Abstract
Most animals from flies to humans count on circadian clocks to synchronize their physiology and behaviors. Daily light cycles are well known environmental cues for setting circadian rhythms. Warmer and cooler temperatures that mimic day and night are also effective in entraining circadian activity in most animals. Even vertebrate organisms can be induced to show circadian responses through exposure to temperature cycles. In poikilothermic animals such as Drosophila, temperature differences of only 2–3°C are sufficient to synchronize locomotor rhythms. However, the molecular sensors that participate in temperature regulation of circadian activity in fruit flies or other animals are enigmatic. It is also unclear whether such detectors are limited to the periphery or may be in the central brain. Here, we showed that Drosophila TRPA1 (transient receptor potential cation channel A1) was necessary for normal activity patterns during temperature cycles. The trpA1 gene was expressed in a subset of pacemaker neurons in the central brain. In response to temperature entrainment, loss of trpA1 impaired activity, and altered expression of the circadian clock protein period (Per) in a subset of pacemaker neurons. These findings underscore a role for a thermoTRP in temperature regulation that extends beyond avoidance of noxious or suboptimal temperatures.
Introduction
Circadian rhythms are self-sustained molecular oscillations, which adjust to environmental changes, such as daily oscillations in light intensities (Reppert and Weaver, 2002; Allada and Chung, 2010; Hardin, 2011; Kwon et al., 2011; Peschel and Helfrich-Forster, 2011). Day/night cycles enable animals to regulate innate behaviors, such as activity patterns. In standard laboratory strains of the fruit fly, Drosophila melanogaster, the animals display activity peaks at dawn and dusk. Once their activity is entrained by the light fluctuations, the flies show anticipatory behavior, which is increased movement before the times that the lights are turned on and off. Another feature of entrainment is that the animals sustain their cyclic behavior in constant darkness.
Over the last 30 years, there has been enormous progress in unraveling the molecular mechanisms underlying the Drosophila light-driven clock (Allada and Chung, 2010; Hardin, 2011; Peschel and Helfrich-Forster, 2011). Light input requires the blue light photoreceptor cryptochrome (Cry), which functions cell-autonomously in pacemaker neurons in the central brain (Emery et al., 1998, 2000; Stanewsky et al., 1998; Krishnan et al., 2001). Changes in circadian activity are controlled by an internal clock that depends on several clock genes and proteins, such as period (Per), which cycle in concentration and in spatial distribution over the circadian cycle in the pacemaker neurons (Konopka and Benzer, 1971; Bargiello et al., 1984; Reddy et al., 1984; Baylies et al., 1987; Siwicki et al., 1988; Zwiebel et al., 1991; Hardin et al., 1992; Curtin et al., 1995).
In addition to light, which is considered to be the strongest Zeitgeber (given time; ZT), it has been long-known that temperature cycles can also synchronize locomotor rhythms in both vertebrate and invertebrate organisms (Roberts, 1962; Helfrich, 1986; Wheeler et al., 1993; Glaser and Stanewsky, 2005, 2007; Boothroyd et al., 2007; Dubruille and Emery, 2008; Buhr et al., 2010; van der Linden et al., 2010). Peripheral neurons participate in temperature entrainment (Glaser and Stanewsky, 2005; Sehadova et al., 2009). In addition, a subset of central pacemaker neurons also contribute to temperature-induced synchronization of locomotor activity and these cells depend on input from the peripheral tissues (Yoshii et al., 2005; Busza et al., 2007; Miyasako et al., 2007; Picot et al., 2009; Sehadova et al., 2009). However, we do not know whether central pacemaker neurons express temperature sensors that function in temperature entrainment.
Members of the TRP family of cations comprise a well established class of temperature detectors (Ramsey et al., 2006; Venkatachalam and Montell, 2007; Damann et al., 2008). Among these channels is Drosophila TRPA1, which functions in the avoidance of noxious tastants (Kang et al., 2010; Kim et al., 2010), insect repellents (Kim et al., 2010), excessively bright light (Xiang et al., 2010), and uncomfortably warm and slightly suboptimal temperatures (Viswanath et al., 2003; Rosenzweig et al., 2005; Kwon et al., 2008; Neely et al., 2011; Shen et al., 2011; Kang et al., 2012; Zhong et al., 2012). Here, we identified a role for Drosophila trpA1 in temperature synchronization of circadian activity. trpA1 was expressed and functioned in a subset of pacemaker neurons in the brain. These data provide evidence that a subset of pacemaker neurons contribute to temperature synchronization of circadian activity through functioning cell-autonomously in temperature sensation.
Materials and Methods
Fly stocks.
We previously described the generation of the trpA11 (Kwon et al., 2008) and trpA1GAL4 (Kim et al., 2010), which we deposited at the Bloomington Stock Center. Both trpA1 mutants were backcrossed into a w1118 background for five generations. The w1118 strain was used as the wild-type control. The tim-GAL4 and cry-GAL4 were contributed by A. Seghal (University of Pennsylvania, Philadelphia, PA) and M. Roshbash (Brandeis University, Waltham, MA), respectively. The w1118 and the UAS-mCD8::GFP flies were from the Bloomington Stock Center.
Immunohistochemistry.
Immunostainings of adult brains were performed as previously described (Lee et al., 2009), except for the addition of the pre-fix step to prepare the data shown in Figures 2, 3, and 6. Briefly, we isolated flies at ZT9 or ZT21 after 5 or 6 d of temperature entrainment (18 h 29°C/6 h 18°C) as mentioned in the figure legends, and fixed the whole flies for 1–2 h with 4% paraformaldehyde in phosphate buffer with 0.1% Triton X-100. We dissected the brains and postfixed them on ice for 45 min by inserting them in 24-well cell culture cluster plates (Costar) containing 940 μl of fix buffer (0.1 m PIPES, pH 6.9, 1 mm EGTA, 1% Triton X-100, 2 mm MgSO4, 150 mm NaCl) and 60 μl of 37% formaldehyde. To perform the immunostainings shown in Figure 4, we used 3- to 7-d-old flies that were entrained using light/dark cycles for 3–5 d. We then dissected the brains at ZT23 and fixed them without a pre-fix. The tissues were washed three times (1× PBS, 0.2% saponin) and blocked overnight at 4°C with 1 ml of Blocking Buffer (1× PBS, 0.2% saponin, 5 mg/ml BSA). The tissues were incubated with the primary antibodies for 24 h at 4°C, washed three times, blocked for 15 min and incubated with the secondary antibodies (Alexa Fluor 488 and Alexa Fluor 568, 1:200; Invitrogen) for 4 h at 4°C, and washed three times. The tissues were transferred into 1.25× PDA solution (37.5% glycerol, 187.5 mm NaCl, 62.5 mm Tris, pH 8.8) and viewed by confocal microscopy (Carl Zeiss LSM510). Primary antibodies were used at the following dilutions: mouse anti-GFP (1:1000, Invitrogen), rabbit anti-GFP (1:1000, Santa Cruz Biotechnology), mouse anti-β-GAL (1:1000, Promega), and rat anti-Per (1:1500; gift from A. Seghal and M. Wu).
Figure 2.
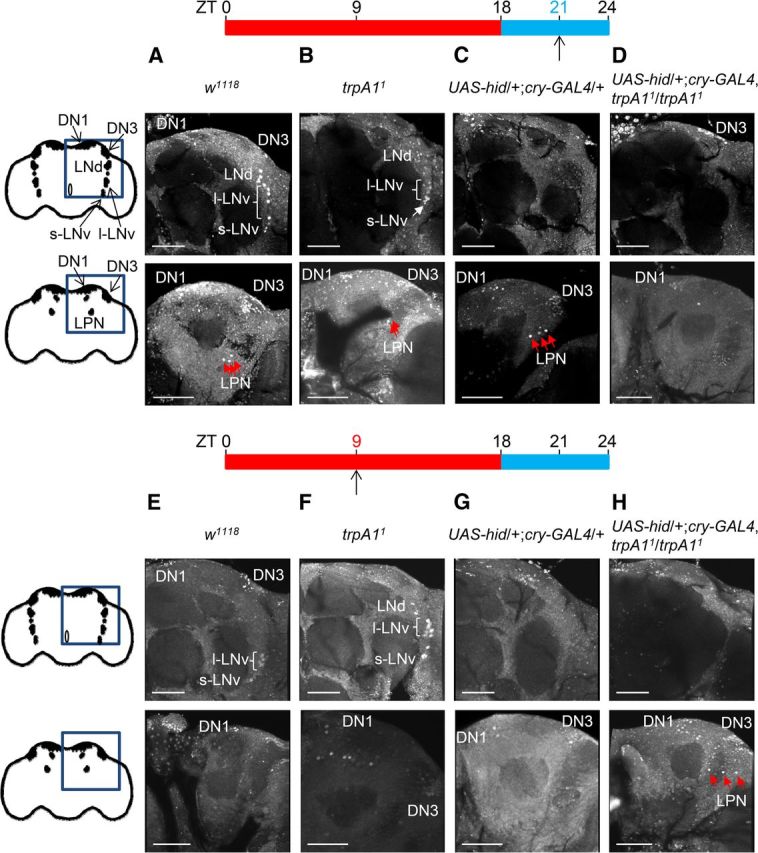
Relative Per levels during a TC cycle at ZT21 and ZT9. Three- to 7-d-old flies were entrained under a TC cycle (29°/18°C) for 6 d and dissected either at ZT21 or ZT9, and stained with anti-Per. The genotypes are indicated. More than 10 hemispheres were examined per genotype. The upper and bottom are anterior and posterior views, respectively. The diagrams represent adult brains. The boxes indicate the brain regions shown. The positions of the clusters of pacemaker neurons are presented. A–D, ZT21. E–H, ZT9. Scale bars, 50 μm.
Figure 3.
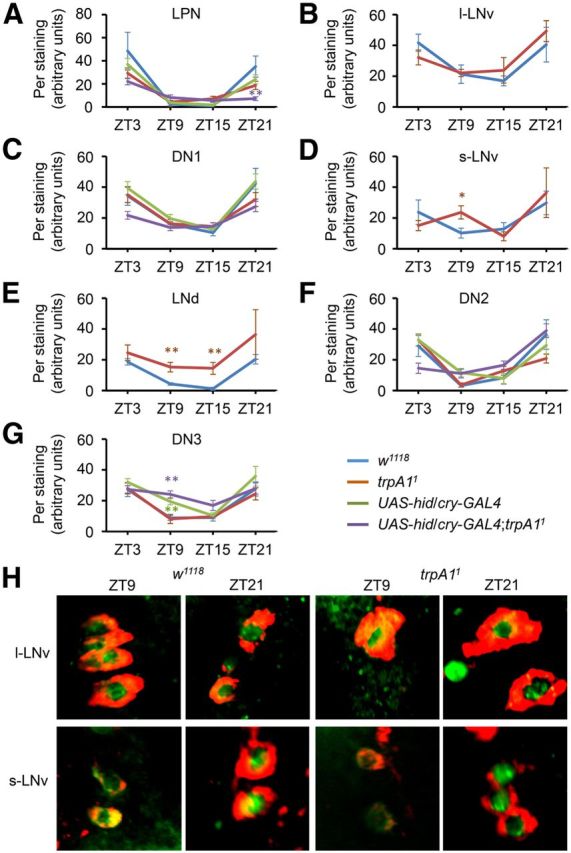
Quantification of Per oscillations in clock neurons. Three- to 7-d-old flies were entrained under a TC cycle (29°/18°C) for 5 d. The brains were then dissected at ZT3, ZT9, ZT15, or ZT21 and stained with anti-Per. A–G, The genotypes and the types of pacemaker neurons are indicated. Data are not indicated for Per levels in l-LNvs, s-LNvs, and LNds since these pacemaker neurons are eliminated in UAS-hid/cry-GAL4;trpA11 (Busza et al., 2007). We confirmed this finding by immunostaining with anti-Per and anti-PDF (data not shown), the latter of which labels only l-LNvs and s-LNvs in wild-type (Renn et al., 1999). The fluorescence intensities were quantified using MacBiophotonics ImageJ software using 3D reconstruction images (10–27 brain hemispheres per sample). The formula for the relative intensity (I) is I = 100(S − B)/B, where S is the fluorescence intensity and B is the background intensity of the region adjacent to the pacemaker neuron (Picot et al., 2009). The error bars indicate SEM. The asterisks indicate significant differences from wild-type (*p < 0.05, **p < 0.01) based on ANOVA and the Scheffé post hoc tests. H, Colabeling with anti-Per (green) and anti-PDF (red) from w1118 and trpA11 l-LNvs, and s-LNvs at ZT9 and ZT21.
Figure 6.
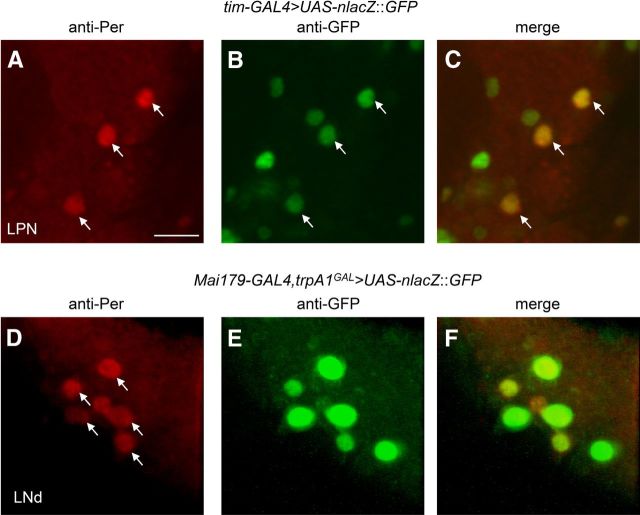
Testing for coexpression of anti-Per with reporters in LPNs and LNds. A–C, The tim-GAL4 reporter was expressed in LPNs. A brain was dissected from UAS-nlacZ::GFP/tim-GAL4 flies at ZT21, after the flies were exposed to 18 h thermophase (29°C)/6 h cryophase (18°C) cycles in the dark for 6 d. The brain was stained with anti-Per and anti-GFP. A, anti-Per (red). The arrows point to LPN neurons that were Per positive. B, anti-GFP (green). C, The merged image from A and B. D–F, Colabeling with anti-Per (red) and anti-GFP (green) from UAS-mCD8::GFP/Mai179-GAL4;trpA1GAL4/+ flies at ZT21 after entraining flies under TC cycles for 6 d in the dark. D, anti-Per (red). A total of six LNds are labeled. The arrows point to LNds that were Per and GFP positive in merged image in F. E, Anti-GFP (green). F, The merged image from D and E. Scale bar, 10 μm.
Figure 4.
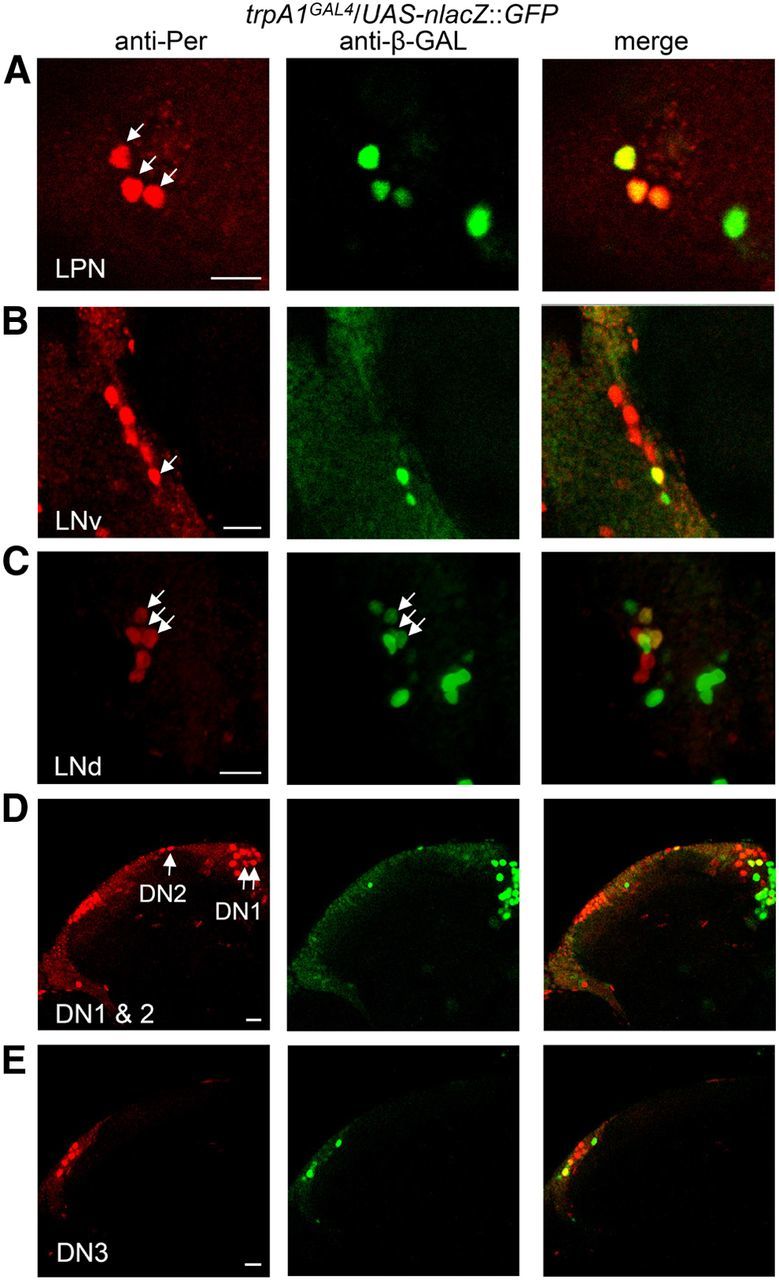
Coexpression of the trpA1-reporter and Per in pacemaker neurons. Brains from 3- to 7-d-old UAS-nlacZ::GFP/+;trpA1GAL4/+ flies immunostained with anti-β-GAL and anti-Per at ZT23 during LD cycles. The right panels are the merged images. A, LPN. B, LNv. C, LNd. These 3D images were reconstituted from confocal stacks. D, DN1 and 2. E, DN3. The arrowheads indicate colabeling of Per and the trpA1 reporter. Scale bar, 10 μm.
Behavioral assays.
For circadian locomotor behavior, we used the Drosophila Activity Monitoring (DAM) system (TriKinetics), which automatically records activity every time a fly crosses an infrared beam. Three- to 7-d-old flies were loaded into tubes that had 1% agarose and 5% sucrose food at one end. We entrained the flies to light/dark (LD) cycles for 3–5 d at 25° or 18°C before changing to the next conditions, such as darkness only (16°, 18°, 25°, or 29°C) or thermophase/cryophase (TC) cycles as indicated in the figures.
The TC entrainments in Figures 1 and 7 were performed as follows. The flies were entrained initially using 12 h light/12 h dark cycles for 4 d at 18°C, followed by 7 d of 18 h thermophase (29°C) and 6 h cryophase (18°C) cycles in constant darkness. The thermophase cycles were advanced 8 h relative to the previous LD cycle. After temperature entrainment, the flies were kept at 18°C in the dark (free run).
Figure 1.
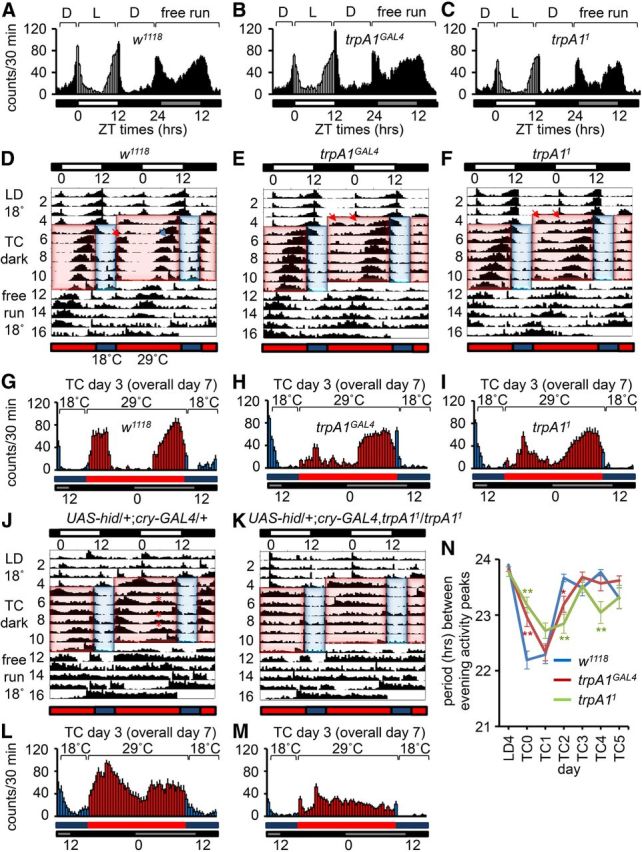
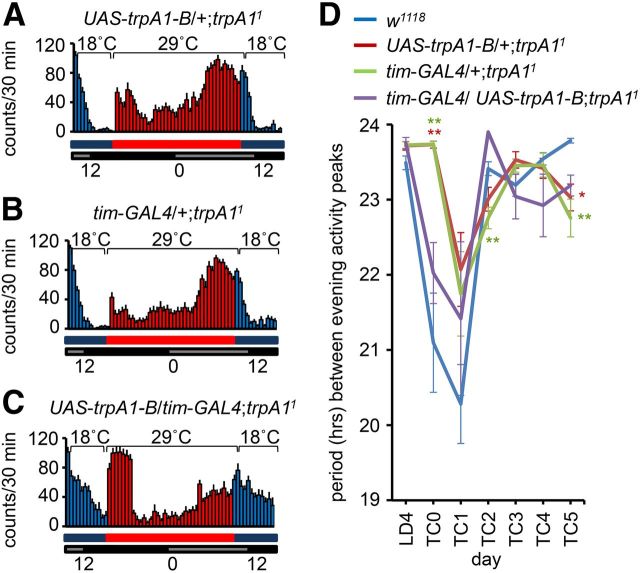
Actograms using LD and TC entrainment. A–C, Average activities of male flies on day 4 under LD conditions (12 h light/dark cycles at 25°C), and day 1 under constant darkness (free run at 25°C). ZT times are indicated. The white, black, and gray bars represent day, night, and subjective day, respectively. A, w1118. B, trpA1GAL4. C, trpA11. D–F, J, K, Average normalized activities for the indicated genotypes. Flies were maintained for 4 d at 18°C <12 h light/dark cycles (LD 18°C) and entrained for 7 d under constant darkness using 18 h (29°C) thermophases/6 h (18°C) cryophases (TC dark). The flies were then kept at a constant temperature (18°C) in the dark (free run 18°C). Except for overall days 1 and 17, the daily activities are presented on two lines. The top white and black bars indicate LD cycles. The bottom red and blue bars indicate TC cycles. The red and blue shading indicates warm and cool phases, respectively. D, w1118. The red and blue arrows on overall day 7 (TC day 3) indicate examples of morning and evening activities, respectively. E, trpA1GAL4. F, trpA11. The two arrows on overall day 5 (TC day 1) indicate small double peaks. G–I, Activities from TC day 3 (overall day 7) from D–F, respectively. G, w1118. H, trpA1GAL4. I, trpA11. J, UAS-hid/+;cry-GAL4/+. Asterisks indicate smaller and broader evening activities. K, UAS-hid/+;cry-GAL4,trpA11/trpA11. L, M, Activities from TC day 3 (overall day 7) from J and K, respectively. N, Periods between evening activity peaks from D–F. The error bars represent SEM. The asterisks indicate significant differences from wild-type (*p < 0.05, **p < 0.01) based on ANOVA and the Scheffé post hoc test.
Figure 7.
Suppression of the trpA1 phenotype by expression of the trpA1-B isoform in pacemaker neurons. Flies were maintained and entrained using the same paradigm as in Figure 1D–K. A–C, Average activities of male flies on TC day 3 (overall day 7) for the indicated genotypes. D, Periods between evening activity peaks from LD4 to TC5. The error bars represent SEMs. The asterisks indicate significant differences from wild-type (*p < 0.05, **p < 0.01) based on ANOVA and the Scheffé post hoc test.
To perform the TC entrainment experiments shown in Figure 5 (antiphase experiments), the flies were entrained using 12 h LD cycles for 3 d at 25°C followed by 6 d of 12 h thermophase (25°C) and 12 h cryophase (16°C) TC cycles in constant darkness. The warm and cold phases were antiphase to the previous LD cycle, such that the first thermophase coincided with the time when the last night cycle during the LD cycle would have occurred. After the temperature entrainment, the flies were maintained at 25°C for the free running analysis.
Figure 5.
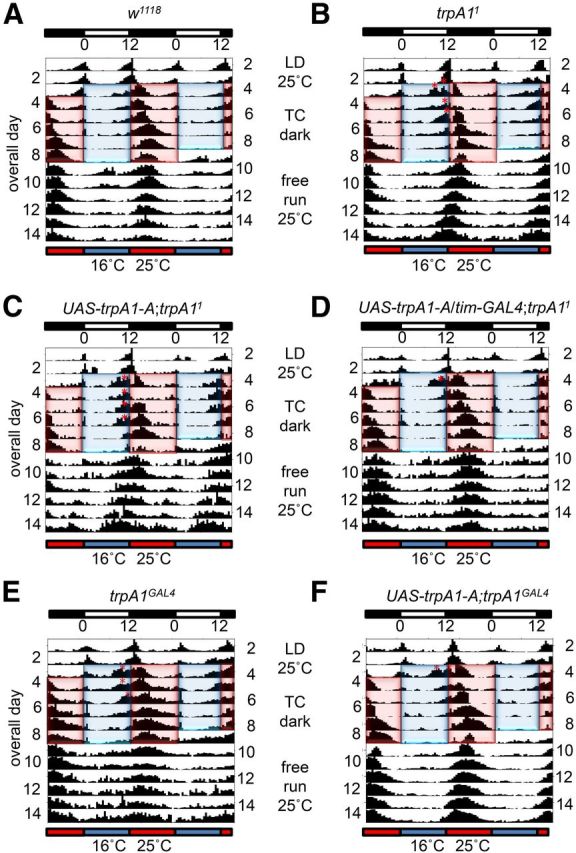
Suppression of the trpA1 phenotype by expression of the wild-type trpA1 transgene in pacemaker neurons. Average actograms using flies that were entrained to 12 h light/dark cycles for 3 d at 25°C followed by 6 d of 12 h thermophase (25°C) and 12 h cryophase (16°C) cycles in constant darkness. The warm and cool cycles, which simulate day and night, were the reverse phase from the night and day during the LD cycles (antiphase paradigm). After the temperature entrainment, the flies were kept at 25°C (free run). The white and black bars at the top indicate the LD cycles and the red and blue bars at the bottom indicate the TC cycles. The thermophases and cryophases are highlighted on the actograms with red and blue boxes, respectively. Asterisks indicate sustained peaks that were not detected in the w1118 actogram. A, w1118. B, trpA11. C, UAS-trpA1-A;trpA11. D, UAS-trpA1-A /tim-GAL4;trpA11. E, trpA1GAL4. F, UAS-trpA1-A;trpA1GAL4.
Data analyses were performed using ClockLab in conjunction with MatLab, after collecting each file using 30 min bins. To determine the period and Fast Fourier Transfer (FFT) values, we analyzed the data obtained during the free runs (d 2–7) using χ2 periodograms and batch analysis. If the τ FFT was not 0.042 (which corresponded to a 24 h period), the FFT values for the individual files were calculated manually. If the value of the FFT was >0.05, this indicated that >5% of the power was at that frequency and considered rhythmic. If the values for the FFT were <0.03, the flies were considered arrhythmic. If the values were between 0.05 and 0.03, the rhythmicities were considered to be borderline.
Results
Loss of trpA1 altered activity during temperature entrainment of circadian rhythm
The Drosophila TRPA1 channel is a candidate for sensing thermal cycles that affect activity rhythms because it participates in thermotaxis (Rosenzweig et al., 2005; Kwon et al., 2008) and is directly activated in a range (>24°C) (Viswanath et al., 2003) that functions in temperature synchronization of circadian rhythms (18°–29°C). This is in contrast to two other thermally-activated Drosophila TRP channels, Pyrexia and Painless, which are activated directly by temperatures with much higher thresholds (40°C and 42°C, respectively) (Tracey et al., 2003; Lee et al., 2005). To determine whether loss of the TRPA1 channel affected activity patterns during temperature entrainment, we analyzed trpA1 mutants using the DAM system (Chiu et al., 2010). Before assaying circadian activity set by TC cycles, we first examined activity entrained by 12 h LD cycles at a constant temperature (25°C). The trpA1+ controls (w1118; referred to as wild-type) have activity peaks at “lights-on” (ZT0) and “lights-off” (ZT12; Fig. 1A) (Allada and Chung, 2010). Flies increase their activity before the morning (lights on) or evening (lights off). This is called “morning anticipation” and “evening anticipation” and is a hallmark of circadian entrainment (Fig. 1A) (Allada and Chung, 2010). The evening anticipation displayed by trpA1 mutants during the LD cycles was similar to the wild-type control (Fig. 1A–C). The trpA1 flies also displayed morning anticipation, although it appeared to be reduced slightly (Fig. 1A–C). After allowing the flies to free run in constant darkness (DD), the morning (dawn) and evening (dusk) peaks remained, and the periodicities were similar to wild-type (Fig. 1A–C; Table 1). During the DD cycles, there also appeared to be reduced morning anticipation in trpA1 mutants (Fig. 1A–C). Thus, with the exception of a reduction in morning anticipation, the trpA1 flies behaved similarly to wild-type during the LD cycle and during the DD free run.
Table 1.
Circadian rhythmicity
| Genotype | Entrain condition | Entrain temp (°C) | Free run (°C) | n | Rhy (%) | Period (h) | FFT |
|---|---|---|---|---|---|---|---|
| Wild-type | LD | 25 | 25 | 77 | 95 | 23.93 ± 0.03 | 0.15 ± 0.01 |
| trpA1GAL4 | LD | 25 | 25 | 59 | 90 | 23.90 ± 0.04 | 0.17 ± 0.01 |
| trpA1GAL4/+ | LD | 25 | 25 | 16 | 100 | 23.83 ± 0.02 | 0.21 ± 0.02 |
| trpA11 | LD | 25 | 25 | 32 | 94 | 23.63 ± 0.05 | 0.15 ± 0.01 |
| Wild-type | LD | 29 | 29 | 16 | 100 | 23.69 ± 0.06 | 0.15 ± 0.01 |
| trpA1GAL4 | LD | 29 | 29 | 16 | 100 | 23.66 ± 0.05 | 0.14 ± 0.01 |
| trpA1GAL4/+ | LD | 29 | 29 | 15 | 100 | 23.68 ± 0.03 | 0.14 ± 0.01 |
| Wild-type | TC | 25/16 | 25 | 45 | 93 | 23.66 ± 0.08 | 0.15 ± 0.01 |
| trpA11 | TC | 25/16 | 25 | 45 | 96 | 23.28 ± 0.04* | 0.13 ± 0.01 |
| trpA1GAL4 | TC | 25/16 | 25 | 36 | 83 | 23.11 ± 0.08* | 0.10 ± 0.01 |
| trpA1GAL4/trpA11 | TC | 25/16 | 25 | 31 | 100 | 23.36 ± 0.07* | 0.15 ± 0.01 |
| UAS-trpA1-A;trpA1GAL4 | TC | 25/16 | 25 | 28 | 96 | 23.64 ± 0.04 | 0.22 ± 0.01 |
| UAS-trpA1-A;trpA11 | TC | 25/16 | 25 | 29 | 97 | 23.19 ± 0.07* | 0.08 ± 0.01 |
| tim-GAL4/+;trpA11 | TC | 25/16 | 25 | 16 | 81 | 23.32 ± 0.12* | 0.10 ± 0.01 |
| tim-GAL4/UAS-trpA1-A; trpA11 | TC | 25/16 | 25 | 16 | 88 | 24.12 ± 0.09 | 0.15 ± 0.02 |
| trpA1GAL4/+ | TC | 25/16 | 25 | 16 | 100 | 23.71 ± 0.06 | 0.20 ± 0.02 |
The flies were exposed to either LD or TC using a 12/12 h paradigm. Data are shown as the period and FFT values (±SEM) after the LD or TC cycles during days 2–7 of the free runs.
*p < 0.05, significant differences from wild-type using ANOVA and the Scheffé post hoc tests. Rhy, Rhythmicity.
To address whether loss of trpA1 impaired temperature synchronization of circadian activity, we switched from a LD cycle at 18°C, to constant darkness and used thermophase (29°C; simulates day) and cryophase (18°C, simulates night) cycles (Busza et al., 2007). To clearly resolve the morning and evening peaks, we initiated the first thermophase cycle 4 h into the final night cycle. We used 18 and 6 h TC cycles, which help separate the morning and evening peaks (Busza et al., 2007). Using this paradigm, wild-type flies reestablish morning and evening peaks within ∼3 d, including pronounced evening anticipation (Fig. 1D,G, red and blue arrows on TC day 3, which was day 7 overall; note that except for day 1, each day of activity is shown on two lines to facilitate comparisons with the previous and following days).
Both trpA1 alleles (trpA1GAL4 and trpA11) (Kwon et al., 2008; Kim et al., 2010) displayed differences from wild-type during the TC cycles. During the first TC cycle the trpA1 mutants exhibited double peaks (Fig. 1E,F, two red arrows on TC day 1, overall day 5). The second peak approximately coincided with the original morning peak established during the LD entrainment, suggesting that this peak persisted. In addition, the morning peaks during the TC cycles were reduced (Fig. 1D–I). The evening activity exhibited by the mutant flies initiated slightly earlier than wild-type (advanced activity), and was broader (Fig. 1D–I). During the TC cycles the trpA1 mutants required an additional day compared with wild-type to establish a constant period between the evening activity peaks (Fig. 1N). By TC day 2, wild-type displayed a period comparable to that during LD4 (Fig. 1N; TC2, 23.67 ± 0.07; LD4, 23.90 ± 0.00). However, the trpA1 mutants did not exhibit a similar period until TC day 3 (Fig. 1N; trpA1GAL4, 23.68 ± 0.09; trpA11, 23.53 ± 0.17).
Because trpA1 mutants still have rhythmic activity induced by temperature entrainment, we tested whether the effect of removing trpA1 would be enhanced by genetically ablating a subset of pacemaker neurons. In the Drosophila brain there are ∼150 pacemaker neurons. In addition to six lateral posterior neurons (LPNs), the other pacemaker neurons sort into two major groups (Allada and Chung, 2010). These include the lateral neurons (LNs), which are located in the anterior cortex between the central brain and optic lobe, and the dorsal neurons (DNs) situated in the cortex associated with the dorsal protocerebrum. The LNs are divided into five small and four large ventrolateral neurons (s-LNvs and l-LNvs, respectively) and six dorsolateral neurons (LNds) in each hemisphere. There are also three groups of DNs (DN1, DN2, and DN3). Several pacemaker neurons have been suggested to participate in temperature entrainment of circadian rhythms. These include LNvs (Busza et al., 2007), DN1s (Zhang et al., 2010), DN2s (Busza et al., 2007; Picot et al., 2009), and LPNs (Busza et al., 2007; Yoshii et al., 2009). In addition, the concentration of Per in LPNs changes strongly in response to TC cycles but not to LD cycles (Busza et al., 2007). With the exception of the LPNs and DN2s, most pacemaker neurons including the LNvs, LNds, and some DN1s and DN3s are removed by expressing the proapoptotic gene UAS-hid under the control of the cry-GAL4 (Busza et al., 2007). Therefore, we compared temperature entrainment of circadian activity in UAS-hid;cry-GAL4 flies in the presence or absence of the trpA11 mutation. As previously described, during a LD cycle, the UAS-hid;cry-GAL4 flies were missing the usual morning and evening peaks of activity, except for brief startle responses when the lights were turned on and off (Stoleru et al., 2004; Busza et al., 2007) (Fig. 1J). When these flies were switched to a TC cycle, they displayed two broad peaks (Fig. 1J,L) (Busza et al., 2007). During the free run at 18°C, 41% of the flies displayed rhythmicity (Table 2).
Table 2.
Circadian rhythmicity
| Genotype | Entrain temp (°C) | Free run (°C) | n | Rhy (%) | Period (h) | FFT |
|---|---|---|---|---|---|---|
| Wild-type | 29/18 | 18 | 29 | 86 | 23.52 ± 0.07 | 0.11 ± 0.01 |
| trpA11 | 29/18 | 18 | 30 | 90 | 23.32 ± 0.07 | 0.15 ± 0.01 |
| trpA1GAL4 | 29/18 | 18 | 30 | 80 | 23.48 ± 0.14 | 0.13 ± 0.01 |
| UAS-hid/cry-GAL4 | 29/18 | 18 | 32 | 41 | 27.12 ± 1.15 | 0.19 ± 0.01 |
| UAS-hid/cry-GAL4;trpA11 | 29/18 | 18 | 32 | 6 | 23.00 ± 0.90 | 0.08 ± 0.02 |
| per01 | 29/18 | 18 | 31 | 23 | 26.50 ± 1.64 | 0.05 ± 0.01 |
| UAS-trpA1-A /cry-GAL4;trpA11 | 29/18 | 18 | 16 | 94 | 22.03 ± 0.13 | 0.09 ± 0.01 |
| UAS-trpA1-B/+;trpA11 | 29/18 | 18 | 32 | 97 | 23.73 ± 0.09 | 0.20 ± 0.01 |
| tim-GAL4/+;trpA11 | 29/18 | 18 | 32 | 88 | 24.14 ± 0.38 | 0.20 ± 0.01 |
| UAS-trpA1-B /tim-GAL4;trpA11 | 29/18 | 18 | 17 | 100 | 23.90 ± 0.10 | 0.19 ± 0.01 |
| Wild-type | 29/18 | 29 | 15 | 73 | 23.33 ± 0.25 | 0.09 ± 0.01 |
| trpA11 | 29/18 | 29 | 16 | 100 | 22.68 ± 0.11* | 0.10 ± 0.01 |
| trpA1GAL4 | 29/18 | 29 | 15 | 67 | 22.72 ± 0.09* | 0.08 ± 0.01 |
| per01 | 29/18 | 29 | 16 | 0 | ||
| UAS-hid/cry-GAL4 | 29/18 | 29 | 16 | 38 | 22.94 ± 0.19 | 0.08 ± 0.01 |
| UAS-hid/cry-GAL4;trpA11 | 29/18 | 29 | 16 | 6 | 22.91 | 0.1221 |
| Wild-type | 25/16 | 16 | 16 | 75 | 23.16 ± 0.20 | 0.07 ± 0.01 |
| trpA11 | 25/16 | 16 | 16 | 100 | 21.88 ± 0.26* | 0.07 ± 0.01 |
| UAS-trpA1-A;trpA11 | 25/16 | 16 | 16 | 81 | 21.78 ± 0.54* | 0.07 ± 0.01 |
| UAS-trpA1-A /tim-GAL4;trpA11 | 25/16 | 16 | 15 | 93 | 22.56 ± 0.21 | 0.08 ± 0.01 |
| UAS-trpA1-A /cry-GAL4;trpA11 | 25/16 | 16 | 16 | 69 | 22.31 ± 0.14* | 0.05 ± 0.01 |
| per01 | 25/16 | 16 | 16 | 0 |
The flies were entrained under 18 h thermophase/6 h cryophase conditions and either 29°/18°C or 25°/16°C TC cycles. The free runs were performed at either 18° or 29°C. The period and FFT values (±SEM) are based on the free runs (days 2–7).
*p < 0.05, significant differences from wild-type, based on ANOVA and the Scheffé post hoc tests.
1No error bars are presented because only one UAS-hid/cry-GAL4;trpA11 fly showed rhythmicity.
Of significance here, when we combined the trpA11 mutation with the UAS-hid;cry-GAL4 transgenes, the activity peaks during the TC cycles were virtually eliminated, and the flies were nearly arrhythmic during the free runs (6% rhythmicity) (Fig. 1K,M; Table 2). We obtained similar results when switched from the TC cycles to a free run using a constant thermophase (29°C) instead of cryophase (Table 2). Because introduction of the trpA1 mutation into the sensitized UAS-hid;cry-GAL4 background virtually eliminated the activity peaks during the TC cycles, and caused the flies to lose nearly all rhythmicity during the free runs, TRPA1 appears to contribute to TC entrainment of circadian rhythm. Even without the sensitized background (UAS-hid;cry-GAL4), loss of trpA1 alone shortened the periodicity during the 29°C free run following 29°/18° TC cycles (18/6 h phases) (Table 2), and during the 16°C free run following 25°/16°C cycles (18/6 h phases) (Table 2).
Increased variation in Per expression in trpA11
In wild-type pacemaker neurons, the concentration of the clock protein Period (Per) (Konopka and Benzer, 1971; Citri et al., 1987) oscillates during the circadian cycle (Siwicki et al., 1988; Yoshii et al., 2005; Allada and Chung, 2010). Per is high at ZT21 (middle of the night) and ZT3 (early morning), and low at ZT9 (middle of the day) and ZT15 (late afternoon (Figs. 2A,E, 3A–G). The pacemaker neurons in the trpA1 mutants displayed oscillations in Per, but the patterns of the oscillations and concentrations of Per among pacemaker neurons were more variable in trpA11, especially in combination with UAS-hid;cry-GAL4. The Per intensities and oscillations in trpA11 brains were similar to wild-type in some pacemaker neurons (e.g., l-LNvs, Figs. 2A,B,E,F, 3B). At ZT9, when Per expression was relatively low in wild-type, Per was elevated in some trpA11 and UAS-hid;cry-GAL4,trpA11 pacemaker neurons (e.g., s-LNvs and LNds, Figs. 2E,F, 3D,E; or DN3, Figs. 2H, 3G). Per expression also tended to be lower at ZT21 in trpA11 LPNs, DN1s, and DN2s, although the differences were not statistically significant using ANOVA (Figs. 2A,B, 3A,C,F). However, in a UAS-hid;cry-GAL4;trpA11 background, Per was virtually undetectable in LPNs at ZT21 (Figs. 2D, 3A), and showed only mild oscillations in Per expression in multiple pacemaker neurons (e.g., LPNs, DN1s, and especially DN3s, Fig. 3A,C,G).
We also examined the subcellular localization of Per. In wild-type pacemaker neurons, with the exception of the l-LNvs, Per was primarily in the cytosol at ZT9, and in the nucleus at ZT21 (Figs. 2A,E, 3H). However, we occasionally detected Per nuclear localization in l-LNvs earlier than s-LNvs (Fig. 3H). At ZT9, Per was distributed in both the cytosol and nucleus of l-LNvs, and primarily in the cytosol of s-LNvs (Figs. 2E, 3H). The subcellular distribution of Per was similar in the mutant pacemaker neurons; however, Per expression in trpA11 tended to be higher and more variable than in wild-type at ZT9 (Figs. 2E,F, 3H). This suggests that the oscillation of Per in trpA11 was slightly advanced, consistent with the behavioral results (Table 2).
Expression of trpA1 in pacemaker neurons contributes to temperature synchronization of locomotor activity
Peripheral neurons participate in temperature entrainment of circadian rhythm (Sehadova et al., 2009). Thus, TRPA1 could promote TC control of circadian activity either by functioning in peripheral neurons that send axons to pacemaker neurons, and/or by acting in pacemaker neurons. To address this question, we first examined the presumptive expression pattern of trpA1 using a GAL4 reporter that we inserted previously at the position of the endogenous trpA1 translation initiation site (Kim et al., 2010) in combination with a UAS-nuclear::lacZ::GFP. The trpA1 reporter was expressed in many pacemaker neurons, which also expressed Per (Fig. 4). These included all three LPNs (Fig. 4A), and 1–3 neurons in other groups of pacemaker neurons (Fig. 4B–E).
To address whether trpA1 has a role in pacemaker neurons for temperature synchronization of activity rhythms (29°/18°C TC cycles), we set out to perform rescue experiments with a transgene that expressed TRPA1-A (Kwon et al., 2010). However, this greatly reduced viability due to activation of the TRPA1 channel at 29°C (Viswanath et al., 2003). Therefore, we used lower temperatures for the TC cycles (25° and 16°C), which had no adverse effects on viability. We entrained the flies with a LD cycle, and initiated the first cryophase (simulates night) when the light cycle would have initiated, as previously described (Glaser and Stanewsky, 2005). This “antiphase paradigm” causes wild-type flies to display a robust single peak in the middle of the thermophase (Glaser and Stanewsky, 2005) (Fig. 5A). During the subsequent free running period at 25°C, the single peak during the thermophase persisted for many days (Fig. 5A).
Using the lower TC temperatures (25°/16°C), the trpA1 mutant phenotype was relatively subtle, indicating that TRPA1 was more important for temperature controlled circadian activity using higher thermophase conditions (29°C). Nevertheless, there were two differences from wild-type, which we used to test for rescue. First, during the first 4 d of the TC cycles, trpA11, or UAS-trpA1-A;trpA11 flies exhibited additional activity near the end of the cryophase cycle, which corresponded approximately with the former evening period under the LD cycles (Fig. 5B,C, red asterisks). Second, during the free run, the circadian cycle was slightly shorter in trpA11 and trpA1GAL4 (23.28 ± 0.04 and 23.11 ± 0.08, respectively) (Table 1) than in w1118 (23.66 ± 0.08; Table 1). To test for rescue, we expressed UAS-trpA1-A under the control of the tim-GAL4, which was expressed in pacemaker neurons (Dubruille and Emery, 2008). The extra peaks in the UAS-trpA1-A;trpA11 control flies during days 2–4 of the TC cycles (Fig. 5C, asterisks) were diminished or eliminated (Fig. 5D), and the length of the circadian cycle during the free run was similar to wild-type (Table 1). We confirmed that the tim-GAL4 was expressed in LPNs, as the tim-GAL4/UAS-nlacZ::GFP was coexpressed with the Per protein (Fig. 6A–C). We also rescued the trpA1GAL4 mutant phenotype using the UAS-trpA1-A;trpA1GAL4 transgenes (Fig. 5E,F; Table 1). Moreover, we performed the lower TC temperatures (25°/16°C) using the 18/6 h paradigm that we used with the higher temperature TC cycles (29°/18°C). Under these latter conditions, the main difference from wild-type was that the periodicity was shorter (wild-type, 23.16 ± 0.20; trpA11, 21.88 ± 0.26) (Table 2). The rescue of this phenotype by expression of UAS-trpA1-A under control of the tim-GAL4 was statistically significant, whereas the suppression resulting from expression of UAS-trpA1-A under control of the cry-GAL4 was not (Table 2).
Recently it was reported that TRPA1 is expressed as at least four isoforms, two of which (TRPA1-B and TRPA1-C) are not activated directly by elevated temperatures (Kang et al., 2012; Zhong et al., 2012). Therefore, to address whether or not TRPA1 might be a direct thermosensor in pacemaker neurons, we expressed UAS-TRPA1-B in trpA11 mutant animals pacemaker neurons under the control of the tim-GAL4, and used the 16 h 29° and 8 h 18°C TC paradigm described above. We found that introduction of TRPA1-B in pacemaker neurons (UAS-trpA1-B/tim-GAL4;trpA11) restored the morning peak (Fig. 7C), which was reduced in trpA1 mutant control flies (Fig. 7A,B). In addition, expression of TRPA1-B in pacemaker neurons reduced the advanced evening activity, and decreased the mid-thermophase activity displayed by the trpA11 mutant animals (Fig. 7A–C). Using this TC paradigm, wild-type flies were capable of establishing the same periodicity between the evening peaks on TC2, as was displayed on LD4 (Figs. 1N, 7D, LD4, 23.49±0.09; TC2, 23.41 ± 0.09). The trpA1 mutant flies required an additional day, and did not show a periodicity similar to LD4 until TC3 (Figs. 1N, 7D, UAS-trpA1-B/+;trpA11 and tim-GAL4/+;trpA11: LD4, 23.72 ± 0.05 and 23.73 ± 0.04, respectively; TC2, 23.01 ± 0.16 and 22.76 ± 0.14, respectively; TC3, 23.53 ± 0.11 and 23.45 ± 0.09, respectively). However, the rescue flies (UAS-trpA1-B/tim-GAL4;trpA11) showed a periodicity comparable to LD4 on TC2 (Fig. 7D, LD4, 23.75 ± 0.07; TC2, 23.90 ± 0.00).
Discussion
Temperature entrainment of the central clock depends at least in part on peripheral sensory neurons, and on the phospholipase C (PLC) encoded by norpA, and a novel glutamate-rich protein referred to Nocte (Glaser and Stanewsky, 2005; Sehadova et al., 2009). However, it was unclear whether TC synchronization of the circadian clock depends exclusively on temperature sensation in peripheral neurons or whether there exist one or more sensors in the pacemaker neurons that promotes temperature-controlled activity patterns. Here, we conclude that a subset of central pacemaker neurons functions cell-autonomously in temperature synchronization of circadian rhythm, and that TRPA1 contributes to this behavior.
TRPA1 may not function in all pacemaker neurons since it appeared to be expressed in just a subset of these cells. The LPNs would appear to constitute one class of pacemaker neurons in which trpA1 is functionally important since trpA1 was expressed in all thee LPNs, and flies with intact LPNs but not other pacemaker neurons (UAS-hid;cry-GAL4) were still capable of TC entrainment of circadian rhythms. Moreover, entrainment was eliminated if the UAS-hid;cry-GAL4 transgenes were combined with a trpA1 mutation. These data are consistent with previous studies suggesting that the LPNs contribute to temperature control of circadian rhythm (Busza et al., 2007; Yoshii et al., 2009). The TRPA1 channel may also function in other pacemaker neurons since the trpA1 reporter was expressed in at least one neuron in most clusters of pacemaker neurons, including LNvs, DN1s, and DN2s, which have also been proposed to contribute to TC entrainment (Busza et al., 2007; Picot et al., 2009; Zhang et al., 2010). We suggest that the trpA1-expressing neuron in each cluster might be the central core sensory neuron that responds to temperature changes, and mediates TC sensitivity in other neurons within the clusters.
In addition to affecting TC-induced activity patterns, mutation of trpA1 also impacted on TC-dependent cycling of Per expression in some pacemaker neurons. Thus, although central brain neurons are not capable of undergoing TC entrainment in the absence of peripheral input (Sehadova et al., 2009), it appears that a temperature sensor in pacemaker neurons promotes normal fluctuations in Per expression. Because we observed alterations in Per cycling in just a subset of pacemaker neurons, it appears that the central neurons are not uniformly sensitive to responding to temperature in a cell autonomous manner.
It has been proposed that separate sets of central neurons are the most critical for light and temperature entrainment, and cry-negative neurons may be the more important temperature sensors (Busza et al., 2007; Miyasako et al., 2007; Picot et al., 2009; Sehadova et al., 2009). Such specialization may limit temperature interference with light entrainment (Sehadova et al., 2009). In this context, it is worth noting that some trpA1-positive central neurons were cry negative. For example, the trpA1 reporter was expressed in three LNds (Fig. 4C), and that cry was expressed in only three of six LNds. The Mai179-GAL4 is expressed in the three Cry-positive LNd cells (Grima et al., 2004; Picot et al., 2007), and five of six LNd cells were labeled with both reporters (Fig. 6D–F), indicating that at least two trpA1-positive cells did not overlap with the Cry-positive LNd cells.
TRPA1 contributes to temperature control of circadian rhythm, but does not appear to be a direct temperature sensor in pacemaker neurons for two reasons. First, loss of TRPA1 impairs 25°/16°C TC cycles, and these temperatures are below the thermal threshold for activation of TRPA1, which is ∼27°C. Second, introduction of a temperature-insensitive TRPA1 isoform (TRPA1-B) (Kang et al., 2012; Zhong et al., 2012) in trpA1 mutant flies restored the morning peaks during 29°/18°C TC cycles, reduced the advanced evening activity, decreased the mid thermophase activity of the mutants and eliminated the need for one additional day to establish a constant period between the evening activity peaks. These data suggest that TRPA1 might function downstream of another temperature sensor. Although the identity of the thermosensor remains to be identified, the activity of TRPA1 might be coupled to the PLC that is encoded by norpA. This is plausible because NORPA contributes to this temperature entrainment pathway (Glaser and Stanewsky, 2005), and TRPA1 acts downstream of NORPA to promote temperature sensation in larvae (Kwon et al., 2008).
TRPA1 cannot define the only pathway that contributes to temperature synchronization of circadian rhythm because mutation of trpA1 alone had a modest effect, although when combined with the UAS-hid;cry-GAL4 transgenes, temperature entrainment was eliminated. If a second cation channel promotes TC synchronization, it may not belong to the TRP superfamily, because no other Drosophila TRP appears to be activated by environmental temperatures in the range that functions in control of circadian rhythms. Given that input from peripheral tissues, such as chordotonal organs, impacts on temperature entrainment (Sehadova et al., 2009), we suggest that thermal sensors exist in both central pacemakers and in peripheral neurons to synchronize activity by TC cycles. It is also plausible that additional thermosensory input is mediated through central brain cells that are not pacemaker neurons. Nevertheless, because TRPA1 is one channel that promotes temperature control of circadian rhythm, our findings demonstrate that the roles of a thermoTRP in a poikilothermic organism are not limited to the avoidance of undesirable temperatures.
Footnotes
This work was supported by a grant to C.M. from the National Institute of General Medical Sciences, NIH (GM085335). Y. L. was supported by the Basic Science Research Program of the National Research Foundation (NRF) of Korea funded by the Ministry of Education, Science, and Technology (2012R1A1A2003727) and the Bio and Medical Technology Development Program of the NRF funded by the Korean government (MEST) (2012M3A9B2052525). We thank A. Seghal, M. Roshbash, M. Wu, and P. Emery for fly stocks and Per antibodies; E. Shin, P. Neela, and J. Tsai for help with the circadian assays; and M. Wu for critical reading of the paper.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 2007;3:e54. doi: 10.1371/journal.pgen.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busza A, Murad A, Emery P. Interactions between circadian neurons control temperature synchronization of Drosophila behavior. J Neurosci. 2007;27:10722–10733. doi: 10.1523/JNEUROSCI.2479-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Low KH, Pike DH, Yildirim E, Edery I. Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp. 2010;43:2157. doi: 10.3791/2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri Y, Colot HV, Jacquier AC, Yu Q, Hall JC, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- Damann N, Voets T, Nilius B. TRPs in our senses. Curr Biol. 2008;18:R880–R889. doi: 10.1016/j.cub.2008.07.063. [DOI] [PubMed] [Google Scholar]
- Dubruille R, Emery P. A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol Neurobiol. 2008;38:129–145. doi: 10.1007/s12035-008-8035-y. [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/S0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Förster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/S0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Glaser FT, Stanewsky R. Temperature synchronization of the Drosophila circadian clock. Curr Biol. 2005;15:1352–1363. doi: 10.1016/j.cub.2005.06.056. [DOI] [PubMed] [Google Scholar]
- Glaser FT, Stanewsky R. Synchronization of the Drosophila circadian clock by temperature cycles. Cold Spring Harb Symp Quant Biol. 2007;72:233–242. doi: 10.1101/sqb.2007.72.046. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci U S A. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich C. Role of the optic lobes in the regulation of the locomotor activity rhythm of Drosophila melanogaster: behavioral analysis of neural mutants. J Neurogenet. 1986;3:321–343. doi: 10.3109/01677068609106857. [DOI] [PubMed] [Google Scholar]
- Kang K, Pulver SR, Panzano VC, Chang EC, Griffith LC, Theobald DL, Garrity PA. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature. 2010;464:597–600. doi: 10.1038/nature08848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Panzano VC, Chang EC, Ni L, Dainis AM, Jenkins AM, Regna K, Muskavitch MA, Garrity PA. Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature. 2012;481:76–80. doi: 10.1038/nature10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lee Y, Akitake B, Woodward OM, Guggino WB, Montell C. Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc Natl Acad Sci U S A. 2010;107:8440–8445. doi: 10.1073/pnas.1001425107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, Hall JC, Hardin PE, Dryer SE. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Kwon I, Choe HK, Son GH, Kim K. Mammalian molecular clocks. Exp Neurobiol. 2011;20:18–28. doi: 10.5607/en.2011.20.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Shim HS, Wang X, Montell C. Control of thermotactic behavior via coupling of a TRP channel to a phospholipase C signaling cascade. Nat Neurosci. 2008;11:871–873. doi: 10.1038/nn.2170. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Kim SH, Ronderos DS, Lee Y, Akitake B, Woodward OM, Guggino WB, Smith DP, Montell C. Drosophila TRPA1 channel is required to avoid the naturally occurring insect repellent citronellal. Curr Biol. 2010;20:1672–1678. doi: 10.1016/j.cub.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Lee J, Bang S, Hyun S, Kang J, Hong ST, Bae E, Kaang BK, Kim J. Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nat Genet. 2005;37:305–310. doi: 10.1038/ng1513. [DOI] [PubMed] [Google Scholar]
- Lee Y, Moon SJ, Montell C. Multiple gustatory receptors required for the caffeine response in Drosophila. Proc Natl Acad Sci U S A. 2009;106:4495–4500. doi: 10.1073/pnas.0811744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasako Y, Umezaki Y, Tomioka K. Separate sets of cerebral clock neurons are responsible for light and temperature entrainment of Drosophila circadian locomotor rhythms. J Biol Rhythms. 2007;22:115–126. doi: 10.1177/0748730407299344. [DOI] [PubMed] [Google Scholar]
- Neely GG, Keene AC, Duchek P, Chang EC, Wang QP, Aksoy YA, Rosenzweig M, Costigan M, Woolf CJ, Garrity PA, Penninger JM. TrpA1 regulates thermal nociception in Drosophila. PLoS One. 2011;6:e24343. doi: 10.1371/journal.pone.0024343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N, Helfrich-Förster C. Setting the clock–by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 2011;585:1435–1442. doi: 10.1016/j.febslet.2011.02.028. [DOI] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5:e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot M, Klarsfeld A, Chélot E, Malpel S, Rouyer F. A role for blind DN2 clock neurons in temperature entrainment of the Drosophila larval brain. J Neurosci. 2009;29:8312–8320. doi: 10.1523/JNEUROSCI.0279-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A PDF neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/S0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Roberts SK. Circadian activity in cockroaches: II. Entrainment and phase shifting. J Cell Comp Physiol Behav. 1962;59:175–186. doi: 10.1002/jcp.1030590210. [DOI] [Google Scholar]
- Rosenzweig M, Brennan KM, Tayler TD, Phelps PO, Patapoutian A, Garrity PA. The Drosophila ortholog of vertebrate TRPA1 regulates thermotaxis. Genes Dev. 2005;19:419–424. doi: 10.1101/gad.1278205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehadova H, Glaser FT, Gentile C, Simoni A, Giesecke A, Albert JT, Stanewsky R. Temperature entrainment of Drosophila's circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron. 2009;64:251–266. doi: 10.1016/j.neuron.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Shen WL, Kwon Y, Adegbola AA, Luo J, Chess A, Montell C. Function of rhodopsin in temperature discrimination in Drosophila. Science. 2011;331:1333–1336. doi: 10.1126/science.1198904. [DOI] [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/S0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Tracey WD, Jr, Wilson RI, Laurent G, Benzer S. Painless: a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/S0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- van der Linden AM, Beverly M, Kadener S, Rodriguez J, Wasserman S, Rosbash M, Sengupta P. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000503. doi: 10.1371/journal.pbio.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanath V, Story GM, Peier AM, Petrus MJ, Lee VM, Hwang SW, Patapoutian A, Jegla T. Opposite thermosensor in fruitfly and mouse. Nature. 2003;423:822–823. doi: 10.1038/423822a. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Heshiki Y, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K. Temperature cycles drive Drosophila circadian oscillation in constant light that otherwise induces behavioural arrhythmicity. Eur J Neurosci. 2005;22:1176–1184. doi: 10.1111/j.1460-9568.2005.04295.x. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Vanin S, Costa R, Helfrich-Förster C. Synergic entrainment of Drosophila's circadian clock by light and temperature. J Biol Rhythms. 2009;24:452–464. doi: 10.1177/0748730409348551. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Bellemer A, Yan H, Ken H, Jessica R, Hwang RY, Pitt GS, Tracey WD. Thermosensory and non-thermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat sensor domains of a thermoTRP channel. Cell Rep. 2012;1:43–55. doi: 10.1016/j.celrep.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiebel LJ, Hardin PE, Liu X, Hall JC, Rosbash M. A posttranscriptional mechanism contributes to circadian cycling of a per-β-galactosidase fusion protein. Proc Natl Acad Sci U S A. 1991;88:3882–3886. doi: 10.1073/pnas.88.9.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]