL needles as previously described in Aybar et al. (2003). For zebrafish embryos, MOs were injected at a concentration of 3 g/liter. Two different MOs against zebrafish Hif-1α were designed: an ATGMOhif-1α that blocks translation, 5′-GTGACAACTCCAGTATCCATTCCTG-3′; and a spice-blocking MO MOSShif-1α, 5′-CAGATTAGTGGACTGACCGGAATTG-3′. For Xenopus, previously characterized MOs were used: MOATGhif-1α was injected at a concentration of 4 g/liter (Nagao et al., 2008), and MOATGtwist was injected at a concentration of 0.8 g/liter (Zhang and Klymkowsky, 2009).
The Hif-GFP construct spans the MO binding site plus 250 bp of the 5′ UTR of Hif and ∼50 bp of the coding region. The Hif mRNA does not include the 5′ UTR and consequently does not hybridize with the Hif-1α MO. All plasmids were linearized, and RNA was transcribed as previously described by Harland and Weintraub (1985), using SP6 or T7 RNA polymerases and the GTP cap analogue (New England Biolabs, Inc.). After DNase treatment, RNA was purified (BD) and resuspended in RNase-free water (Ambion).
Immunostaining, Western blot, RT-PCR, and quantitative PCR
SDS-PAGE and blotting were performed using NuPAGE Bis-Tris Gels (Novex; Invitrogen) following the manufacturer’s instructions, and polyvinylidene difluoride membrane (GE Healthcare) was used for transfer blotting. Samples were taken at the appropriate stages and homogenized with buffer containing antiphosphorylation reagent (Sigma-Aldrich) and protease inhibitor cocktail (Roche). Antibodies for HIF-1α and MAPK (Cell Signaling Technology) were used at 1:2,000 in 4% BSA in PBS-Twin. After three washes, anti–rabbit IgG (high and light immunoglobulin chains) HRP conjugate (Jackson ImmunoResearch Laboratories, Inc.) was applied as a secondary antibody at 1:25,000. Signal was visualized with luminescent HRP substrate and exposed to film (Fujifilm). For E-cadherin immunostaining, a 5D3 antibody (Developmental Studies Hybridoma Bank) was used at 1:200, and a secondary antibody conjugated to Alexa Fluor 647 was used at 1:500 (Invitrogen). Confocal imaging was performed at room temperature by using a confocal microscope (FluoView FV1000; Olympus), and emission was captured using a charge-coupled device camera (DP73; Olympus) with a U Plan Apochromat 60×/1.35 NA oil immersion objective. Camera, filter wheels, and shutters were controlled by FluoView Software (Olympus).
For the RT-PCR experiments, total RNA was isolated from whole embryos or embryonic tissues after microdissection and cDNA were synthesized as previously described (Aybar et al., 2003). The primers used are listed in Table 1. PCR amplification with these primers was performed over 23–30 cycles, and the PCR products were analyzed on 1.5% agarose gels. RNA was treated with DNase (QIAGEN) to avoid DNA contamination.
Table 1.
Primers used for quantitative PCR analysis.
| Name | Sequences | Citation |
| Hif-1α cDNA forward | 5′-TGGTATCGATCAGGATCAGGAATGGATACTGGAGTTG-3′ | New |
| Hif-1α cDNA reverse | 5′-TGGTATCGATCAGGATCAGGAATGGATACTGGAGTTG-3′ | New |
| β-Actin forward | 5′-TTCTGGTCGGTACTACTGGTATTGTG-3′ | New |
| β-Actin reverse | 5′-ATCTTCATCAGGTAGTCTGTCAGGT-3′ | New |
| Intron 2-3 forward | 5′-CGCATGAGGAAGCTGCTC-3′ | New |
| Intron 2-3 reverse | 5′-GCTCATTCTCCTCCTTCTCC-3′ | New |
| vegf forward | 5′-GAGAGCCAGCGACTCACCGCAACAC-3′ | New |
| vegf reverse | 5′-GTTCGCTCGATCATCATCTTGGC-3′ | New |
| aldolase forward | 5′-CGCAGATGAGTCCACAGGTA-3′ | New |
| aldolase reverse | 5′-TGTCAACCTTGATTCCCACA-3′ | New |
| E-cadherin forward | 5′-CGAAGATGTAAACGAAGCC-3′ | Nandadasa et al., 2009 |
| E-cadherin reverse | 5′-GCCATTTCCAGTGACAATC-3′ | Nandadasa et al., 2009 |
| twist forward | 5′-GGGAGAAAATGATGCAGGAA-3′ | Hong and Saint-Jeannet, 2007 |
| twist reverse | 5′-TCACTGAGATCGGACTGTCG-3′ | Hong and Saint-Jeannet, 2007 |
| snail1 forward | 5′-TCACAAAGGCAGTGCTTCAC-3′ | Hong and Saint-Jeannet, 2007 |
| snail1 reverse | 5′-TGTTCTCTGTGCCAACTGC-3′ | Hong and Saint-Jeannet, 2007 |
| snail2 forward | 5′-CATGGGAATAAGTGAAACCA-3′ | Hong and Saint-Jeannet, 2007 |
| snail2 reverse | 5′-AGGCACGTGAAGGGTAGAGA-3′ | Hong and Saint-Jeannet, 2007 |
| cxcr4 forward | 5′-GGCTATCAAAAGAAATCCAGG-3′ | New |
| cxcr4 reverse | 5′-GCAGGAATCTAAACCCAAACAGTC-3′ | New |
| sdf-1 forward | 5′-CACAGCTCCAGCCACAACATG-3′ | New |
| sdf-1 reverse | 5′-GCCAGAACACTAACAAAGAAATTA-3′ | New |
| ef-1 forward | 5′-ACCCTCCTCTTGGTCGTTT-3′ | Hong and Saint-Jeannet, 2007 |
| ef-1 reverse | 5′-TTTGGTTTTCGCTGCTTTCT-3′ | Hong and Saint-Jeannet, 2007 |
For quantitative PCR, total RNAs were extracted from Xenopus embryos or neural crest explants by using an RNeasy isolation kit (QIAGEN) according to the manufacturer’s instructions. In brief, samples were disrupted and lysed by using buffer RLT, homogenized samples were mixed with an appropriate volume of 70% ethanol and placed in a spin column, and then, the samples were washed with RW1 buffer and treated with DNase I to avoid contamination with genomic DNA. Then, the column was washed with RPE buffer, and finally, the RNA was eluted by adding 50 µl RNase-free water. cDNA was synthesized by using a reverse transcription kit (Improm II; Promega). The amount of RNA isolated from tissues and cDNA synthesized was quantified by using a spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific). Real-time PCR was performed using the primers shown in Table 1 and the SYBR green quantitative PCR master mix (Brilliant II; Agilent Technologies) on the Mx3000 LightCycler system (Agilent Technologies). The reaction mixture consisted of 12.5 µl Brilliant II SYBR green quantitative PCR master mix, 500 nM forward and reverse primers, and 100 ng cDNA template. Cycling conditions were as follows: denaturation at 95°C (5 s), annealing at 55°C (10 s), and extension at 72°C (30 s). The amplification of a single specific product was confirmed by melting curve analysis. Control without template was included on each reaction, and a standard curve of serial dilution points (in steps of 10-fold) was set for each primer. In each reaction, elongation factor-1 (EF-1α) was used as an internal reference to normalize the cycle threshold values obtained from each primer (Hong and Saint-Jeannet, 2007). The histogram bars in the figures are representative cases of at least three independent experiments.
In vitro culture of neural crest, chemotaxis assay, and quantification
Xenopus cranial neural crest were dissected and cultured on fibronectin as previously described (DeSimone et al., 2005). In brief, Petri dishes were covered with a solution of 50 µg/ml fibronectin (Sigma-Aldrich), incubated overnight, and then, washed with PBS and blocked with 1% BSA (Sigma-Aldrich) for 2 h at room temperature. Neural crests were dissected from embryos at stage 16 as described in the Zebrafish and Xenopus embryos, micromanipulation and whole-mount in situ hybridization section, transferred to the fibronectin-covered dish in Danilchick’s solution (DeSimone et al., 2005), and cultured at room temperature for several hours. A new chemotaxis assay was used to test the ability of neural crest cells to respond to SDF-1 (Theveneau and Mayor, 2011; Theveneau et al., 2010). In brief, acrylic Heparin beads (Sigma-Aldrich or Adar Biotech) were incubated overnight in 1 µg/ml Sdf-1 (Sigma-Aldrich), washed in PBS, and fixed with silicone grease to a fibronectin-covered dish. Neural crest cells were placed at 50 µm from the beads, and time lapse and tracking of migrating neural crest cells were performed as previously described (Carmona-Fontaine et al., 2008; Matthews et al., 2008). Tracking was made using ImageJ Manual Tracking plug-in (National Institutes of Health) as described in Theveneau et al. (2010). The tracks of each individual cells were represented in a graph with the origin at 0, 0 using MATLAB (MathWorks) or ImageJ Chemotaxis Tool plug-in, as described in Theveneau et al. (2010). The chemotaxis index, defined as the ratio of the cell displacement in the direction of the gradient and its total traveled distance, was determined as follows. First, the time-lapse videos were orientated with the source of the chemoattractant to the top of the frame; therefore, the direction of the chemotactic gradient was parallel to the y axis. Second, cells were manually tracked, and the forward migration index in the y direction was determined using the ImageJ Chemotaxis plugin. The data shown are the mean of the chemotaxis indices from at least three independent experiments. Cell dispersion was quantified using the Delaunay triangulation (Carmona-Fontaine et al., 2011). Neural crest time-lapse videos were carried on at room temperature in an upright microscope (DMX6000; Leica) equipped with a motorized stage (Prior Scientific) and a camera (DFL420; Leica). N Plan Apochromat 10×/0.25 NA PH1 or HCX Apochromat L20×/1.00 NA water immersion objectives (Leica) were used. Camera, filter wheels, stage, and shutters were controlled by LAS EZ software (Leica). Frames were captured every 5 min during ≥5 h.
Hypoxia treatments
Xenopus embryos and neural crest cultured in vitro were cultured for different period of time under hypoxic conditions. A mixture of 1% O2 and 99% N2 was permanently flushed through the hypoxic chamber (InVivo2 1000; Ruskinn Technology). For analysis of gene expression, embryos were fixed, or the mRNA was extracted immediately after the hypoxic treatment. For time-lapse analysis, cells that were cultured under hypoxia were sealed in glass containers where the hypoxic conditions were maintained for several hours.
Time-lapse microscopy and live-cell imaging
Transgenic zebrafish embryos, Tg(Sox10:mRFP) (which have fluorescently labeled pre- and postmigratory neural crest cells membranes; Kirby et al., 2006), were softly anesthetized with tricaine and mounted for lateral imaging in 1.5% low melting agarose prepared in E3 solution. Images were captured at room temperature in a confocal microscope (LSM 710 NLO; Carl Zeiss) equipped with a motorized stage and a camera (AxioCam MRc5; Carl Zeiss). A Plan Apochromat 20×/0.80 NA water immersion objective was used. Camera, filter wheels, stage, and shutters were controlled by ZEN software (Carl Zeiss). Z stacks were captured every 5 min, and maximum projection images were created using ImageJ software. Images were further processed using ImageJ and/or Photoshop (Adobe). Directionality of migration was measured as persistence, which corresponds to the linear displacement of a cell divided by the total distance traveled (Matthews et al., 2008). Chemotaxis index reflects how straight a cells moves in the direction of the chemoattractant and was measured as previously described in Theveneau et al. (2010).
Online supplemental material
Fig. S1 shows that specificity and the efficiency of two zebrafish antisense MOs (ATGMOhif-1α and SSMOhif-1α). Fig. S2 show the effect of blocking Hif-1α in zebrafish embryo on neural crest, mesoderm, and placode markers. Video 1 shows that Hif-1α is required for normal migration of zebrafish neural crest. Video 2 shows that Hif-1α is required for SDF-1 chemotaxis in a Cxcr4-dependent manner. Video 3 shows that activation of Hif-1α promotes neural crest cell dispersion and blocks chemotaxis. Video 4 shows that Hif-1α is required for normal neural crest cell dispersion. Video 5 shows that Hif-1α controls neural crest dispersion in a Twist-dependent manner. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212100/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Chenbei Chang (University of Alabama at Birmingham, Birmingham, AL) for the XTwist clone. We also thank C. Stern, A. Nieto, E. Theveneau, and J. Leslie for comments on the manuscript.
This study was supported by grants to R. Mayor from the Medical Research Council, Biotechnology and Biological Sciences Research Council, and Welcome Trust; to A.E. Reyes from the Fondo de Investigacion Cienntifica y Tecnologica (FONDECYT; 1120552), Consejo Nacional de Investigacion Cientifica y Tecnologica (CONICYT)/Fondo de Financiamiento de Centros de Excelencia en Investigación (15110027), and Universidad Andres Bello (UNAB; DI-17-12/R); and to E.H. Barriga from FONDECYT (24121369) and UNAB (DI-36-12/I). E.H. Barriga is a CONICYT scholarship holder, and he has been supported by The Company of Biologists and Boehringer Ingelheim Fonds.
Author contributions: Preliminary zebrafish experiments were performed by E.H. Barriga as a student at the Marine Biology Laboratory Embryology course (Woods Hole, MA).
Footnotes
Abbreviations used in this paper:
- EMT
- epithelial to mesenchymal transition
- FDX
- fluorescein dextran
- HIF
- hypoxia-inducible transcription factor
- MO
- morpholino
- SS
- splice site
References
- Akimenko M.A., Ekker M., Wegner J., Lin W., Westerfield M. 1994. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14:3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfandari D., Cousin H., Gaultier A., Hoffstrom B.G., DeSimone D.W. 2003. Integrin alpha5beta1 supports the migration of Xenopus cranial neural crest on fibronectin. Dev. Biol. 260:449–464 10.1016/S0012-1606(03)00277-X [DOI] [PubMed] [Google Scholar]
- Aybar M.J., Nieto M.A., Mayor R. 2003. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 130:483–494 10.1242/dev.00238 [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Nieto M.A. 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 132:3151–3161 10.1242/dev.01907 [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Holzschuh J., Driever W., Knapik E.W. 2004. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development. 131:1463–1477 10.1242/dev.01033 [DOI] [PubMed] [Google Scholar]
- Betancur P., Bronner-Fraser M., Sauka-Spengler T. 2010. Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 26:581–603 10.1146/annurev.cellbio.042308.113245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bildsoe H., Loebel D.A., Jones V.J., Chen Y.T., Behringer R.R., Tam P.P. 2009. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev. Biol. 331:176–188 10.1016/j.ydbio.2009.04.034 [DOI] [PubMed] [Google Scholar]
- Breau M.A., Pietri T., Stemmler M.P., Thiery J.P., Weston J.A. 2008. A nonneural epithelial domain of embryonic cranial neural folds gives rise to ectomesenchyme. Proc. Natl. Acad. Sci. USA. 105:7750–7755 10.1073/pnas.0711344105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacheux V., Dastot-Le Moal F., Kääriäinen H., Bondurand N., Rintala R., Boissier B., Wilson M., Mowat D., Goossens M. 2001. Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum. Mol. Genet. 10:1503–1510 10.1093/hmg/10.14.1503 [DOI] [PubMed] [Google Scholar]
- Cairns R.A., Harris I.S., Mak T.W. 2011. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 11:85–95 10.1038/nrc2981 [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C., Matthews H.K., Kuriyama S., Moreno M., Dunn G.A., Parsons M., Stern C.D., Mayor R. 2008. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 456:957–961 10.1038/nature07441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C., Theveneau E., Tzekou A., Tada M., Woods M., Page K.M., Parsons M., Lambris J.D., Mayor R. 2011. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev. Cell. 21:1026–1037 10.1016/j.devcel.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q., Jurisica I., Do T., Hedley D.W. 2011. Hypoxia predicts aggressive growth and spontaneous metastasis formation from orthotopically grown primary xenografts of human pancreatic cancer. Cancer Res. 71:3110–3120 10.1158/0008-5472.CAN-10-4049 [DOI] [PubMed] [Google Scholar]
- Chen Z.F., Behringer R.R. 1995. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9:686–699 10.1101/gad.9.6.686 [DOI] [PubMed] [Google Scholar]
- Chong S.W., Emelyanov A., Gong Z., Korzh V. 2001. Expression pattern of two zebrafish genes, cxcr4a and cxcr4b. Mech. Dev. 109:347–354 10.1016/S0925-4773(01)00520-2 [DOI] [PubMed] [Google Scholar]
- Cockman M.E., Masson N., Mole D.R., Jaakkola P., Chang G.W., Clifford S.C., Maher E.R., Pugh C.W., Ratcliffe P.J., Maxwell P.H. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275:25733–25741 10.1074/jbc.M002740200 [DOI] [PubMed] [Google Scholar]
- Compernolle V., Brusselmans K., Franco D., Moorman A., Dewerchin M., Collen D., Carmeliet P. 2003. Cardia bifida, defective heart development and abnormal neural crest migration in embryos lacking hypoxia-inducible factor-1alpha. Cardiovasc. Res. 60:569–579 10.1016/j.cardiores.2003.07.003 [DOI] [PubMed] [Google Scholar]
- Dady A., Blavet C., Duband J.L. 2012. Timing and kinetics of E- to N-cadherin switch during neurulation in the avian embryo. Dev. Dyn. 241:1333–1349 10.1002/dvdy.23813 [DOI] [PubMed] [Google Scholar]
- de Beaucourt A., Coumailleau P. 2007. Molecular cloning and characterization of the Xenopus hypoxia-inducible factor 1alpha (xHIF1alpha). J. Cell. Biochem. 102:1542–1552 10.1002/jcb.21376 [DOI] [PubMed] [Google Scholar]
- De Calisto J., Araya C., Marchant L., Riaz C.F., Mayor R. 2005. Essential role of non-canonical Wnt signalling in neural crest migration. Development. 132:2587–2597 10.1242/dev.01857 [DOI] [PubMed] [Google Scholar]
- DeSimone D.W., Davidson L., Marsden M., Alfandari D. 2005. The Xenopus embryo as a model system for studies of cell migration. Methods Mol. Biol. 294:235–245 [DOI] [PubMed] [Google Scholar]
- Erler J.T., Cawthorne C.J., Williams K.J., Koritzinsky M., Wouters B.G., Wilson C., Miller C., Demonacos C., Stratford I.J., Dive C. 2004. Hypoxia-mediated down-regulation of Bid and Bax in tumors occurs via hypoxia-inducible factor 1-dependent and -independent mechanisms and contributes to drug resistance. Mol. Cell. Biol. 24:2875–2889 10.1128/MCB.24.7.2875-2889.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A.J., Russell R.C., Roche O., Burry T.N., Fish J.E., Chow V.W., Kim W.Y., Saravanan A., Maynard M.A., Gervais M.L., et al. 2007. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol. Cell. Biol. 27:157–169 10.1128/MCB.00892-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta J.L., Glavic A., de la Calle-Mustienes E., Modolell J., Mayor R. 1998. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development at the neural plate. EMBO J. 17:181–190 10.1093/emboj/17.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gort E.H., van Haaften G., Verlaan I., Groot A.J., Plasterk R.H., Shvarts A., Suijkerbuijk K.P., van Laar T., van der Wall E., Raman V., et al. 2008. The TWIST1 oncogene is a direct target of hypoxia-inducible factor-2alpha. Oncogene. 27:1501–1510 10.1038/sj.onc.1210795 [DOI] [PubMed] [Google Scholar]
- Graeber T.G., Osmanian C., Jacks T., Housman D.E., Koch C.J., Lowe S.W., Giaccia A.J. 1996. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 379:88–91 10.1038/379088a0 [DOI] [PubMed] [Google Scholar]
- Gray M.J., Zhang J., Ellis L.M., Semenza G.L., Evans D.B., Watowich S.S., Gallick G.E. 2005. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 24:3110–3120 10.1038/sj.onc.1208513 [DOI] [PubMed] [Google Scholar]
- Gupta G.P., Massagué J. 2006. Cancer metastasis: building a framework. Cell. 127:679–695 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Harland R.M. 1991. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36:685–695 10.1016/S0091-679X(08)60307-6 [DOI] [PubMed] [Google Scholar]
- Harland R., Weintraub H. 1985. Translation of mRNA injected into Xenopus oocytes is specifically inhibited by antisense RNA. J. Cell Biol. 101:1094–1099 10.1083/jcb.101.3.1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A.L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer. 2:38–47 10.1038/nrc704 [DOI] [PubMed] [Google Scholar]
- Hendrix M.J., Seftor E.A., Seftor R.E., Kasemeier-Kulesa J., Kulesa P.M., Postovit L.M. 2007. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat. Rev. Cancer. 7:246–255 10.1038/nrc2108 [DOI] [PubMed] [Google Scholar]
- Hill R.P., Marie-Egyptienne D.T., Hedley D.W. 2009. Cancer stem cells, hypoxia and metastasis. Semin. Radiat. Oncol. 19:106–111 10.1016/j.semradonc.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Hong C.S., Saint-Jeannet J.P. 2007. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell. 18:2192–2202 10.1091/mbc.E06-11-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood N.D., Pluck A., Gurdon J.B. 1989. A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell. 59:893–903 10.1016/0092-8674(89)90612-0 [DOI] [PubMed] [Google Scholar]
- Ishii M., Merrill A.E., Chan Y.S., Gitelman I., Rice D.P., Sucov H.M., Maxson R.E., Jr 2003. Msx2 and Twist cooperatively control the development of the neural crest-derived skeletogenic mesenchyme of the murine skull vault. Development. 130:6131–6142 10.1242/dev.00793 [DOI] [PubMed] [Google Scholar]
- Kamura T., Sato S., Iwai K., Czyzyk-Krzeska M., Conaway R.C., Conaway J.W. 2000. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA. 97:10430–10435 10.1073/pnas.190332597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh R.N., Dutton K., Medlin J., Eisen J.S. 2000. Expression of zebrafish fkd6 in neural crest-derived glia. Mech. Dev. 93:161–164 10.1016/S0925-4773(00)00250-1 [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kioi M., Vogel H., Schultz G., Hoffman R.M., Harsh G.R., Brown J.M. 2010. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J. Clin. Invest. 120:694–705 10.1172/JCI40283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby B.B., Takada N., Latimer A.J., Shin J., Carney T.J., Kelsh R.N., Appel B. 2006. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat. Neurosci. 9:1506–1511 10.1038/nn1803 [DOI] [PubMed] [Google Scholar]
- Knight R.D., Nair S., Nelson S.S., Afshar A., Javidan Y., Geisler R., Rauch G.J., Schilling T.F. 2003. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 130:5755–5768 10.1242/dev.00575 [DOI] [PubMed] [Google Scholar]
- Koh M.Y., Powis G. 2009. HAF: the new player in oxygen-independent HIF-1alpha degradation. Cell Cycle. 8:1359–1366 10.4161/cc.8.9.8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucia M., Reca R., Miekus K., Wanzeck J., Wojakowski W., Janowska-Wieczorek A., Ratajczak J., Ratajczak M.Z. 2005. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 23:879–894 10.1634/stemcells.2004-0342 [DOI] [PubMed] [Google Scholar]
- Kulesa P.M., Kasemeier-Kulesa J.C., Teddy J.M., Margaryan N.V., Seftor E.A., Seftor R.E., Hendrix M.J. 2006. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proc. Natl. Acad. Sci. USA. 103:3752–3757 10.1073/pnas.0506977103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama S., Mayor R. 2008. Molecular analysis of neural crest migration. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:1349–1362 10.1098/rstb.2007.2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker C., Bronner-Fraser M., Mayor R. 2000. Relationship between gene expression domains of Xsnail, Xslug, and Xtwist and cell movement in the prospective neural crest of Xenopus. Dev. Biol. 224:215–225 10.1006/dbio.2000.9723 [DOI] [PubMed] [Google Scholar]
- Locascio A., Manzanares M., Blanco M.J., Nieto M.A. 2002. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc. Natl. Acad. Sci. USA. 99:16841–16846 10.1073/pnas.262525399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., An M., Arduini B.L., Henion P.D. 2001. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev. Dyn. 220:169–174 [DOI] [PubMed] [Google Scholar]
- Mancilla A., Mayor R. 1996. Neural crest formation in Xenopus laevis: mechanisms of Xslug induction. Dev. Biol. 177:580–589 10.1006/dbio.1996.0187 [DOI] [PubMed] [Google Scholar]
- Matthews H.K., Marchant L., Carmona-Fontaine C., Kuriyama S., Larraín J., Holt M.R., Parsons M., Mayor R. 2008. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development. 135:1771–1780 10.1242/dev.017350 [DOI] [PubMed] [Google Scholar]
- Mayor R., Morgan R., Sargent M.G. 1995. Induction of the prospective neural crest of Xenopus. Development. 121:767–777 [DOI] [PubMed] [Google Scholar]
- Moepps B., Braun M., Knöpfle K., Dillinger K., Knöchel W., Gierschik P. 2000. Characterization of a Xenopus laevis CXC chemokine receptor 4: implications for hematopoietic cell development in the vertebrate embryo. Eur. J. Immunol. 30:2924–2934 [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Csete M., Groves A.K., Melega W., Wold B., Anderson D.J. 2000. Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J. Neurosci. 20:7370–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K., Taniyama Y., Kietzmann T., Doi T., Komuro I., Morishita R. 2008. HIF-1alpha signaling upstream of NKX2.5 is required for cardiac development in Xenopus. J. Biol. Chem. 283:11841–11849 10.1074/jbc.M702563200 [DOI] [PubMed] [Google Scholar]
- Nandadasa S., Tao Q., Menon N.R., Heasman J., Wylie C. 2009. N- and E-cadherins in Xenopus are specifically required in the neural and non-neural ectoderm, respectively, for F-actin assembly and morphogenetic movements. Development. 136:1327–1338 10.1242/dev.031203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P.D., Faber J. 1967. Normal Table of Xenopus laevis (Daudin): A systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Second edition North-Holland Pub. Co, Amsterdam: 260 pp [Google Scholar]
- Ocaña O.H., Córcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S., Barrallo-Gimeno A., Cano A., Nieto M.A. 2012. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 22:709–724 10.1016/j.ccr.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Ohh M., Park C.W., Ivan M., Hoffman M.A., Kim T.Y., Huang L.E., Pavletich N., Chau V., Kaelin W.G. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423–427 10.1038/35017054 [DOI] [PubMed] [Google Scholar]
- Olesnicky Killian E.C., Birkholz D.A., Artinger K.B. 2009. A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev. Biol. 333:161–172 10.1016/j.ydbio.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M.S., Loebel D.A., O’Rourke M.P., Wong N., Tsoi B., Tam P.P. 2004. Twist is required for patterning the cranial nerves and maintaining the viability of mesodermal cells. Dev. Dyn. 230:216–228 10.1002/dvdy.20047 [DOI] [PubMed] [Google Scholar]
- Peinado H., Olmeda D., Cano A. 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 7:415–428 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- Pennacchietti S., Michieli P., Galluzzo M., Mazzone M., Giordano S., Comoglio P.M. 2003. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 3:347–361 10.1016/S1535-6108(03)00085-0 [DOI] [PubMed] [Google Scholar]
- Rojas D.A., Perez-Munizaga D.A., Centanin L., Antonelli M., Wappner P., Allende M.L., Reyes A.E. 2007. Cloning of hif-1alpha and hif-2alpha and mRNA expression pattern during development in zebrafish. Gene Expr. Patterns. 7:339–345 10.1016/j.modgep.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Rouschop K.M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J.W., et al. 2010. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120:127–141 10.1172/JCI40027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker S., Ho R.K., Herrmann B.G., Nüsslein-Volhard C. 1992. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development. 116:1021–1032 [DOI] [PubMed] [Google Scholar]
- Sefton M., Sánchez S., Nieto M.A. 1998. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 125:3111–3121 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. 2000. HIF-1: using two hands to flip the angiogenic switch. Cancer Metastasis Rev. 19:59–65 10.1023/A:1026544214667 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. 2002. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8(Suppl.):S62–S67 10.1016/S1471-4914(02)02317-1 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. 2007. Hypoxia and cancer. Cancer Metastasis Rev. 26:223–224 10.1007/s10555-007-9058-y [DOI] [PubMed] [Google Scholar]
- Semenza G.L. 2010. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 29:625–634 10.1038/onc.2009.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G.L., Nejfelt M.K., Chi S.M., Antonarakis S.E. 1991. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc. Natl. Acad. Sci. USA. 88:5680–5684 10.1073/pnas.88.13.5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo K., O’Rourke M.P., Khoo P.L., Steiner K.A., Wong N., Behringer R.R., Tam P.P. 2002. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev. Biol. 247:251–270 10.1006/dbio.2002.0699 [DOI] [PubMed] [Google Scholar]
- Stachel S.E., Grunwald D.J., Myers P.Z. 1993. Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development. 117:1261–1274 [DOI] [PubMed] [Google Scholar]
- Staller P., Sulitkova J., Lisztwan J., Moch H., Oakeley E.J., Krek W. 2003. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 425:307–311 10.1038/nature01874 [DOI] [PubMed] [Google Scholar]
- Steventon B., Carmona-Fontaine C., Mayor R. 2005. Genetic network during neural crest induction: from cell specification to cell survival. Semin. Cell Dev. Biol. 16:647–654 10.1016/j.semcdb.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Stoetzel C., Weber B., Bourgeois P., Bolcato-Bellemin A.L., Perrin-Schmitt F. 1995. Dorso-ventral and rostro-caudal sequential expression of M-twist in the postimplantation murine embryo. Mech. Dev. 51:251–263 10.1016/0925-4773(95)00369-X [DOI] [PubMed] [Google Scholar]
- Studer L., Csete M., Lee S.H., Kabbani N., Walikonis J., Wold B., McKay R. 2000. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J. Neurosci. 20:7377–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K., Makino Y., Pereira T., Poellinger L. 2000. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19:4298–4309 10.1093/emboj/19.16.4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E., Mayor R. 2010. Integrating chemotaxis and contact-inhibition during collective cell migration: Small GTPases at work. Small GTPases. 1:113–117 10.4161/sgtp.1.2.13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E., Mayor R. 2011. Collective cell migration of the cephalic neural crest: the art of integrating information. Genesis. 49:164–176 10.1002/dvg.20700 [DOI] [PubMed] [Google Scholar]
- Theveneau E., Marchant L., Kuriyama S., Gull M., Moepps B., Parsons M., Mayor R. 2010. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell. 19:39–53 10.1016/j.devcel.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Tsai J.H., Donaher J.L., Murphy D.A., Chau S., Yang J. 2012. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 22:725–736 10.1016/j.ccr.2012.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ohh M. 2010. Oxygen-mediated endocytosis in cancer. J. Cell. Mol. Med. 14:496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E.S., Allende M.L., Kelly C.S., Abdelhamid A., Murakami T., Andermann P., Doerre O.G., Grunwald D.J., Riggleman B. 1996. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 122:271–280 [DOI] [PubMed] [Google Scholar]
- Westerfield M. 2000. The Zebrafish Book: a guide for the laboratory use of zebrafish (Brachydanio rerio). Fourth edition University of Oregon Press, Eugene, OR [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Yang M.H., Wu M.Z., Chiou S.H., Chen P.M., Chang S.Y., Liu C.J., Teng S.C., Wu K.J. 2008. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 10:295–305 10.1038/ncb1691 [DOI] [PubMed] [Google Scholar]
- Zhang C., Klymkowsky M.W. 2009. Unexpected functional redundancy between Twist and Slug (Snail2) and their feedback regulation of NF-kappaB via Nodal and Cerberus. Dev. Biol. 331:340–349 10.1016/j.ydbio.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., Chiles K., Feldser D., Laughner E., Hanrahan C., Georgescu M.M., Simons J.W., Semenza G.L. 2000. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60:1541–1545 [PubMed] [Google Scholar]
- Zhou J., Brüne B. 2006. Cytokines and hormones in the regulation of hypoxia inducible factor-1alpha (HIF-1alpha). Cardiovasc. Hematol. Agents Med. Chem. 4:189–197 10.2174/187152506777698344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.