Abstract
Riboswitches are mRNA elements that specifically bind cellular metabolites and control gene expression by modifying their structure. As riboswitches often control essential genes in pathogenic bacteria, riboswitches have been proposed as new targets for antibiotics. High-throughput screening provides a powerful approach to identify riboswitch ligand analogs that could act as powerful antibacterial drugs. Biochemical assays have already been used to find riboswitch-binding analogs, but those methods do take into account the transcriptional context for riboswitch regulation. As the importance of co-transcriptional ligand binding has been shown for several riboswitches, it is vital to develop an assay that screens riboswitch-binding analogs during the transcriptional process. Here, we describe the development of a dual molecular beacon system monitoring the transcriptional regulation activity of the Bacillus subtilis pbuE adenine riboswitch. This system relies on two molecular beacons that enable the monitoring of transcription efficiency, as well as the regulatory activity of the riboswitch. Different analogs were tested using our system, and a good correlation was observed between riboswitch activity and reported metabolite affinities. This method is specific, reliable and could be applied at the high-throughput level for the identification of new potential antibiotics targeting any riboswitch-regulating gene expression at the mRNA level.
INTRODUCTION
Riboswitches are gene regulatory elements located within mRNA untranslated regions that modulate gene expression through the specific binding of small metabolites (1). The ligand-binding activity of riboswitches is performed by the aptamer domain consisting of a defined structure involved with most of the ligand atomic groups, ensuring high-binding affinity and specificity of recognition (2). The formation of the metabolite–aptamer complex is coupled to the modulation of the downstream expression platform that controls gene expression through various mechanisms, such as transcription termination, translation initiation and mRNA splicing (3). The versatility of riboswitches is well illustrated by purine riboswitches. They comprise the adenine- and guanine-sensing variants, which have been shown to activate or repress gene expression through metabolite binding, respectively (4,5). Although the guanine-specific riboswitch regulates only at the level of transcription by modulating the formation of a transcription terminator, the adenine riboswitch is involved in both the control of transcription elongation and in the modulation of translation initiation by sequestering the Shine–Dalgarno and AUG start codon sequences (6–8).
As pathogenic bacteria are increasingly found to acquire resistance to a large variety of antibiotics (9), riboswitches have been considered to be promising targets for the development of novel antibacterial drugs (10–12). The large-scale exploration of new riboswitch-targeting analogs requires the development of novel assays easily applicable to high-throughput screening approaches. Although initial efforts using high-throughput screens have been put forward by engineering allosteric ribozymes having the ability to be regulated via molecular effectors (13), recent studies have reported the use of the naturally occurring glmS ribozyme that can be specifically activated by the binding of glucosamine-6-phosphate (GlcN6P) (14,15). However, for most other riboswitches that are putative candidates as drug targets but do not have ribozyme activity, it may be challenging to identify an antimicrobial compound that is specific towards the targeted riboswitch rather than to the ribozyme-derived riboswitch construct.
In this work, we report the development of a straightforward detection assay based on molecular beacons that enables the identification of drug compounds targeting any riboswitch that regulates expression at the mRNA level. Molecular beacons are nucleic acid molecules possessing a stem–loop structure in which the loop portion of the beacon is complementary to a pre-determined sequence in a target mRNA (16). These fluorescent reporters contain a fluorophore and a quencher that are attached to both extremities of the molecule. Although a beacon does not fluoresce when folded into the stem–loop conformation, it emits fluorescence when hybridized to its target mRNA because the fluorophore and the quencher are too far apart from each other. Recent studies have used various types of molecular beacons, including DNA (16), peptidyl nucleic acid (17) and 2′-O-methylribonucleotide (2′-O-Me) backbone beacons (18). Although DNA beacons have been shown to be poor reporters of transcription reactions because they are substrates for RNA polymerases (RNAP), 2′-O-Me beacons (OMB) have been demonstrated to be appropriate reporters of transcription reactions (18). The assay described here exploits the high specificity and stability of OMB molecules to follow the transcriptional regulatory activity of the pbuE adenine riboswitch from Bacillus subtilis in presence of various purine compounds.
MATERIALS AND METHODS
Molecular beacons
The sequence of 2′-O-methyl molecular beacons are DABCYL-GCGUUUUUUUUUUUUCGC–Fluorescein (5′-OMB) and DABCYL-GCGUUGUUAAUUUUUCGC-5′–TMR (3′-OMB), respectively, for the 5′ and 3′ molecular beacons.
Synthesis of RNA molecules
Templates for transcription were made by polymerase chain reaction from synthetic DNA oligonucleotides. Synthetic DNA templates included an xpt-pbuX bacterial promoter (19), followed by the B. subtilis pbuE riboswitch sequence, including 25 nt after the terminator stem. Where indicated, the T7 RNAP promoter (20) was used instead of the xpt-pbuX promoter. DNA sequences are shown for wild-type and mutant constructs. The 5′-OMB and 3′-OMB target sites are underlined.
WT
CAGCCTATGCAAGAGATTAGAATCTTGATATAATTTATTACATGTGGTACACTCATCAACGGAAACGCAAAAAAAAAAAAGCGATTATCACTTGTATAACCTCAATAATATGGTTTGAGGGTGTCTACCAGGAACCGTAAAATCCTGATTACAAAATTTGTTTATGACATTTTTTGTAATCAGGATTTTTTTTATTTATCGCAACAATTAAAAAGCG
ONMutant
CAGCCTATGCAAGAGATTAGAATCTTGATATAATTTATTACATGTGGTACACTCATCAACGGAAACGCAAAAAAAAAAAAGCGATTATCACAACAATAACCTCAATAATATGGTTTGAGGGTGTCTACCAGGAACCGTAAAATCCTGATTTGTTAATTTGTTTATGACATTTTTTGTAATCAGGATTTTTTTTATTTATCGCAACAATTAAAAAGCG
OFFMutant
CAGCCTATGCAAGAGATTAGAATCTTGATATAATTTATTACATGTGGTACACTCATCAACGGAAACGCAAAAAAAAAAAAGCGATTATCACTTGTATAACCTCAATAATATCCTTTGAGGGTGTCTACCAGGAACCGTAAAATCCTGATTACCAAAATTTGTTTATGACATTTTTTGTAATCAGGATTTTTTTTATTTATCGCAACAATTAAAAAGCG
Single-rounds in vitro transcription assays
DNA templates for single-round in vitro transcriptions were prepared as described previously (8). Briefly, the xpt-pbuX promoter sequence was used to generate a transcription start site ∼46-nt upstream of the aptamer domain, and transcription reactions were performed using Escherichia coli RNA polymerase from Epicenter Biotechnologies as previously described (8). Transcription products were migrated on a 10% polyacrylamide gel at 18 W for 2 h in 1× TBE (90 mM Tris-borate, pH 8.3, 2 mM EDTA) buffer. Experiments were conducted at least three times, and all exhibited similar uncertainties (<10%). Additional mutations introduced are indicated in the text.
Native gel electrophoresis
[5′-32P]-labeled adenine riboswitches were incubated in absence or in presence of molecular beacons at 70°C for 3 min and allowed to slowly cool to room temperature to ensure hybridization. Samples were separated in 1× TB (90 mM Tris-borate, pH 8.3) and 1 mM MgCl2 in 8% acrylamide:bisacrylamide (29:1) gel in Tris–borate buffer containing 1 mM MgCl2 at room temperature at 150 V for 8 h with the running buffer circulated during the electrophoresis.
Fluorescence analysis
Fluorescence spectroscopy was performed on a Quanta Master fluorometer, and spectra were corrected for lamp fluctuations and instrument variations as described previously (21). All data were collected at 37°C in a buffer containing 40 mM Tris, pH 7.5, 20 mM MgCl2 and 10 mM DTT in a final volume of 100 µl. Each reaction is composed of 5 pmoles template, 65 µM rNTP and 10 pmoles of each 5′-OMB and 3′-OMB. The solution was kept at 37°C for 5 min before 5 U of E. coli RNAP were added. Tetramethyl-rhodamine (TMR) was excited at 558 nm, and the emission values were collected from 568 to 595 nm. Fluorescein was excited at 494 nm, and the emission values were collected from 505 to 530 nm. For each time point, the initial fluorescence intensity (I0) is subtracted to the fluorescence intensity of both 5′-OMB and 3′-OMB (I) and divided by I0, given the equation {(I − I0)/I0}. The riboswitch activity is measured by normalizing the fluorescence intensity of the 3′-OMB with the corresponding 5′-OMB value. Experiments were performed in absence or presence of 10 µM ligand. Experiments were performed at least three times, and all exhibited similar uncertainties (<5%).
RESULTS
General strategy for monitoring riboswitch activity using molecular beacons
The B. subtilis adenine riboswitch modulates transcription elongation of the pbuE gene encoding a purine efflux pump involved in the export of adenine (22,23). In presence of low adenine concentrations, the riboswitch adopts an OFF state-inhibiting gene expression by inducing the formation of a transcription terminator (Figure 1A). However, in presence of high-adenine concentrations, the riboswitch folds into an ON state that allows transcription elongation and synthesis of a full-length pbuE transcript. Although the riboswitch domain is transcribed independently of the adenine concentration, the elongated mRNA is only produced when the intracellular concentration of adenine is high (5).
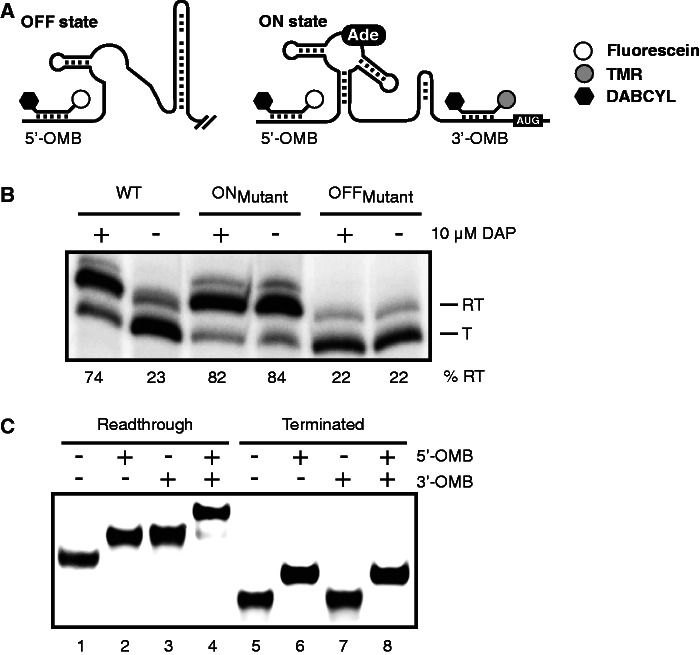
Figure 1.
Strategy for monitoring riboswitch activity using molecular beacons. (A) Schematic representation of the pbuE riboswitch regulation, including hybridization sites for both molecular beacons. In absence of adenine, the B. subtilis pbuE riboswitch folds in the OFF state and forms a terminator stem that prematurely stops transcription. However, on adenine (Ade) binding, the terminator stem is destabilized, and transcription elongation is allowed (ON state). Although the 5′ molecular beacon (5′-OMB) can hybridize to both OFF and ON state transcripts, the 3′ beacon (3′-OMB) is only able to bind to the elongated ON state transcript. Both molecular beacons contain a quencher (DABCYL, black hexagon) at the 3′ extremity, preventing fluorescence emission when beacons are not hybridized to the riboswitch. TMR and fluorescein are represented by gray and white circles, respectively. (B) Single-round in vitro transcription assays performed in absence (−) and in presence (+) of 10 µM DAP for the wild-type (WT), ON and OFF state mutants. Readthrough (RT) and terminated (T) transcripts are shown on the right of the gel. Percentages of termination are indicated later in the text the gel. (C) EMSA performed on the pbuE riboswitch elongated (lanes 1–4) and terminated (lanes 5–8) transcripts in complex with indicated molecular beacons. Free and bound riboswitch transcripts were separated on polyacrylamide gel in native conditions.
We have designed a reporter system in which two OMBs are used to monitor the promoter efficiency, as well as the control of the pbuE riboswitch transcription elongation (Figure 1A). We first designed a molecular beacon (5′-OMB) that binds upstream of the aptamer domain so that the promoter transcription activity could be monitored. A second molecular beacon (3′-OMB) was engineered to hybridize downstream of the transcription terminator; therefore, it only detects the elongated riboswitch mRNA. Although the 5′-OMB reporter was labeled with fluorescein and a DABCYL quencher, tetramethylrhodamine (TMR) and DABCYL were used for the 3′-OMB molecule. Riboswitch target sequences were modified to allow efficient binding of molecular beacons without affecting riboswitch activity. By using such a dual molecular beacon system, it is possible to simultaneously monitor both transcription efficiency and riboswitch regulation control.
The transcriptional regulation activity of the pbuE riboswitch was first assessed using single-round in vitro transcription assays (19). In these assays, we used a DNA template, including the B. subtilis xpt promoter fused to the pbuE riboswitch to allow transcription of a readthrough product terminating 40 nt after the AUG start codon. In our experimental conditions, a low readthrough efficiency of 23% was obtained when performing a transcription reaction in absence of ligand (Figure 1B; WT). However, in presence of 10 µM 2,6-diaminopurine (DAP), which is known to bind the pbuE riboswitch with high affinity (5), the readthrough efficiency was increased to a high level of 84%, consistent with ligand binding, promoting pbuE transcription elongation in vivo (5,8,21). As expected, the introduction of mutations in the P1 stem stabilizing the riboswitch ON state (ONMutant) promoted a high level of mRNA elongation, regardless of the presence of the ligand (>80% readthrough efficiency; Figure 1B). In contrast, destabilization of the P1 stem to favor the OFF state (OFFMutant) resulted in the constitutive production of a high fraction of prematurely terminated mRNA species (22% readthrough efficiency, Figure 1B). Thus, our data are in agreement with the pbuE riboswitch transcription modulation being controlled by a ligand-induced conformational change.
To determine whether both molecular beacons can efficiently bind to the pbuE transcript, RNA molecules corresponding either to the readthrough or terminated mRNA were transcribed using the T7 RNAP and were assessed for their ability to bind molecular beacons using electrophoretic mobility shift assay (EMSA) (Figure 1C). When transcripts corresponding to the readthrough species were migrated in native conditions in presence of either 5′-OMB or 3′-OMB molecules, a similar gel retardation was observed in both cases, suggesting the formation of a transcript-beacon complex (Figure 1C, lanes 2 and 3, respectively). Furthermore, a slower migrating complex was observed when both beacons were present, indicating that riboswitch transcripts hybridized simultaneously to both OMBs (Figure 1C, lane 4). When the EMSA assay was repeated using the terminated transcript (Figure 1C, lanes 5–8), complex formation was observed only when the 5′-OMB was present (Figure 1C, lanes 6 and 8), agreeing with the absence of a 3′-OMB target sequence in the terminated transcript. These results suggest that both beacons can efficiently bind to their respective target sequence in the context of the adenine riboswitch.
Fluorescence detection of pbuE riboswitch transcription
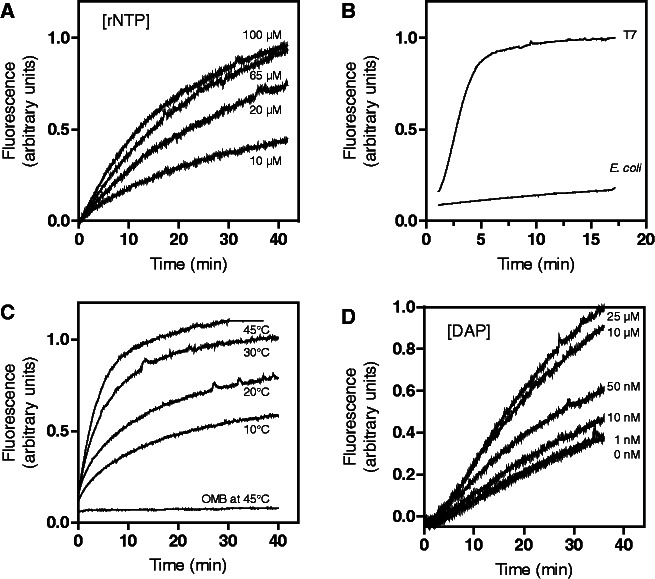
Using the transcription conditions previously established (Figure 1B), we used the 5′-OMB reporter to monitor E. coli RNAP transcription activity. The fraction of bound 5′-OMB was obtained by exciting fluorescein at 494 nm and by measuring the emitted fluorescence at 517 nm at 1-s intervals. When the transcription was performed using 10 µM rNTP, the 5′-OMB fluorescence emission was observed to increase steadily over the course of the transcription reaction (Figure 2A), consistent with an accumulation of pbuE transcripts. As we previously observed that the rNTP concentration influences the rate of transcription (8), we repeated the experiment using a different rNTP concentration to determine whether the fluorescence increase is proportional to the transcriptional activity. As expected, the 5′-OMB fluorescence signal increased proportionally to the concentration of rNTPs (Figure 2A), consistent with fluorescence emission being related to transcription efficiency. However, signal saturation was obtained in presence of 65 µM rNTP, suggesting either that the maximal rate of transcription was attained, or that the binding of the molecular beacon was limiting. To distinguish between these alternatives, we repeated the transcription reaction assay using T7 RNAP, which is known to exhibit ∼10–20 times faster transcription rates (24). When using T7 RNAP, the 5′-OMB fluorescence signal was dramatically increased over the first 5 min of the reaction and then reached a plateau for the rest of the time course (Figure 2B). Thus, because our assay can monitor the higher transcriptional activity of T7 RNAP, it suggests that the 5′-OMB reporter can readily detect the transcriptional activity of the E. coli RNAP. The signal saturation obtained when increasing rNTP concentrations also suggests that the transcriptional activity does not significantly change at rNTP concentrations higher than 65 µM.
Figure 2.
Characterization of 5′-OMB and 3′-OMB reporter activities in the context of the pbuE riboswitch. (A) 5′-OMB fluorescence intensity monitored as a function of time by varying the rNTP concentrations to 10, 20, 65 and 100 µM. (B) The 5′-OMB fluorescence emission recorded when transcribing the pbuE riboswitch using either T7 or E. coli RNA polymerase. The fluorescence intensity of the 5′-OMB is represented as a function of the transcription reaction time. (C) The 5′-OMB fluorescence intensity measured at various temperatures in presence of its target sequence. As a control, the 5′-OMB was incubated alone at 45°C and showed no significant fluorescence emission because of temperature destabilization. (D) Effect of DAP concentration on the pbuE riboswitch transcriptional regulation monitored using the 3′-OMB fluorescence emission. DAP was added to the transcription reaction at concentrations ranging from 0 to 25 µM, and the fluorescence emission of the 3′-OMB was measured as a function of time.
Although molecular beacons are designed to fluoresce only when bound to their target, it is possible that unbound beacons may emit fluorescence because of an equilibrium existing between open (fluorescent) and closed (non-fluorescent) conformations (16). For example, given that the transcription reaction is performed at 37°C, the partial melting of the unbound beacon could give rise to fluorescence emission. To rule out this possibility, the 5′-OMB was incubated at 45°C in absence of the target transcript, and its fluorescence was recorded over time (Figure 2C). No significant fluorescence emission was detected, suggesting that the beacon remains folded in its non-fluorescent conformation. In contrast, significant fluorescence emission was observed when incubating the beacon in presence of the target transcript at all tested temperatures (Figure 2C), consistent with the hybridization of 5′-OMB being required for fluorescence emission.
The riboswitch transcriptional control was next monitored by measuring the fluorescence signal of the 3′-OMB as a function of DAP concentration (Figure 2D). As expected from single-round transcription assays (Figure 1B), we found that the 3′-OMB fluorescence signal was increased proportionally to DAP concentrations (Figure 2D). No significant 3′-OMB fluorescence increase was observed >10 µM DAP, suggesting that ligand saturation was attained in these conditions. This result agrees well with a previous single-round in vitro transcription study in which a T50 value of ∼0.5 µM was reported for DAP (8). Taken together, these data suggest that our molecular beacon assay can be used to accurately monitor the pbuE transcription efficiency, as well as the ligand-dependent riboswitch transcriptional regulatory control. For all of the following experiments, transcription reactions were performed at 37°C in presence of 10 µM DAP and 65 µM rNTP, which represents the lowest concentrations showing the highest riboswitch transcriptional control (Figure 2A).
Monitoring pbuE riboswitch activity using a dual molecular beacon assay
We next modified our method to monitor both molecular beacons within a single experiment to ensure a more robust and reliable assay across experimental conditions. In these assays, the ratio of bound 5′-OMB and 3′-OMB were obtained by selectively exciting fluorescein at 494 nm and collecting the emitted fluorescence from 505 to 530 nm, and by exciting TMR at 558 nm and measuring the emitted fluorescence signal from 568 to 595 nm. The sensitivity of the method was first determined by varying the concentration of DNA template from 2 to 20 pmoles (Figures 3A and 3B). From the emission spectra, we observed that the 5′-OMB fluorescence increased proportionally to concentration of the template (Figure 3A). Furthermore, when analyzing the 3′-OMB fluorescence, a signal increase was also observed as a function of the concentration of the template (Figure 3B), even if the transcription reaction was performed in absence of ligand. These results are expected according to our single-round transcription data because a small fraction of elongated RNA is still produced in these conditions (23% readthrough product; Figure 1B).
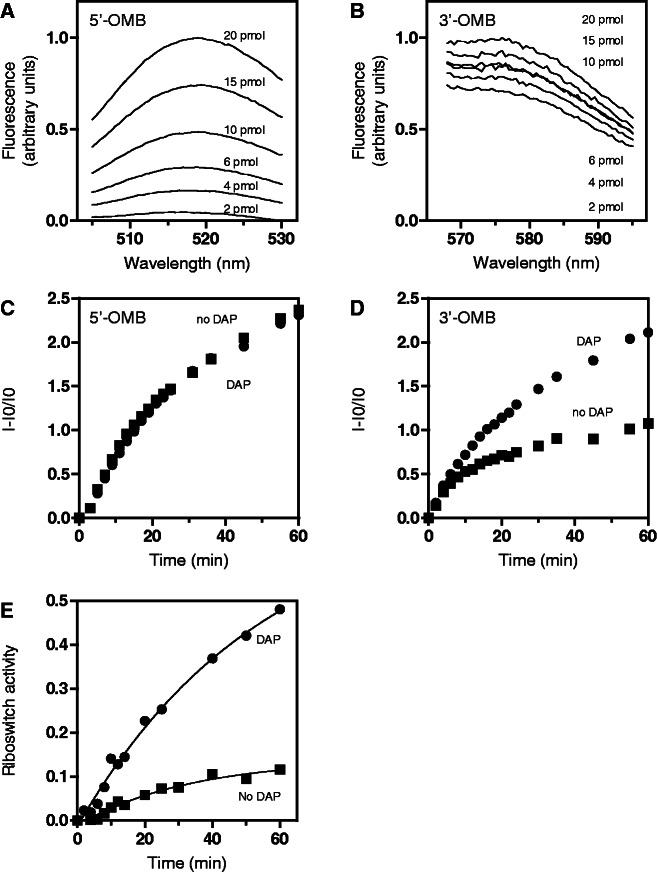
Figure 3.
Monitoring riboswitch activity using a dual molecular beacon assay. (A) 5′-OMB fluorescence emission monitored from 505 to 530 nm with increasing concentrations of DNA template ranging from 2 to 20 pmoles. (B) Fluorescence emission of 3′-OMB measured from 565 to 595 nm using increasing DNA template concentrations from 2 to 20 pmoles. (C) Transcription reaction monitored using 5′-OMB fluorescence. The fraction of 5′-OMB fluorescence emission was determined in absence (squares) and in presence (circles) of 10 µM DAP. (D) Transcriptional control of the riboswitch monitored using 3′-OMB fluorescence. The fraction of 3′-OMB fluorescence was determined for each time point in absence (squares) and in presence (circles) of 10 µM DAP. (E) Normalized pbuE riboswitch activity obtained in absence (squares) and in presence (circles) of 10 µM DAP. The riboswitch activity is normalized by using the 5′-OMB fluorescence emission, which is not dependent on the riboswitch transcriptional control.
Next, we monitored the transcription reaction as a function of DAP when using both beacons. Because the 5′-OMB target sequence is located upstream of the transcription terminator (Figure 1A), the 5′-OMB fluorescence should not be modulated by the riboswitch transcriptional control and thus could be used as an ‘internal reporter’ to normalize the 3′-OMB fluorescence signal. In agreement with this, no signal variation was observed when monitoring the 5′-OMB fluorescence in absence or presence of DAP (Figure 3C). In contrast, a significant fluorescence increase was detected for the 3′-OMB in presence of DAP (Figure 3D), consistent with the riboswitch-promoting transcription readthrough in presence of ligand (Figure 1B). By combining data obtained for both 5′-OMB and 3′-OMB beacons (Figures 3C and D), a plot of transcription reactions was generated in which the 3′-OMB fluorescence signal is normalized to provide a direct measure of the ligand-induced riboswitch transcriptional control (Figure 3E). In our conditions, we observed that the presence of 10 µM DAP resulted in an ∼5-fold increase in transcription readthrough (Figure 3E; compare time points at 60 min), in agreement with single-round transcription reactions (Figure 1B). Control experiments were also performed to establish the sensitivity and robustness of the fluorescent reporters in the context of the transcription reaction. As expected, when using ONMutant and OFFMutant templates, we obtained elevated and reduced riboswitch transcriptional activities for the ONMutant and OFFMutant constructs, respectively (Figure 4A). Our results suggest that the extent of transcription elongation is slightly higher for the ONMutant (Figure 4A) than for the wild-type riboswitch in presence of DAP (Figure 3E), in agreement with our previous study (8).
Figure 4.
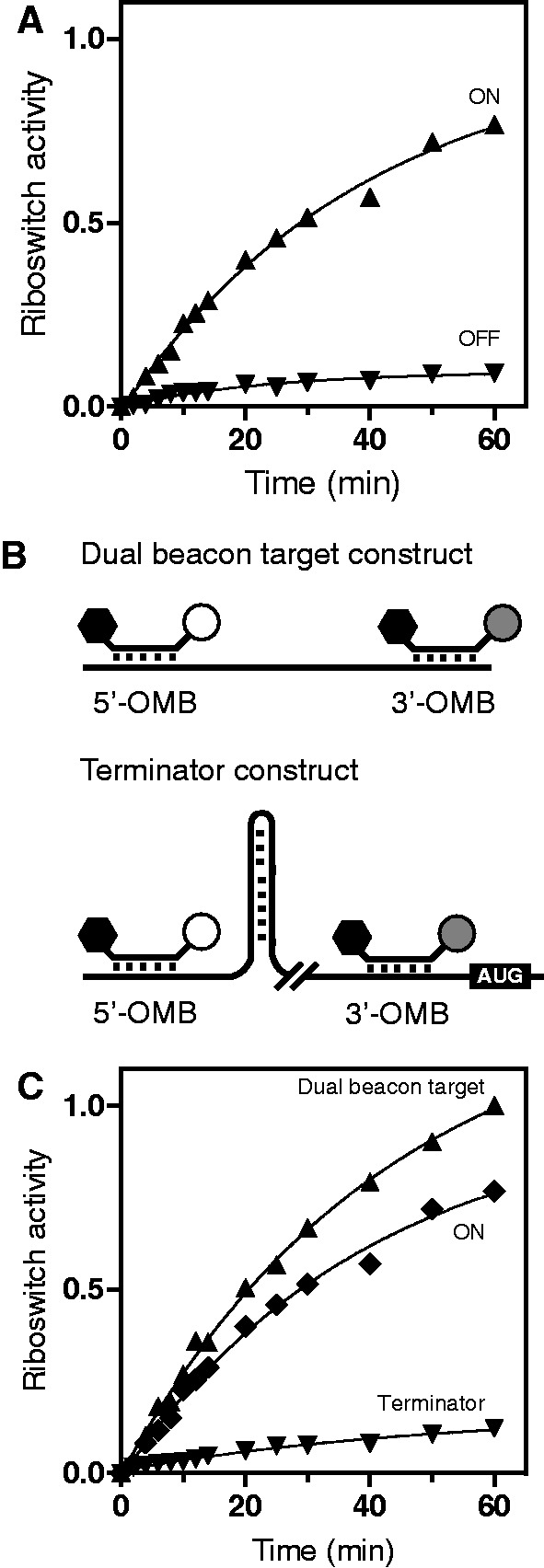
Characterization of the molecular beacon detection activity. (A) Normalized transcriptional activity of ONMutant (triangles) and OFFMutant (inverted triangles) riboswitch constructs. (B) Schematic representations of the dual beacon target and terminator constructs. Although both molecular beacons can bind to the dual beacon target transcript, only the 5′-OMB binds the terminator transcript when premature termination occurs. (C) Riboswitch activity measured for both the dual beacon target and terminator transcripts. The ONMutant data are added for comparison.
We next engineered a dual beacon target construct containing both 5′-OMB and 3′-OMB-binding sites (Figure 4A). Importantly, this construct lacks the metabolite-binding aptamer domain and was engineered to estimate the maximum signal that can be obtained in conditions where all transcribed riboswitch mRNAs are in their elongated form, thus providing binding sites for both OMBs. Using this construct, we observed an increase of the fluorescence signal which was higher than what was obtained using the riboswitch ONMutant template (Figure 4B). This result indicates that even in the context of the ONMutant riboswitch (Figure 4B), where the P1 stem is stabilized, there is still a fraction of premature transcription termination. To estimate the maximum efficiency of premature termination in our conditions, we designed an additional template control lacking the aptamer domain in which the transcription terminator of the riboswitch was inserted between both 5′-OMB and 3′-OMB target sequences (Figure 4A; terminator construct). Using this control, transcription reactions yielded a small fraction of transcripts exhibiting 3′-OMB–binding site (Figure 4B), which is reminiscent of what we obtained for the wild-type riboswitch in absence of ligand (Figure 3E) or for the OFFMutant construct (Figure 4A). These data allowed us to determine the dynamic range of the adenine riboswitch regulation usable to screen riboswitch analogs. Together, our experimental controls indicate that our fluorescent assay is robust and sensitive to detect riboswitch conformational states.
Low-throughput analysis of the ligand-induced pbuE riboswitch transcriptional activity
To demonstrate the suitability of our system for the search of riboswitch-binding small molecules, we performed a low-throughput analysis of various purine-related compounds that have been previously tested to bind purine riboswitch aptamers (4,5). The concentration of reagents added to our assay was optimized to use a minimal ligand concentration giving a reproducible and significant fluorescence signal. Individual experiments were monitored at the beginning and after 1 h of transcription, and results were normalized according to the dual beacon target construct (Figure 5). Without ligand, normalized transcription efficiencies were low, whereas reactions performed in presence of DAP, adenine or 2-aminopurine (2AP) yielded transcription elongation ratios that were proportional to their determined dissociation constants (5). This result is in agreement with the ability of the adenine riboswitch to strongly discriminate against guanine-related compounds (5). Indeed, a small increase in transcription elongation was observed in presence of guanine, 9-methylguanine (9-Me-Gua) or O6-thioguanine (Thio-Gua). Taken together, our results strongly indicate that our molecular beacon assay is functional, specific and could be used to evaluate ligand affinities of any transcriptionally acting riboswitches on a high-throughput scale.
Figure 5.
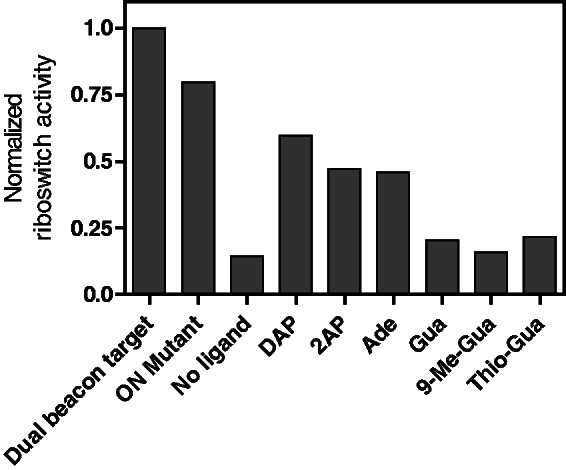
Low-throughput analysis of adenine- and guanine-related analogs using the dual molecular beacon assay. Transcription reactions of the pbuE adenine riboswitch were performed in absence of ligand or in presence of DAP, 2AP, adenine (Ade), guanine (Gua), 9-methylguanine (9-Me-Gua) and O6-thioguanine (Thio-Gua). Only DAP, 2AP and adenine allow significant transcription elongation. Control experiments were performed with the dual beacon target and the ONMutant transcripts, both enabling transcription elongation without added ligand.
DISCUSSION
The system developed herein addresses the need for a screening assay able to identify new riboswitch analogs that could potentially be used as antimicrobial agents (11). This method is the first one designed to directly measure the riboswitch transcriptional regulatory activity using fluorescence detection, thus substantially facilitating its implementation in a high-throughput context. Although RNA misfolding could influence the outcome of the assay, it should not occur in a ligand-dependent fashion and, therefore, should not introduce a bias in the detection of binding analogs. The development of such a system strongly depends on the efficient binding of molecular beacons to accurately report on the riboswitch transcriptional control (Figure 3). However, because it is possible to alter the molecular beacon sequences to achieve strong and specific binding, we do not anticipate this step to be problematic in implementing our approach for other riboswitch classes. Importantly, among all parameters that could influence the identification of potential ligand-binding analogs, we found that the inefficiency of transcriptional control—or ‘leakiness’—was the most important in our detection assays. For example, addition of DAP resulted in an ∼5-fold transcription activation (Figures 3E and 5), which provides enough dynamic range to assess various ligands, such as 2-AP and adenine. A second important parameter to consider is the concentration of the compound used in the transcription reaction. Indeed, because large concentrations of compounds are used in our assays (10 µM), it is expected that low-binding affinity metabolites, such as guanine and related compounds, may be detected (Figure 5). However, more stringent conditions can readily be applied by decreasing the ligand concentration to detect compounds having higher binding affinity.
Fluorescent nucleic acids have already been described as tools to measure riboswitch ligand-binding affinities. For example, two recent studies have used the ligand-dependent glmS ribozyme cleavage activity to identify new potential ligands (14,15). In both cases, fluorescent versions of the RNA were engineered in which ligand-induced ribozyme cleavage would alter the fluorescence signal. As the modulation is directly related to the ligand-induced ribozyme activity, analogs exhibiting high affinity towards the ribozyme can easily be detected. Another method recently described uses a synthetic aptamer responsive to the aminoglycoside antibiotic tobramycin (25). The system consists of a fluorescein-labeled aptamer exhibiting a sequence complementary to a DNA oligonucleotide attached to magnetic beads. On ligand binding, the aptamer undergoes structural changes that prevent its pairing to DNA. After purification of RNA bound to the magnetic beads, the binding specificity for various molecules is evaluated by measuring the fluorescence emission. Finally, Sanbonmatsu and coworkers monitored the SAM-I riboswitch-binding activity by designing a trans-acting version of the riboswitch in which the terminator helix is formed intermolecularly by the hybridization of the aptamer and of an RNA oligonucleotide containing a 2-AP fluorescent base (26). In absence of the ligand, the SAM-I aptamer is free to interact with the 2-AP–containing strand to form the riboswitch ON state, which does not allow 2-AP fluorescence. However, on SAM binding, the aptamer is stabilized and does not efficiently interact with the fluorescent strand, thus effectively allowing fluorescence to be emitted from 2-AP. Although this construct was used to measure the SAM-binding affinity for many aptamer variants, it could also be used to screen for potential ligand analogs binding to the SAM-I riboswitch.
Even though the techniques described earlier in the text can monitor the ligand-binding activity of riboswitches in a high-throughput fashion, they do not address the transcriptional context of the riboswitch regulation activity. For instance, a recent study on the B. subtilis flavin mononucleotide (FMN) riboswitch has shown that the riboswitch relies on RNAP transcription-pausing sites to allow more time for ligand binding (27). Moreover, it was recently demonstrated that two adenine-sensing riboswitches differ by using different regulation regimes (8). Although the transcriptionally acting B. subtilis pbuE riboswitch was also shown to operate under a kinetic regime, the translationally acting Vibrio vulnificus add riboswitch was found to use a different regulation mechanism that seems to be thermodynamically driven and where ligand binding can occur post-transcriptionally. Furthermore, the btuB, mgtA and ribB riboswitches from E. coli were also shown to depend on the transcriptional process (28,29), suggesting that other riboswitches are likely to rely on the transcriptional context to regulate gene expression. Therefore, because riboswitch genetic regulation cannot always be predicted from thermodynamic characteristics derived from the aptamer–ligand complex, it is important to seek alternative high-throughput analogs screening methods that take into account the riboswitch regulatory context.
The dual molecular beacon assay described here is robust and can be fully automated to screen compounds binding to any riboswitch-controlling expression at the transcription level. The main advantage of our method is that it directly measures the ligand effect on riboswitch transcriptional regulation activity. Indeed, this system facilitates the identification of molecules that bind to the riboswitch aptamer and induce the structural switch in the expression platform. Because this assay relies on transcription, it is obvious that any compound like rifampicin that inhibits transcription would not be good candidates owing to their likely large antibiotic spectrum (18). However, because the 5′-OMB signal monitors the level of transcription, such compounds will be readily detected and avoided. Although the described method is useful and informative, the molecular beacon assay has its own limitations. First, some inherent physical characteristics of screened analogs could potentially affect data analysis and lead to misinterpretation. Precisely, any fluorescent analogs could affect the fluorescence signal of the molecular beacons if both signals are emitted in the same range of wavelengths. To avoid such false positives, it is advisable to perform control transcription reactions using an OFFMutant construct, which should not show any ligand-dependent fluorescence increase. One possible solution for those cases would be to label the problematic molecular beacon using alternative dyes. Second, the insolubility of compounds in the reaction buffer could also be a problem for this assay. For those molecules, different buffer conditions should be tested to promote their solubility.
In summary, we have used the adenine-sensing riboswitch as a model to validate a novel screening assay using molecular beacons. However, this system could be used with any riboswitch-regulating gene expression at the transcriptional level. This system is also amenable to high-throughput approaches and is promising for the discovery of new potential inhibitors of riboswitch-regulated genes. Moreover, similar approaches to the one described here could be developed to screen molecules against translational riboswitches. A simple method would use a molecular beacon complementary to the ribosome binding site (RBS) sequence. On metabolite recognition by the aptamer domain, the RBS would be sequestered and inaccessible for binding the molecular beacon. However, the absence of ligand binding would result in the binding of the molecular beacon and fluorescence emission. This general approach could provide a high-throughput platform for the discovery of novel riboswitch-binding analogs exhibiting powerful antimicrobial properties.
FUNDING
National Sciences and Engineering Research Council of Canada (NSERC); Doctoral fellowship from NSERC (to J.F.L.); Canadian Institute for Health Research (CIHR) New Investigator scholar (to D.A.L.). Funding for open access charge: NSERC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank members of the Lafontaine laboratory for discussion and Dr Alain Lavigueur for critical reading of the manuscript.
REFERENCES
- 1.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 3.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012;4:1–15. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 5.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 6.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-Riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 8.Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talbot GH, Bradley J, Edwards JE, Jr, Gilbert D, Scheld M, Bartlett JG. Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the Infectious Diseases Society of America. Clin. Infect. Dis. 2006;42:657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 10.Mulhbacher J, St-Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Curr. Opin. Pharmacol. 2010;10:551–556. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 12.Theuretzbacher U, Toney JH. Nature's clarion call of antibacterial resistance: are we listening? Curr. Opin. Investig. Drugs. 2006;7:158–166. [PubMed] [Google Scholar]
- 13.Breaker RR. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 14.Mayer G, Famulok M. High-throughput-compatible assay for glmS riboswitch metabolite dependence. Chembiochem. 2006;7:602–604. doi: 10.1002/cbic.200500490. [DOI] [PubMed] [Google Scholar]
- 15.Blount K, Puskarz I, Penchovsky R, Breaker R. Development and application of a high-throughput assay for glmS riboswitch activators. RNA Biol. 2006;3:77–81. doi: 10.4161/rna.3.2.3102. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 18.Marras SA, Gold B, Kramer FR, Smith I, Tyagi S. Real-time measurement of in vitro transcription. Nucleic Acids Res. 2004;32:e72. doi: 10.1093/nar/gnh068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tremblay R, Lemay JF, Blouin S, Mulhbacher J, Bonneau E, Legault P, Dupont P, Penedo JC, Lafontaine DA. Constitutive regulatory activity of an evolutionary-excluded riboswitch variant. J. Biol. Chem. 2011;286:27406–27415. doi: 10.1074/jbc.M111.229047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Johansen LE, Nygaard P, Lassen C, Agerso Y, Saxild HH. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL) J. Bacteriol. 2003;185:5200–5209. doi: 10.1128/JB.185.17.5200-5209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nygaard P, Saxild HH. The purine efflux pump PbuE in Bacillus subtilis modulates expression of the PurR and G-box (XptR) regulons by adjusting the purine base pool size. J. Bacteriol. 2005;187:791–794. doi: 10.1128/JB.187.2.791-794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 25.Vandenengel JE, Morse DP. Mutational analysis of a signaling aptamer suggests a mechanism for ligand-triggered structure-switching. Biochem. Biophys. Res. Commun. 2009;378:51–56. doi: 10.1016/j.bbrc.2008.10.180. [DOI] [PubMed] [Google Scholar]
- 26.Hennelly SP, Sanbonmatsu KY. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 2011;39:2416–2431. doi: 10.1093/nar/gkq1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 28.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc. Natl Acad. Sci. USA. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perdrizet GA, II, Artsimovitch I, Furman R, Sosnick TR, Pan T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl Acad. Sci. USA. 2012;109:3323–3328. doi: 10.1073/pnas.1113086109. [DOI] [PMC free article] [PubMed] [Google Scholar]