Abstract
Runs of homozygosity (ROHs) are a class of important but poorly studied genomic variations and may be involved in individual susceptibility to diseases. To better understand ROH and its relationship with lung cancer, we performed a genome-wide ROH analysis of a subset of a previous genome-wide case-control study (1,473 cases and 1,962 controls) in a Han Chinese population. ROHs were classified into two classes, based on lengths, intermediate and long ROHs, to evaluate their association with lung cancer risk using existing genome-wide single nucleotide polymorphism (SNP) data. We found that the overall level of intermediate ROHs was significantly associated with a decreased risk of lung cancer (odds ratio = 0.63; 95% confidence interval: 0.51-0.77; P = 4.78×10−6 ), while the long ROHs seemed to be a risk factor of lung cancer. We also identified one ROH region at 14q23.1 that was consistently associated with lung cancer risk in the study. These results indicated that ROHs may be a new class of variation which may be associated with lung cancer risk, and genetic variants at 14q23.1 may be involved in the development of lung cancer.
Keywords: lung cancer, runs of homozygosity (ROHs), genome-wide association study
INTRODUCTION
Lung cancer is the leading cause of cancer death worldwide; its incidence and mortality rates have been increasing rapidly in the last three decades[1]. Although lung cancer is largely caused by tobacco smoking, there is accumulating evidence for the role of genetic factors in the occurrence of the disease[2]. Until now, several genome-wide association studies (GWAS) have captured a few polymorphic variations at 3q28 (TP63), 5p15.33 (TERT/CLPTM1L), 5q32 (PPP2R2B-STK32A-DPYSL3), 6p21.32 (BTNL2), 6p21.33 (BAT3/MSH5), 9p21 (CDKN2A,CDKN2B), 10p14 (GATA3), 12p13.33 (RAD52), 15q25.1 (CHRNA5/CHRNA3/CHRNB4), 13q12.12 (MIPEP-TNFRSF19), 17q24.2 (BPTF), 20q13.2 (CYP24A1) and 22q12.2 (MTMR-HORMAD2-LIF) that are associated with lung cancer risk[3]–[11]. However, these findings are not sufficient to explain the heritability of lung cancer. Thus, further studies are needed to discover more regions/variants related to lung cancer risk.
A run of homozygosity (ROH) is defined as a continuous or uninterrupted stretch of a genomic sequence without heterozygosity in the diploid state. In general, very long ROHs are the by-product of recent inbreeding or chromosome abnormity, whereas the derivation of relatively shorter ROHs is still disputable. It is also hypothesized that ROH may be a result of linkage disequilibrium (LD)[12],[13]. However, increased LD in the vicinity of a given variant is neither necessary nor sufficient for a series of variants to be included in an ROH[14]. Emerging evidence has supported that ROH may represent a novel type of independent characteristic of the genome[15]. With the development of the genotyping technology of single nucleotide polymorphism (SNP), it has become more feasible to carry out ROH studies. Several studies have reported that ROHs are widely but not randomly distributed in the outbred human genomes[12],[14],[16], and have been implicated in multiple complex diseases[12],[17]–[24]. In the current study, we attempted to clarify the association of ROHs with the development of lung cancer by using a case-control study including 1,473 cases and 1,962 cancer-free controls of Han Chinese.
SUBJECTS AND METHODS
Subjects
Demographic and clinical information is summarized in Table 1 and has been described elsewhere[9]. Subjects which were included in this study consisted of 1,473 cases and 1962 controls. The cases that were histopathologically or cytologically confirmed as lung cancer were recruited from local hospitals. All cancer-free control subjects were selected from individuals receiving routine physical examinations at local hospitals or those participating in our community-based screening of non-communicable diseases. All subjects were unrelated ethnic Han Chinese. At recruitment, informed consent was obtained from each subject, and this study protocol was approved by the local institutional review boards of authors' affiliated institutions.
Table 1. Summary of Class A and B ROHs at 14q23.1 and its association with lung cancer risk.
| Variable | Case | Control |
| Age (Mean±SD) | 60.08±10.30 | 59.35±9.74 |
| Gender (%) | ||
| Male | 1057 | 1214 |
| Female | 416 | 748 |
| Smoking status (%) | ||
| Current smokers | 741 | 636 |
| Former smokers | 168 | 83 |
| Never smokers | 564 | 1243 |
| Smoking levels (Mean±S.D.) | 41.43±26.86 | 30.96±20.28 |
| ≤ 25 (Pack-years) | 254 | 327 |
| > 25 (Pack-years) | 655 | 392 |
| Histology | ||
| Squamous cell carcinoma | 421 | |
| Adenocarcinoma | 896 | |
| Small cell carcinoma | 129 | |
| Others * | 27 |
*Other includes large cell lung cancer and mixed cell carcinoma.
Genome-wide scan and ROH calling
Genome-wide scan was conducted using Affymetrix Genome-Wide Human SNP Array 6.0 chips. Systematic quality control procedure was used to filter out both unqualified samples and SNPs based on predefined criteria[9]. Briefly, SNPs were excluded if: (i) they were not mapped on autosomal chromosomes; (ii) they had a call rate < 95%; or (iii) they had a minor allele frequency (MAF) < 0.05. Samples with low call rates (< 95%), ambiguous gender or familial relationships (PI_HAT > 0.25), (outlying values) in the principal component analysis, or extreme heterozygote rate (> 6 standard deviations away from nearest neighbor) were removed. Finally, 591,370 SNPs from 1,473 cases and 1,962 controls were used for ROH detection.
ROHs were determined using the command “--homozyg” implemented in PLINK v1.07[25], by setting the minimum length of ROH at 0.50 Mb, the minimum number of SNPs per ROH at 50 and gap threshold between two ROHs at 0.10 Mb. The minimum length of ROH was selected in order to exclude some short copy number variations (CNVs) and false ROHs formed by chance[16]. A ROH was broken into two if a gap was found more than 100 kb between adjacent homozygous SNPs. To show a full view of ROH burden, we set the minimum number of SNPs in an ROH at 50, 60, 70, 80, 90 and 100 and performed detection, respectively.
Statistical analysis
For each individual, FROH, defined as the proportion of the autosomal genome in ROHs above a specified length threshold (the total length of all their ROHs in the autosome divided by the total SNP-mappable autosomal distance)[16],[21], was used as a predictor of case-control status in ROH description and burden analysis.
Mclust from the mclust package (v.4) in R was used to run unsupervised Gaussian fitting of the ROH length distribution. According to Pemberton et al.[26], we divided ROHs into two groups (Class A and B as one group, and Class C as another) depending on their length. For each group, overlapping pools between individuals were defined in case and control separately via a program coded by R based on the algorithm from command “-homozyg-group” implemented in PLINK v1.07, which calculated overlapping ROH number in each SNPs and considered regions with peak overlapping ROH number as pools. Considering that there might be difference between ROH structure in cases and controls, pools identified either in case or controls were involved in subsequent analysis. While analyzing, the status of each pool was coded as 0 for no ROH, 1 for class A or class B ROH, and 2 for class C ROH (dummy variables). Pools with frequency greater than 20% in either the case or control groups were considered to be “hotspot” and were further analyzed in the study.
The relationship of FROH with lung cancer risk was analyzed by using logistic regression to assess the burden of ROHs on lung cancer. The association of each hotspot with lung cancer risk was also evaluated by using logistic regression with adjustment for age, gender and pack-years of smoking as top principle component. Population structure was evaluated by principle component analysis (PCA) using SNPs in the software package EIGENSTRAT 3.0[21],[27]. R v2.15.1 was used for general statistical analysis[28].
RESULTS
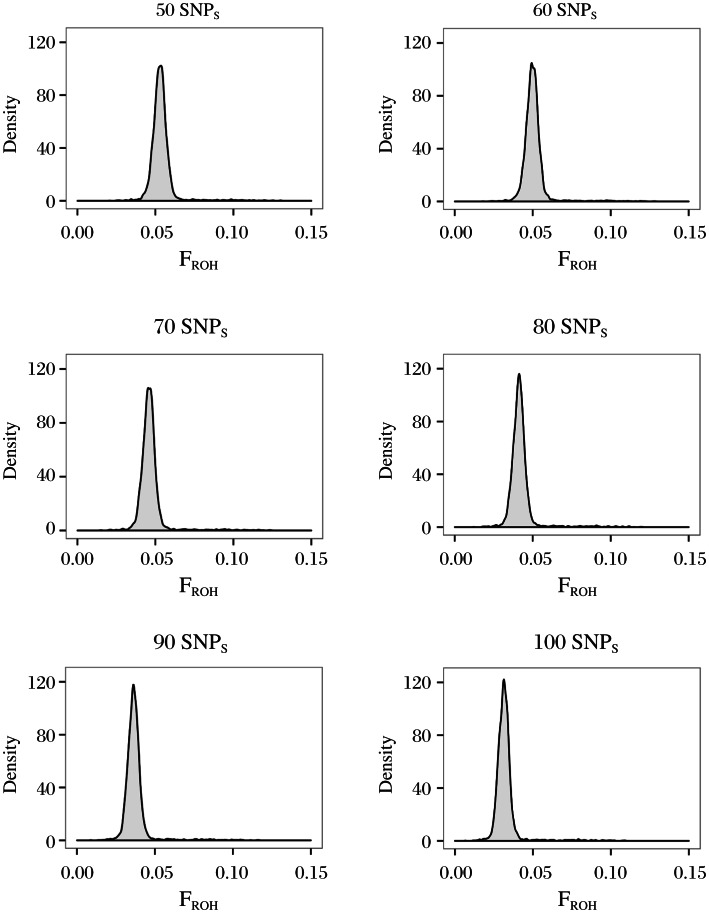
FROH of ROHs was defined as a SNP number of more than 50 approximates to a normal distribution except for some individuals with extremely large FROH (Fig. 1A to 1F). To extensively evaluate ROHs across the entire genome, we set 50 as a default value to define ROHs in subsequent analysis. FROH distribution suggested that there were multiple components (Fig. 1A); therefore, we classified ROHs into three classes (class A: 500Kb-689.346Kb, class B: 689.346Kb-1548.887Kb, and class C: over 1548.887Kb) according to the cluster method based on Gaussian mixture model (see Method).
Fig. 1. Distribution of runs of homozygosity (ROHs) at different single nucleotide polymorphism (SNP) thresholds.
X axis represents FROH of each individual. Y axis represents the number of individuals with specific FROH.
We evaluated the ROH burden on lung cancer using FROH. As shown in Table 2, overall moderate ROH level (FROH) was significantly associated with a decreased risk of lung cancer (OR = 0.63, 95% CI: 0.51-0.77, P = 4.78×10−6). In contrast, long FROH levels was significantly associated with an increased risk of lung cancer (OR = 1.13, 95% CI: 1.01-1.26, P = 0.030). As individuals were divided into 4 ROH levels according to the quartile of FROH in controls, logistic regression analysis showed that the high levels of moderate ROH were consistently associated with a decreased risk of lung cancer (trend OR = 0.85, 95% CI: 0.79-0.91, P = 3.33×10−5) as compared to low levels while the association was not observed with the high levels of long ROH (trend OR = 1.06, 95% CI: 0.99-1.14, P = 0.08).
Table 2. Distribution of FROH for overall ROHs between lung cancer cases and controls and its association with lung cancer risk.
| Class A&B |
Class C |
|||||||
| ROH group | Case/Control | OR (95%CI) | P | ROH group | Case/Control | OR (95%CI) | P | |
| ≤4.78% | 399/459 | 1 | ≤0.155% | 322/456 | 1 | |||
| 4.78%-5.01% | 397/462 | 0.90 (0.72;1.12) | 3.29×10−1 | 0.155%-0.246% | 348/511 | 0.92 (0.73;1.15) | 4.52×10−1 | |
| 5.01%-5.26% | 341/517 | 0.72 (0.57;0.89) | 2.75×10−3 | 0.246%-0.353% | 384/474 | 1.17 (0.93;1.46) | 1.73×10−1 | |
| > 5.26% | 336/523 | 0.63 (0.51;0.78) | 3.33×10−5 | > 0.353% | 384/475 | 1.13 (0.90;1.41) | 2.86×10−1 | |
| TREND | 1,473/1,962 | 0.85 (0.79;0.91) | 4.46×10−6 | TREND | 1,473/1,962 | 1.06 (0.99;1.14) | 8.55×10−2 | |
ROH: Runs of homozygosity.
To identify specific regions associated with lung cancer risk, we performed a genetic association analysis of ROH hotspots. Totally 3,288 and 3,657 ROH hotspots detected from the case and control groups, respectively, were separately evaluated for an association with lung cancer risk. Several intermediate ROH pools at 14q23.1 were found to be significantly associated with lung cancer risk (PFDR < 0.05, OR < 1) after multiple test correction (FDR) (Table 3), and interestingly, long ROH pools at the same location exerted reverse effects on lung cancer (P < 0.05, OR > 1).
Table 3. Summary of Class A and B ROHs at 14q23.1 and its association with lung cancer risk.
| Pools* | Case |
Control |
P | PFDR | OR (95%CI) | |||
| ROH | ROH free | ROH | ROH free | |||||
| chr14:58952891-58963188 | 254 | 1219 | 438 | 1524 | 4.13×10−5 | 4.15×10−2 | 0.67 (0.55,0.81) | |
| chr14:59160264-59160264 | 281 | 1192 | 472 | 1490 | 4.47×10−5 | 4.15×10−2 | 0.68 (0.56,0.82) | |
| chr14:59121306-59143688 | 288 | 1185 | 487 | 1475 | 5.44×10−5 | 4.15×10−2 | 0.68 (0.57,0.82) | |
| chr14:59116443-59116443 | 287 | 1186 | 480 | 1482 | 6.68×10−5 | 4.15×10−2 | 0.69 (0.57,0.83) | |
| chr14:59145071-59145071 | 286 | 1187 | 478 | 1484 | 7.04×10−5 | 4.15×10−2 | 0.69 (0.57,0.83) | |
| chr14:59098920-59098920 | 279 | 1194 | 468 | 1494 | 7.72×10−5 | 4.15×10−2 | 0.69 (0.57,0.83) | |
| chr14:59163968-59163968 | 246 | 1227 | 416 | 1546 | 7.94×10−5 | 4.15×10−2 | 0.67 (0.55,0.82) | |
| chr14:59174889-59174889 | 252 | 1221 | 425 | 1537 | 1.07×10−4 | 4.90×10−2 | 0.68 (0.56,0.83) | |
*Position of pools were based on Hg17 (Human May 2004 Assembly). ROH: Runs of homozygosity.
DISCUSSION
SNPs are considered as the major source of genetic diversity in humans and have been extensively implicated in multiple diseases and traits. Although GWAS has successfully established the link between SNPs and phenotypes, the identified loci can only explain a small fraction of the risk of diseases or the variance of traits. In addition to SNPs, other types of genetic variants may also contribute to the individual risk of disease either as causal variants or as proxies for causal variants. In the current study, we performed a genome-wide survey on ROHs, a mysterious type of genomic variant, utilizing SNP data from genome SNP scanning chips, evaluated the overall ROH levels on lung cancer and conducted genome-wide association of ROHs with lung cancer risk. We found a significantly decreased overall ROH level among lung cancer cases and identified a ROH region at 14q23.1 that was consistently associated with lung cancer risk. This study has made an important effort to investigate the role of ROHs in a specific disease and provided a proof-of-principle approach that can be used in further ROH studies of other diseases using existing GWAS data.
In this study, FROH of ROHs approximated to a normal distribution, which is consistent with the results reported in previous studies[21],[26]. According to the right-skewed distribution of FROH for overall ROHs, the boundaries of each component were defined at 689.346 kb and 1548.887 kb, similar to the cutoffs (0.5 Mb and 1.5 Mb) reported by McQuillan et al.[16]. Moreover, we found that the similar FROH distributions of class A and class B ROHs, which, however, were significantly different from that of class C. Class C ROHs, representing long ROHs, are very rare in Han Chinese and may be caused by recent inbreeding[16],[26],[29]. According to the simulation analyses in previous studies[26],[29], a large sample size (e.g., 12,000-65,000) is needed for an adequate statistical power as detecting the associations of long ROHs with phenotypes. In the current study, we paid attention primarily to class A and B ROHs that have a relatively high frequency. These ROHs are probably the consequence of ancestry inbreeding and positive selection and act as a stable genomic structure across populations[16],[26]. Intriguingly, we obtained consistent results that long ROHs (class C), accompanied by some deleterious recessive variants passed down through recent inbreeding, contributed to the development of cancer. Furthermore, we found a protective effect of intermediate ROHs.
On the basis of candidate gene/region approach, ROHs have been reported to be associated with several diseases, including schizophrenia, late-onset Alzheimer's disease, cancer and etc.[22],[31]–[33]. In the current study, we identified several pools associated with lung cancer risk in the region (chr14:59,835,003-60,105,136) at 14q23.1 including genes DAAM1, GRP135, c14orf149, JKAMP and RTN1. Gain of copy numbers on this region was associated with shorter lung cancer survival[34]. The phenomenon of ROH increase in this region may indicate the presence of a number of deleterious heterozygosity mutation, suggesting that it is worthy of doing some further studies in this region.
DAAM1 is one important participant in the planar cell polarity (PCP) pathway, which is one of the key sub-pathways of the Wnt signaling pathway. Bounded to both DVL and Rho, DAAM1 is thought to function as a scaffolding protein and mediates Wnt-induced DBL-Rho complex formation[35]. DAAM1 is also considered as a form in homology (FH) protein that is expressed in complementary patterns in lungs[36], and DAAM1 protein is enriched at the apical surface of airways[37]. However, there was no further data and research on the relationship between DAAM1 and the lungs. Recently, two studies found that DAAM1 was involved in regulating heart and kidney morphogenesis[38],[39]. In our study, the protective region in 14q23.1 included DAAM1 upstream position and some parts of DAAM1, within which there were some eQTL SNPs of DAAM1 that may regulate the expression of DAAM1.
JKAMP, also called JNK1-associated membrane protein (JAMP), was reported to be associated with JNK1. Its association with JNK1 outcompetes JNK1 association with MKP5, resulting in increased and prolonged JNK1 activity following stress[40]. JNK-deficient mice exhibited delayed epithelial development in the lungs[41]. Furthermore, one study indicated that JNK was activated in a subset of NSCLC biopsy samples and promoted ontogenesis of the bronchial epithelium[42].
RTN1 was regarded as neuroendocrine-specific proteins and identified by screening the expression library of the small-cell lung cancer (SCLC) NCI-H82 cell line with antibodies to the previously identified proteins[43]. The gene can produce 3 different transcripts with identical 3-prime ends but unique amino termini. The B transcript was found only in the NCI-H82 cell lines, while A and C transcripts were found in 18 different SCLC lines but not in any of the 11 non-endocrine NSCLCs[44].
In summary, our results indicated that the overall intermediate ROH levels were low in lung cancer cases and may be implicated with susceptibility to lung cancer. We also identified one intermediate ROH region at 14q23.1 that was associated with the risk of lung cancer in Han Chinese population by using genome-wide ROH association analysis. It is important to further elucidate the genetic determinants of lung cancer. However, the exact mechanisms through which this region leads to the development of lung cancer are still unclear and need to be further investigated. Nevertheless, this study represents a pioneering effort to explore the role of intermediate ROHs in a specific disease on a genome-wide scale.
Footnotes
This work was supported in part the by National Natural Science Foundation of China (81230067, 81270044 and 30901233); Doctoral Fund of Ministry of Education of China (20093234110001), New Century Excellent Talents in University (NCET-10-0178), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12:399–408. doi: 10.1016/S1470-2045(10)70126-1. [DOI] [PubMed] [Google Scholar]
- 3.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 4.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15. 33. Nat Genet. 2008;40:1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, et al. Common 5p15. 33 and 6p21. 33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–9. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. The American Journal of Human Genetics. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, et al. The 5p15. 33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6:e1001051. doi: 10.1371/journal.pgen.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893–6. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12. 12 and 22q12. 2 in Han Chinese. Nat Genet. 2011;43:792–6. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 10.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900–3. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 11.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–95. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- 13.Curtis D, Vine A, Knight J. Study of regions of extended homozygosity provides a powerful method to explore haplotype structure of human populations. Ann Hum Genet. 2008;72:261–78. doi: 10.1111/j.1469-1809.2007.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nothnagel M, Lu TT, Kayser M, Krawczak M. Genomic and geographic distribution of SNP-defined runs of homozygosity in Europeans. Hum Mol Genet. 2010;19:2927–35. doi: 10.1093/hmg/ddq198. [DOI] [PubMed] [Google Scholar]
- 15.Ku CS, Naidoo N, Teo SM, Pawitan Y. Regions of homozygosity and their impact on complex diseases and traits. Hum Genet. 2011;129:1–15. doi: 10.1007/s00439-010-0920-6. [DOI] [PubMed] [Google Scholar]
- 16.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–72. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spain SL, Cazier JB, Houlston R, Carvajal-Carmona L, Tomlinson I. Colorectal cancer risk is not associated with increased levels of homozygosity in a population from the United Kingdom. Cancer Res. 2009;69:7422. doi: 10.1158/0008-5472.CAN-09-0659. [DOI] [PubMed] [Google Scholar]
- 18.Vine AE, McQuillin A, Bass NJ, Pereira A, Kandaswamy R, Robinson M, et al. No evidence for excess runs of homozygosity in bipolar disorder. Psychiatr Genet. 2009;19:165. doi: 10.1097/YPG.0b013e32832a4faa. [DOI] [PubMed] [Google Scholar]
- 19.Enciso-Mora V, Hosking FJ, Houlston RS. Risk of breast and prostate cancer is not associated with increased homozygosity in outbred populations. Eur J Hum Genet. 2010;18:909–14. doi: 10.1038/ejhg.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosking FJ, Papaemmanuil E, Sheridan E, Kinsey SE, Lightfoot T, Roman E, et al. Genome-wide homozygosity signatures and childhood acute lymphoblastic leukemia risk. Blood. 2010;115:4472–7. doi: 10.1182/blood-2009-09-244483. [DOI] [PubMed] [Google Scholar]
- 21.Keller MC, Simonson MA, Ripke S, Neale BM, Gejman PV, Howrigan DP, et al. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 2012;8:e1002656. doi: 10.1371/journal.pgen.1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orloff MS, Zhang L, Bebek G, Eng C. Integrative genomic analysis reveals extended germline homozygosity with lung cancer risk in the PLCO cohort. PLoS One. 2012;7:e31975. doi: 10.1371/journal.pone.0031975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siraj AK, Khalak HG, Sultana M, Al-Rasheed M, Bavi P, Al-Sanea N, et al. Colorectal cancer risk is not associated with increased levels of homozygosity in Saudi Arabia. Genet Med. 2012 Apr 5; doi: 10.1038/gim.2012.27. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Yang HC, Chang LC, Liang YJ, Lin CH, Wang PL. A genome-wide homozygosity association study identifies runs of homozygosity associated with rheumatoid rrthritis in the human major histocompatibility complex. PLoS One. 2012;7:e34840. doi: 10.1371/journal.pone.0034840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91:275–92. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996:299–314. [Google Scholar]
- 29.Keller MC, Visscher PM, Goddard ME. Quantification of Inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics. 2011;189:237–49. doi: 10.1534/genetics.111.130922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nalls MA, Simon-Sanchez J, Gibbs JR, Paisan-Ruiz C, Bras JT, Tanaka T, et al. Measures of autozygosity in decline: globalization, urbanization, and its implications for medical genetics. PLoS Genet. 2009;5:e1000415. doi: 10.1371/journal.pgen.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A. 2007;104:19942–7. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assié G, LaFramboise T, Platzer P, Eng C. Frequency of germline genomic homozygosity associated with cancer cases. JAMA. 2008;299:1437–45. doi: 10.1001/jama.299.12.1437. [DOI] [PubMed] [Google Scholar]
- 33.Nalls M, Guerreiro R, Simon-Sanchez J, Bras J, Traynor B, Gibbs J, et al. Extended tracts of homozygosity identify novel candidate genes associated with late-onset Alzheimer's disease. Neurogenetics. 2009;10:183–90. doi: 10.1007/s10048-009-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Micke P, Edlund K, Holmberg L, Kultima HG, Mansouri L, Ekman S, et al. Gene copy number aberrations are associated with survival in histologic subgroups of non-small cell lung cancer. J Thorac Oncol. 2011;6:1833–40. doi: 10.1097/JTO.0b013e3182295917. [DOI] [PubMed] [Google Scholar]
- 35.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–54. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 36.Nakaya M, Habas R, Biris K, Dunty WC, Kato Y, He X, et al. Identification and comparative expression analyses of Daam genes in mouse and Xenopus. Gene Expr Patterns. 2004;5:97–105. doi: 10.1016/j.modgep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Yates LL, Dean CH. Planar polarity: A new player in both lung development and disease. Organogenesis. 2011;7:209–16. doi: 10.4161/org.7.3.18462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D, Hallett MA, Zhu W, Rubart M, Liu Y, Yang Z, et al. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development. 2011;138:303–15. doi: 10.1242/dev.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller RK, de la Torre Canny SG, Jang CW, Cho K, Ji H, Wagner DS, et al. Pronephric tubulogenesis requires Daam1-mediated planar cell polarity signaling. J Am Soc Nephrol. 2011;22:1654–64. doi: 10.1681/ASN.2010101086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadoya T, Khurana A, Tcherpakov M, Bromberg KD, Didier C, Broday L, et al. JAMP, a Jun N-terminal kinase 1 (JNK1)-associated membrane protein, regulates duration of JNK activity. Mol Cell Biol. 2005;25:8619–30. doi: 10.1128/MCB.25.19.8619-8630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weston CR, Wong A, Hall JP, Goad MEP, Flavell RA, Davis RJ. The c-Jun NH2-terminal kinase is essential for epidermal growth factor expression during epidermal morphogenesis. Proc Natl Acad Sci USA. 2004;101:14114–9. doi: 10.1073/pnas.0406061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khatlani T, Wislez M, Sun M, Srinivas H, Iwanaga K, Ma L, et al. c-Jun N-terminal kinase is activated in non-small-cell lung cancer and promotes neoplastic transformation in human bronchial epithelial cells. Oncogene. 2006;26:2658–66. doi: 10.1038/sj.onc.1210050. [DOI] [PubMed] [Google Scholar]
- 43.Roebroek A, Van De Velde H, Van Bokhoven A, Broers J, Ramaekers F, Van de Ven W. Cloning and expression of alternative transcripts of a novel neuroendocrine-specific gene and identification of its 135-kDa translational product. J Biol Chem. 1993;268:13439–47. [PubMed] [Google Scholar]
- 44.van de Velde HJK, Senden NHM, Roskams TAD, Broers JLV, Ramaekers FCS, Roebroek AJM, et al. NSP-encoded reticulons are neuroendocrine markers of a novel category in human lung cancer diagnosis. Cancer Res. 1994;54:4769–76. [PubMed] [Google Scholar]