Abstract
The 25S rRNA of yeast contains several base modifications in the functionally important regions. The enzymes responsible for most of these base modifications remained unknown. Recently, we identified Rrp8 as a methyltransferase involved in m1A645 modification of 25S rRNA. Here, we discovered a previously uncharacterized gene YBR141C to be responsible for second m1A2142 modification of helix 65 of 25S rRNA. The gene was identified by reversed phase–HPLC screening of all deletion mutants of putative RNA methyltransferase and was confirmed by gene complementation and phenotypic characterization. Because of the function of its encoded protein, YBR141C was named BMT2 (base methyltransferase of 25S RNA). Helix 65 belongs to domain IV, which accounts for most of the intersubunit surface of the large subunit. The 3D structure prediction of Bmt2 supported it to be an Ado Met methyltransferase belonging to Rossmann fold superfamily. In addition, we demonstrated that the substitution of G180R in the S-adenosyl-l-methionine–binding motif drastically reduces the catalytic function of the protein in vivo. Furthermore, we analysed the significance of m1A2142 modification in ribosome synthesis and translation. Intriguingly, the loss of m1A2142 modification confers anisomycin and peroxide sensitivity to the cells. Our results underline the importance of RNA modifications in cellular physiology.
INTRODUCTION
The non-coding RNAs (ncRNAs) undergo extensive modification to extend their topological potential, which otherwise is limited by four bases (1). These four bases are sufficient to carry the genetic information. However, when it comes to the extra-hereditary functions like catalysis and gene regulation, the nucleic acids pool is insufficient compared with amino acids pool. The chemical modifications provide RNA the necessary complexity and flexibility to perform more sophisticated processes like translation and gene regulation (2). The RNA is broadly subjected to two types of modification, either methylation of sugar moieties and bases or isomerization of uridine to pseudouridine.
There are >100 structurally distinct ribonucleosides that have been identified in all three domains of life (http://rna-mdb.cas.albany.edu/RNAmods/) (3). Many modified nucleosides are conserved throughout bacteria, archaea and eukaryotes, whereas some are unique to each branch of life (4). Most of RNA modifications are not essential for life, which parallels the observation that many well-characterized protein and DNA modifications are also not essential for life (2). Instead, increasing evidence indicates that RNA modifications can play regulatory roles in cells, especially in response to stress conditions (2). Recent studies, especially with the tRNA and mRNA modification, have accentuated the significance of these modifications in the gene regulation and stress signalling (3,5).The regulatory RNAs, including microRNA (miRNA), Piwi-interacting RNA (piRNA) and small interfering RNA (siRNA), have also been discovered to contain modified nucleosides (6,7). The mRNAs, which were until known to contain only m7G nucleosides, have been recently demonstrated to contain other modified bases like m6A (8–10).
To further analyse and explore the function of rRNA modification in the cellular physiology, it is important to identify the respective RNA-modification enzymes. In contrast to 2′-O-ribose methylations and pseudouridylation, which are performed by C/D and H/ACA snoRNPs, respectively, many of the RNA-modification enzymes of eukaryotes responsible for catalysing the base modification are still unknown. Most of the currently known base modification enzymes are essential; however, others escape from genetic analysis, as deletion mutants only provide moderate phenotypes. Therefore, identification of base methyltransferase requires sophisticated genetic and biochemical analysis.
RNA base modifications are the result of processing of corresponding primary transcripts by snoRNA-independent enzymes. A number of enzymes and pathways that catalyse post-transcriptional RNA modifications have been studied for many years and extensively reviewed previously (2). RNA methyltransferases catalyse the transfer of the reactive methyl group of the S-adenosyl-l-methionine (SAM) to an acceptor residue in rRNA. Five classes of methyltransferases with structurally distinct folds have been described to perform the methyltransferase reaction (11). All known rRNA methyltransferases belong to class I or class IV. The majority of the known RNA methyltransferases belonging to class I are characterized by a Rossmann-like fold SAM-binding domain (11). The methyltransferases belongings to class IV are characterized by an α/β knot structure or SPOUT domain. Recently, the SPOUT domain methyltransferases have also been shown to methylate ribosomal proteins (12).
Ribosomes are supramolecular complexes of RNA and proteins, responsible for translation of genetic information. Ribosomes consist of two subunits, a small 40S and a large 60S subunit, named according to their sedimentation coefficient. The 40S subunit decodes the genetic information carried on an mRNA transcript and the larger 60S subunit catalyses the formation of peptide bond. Ribosomes are also known as ‘ribozymes’, as the decoding and peptidyl transferase functions are carried out by the non-coding RNAs (ncRNAs) they harbour (13,14). The 18S and 25S rRNA of small and large subunits has been reported to undergo different chemical modifications to optimize their role in translation of genetic information, which demands high fidelity and structural complexity. The clustering of these modifications in highly conserved areas of the ribosome like peptidyl transfer sites and intersubunit bridges emphasizes their importance in the functioning of ribosomes (15,16).
The 18S rRNA primarily contains the pseudouridylation and ribose methylation catalysed by H/ACA snoRNPs and C/D box snoRNPs, respectively. Apart from these modifications, there are also three base modifications present in the 18S rRNA, the m7G catalysed by Bud23, the m26A catalysed by Dim1 and m1Ψ catalysed by Nep1 (17–19). On the other hand, apart from 2′-O-ribose methylation and pseudouridylation, the 25S rRNA has been reported to contain more diversity in terms of base modifications. The 25S rRNA contains seven modified nucleosides: two m1A, one m5C, two m5U and two m3U (15,20–23). We recently discovered Rrp8 to be one of the two m1A modification enzyme, catalysing the transfer of methyl group to the N1 of adenine at position 645 in helix 25.1 of Saccharomyces cerevisiae (24).
In the present study, we reported the identification of a new gene YBR141C [BMT2, (base methyltransferase of 25S RNA)] responsible for m1A2142 base modification of helix 65 in 25S rRNA of S. cerevisiae. The gene was identified by screening of all deletion mutants of putative RNA methyltransferase and was confirmed by gene complementation and phenotypic characterization. Furthermore, we analysed the significance of the modification in growth, antibiotic sensitivity and translation. Interestingly, the deletion mutant of Δybr141c was previously described to confer a cold resistance, an extended hibernating life span and peroxide sensitivity (25). In our present study, we corroborated the previously described phenotypes of the YBR141C and provided evidences for the role of m1A modifications in conferring these phenotypes.
MATERIALS AND METHODS
Yeast strains and plasmids
The strains used in the present study are listed in Supplementary Table S1. The plasmids were constructed using Gap repair as described previously (26,27) and are listed in Supplementary Table S2.The PCR primers used for the construction of the plasmids are listed in Table S3. The rDNA point mutants were constructed as described previously (18). The substitutions of amino acids were performed by PCR-mediated site-directed mutagenesis using the primers listed in Supplementary Table S2. A detailed protocol for construction of all plasmids will be provided on request.
Growth conditions and yeast media
Yeast strains were grown at 30°C in YPD medium (1% yeast extract, 2% peptone and 2–4% glucose) or in synthetic dropout medium (0.5% ammonium sulphate, 0.17% yeast nitrogen base and 2–4% glucose). For serial dilution growth assays, yeast cells were grown overnight in YPD medium and diluted to an OD600 of 1 followed by 1:10 serial dilutions. From the diluted cultures, 5 µl was spotted onto YPD plates and incubated at 30°C or 19°C. For the antibiotic analysis, 5 µl of paromomycin solution (200 mg/ml) and anisomycin (20 µg/ml) was spotted on filter discs, which were then placed on YPD plates containing the strains to be tested. The H2O2 sensitivity analysis was performed exactly as described previously (28). The overnight culture of the yeast was inoculated to a starting OD600 of 0.1 with YPD. The 9.79 M stock solution of H2O2 was diluted with phosphate-buffered saline (1.5 mM KH2PO4, 2.7 mM Na2HPO4 and 155.1 mM NaCl, pH 7.2), and culture was exposed to 0 and 5 mM H2O2 for 2 h at 30°C. After growth, the cultures were diluted 1000 times with 1× phosphate-buffered saline, and 100 µl of each was plated in duplicates on YPD and incubated for 48 h at 30°C. Colonies were then counted, and the cell survival rates were determined based on the comparison with the number of colonies formed by strain when not exposed to H2O2.
Sucrose gradient analysis
The ribosomal subunits were separated using sucrose gradient centrifugation. The yeast strains were grown in YPD medium (100 ml) at 30°C to early logarithmic phase (OD600 = 1.0) and harvested at 4°C into two 50-ml falcon tubes. The cells were washed twice with 10 ml of buffer B (50 mM Tris–HCl, pH 7.4, 50 mM NaCl and 1 mM freshly added DTT). The washed cells were then suspended in 0.5 ml of buffer B and were lysed by vortexing with equal volume of glass beads. After the lysis, 500 µl of buffer B was added to the lysates. Two centrifugation steps at 3000 g followed by 10 000 g for 15 min, respectively, were used to clarify the lysates. Equivalent amounts of absorbing material were layered on a 20–50% (w/v) sucrose gradient in buffer B. The gradient was made using Gradient Master 107 (Biocomp). The samples were then centrifuged at 24 500 g for 17 h at 4°C in an SW40 rotor using Beckman ultracentrifuge (L-70: Beckman). The gradients were fractionated in an ISCO density gradient fractionar, and the absorbance profile at 254 nm was analysed in ISCO UA-5 absorbance monitor.
The polysome profiles were performed on 10–50% gradients in polysome buffer A (20 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA and 1 mM DTT). Before processing the cell for lysate formation, the cycloheximide was added to the final concentration of 100 µg/ml to a 100 ml YPD culture (OD600 = 1.0). The cells were then washed and processed exactly as described for subunit profiles. The 10 OD254 units of the cell lysates were layered on the gradients and were then subjected to ultracentrifugation in an SW40 Ti rotor (Beckman Coulter, Inc.) for 17 h at 19 000 g and 4°C.
RNA extraction and northern hybridization
For northern blot analysis, RNA was prepared by phenol/chloroform extraction as previously described (29). Ten micrograms of total RNA was separated on a 1% agarose gel in 1× TAE supplemented with 6.66% formaldehyde and transferred to a positively charged nylon membrane (Hybond N+, GE Healthcare) using capillary blotting. Fifty picomoles of the corresponding oligonucleotides (Supplementary Table S3) was radioactively labelled at the 5′-end using 6 µl of γ-[32P]ATP (∼3.3 pmol/µl, Hartmann-Analytics) and 1 µl of T4 polynucleotide kinase (Roche) in the supplied buffer for 1 h at 37°C and purified with G-25 column. Hybridization was done in 15 ml of hybridization buffer (GE Healthcare) overnight at 42°C, and signals were visualized by phosphoimaging using a Typhoon 9400 (GE Healthcare).
Preparation of 25S ribosomal RNA
After the sucrose gradient centrifugation, the fractions corresponding to 60S subunits were collected with the Density Gradient Fractionation System (Teledyne Isco) and precipitated with 2.5 volume of 100% ethanol at −20°C for 16 h. Precipitated 60S subunits were dissolved in water, and 25S rRNA was purified using the RNeasy Kit (QIAGEN).
Reversed phase–high performance liquid chromatography
For reversed phase (RP)–HPLC analysis of base modifications, 70 µg of 25S rRNA was digested to nucleosides by nuclease P1 and alkaline phosphatase. The hydrolysate was analysed by HPLC according to the method described earlier in the text (30). Nucleosides were analysed by RP–HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 µm) equipped with a pre-column (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. For mass spectrometry analysis, nucleosides were collected from four HPLC experiments and desalted twice with a Zorbax Eclipse XDB-C18 column (Agilent; 4.6 × 150 mm, 5 µm) using 5 mM ammonium acetate, pH 6.0, with a flow rate of 0.5 ml/min. After buffer evaporation, samples were resolved in water and applied to MALDI mass spectrometry on a VG Tofspec (Fisons Instruments) in the negative ion mode.
Mung bean nuclease protection assay
The nuclease protection assay was performed exactly as described previously. Specific sequence of the 25S rRNA was isolated by hybridization to complementary (25S-645 and 25S-2142) deoxyoligonucleotides following a protocol previously described with slight modifications (31). Thousand picomoles of the synthetic deoxyoligonucleotides complementary to C633–G680 or C2118–G2166 of yeast 25S rRNA was incubated with 100 pmol of total rRNA and 1.5 µl of DMSO in 0.3 volumes of hybridization buffer (250 mM HEPES and 500 mM KCl, pH 7.0). After hybridization, mung bean nuclease and 0.02 µg/µl RNase A (Sigma-Aldrich) were added to start the digestion. Before the separation of the samples on a 13% polyacrylamide gel containing 7 M urea, they were extracted once with phenol/chloroform and precipitated as described. Bands were extracted using the D-Tube™ Dialyzers according to the manufacturer’s protocol for electroelution (Novagen®).
Primer extension
Primer extension analysis was carried out following the published protocol with some modifications (32). Ten picomoles of the DNA primer GCACTGGGCAGAAATCACATTGCG, complementary to the positions 2178–2201 of 25S rRNA, was 32P-5′-terminally labelled by incubation in the final volume of 40 µl with 50 µCi γ- [32P] ATP and 20 U of polynucleotide kinase in a 2 µl polynucleotide kinase buffer (Fermentas). Reaction was incubated at 37°C for 30 min and then stopped by incubation for 2 min at 90°C. The reaction mixture was then purified using Roche columns (11814419001) to get rid of free γ- [32P] ATP.
An extension reaction premix contained 6 µl of water, 2 µl of dNTP mix (10 µM dGTP, 10 µM dATP, 10 µM dTTP and 10 µM dCTP), 6 µl of RNase-free water and 1 µl (10 U) of Superscript III reverse transcriptase (Invitrogen) per reaction. Fourteen microlitres of the pre-mix was added to the reaction tube with the annealed primer/RNA complex. Samples were incubated at 42°C for 30 min. After incubation, the RNA was precipitated using 3 M sodium acetate, pH 5.2, and 100% ethanol, followed by a washing step with 70% ethanol. After complete removal of supernatant and air-drying of the tubes for 2–3 min, the pellets were resuspended in 6 µl of formamide loading dye (Sambrook et al. 1989). We loaded 1 µl of the sample on to the sequencing gel (Model S2, Biometra) and let it run till the phenol blue dye reached the bottom of the gel. The gel was transferred to Whatman 3MM paper, dried and exposed on a phosphoimager screen. The screen was scanned using Typhoon 9400 (GE) using the Storage Phosphor acquisition mode with Red (633 nm) laser.
Western blot analysis of Bmt2-7xHis
Cell extract from the Δbmt2 strain harbouring a plasmid pSH20 carrying heptahistidine-tagged Bmt2 was prepared using glass beads. Fifty micrograms of total protein from each sample was separated with 12% sodium dodecyl sulphate polyacrylamide gel and blotted on a PVDF membrane (Millipore). The membrane was blocked with 5% non-fat dry milk, and the tagged protein was detected with anti-His monoclonal antibody (Roche; 1:1000 dilution) followed by anti-mouse IgG-conjugated horseradish peroxidase (Bio-Rad; 1:10 000 dilution).
Protein localization
The plasmid pSH18 encoding GFP–Bmt2 fusion protein was constructed using pUG35 plasmid (EUROSCARF). The plasmid pSH18 was then transformed in a strain containing a gene encoding for ScNop56-mRFP. The transformants carrying plasmid were grown to mid-logarithmic phase in synthetic medium lacking uracil at 30°C. The GFP-fused Bmt2 was visualized using a Leica TCS SP5. The RFP-fused Nop56 was used as reference for nucleolar localization.
Bioinformatic analysis
The 3D structure prediction was carried out with amino acid sequence of Bmt2 using a recent protocol (33). The software used is available as a web-based tool. The submitted sequence (query) was scanned against the non-redundant sequence database. Five iterations of PSI-BLAST were used to gather close and remote sequence homologues. A profile from the multiple alignments was then constructed, which was followed by secondary structure prediction using Phyre:Psi-Pred13, SSPro14 and Jnet15 (34–36). The output of each program was predicted in three-states: α-helix (H), β-strand (E for extended) and coil (C). A confidence value for each position of the query for each of three secondary structures was assigned. All these confidence values were then averaged, and a final prediction value was displayed under the individual prediction. DISOPRED software program was run to calculate a two-state prediction of which regions of the query were most likely to be structurally ordered (o) and which disordered (d) (37).
This profile and secondary structure was then scanned against the fold library using a profile–profile alignment algorithm (38). This alignment process generated a score that was then used for ranking the alignments. E-values were then generated using these scores. Full 3D models of the query were generated using the top 10 highest scoring alignments. The missing or inserted regions were repaired using a loop library and reconstruction procedure. Finally, side chains were placed on the model using a fast graph-based algorithm and side chain rotamer library.
RESULTS
Mutants screening for the identification of methyltransferase, catalysing m1A2142 modification
The 25S rRNA undergoes seven base modifications apart from pseudouridylations and ribose methylations. These modifications are performed by snoRNA-independent enzymes, most of which remained elusive. We recently identified Rrp8 as a methyltransferase responsible for one of the two m1A base modifications (24). To find the enzyme or enzymes responsible for m1A2142 modification, we biochemically analysed deletion mutants of all uncharacterized RNA methyltransferases (39).
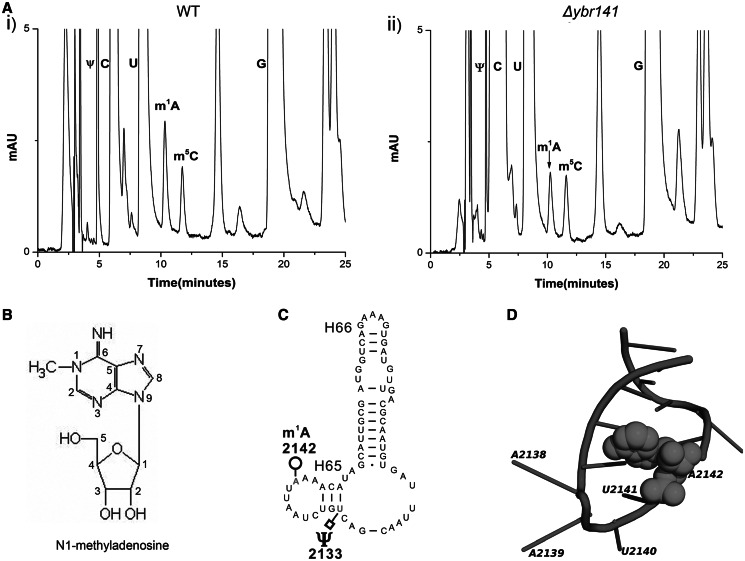
The 25S rRNAs from these mutants were isolated and processed for analysis of m1A modification by RP–HPLC as described in ‘Materials and Methods’ section. Intriguingly, the deletion mutant of YBR141C (Δybr141c) showed a similar reduction in the m1A peak as observed previously for rrp8ΔC mutant, where compared with wild-type, the peak corresponding to m1A reduces to half [Figure 1A (ii)].
Figure 1.
Identification of the m1A2142 methyltransferase of 25S rRNA in S. cerevisiae. The 25S rRNA from the mutant and the isogenic wild-type was digested to nucleosides using P1 nuclease and alkaline phosphatase. Nucleosides from the wild-type and the Δybr141 mutant were analysed by RP–HPLC. (A) Chromatogram from the wild-type (i) and Δybr141 mutant (ii). The peak corresponding to the m1A with a retention time of ∼10 min reduces to half in the Δybr141 mutant. (B) Chemical structure of the N1-methyl adenosine (m1A). (C) Secondary structure of Helix 65 with the m1A and pseudouracil (Ψ) modifications (http://www.rna.icmb.utexas.edu/). (D) Cartoon representing the helix 65 of 25S rRNA. The modified base A2142 is shown as spheres. The cartoon was made using PyMol software (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC) with PDB file 3U5D.pdb.
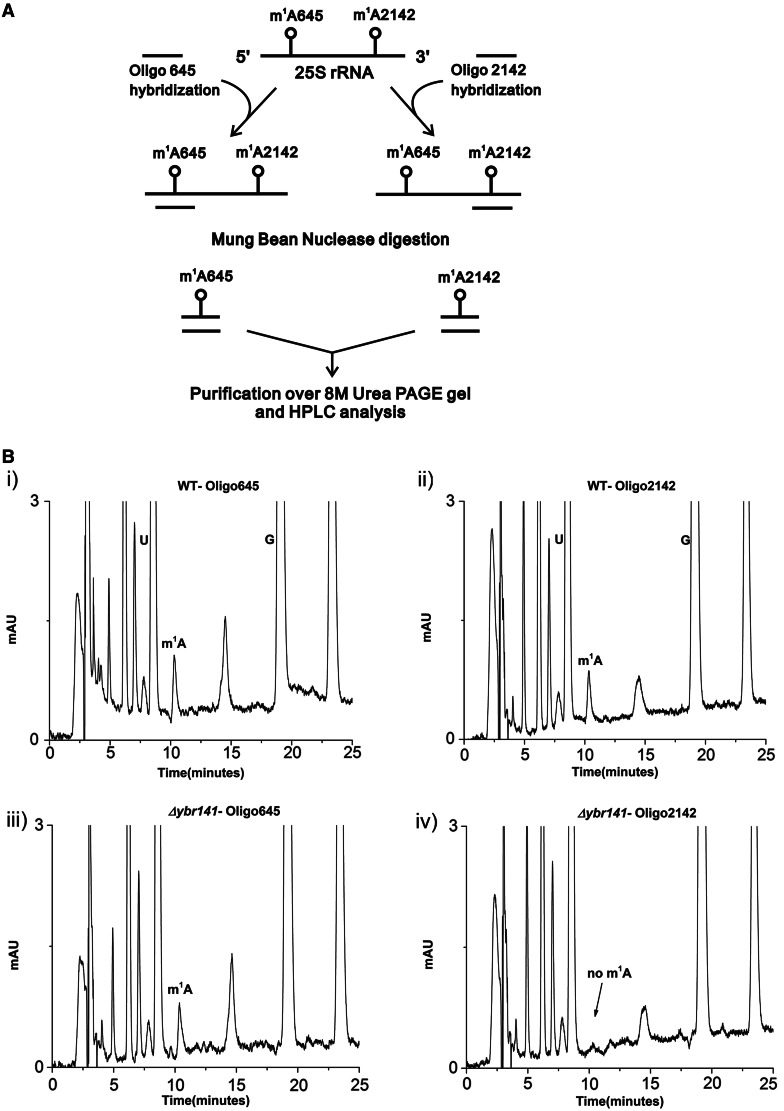
To determine the nucleotide position at which the m1A modification was missing in the Δybr141c mutant, a mung bean nuclease protection assay was performed. Mung bean nuclease is a single-strand–specific endonuclease, which degrades single-strand extension from the ends of RNA and DNA. However, the double-stranded nucleotides are protected against the endonucleolytic cleavage, and it is this property of the nuclease that was exploited to discover the position of modified base in a Δybr141c mutant. Synthetic oligonucleotides complementary to nucleotides 633–680 (Oligo-645) and 2118−2166 (Oligo-2142) were designed corresponding to the position 645 and 2142 (24) (Figure 2A). The 25S rRNA isolated from Δybr141c and wild-type cells was hybridized to these oligonucleotides and subjected to nuclease digestion. Two independent experiments using the oligonucleotides corresponding to two different m1A positions were performed. Now, as only these fragments were protected because of hybridization or double-stranded conformation, both m1A modifications could be separated and addressed independently (Figure 2A). These protected RNA–DNA hybrid fragments were then separated from the rest of the debris and from each other using 8 M-urea PAGE gels. The protected RNA fragments were eluted from the gels using D-Tube™ Dialyzers according to the manufacturer’s protocol by electroelution (Novagen®).
Figure 2.
Mung bean nuclease protection assay. (A) Schematic representation of the Mung bean nuclease protection assay used in the present study for the identification of the position of modified m1A nucleoside in the Δybr141 mutant. (B) RP–HPLC chromatogram of the nucleosides derived from the protected RNA fragments. Specific sequences of the 25S rRNA from wild-type [B (i) helix 25.1] and [B (ii) helix 65] and Δybr141mutant [B (iii) helix 25.1 and B (iv) helix 65] were isolated by hybridization to complementary deoxyoligonucleotides (Oligo645 and Oligo2142) followed by mung bean digestion. The RP–HPLC analyses with these fragments were then performed. The peak corresponding to m1A from helix 65 disappears in Δybr141 mutant.
The eluted fragments were next analysed for the status of m1A modification. As expected, in the wild-type cells the m1A modification was present in both fragments corresponding to the m1A645 and m1A2142 [Figure 2B (i and ii)]. However, in the Δybr141c mutant, the m1A modification from the m1A2142 fragment was missing highlighting the involvement of Ybr141 in m1A2142 modification [Figure 2B (iii and iv)].
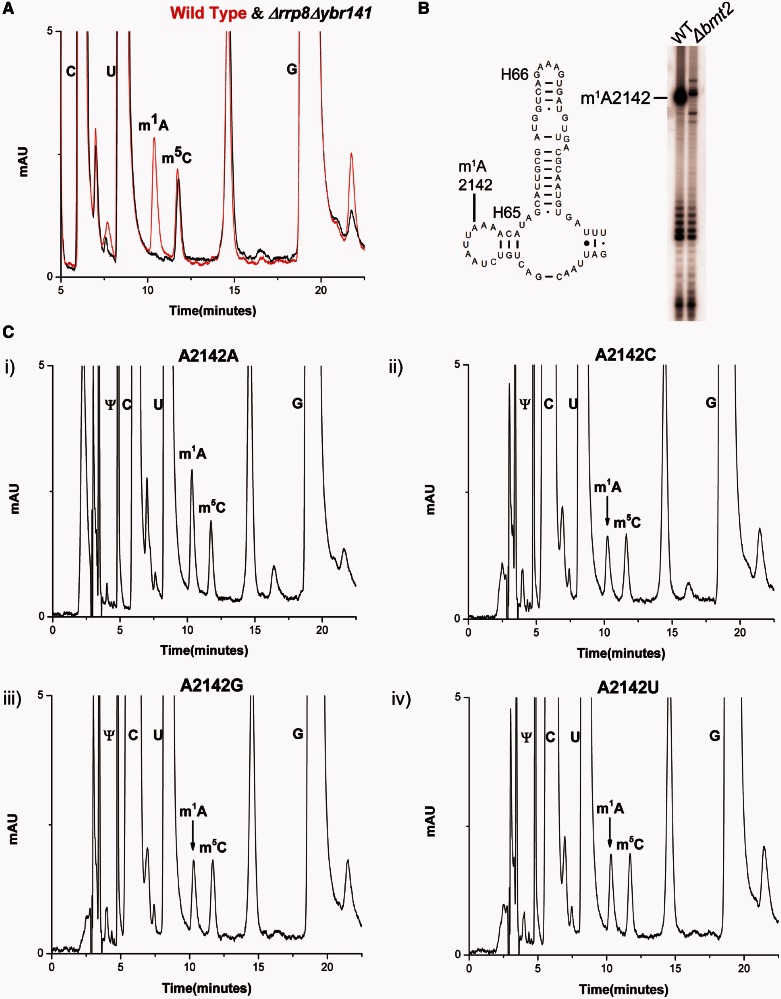
To further augment the role of Ybr141 in m1A2142 modification, a Δybr141cΔrrp8 double mutant strain (BY.PK1033-6D) was constructed. The 25S rRNA from the double mutant was isolated and processed for the m1A modification analysis using the RP–HPLC. As expected and illustrated in the Figure 3A, the peak corresponding to the m1A modification disappeared completely from the 25S rRNA of the double mutant, confirming again Rrp8 and Ybr141 involvement in m1A645 and m1A2142 modifications, respectively (24).
Figure 3.
RP–HPLC analysis of 25S rRNA nucleosides from the double mutant Δrrp8Δbmt2 and rRNA point mutants. (A) Overlaid RP–HPLC chromatogram of the nucleosides derived from the 25S rRNA of Δrrp8Δbmt2 mutant (black) and wild-type (red). The Δrrp8Δbmt2 contains no m1A nucleosides, validating the specific involvement of these two methyltransferases in the m1A modification of 25S rRNA at position 645 and 2142. (B) Primer extension gel. The 25S rRNA from the wild-type and Δbmt2 mutant was isolated using sucrose gradient centrifugation and was analysed for the m1A modification using primer extension with the primer 25S-A2142. The N1 methylation of adenine blocks the Watson–Crick pairing and leads to a strong stop. A strong stop signal indicates the presence of methylation at position 2142. A strong stop at a position 2142 was observed from the 25S rRNA of the wild-type, whereas this stop was missing from the 25S rRNA of Δbmt2 mutant, assigning the position of the modification to A2142. (C) RP–HPLC chromatogram from the rDNA point mutants corresponding to A2142 of 25S rRNA. The A2142 of 25S rRNA was exchanged with U, C and G in a plasmid-borne copy of 35S rDNA transcribed under the native promoter in a strain where the genomic rDNA was deleted. The mutated 25S rRNA was expressed from plasmids pPK622, pPK623 and pPK624 corresponding to A2142C, A2142U, A2142G mutants, respectively. The wild-type 25S rRNA was expressed from the pAV164 plasmid. The peak corresponding to m1A reduces to half in the chromatogram of A2142C (ii), A2142G (iii) and A2142T (iv) compared with corresponding wild-type A2142 (iv).
The 25S rRNA point mutants and primer extension
To corroborate the position of m1A2142 modification and analyse the significance of the modification at this position, rRNA point mutants were created, where the A2142 of 25S rRNA was exchanged with U, C and G.For this analysis, we used a plasmid-borne copy of 35S rDNA transcribed under the native promoter in a strain where the genomic rDNA was deleted. The mutated 25S rRNA was expressed from plasmids pPK622, pPK623 and pPK624 corresponding to A2142C, A2142U, A2142G mutants, respectively. The wild-type 25S rRNA was expressed from the pAV164 plasmid. The 25S rRNAs from these mutants were isolated as described previously and processed for the RP–HPLC analysis. As evident in the Figure 3C, the amount of m1A peak reduces to half when the base A2142 was changed to C, G or U, thus validating the position of the modification. Primer extension analysis using the 25S rRNA from the wild-type and Δybr141c mutant strain was also performed. The methylation of N1 of adenine disrupts the Watson–Crick base pairing and, therefore, should result in a strong stop in the primer extension analysis (40,41). As evident in the Figure 3B, this was indeed the case. A strong stop at position 2142 was observed in case of 25S rRNA of wild-type, whereas this stop was missing in the 25S rRNA derived from the Δybr141c mutant strain.
Once, it became obvious that Ybr141 is the protein involved in the base modification at A2142 of 25S rRNA, we decided to name the gene YBR141c as BMT2.
Bmt2 is predicted to be a Rossmann-like fold methyltransferase
The amino acid sequence comparisons have already predicted Bmt2 to be a Rossmann-like fold methyltransferase, which is characterized by a central seven-stranded β-sheet that is flanked by three helices on each side. Class I methyltransferases modify a wide variety of substrates, including nucleic acids and proteins. Additional recognition domains in many of these enzymes are responsible for substrate specificity. HHpred analysis for the comparison at family level revealed that Bmt2 is indeed related to a large RFM superfamily characterized by proteins whose catalytic domains comprise a seven-stranded β-sheets surrounded by helices.
We also performed further bioinformatic analysis for the 3D structure prediction of Bmt2 using a recently described protocol of Kelley and Sternberg (33). The 3D structure was constructed with sequence coverage of 52%, where 174 residues (52% of Bmt2 amino acid sequence) were modelled with 99.3% confidence by the single highest scoring template. Supplementary Figure S1 shows the cartoon of the predicted 3D structure of Bmt2. This model further supported Bmt2 to be an Ado Met methyltransferase with the characteristic β-sheets surrounded by helices belonging to Rossmann fold superfamily.
As far as the conservation of the Bmt2 is concerned, our searches of the non-redundant sequence database at the NCBI using PSI-BLAST revealed that the Bmt2 orthologues are conserved in members of lower eukaryotes. The members of archaea and bacteria did not show any amino acid sequence conservation with respect to the Bmt2 (data not shown).
Cellular localization and pre-ribosomal association of Bmt2 (Ybr141)
Bmt2 was previously shown in a high-throughput analysis to localize in the nucleolus (42). To validate the nucleolar localization, a plasmid pSH18 expressing Bmt2–GFP fusion was transformed into a strain where Nop56, a nucleolar protein, was expressed as a RFP fusion protein. The cells expressing the fusion protein were visualized with the help of a confocal Leica TCS SP5 microscope. Bmt2 was localized in the nucleolus, indeed, which was in complete consent with the previous study (Supplementary Figure S2A).
Once, the nucleolar localization was confirmed, we next analysed any ribosomal/pre-ribosomal association of Bmt2. A plasmid pSH20 was constructed, where the Bmt2 was fused C-terminally to a heptahistidine tag. The fusion protein was placed and expressed under a strong TDH3 promoter. This plasmid was then introduced into a Δbmt2 strain and polysome profile from the transformed strain was followed. The proteins from the collected fractions were precipitated, and western blot analysis demonstrated that the Bmt2 was associated with the ribosomal subunits (Supplementary Figure S2B). Because of its expression under a strong promoter, there seemed to be diffusion or an unspecific association of the Bmt2 with the 40S fractions.
A C-terminal genomic tagging of Bmt2 with HA epitope was also performed, where the fusion protein was expressed under a native promoter. However, the lower expression level of the fusion protein made it difficult to detect a significant signal above the background. Nevertheless, the nucleolar localization along with the polysome profiles from the pSH20 plasmid carrying heptahistidine-tagged Bmt2 exhibited clearly that Bmt2 associates with the pre-ribosomes to perform the modification of m1A2142 of 25S rRNA. Interestingly, it was shown previously that both m1A modification at position 645 and 2142 are performed early during the 25S rRNA biogenesis (22). This is also in line with the position of these modifications in the mature ribosomes. Both m1A modification are buried deep into the ribosomal structure and, therefore, should be performed early during the 25S rRNA biogenesis, as these positions may not be accessible to the methyltransferases in the mature 60S subunit. The nucleolar localization and ribosomal association of the Bmt2 further established this notion.
Episomally expressed Bmt2 catalyses m1A modification of 25S rRNA in vivo
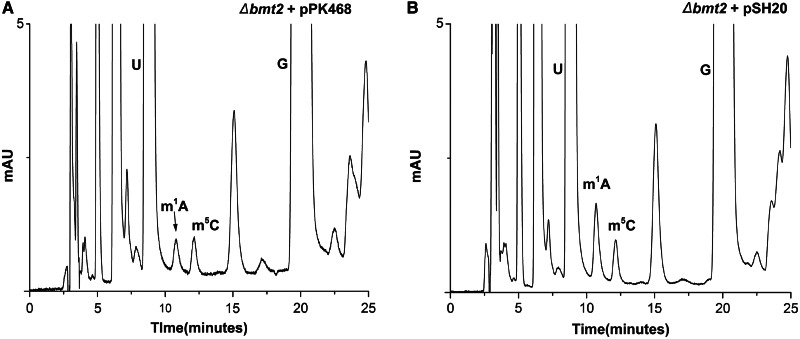
To confirm the functionality of the Bmt2 expressed from plasmid pSH20, we made RP–HPLC analysis with the mutant Δbmt2 strain carrying pSH20 and pPK468 (empty) plasmid. As seen in Figure 4, the episomally expressed Bmt2 from pSH20 plasmid was able to perform the m1A2142 modification in vivo and leads to an increase in m1A peak area as compared with the Δbmt2 mutant with an empty pPK468 plasmid. This clearly demonstrated the functionality of episomally expressed Bmt2 (pSH20) and its involvement in performing the methylation of A2142.
Figure 4.
In vivo methylation of m1A2142 by episomally expressed recombinant Bmt2. To confirm the functionality of episomally expressed Bmt2 and analyse the function of Bmt2 in methylating 25S rRNA at position 2142, the plasmid pSH20 and pPK468 was transformed in the Δbmt2 strain. The 25S rRNA was isolated from the strains carrying these plasmids and processed for HPLC analysis. (A) RP–HPLC chromatogram of the Δbmt2 strain carrying pPK468 plasmid. (B) RP–HPLC chromatogram of the Δbmt2 strain harbouring pSH20 plasmid. It became apparent from the chromatograms that the recombinant Bmt2 expressed from pSH20 plasmid was able to methylate the N1 of A2142 of 25S rRNA, as the area corresponding to the m1A peak increases to almost double amount compared with strain carrying pPK468.
G180R mutation in SAM-binding motif of Bmt2 abolishes its catalytic function
To further corroborate the involvement of Bmt2 in performing the N1-methylation of adenine 2142, we created point mutation in the glycine180 (G180) residue of highly conserved SAM-binding motif of Bmt2. The glycine 180 of Bmt2 corresponds to a highly conserved amino acid in the motif 1 of SAM-binding domain, and a replacement of this conserved glycine with arginine residue was previously shown to be essential for the catalytic functions for Hmt1 and Rrpr8 (24). Moreover, the bioinformatics analysis with the software program (3DLigandSite) also predicted glycine 180 of Bmt2 to be involved in SAM binding.
The point mutation was created on plasmid pSH20 carrying wild-type Bmt2 with heptahistidine tag, as the expression of mutant allele could be easily analysed with a western blot. The plasmid pSH20-G180R carrying the point mutant was then transformed into the Δbmt2 strain, and western blot signal as seen in Supplementary Figure S3E showed that the mutant protein was expressed just like the wild-type protein. We next checked the functionality of the point mutant protein in performing the m1A2142 methylation. The 25S rRNA from the Δbmt2 strain carrying the mutant bmt2-G180R was isolated, and an RP–HPLC analysis was made. As seen in Supplementary Figure S3A, the G180R substitution drastically reduced the catalytic function of the Bmt2 and caused reduction of m1A2142 modification, observed as a significant decrease in the m1A peak in the RP–HPLC chromatogram. Intriguingly, compared with the Δbmt2 deletion mutant, the Δbmt2 mutant strain with pSH20-G180R carried slightly more amount of m1A peak as observed in the Supplementary Figure S3A. Therefore, to analyse any residual activity of the mutant protein, an RP–HPLC analysis of the 25S rRNA from Δbmt2Δrrp8 double mutant with pSH20-G180R was performed. Interestingly, as seen in the Supplementary Figure S3B, we could observe a mild residual activity of mutant protein. The mutant protein bmt2-G180R was able to perform a slight amount of m1A modification in the 25S rRNA. Interestingly during our previous analysis with the Rrp8 point mutant G209R, we observed a similar outcome, where the exchange of this conserved glycine residue (G209R) in the motif 1 of SAM-binding domain affected the SAM binding and catalytic activity of the protein in a similar fashion (24).
Similarly, we also created the point mutations in the N-terminal domain of the protein where we substituted G79R and D116A. These mutants were also expressed like the wild-type (Supplementary Figure S3D). Interestingly, the substitution G79R did not disturb the catalytic function of the protein, whereas the exchange of D116A completely abolished the catalytic function of the protein (Supplementary Figure S3C and D). Taken together, our results demonstrated that Bmt2 methylate m1A of 25S rRNA and the composition of SAM-binding motif are essential for performing the modification.
Growth analysis and antibiotic sensitivity
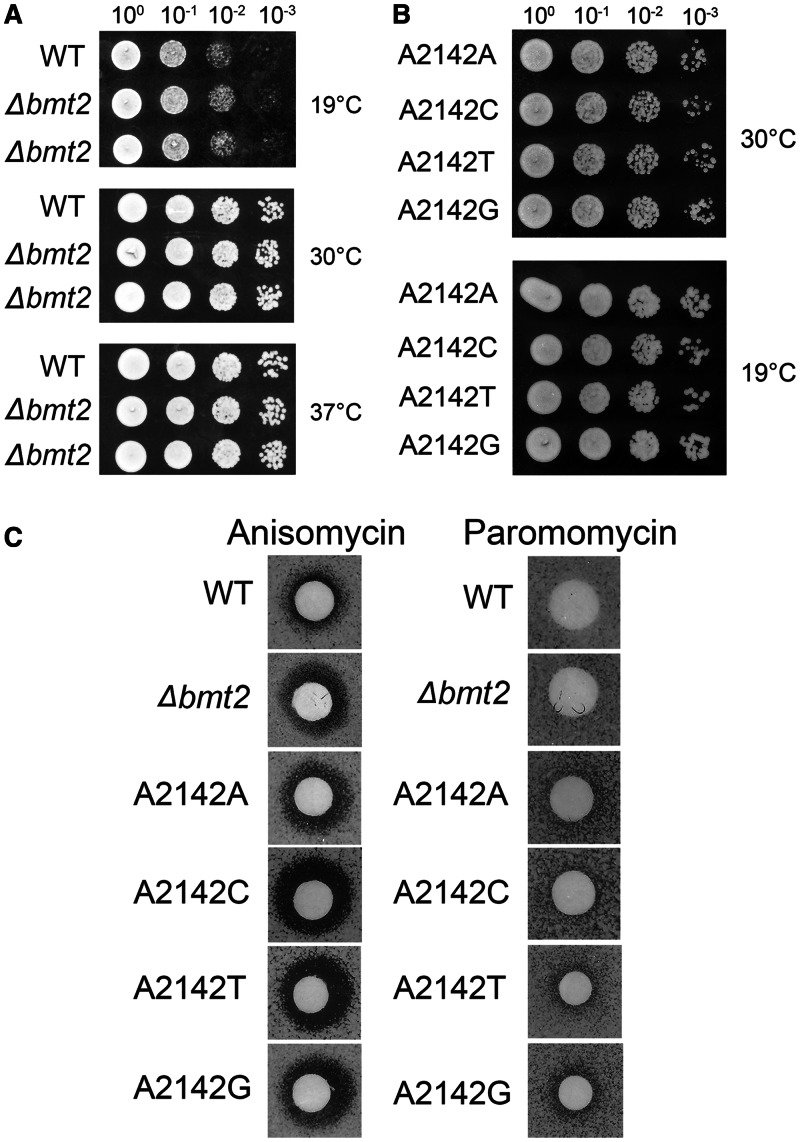
Once, the enzyme responsible for m1A2142 was identified, the effect of lack of this modification on the growth of cells could be analysed. The rDNA point mutants could also be used as a significant control for the analysis. Surprisingly, as known for other modifications of the rRNA, the deletion of the Bmt2 and rDNA point mutants had also no growth phenotype when compared with the wild-type at different temperatures 19°C, 30°C and 37°C on YPD media (Figure 5A and B).
Figure 5.
Growth analysis and antibiotic sensitivity of the Δbmt2 and rDNA point mutants. (A) Ten-fold serial dilutions of the wild-type and Δbmt2 strains were spotted onto solid YPD plates and were incubated at different temperatures. (B) Ten-fold serial dilutions of the strains carrying 25S rRNA point mutants corresponding to A2142 of 25S rRNA. The A2142 of 25S rRNA was exchanged with U, C and G in a plasmid-borne copy of 35S rDNA transcribed under the native promoter in a strain where the genomic rDNA was deleted. The mutated 25S rRNA was expressed from plasmids pPK622, pPK623 and pPK624 corresponding to A2142C, A2142U, A2142G mutants, respectively. The wild-type 25S rRNA was expressed from the pAV164 plasmid. These strains were spotted onto solid YPD plates and were incubated at different temperatures. (C) Anisomycin and paromomycin sensitivity tests were performed by spotting 5 μl of anisomycin (5 µg/ml) and paromomycin solution (200 mg/ml) on filter discs, which were then applied on YPD plates containing the strains indicated.
We previously showed that lack of m1A645 modification makes the cells susceptible to paromomycin, which was also evident with the rDNA mutants where the nucleotide base at 645 position of 25S rRNA was changed from A to U (24). Therefore, to analyse any structural rearrangement, which might lead to antibiotic sensitivity because of lack of m1A2142 modification, we analysed the Δbmt2 mutant cells along with the rDNA mutants of helix 65 for the paromomycin and anisomycin antibiotic sensitivity. Intriguingly, the loss of m1A2142 leads to a mild anisomycin sensitivity, which was more pronounced in the rDNA point mutants (Figure 5C). In contrast, the cells did not show any paromomycin sensitivity (Figure 5C). The antibiotic sensitivity analysis is an interesting tool to analyse the ribosomal biochemistry, especially alteration in structural rearrangement. The anisomycin sensitivity illustrated that the absence of this modification influences the conformation of the helix 65 and ribosomal RNA.
The helix 65 along with the m1A2142 modifications also contains two other pseudouridylations at position 2129 and 2133 that are catalysed by Snr3 ribonucleoprotein complex. To analyse the influence of these modifications on growth and antibiotic sensitivity, we constructed a deletion mutant of SNR3, in which both pseudouridylations were abolished. Surprisingly, we observed minor growth defect and anisomycin sensitivity for the double mutant (data not shown).
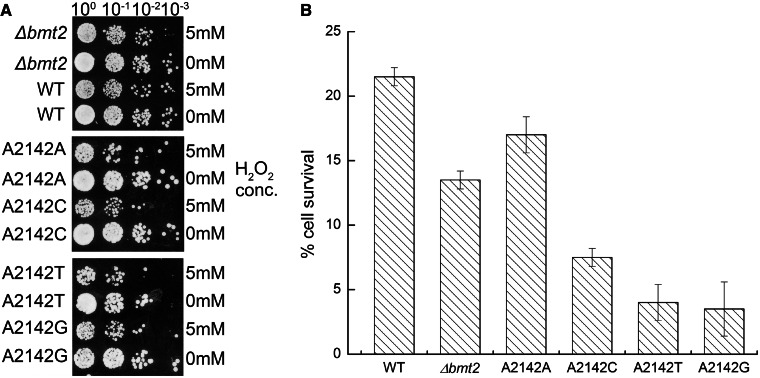
Absence of m1A 2142 from helix 65 makes the cell susceptible to hydrogen peroxide
Interestingly, it was recently shown that the deletion of BMT2 leads to an extended hibernating life span and peroxide sensitivity (25). We also performed H2O2 sensitivity analysis with the Δbmt2 mutant along with the rDNA point mutants. Intriguingly, as evident from the Figure 6A, we could also observe that the Δbmt2 mutant is indeed sensitive to the hydrogen peroxide. We also quantified the sensitivity by calculating the percentage of cell survival on exposure to a lethal concentration of hydrogen peroxide (5 mM) as described earlier in the text (28). The Δbmt2 mutant cells were more prone to peroxide as compared with the isogenic wild–type cells. The cell survival rate on H2O2 exposure was ∼40% lesser in the Δbmt2 mutant as compared with wild-type (Figure 6B). Interestingly, the rDNA mutant exhibited even stronger sensitivity to the peroxide (Figure 6B). Our results clearly showed that the loss of the m1A modification at helix 65 makes the cells more susceptible to hydrogen peroxide.
Figure 6.
Hydrogen peroxide sensitivity analysis. The Δbmt2 strain and the 25S rRNA point mutants along with their corresponding isogenic wild-type were exposed to 5 mM of H2O2 for 2 h. (A) Seven microlitres of undiluted and 10-fold serial dilutions from these exposed strains were then spotted onto YPD plates, and growth was scored after 2 days at 30°C. (B) The ability of these strains to grow on YPD was evaluated by plating 100 μl of a 1:1000 dilution and counting the surviving individual colonies. The bars in the histogram depict the results from three independent experiments, with the error bars indicating the standard deviation.
Influence of the lack of m1A2142 modification on ribosomal processing and translation
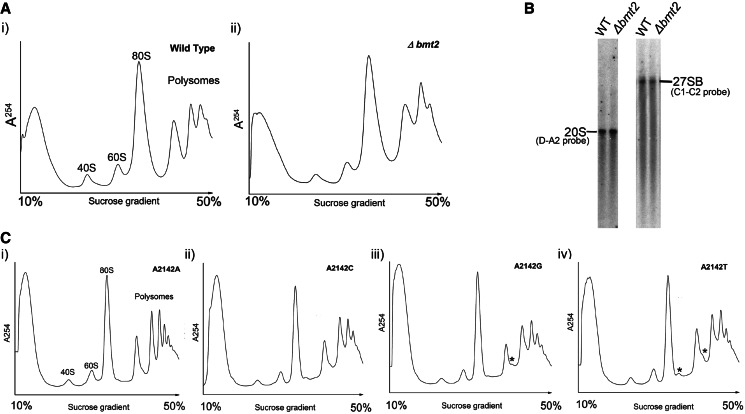
Previous studies have demonstrated that the RNA-modification enzymes play an important role in the processing of the ribosome biogenesis, where deletion or depletion of the enzymes causes serious defects in the final 40S and 60S amounts (24). Therefore, to analyse any such differences in the amount of 40S and 60S subunits in the Δbmt2 mutants, we performed subunit analysis and compared the amount of the 40S and 60S with the wild-type as described in ‘Materials and Methods’ section. We did not observe any significant differences between the amounts of ribosomal subunits in the Δbmt2 mutant compared with wild-type (data not shown), which was also in accordance with our northern blot analysis, where we did not observe any processing or precursors defects (Figure 7B).
Figure 7.
Polysome profile analysis of the Δbmt2 and 25S rRNA point mutants. Polysome profile analyses were performed to detect the translational status of Δbmt2 (A, i) in comparison with its isogenic wild-type (A, ii). (B) Northern blot analysis of the Δbmt2 mutant. Ten micrograms of total RNA was separated on a 1% agarose gel in 1×TAE supplemented with 6.66% formaldehyde and transferred to a positively charged nylon membrane. The membrane was hybridized with radioactively labelled probes ITS1 (D-A2) and ITS2 (C2–C1). (C) Polysome profile analysis of the 25S rRNA point mutants where A2142 of 25S rRNA was exchanged with U, C and G in a plasmid-borne copy of 35S rDNA transcribed under the native promoter in a strain where the genomic rDNA was deleted. The mutated 25S rRNA was expressed from plasmids pPK622, pPK623 and pPK624 corresponding to A2142C, A2142U, A2142G mutants, respectively. The wild-type 25S rRNA was expressed from the pAV164 plasmid. Half-mers formations are indicated by asterisk.
In several cases like Rrp8, Bud 23 and Spb1, the rRNA modification plays an important role in the translation, where deletion leads to a decrease in the translational potential of cells observed by reduction in the polysome fractions or leading to a subunit joining defects observed as half-mer formation (24,43,44). To find any such translational defect, the polysome profiles from the Δbmt2 mutant along with the rDNA point mutants were made and compared with the wild-type (Figure 7A and C). Interestingly, the exchange of adenine at position 2142 to other bases, which eventually leads to lack of m1A modification in the 25S rRNA, leads to the formation of half-mers. Now, as there were no significant differences in the amount of 60S subunits in the rDNA point mutants, the half-mers formation in these mutants seemed to be the result of subunit joining defects (Figure 7C). As the helix 65 comprises the intersubunit region of 60S subunit, the appearances of half-mers suggest that the exchange of the adenine to other bases alters the framework of the rDNA, which then influences the subunit joining. The anisomycin sensitivity also supports this notion. Furthermore, it was recently shown that in Escherichia Coli, the Helix 65 and 66 mutants also promoted subunit association defects (45). Surprisingly, no such half-mers formation was observed in the polysome profiles from the Δbmt2 mutant, which is also in accordance with mild anisomycin sensitivity. Interestingly, we recently demonstrated a similar case with the Δrrp8 mutant, where the half-mers formation and stronger antibiotic sensitivity were observed only in the rDNA point mutants of helix 25.1 (24). Nevertheless, with the rDNA point mutants, the phenotypes corresponding to the modification are highlighted more strongly than the deletion mutant of the corresponding enzyme. This can be because of either the growth rate, which is slower in the mutants, as the rDNA is expressed from the plasmid, or the lack of different multiple copies of rDNA in the plasmid-borne expression system, where only mutated plasmid is the sole source of rRNA. Interestingly, with the rDNA point mutants, we could not only address the modification but could also emphasize the role of helix 65 in the subunit joining and translation.
Investigation of any regulatory role of m1A modifications
RNA modifications have been recently shown to play a regulatory role, where the modifications, especially methylation, vary on different growth conditions (46). Methylation is a reversible modification, and it was recently demonstrated for the m6A modification of mRNAs that the amount of this modification alters depending on the growth phase of the cells and during different stress conditions (46). To analyse any such role played by the m1A modifications of 25S rRNA during the different growth phases, we isolated the 25S rRNA as described earlier in the text from the cells at different growth phases [early exponential (OD600 − 1), at the end of first growth phase (OD600 − 20) and stationary phase (OD600 − 43)]. These stationary phase cultures provided an interesting prospect to analyse 25S rRNA from the cultures where the cells experience nutrient starvation, especially with respect to the carbon source. The cell cultures derived from the first growth phase cells (OD600 − 20) represented the cells experiencing glucose starvation, whereas the cultures derived from the stationary phase cells (OD600 − 43) represented the cells experiencing complete nutrient starvation.
The cells were inoculated in a 1 l of YPD medium with a starting OD600 − 0.2, and an equivalent number of cells for different growth phases as described earlier in the text were harvested. The 25S rRNA from these cells was then isolated and processed for the HPLC analysis. As seen in Supplementary Figure S4, the amount of m1A modifications did not display any significant differences for the different growth phases, demonstrating that the amount of m1A modifications does not alter at the end of first growth phase and stationary phase and remains same as at early exponential phase.
DISCUSSION
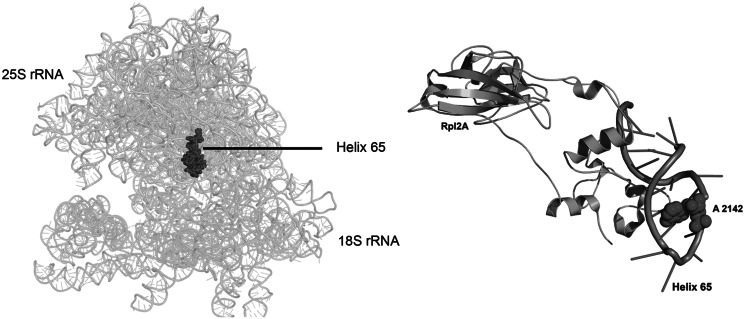
The presence of the base modifications in 25S rRNA was documented as early as in 1973 by Planta’s laboratory (20,22). However, the enzymes involved in these modifications remained elusive for a long time. In the present study, we unravelled a previously uncharacterized methyltransferase, Bmt2, to be involved in the base modification of 25S rRNA at position 2142. After Rrp8, Bmt2 is the second m1A methyltransferase that is involved in the base modification of helix 65 of 25S rRNA in S. cerevisiae. H65 belongs to domain IV, which accounts for most of the intersubunit surface of the large subunit (Figure 8A). H65 is also exposed to the subunit surface on the intersubunit side. Interestingly, L2, a highly conserved protein, has also been shown to make physical contact with the helix 65, especially with its SH3-β barrel globular domain (47) (Figure 8B). The domain IV of 25S rRNA plays a significant role in the translation. This is well supported by a number of studies that have highlighted the importance of this region and the modifications they harbour for the optimal function of the ribosomes (1,47,48). Our analysis with the 25S rRNA lacking m1A modification of helix 65 also confirmed the importance of domain IV of 25S rRNA in subunit joining and antibiotic sensitivity.
Figure 8.
Location of helix 65 in mature 80S ribosome. A 3D cartoon of the rRNA structure of ribosome, emphasizing the helix 65 in the spheres. (A) The helix 65 belongs to domain IV of the 25S rRNA, which accounts for most of the intersubunit surface of the large subunit. (B) Cartoon representing the vicinity of Rpl2A with Helix 65. The A2142 is shown as spheres. All the pictures were made using PyMol software (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.). The PDB files 3U5B, 3U5C, 3U5D and 3U5E were used for the representation of ribosome structure.
Methylation is a predominant post-transcriptional modification found in rRNA (4). Base methylation promotes base-stacking by increasing the hydrophobicity and expanding the polarizability (49). Methylation also influences the structure by increasing steric hindrance, blocking hydrogen bonds [e.g. at Watson–Crick positions and providing positive charge to the nucleosides which in turn effect the hydrogen bonding (40,50)]. m1A is a conserved modification in the TΨC loop and helix 25.1 of eukaryotic tRNAs and 25/28S rRNA, respectively (4). The presence of the methyl group at the N1 position disrupts the Watson–Crick interface of adenine, but it still allows formation of a Hoogsteen base pair with uridine or thymine (2). The m1A58 modification has been shown to play a significant role in maintaining the tRNA tertiary structure, which helps in stabilizing initiator-tRNAmet in yeast; thus, it is an essential modification (51).
As far as 25S rRNA is concerned, we recently showed that m1A645 seems to play an important structural role in the 60S assembly. In contrast, the m1A2142 does not interfere with 60S assembly, as we could not observe any 60S biogenesis defect in the Δbmt2 mutant. However, as shown previously, we could show that the Δbmt2 mutant exhibits peroxide sensitivity. The base modifications, especially for tRNAs, have also been observed to display peroxide sensitivity. The modification levels of 2′-O-methyl cytidine 5-methyl cytidine (m5C) and N-2,2-dimethyl guanosine (m22G) alter on treatment of hydrogen peroxide, and the yeast cells lacking the enzymes responsible for the biogenesis of these three modifications were demonstrated to have hydrogen peroxide hypersensitivity (52). These observations with the tRNA base modification and the m1A 2142 indicate that the biosynthetic pathways of these modifications might be involved in the cellular response towards oxidative stress stimulated by hydrogen peroxide. However, future studies are required to elucidate the specific role of these modifications in oxidative stress. Recent studies have elaborated yet unknown role of ribosomes and the factors associated with ribosomes in oxidative stress. Proteins like Fap7, which were identified as a protein involved in oxidative stress, have been recently shown to be involved in 40S biogenesis (53, 54). Interestingly, the antisense strand of 25S rRNA also carries a mitochondrial protein Tar1, which is the most abundant protein-coding gene in yeast and has been reported to be essential for the mitochondrial function (55). Although the clue why this results in increased hydrogen peroxide sensitivity remains still obscure, the interference of ribosome biogenesis and ribosome function with oxidative stress becomes more and more obvious, and future studies are required to elucidate this phenomenon.
RP–HPLC has been instrumental in identifying the nucleoside modification. Interestingly, retention time also helps in providing insights into the physical properties of the modified bases. Generally, the alkylation provides hydrophobicity to the molecules, but as observed before, we could also observe an increase in polarity of the adenosine once it is modified at first nitrogen atom (51). This increased polarity of m1A by virtue of methyl group could be seen as faster elution at ∼10 min on C-18 Supelcosil column compared with elution of adenosine at 35 min. Intriguingly, this also depends on the nitrogen atom selected for alkylation, as methylation at N6 position results in increased hydrophobicity compared with adenosine, and this could be observed as late elution at ∼45 min on the column (data not shown). These modifications can, therefore, provide specific physical properties to the RNA, which then allow RNA to have a conformation that apparently stabilizes vital transient interactions for the optimum functionality of ribosomes in translating genetic information.
Ribosome biogenesis is a complex multi-step process, where the modifications are introduced on a newly transcribed rRNA before its assembly into subunits (56). However, the synthesis of other modifications requires specific conformations of pre-RNA available only in fully or partial assembled subunits (57). In the present study, we also provide some evidences for the site and timing of the modifications. The nucleolar localization of the protein along with its pre-ribosomal association made it apparent that the m1A2142 modification is performed in the nucleolus, during early 60S maturation phase. This is further supported by the 80S structure, as the A2142 of helix 65 is buried deep in the structure and will not be accessible in the mature 60S ribosome. Interestingly, the Planta’s group could also assign a similar chronology to the modification almost 50 years ago (22).
The amino acid sequence analysis of the Bmt2 has a characteristic C-terminal motif of Class I methyltransferases (Rossmann-like fold protein family). Interestingly, the in silico 3D model of Bmt2 further illustrated that the protein belongs to the Rossmann-like fold protein family. Furthermore, in the present study, we also demonstrated that the substitution of highly conserved glycine180 residue in the SAM-binding motif of Bmt2 with arginine abolishes the catalytic function of the protein. Proteins with highly significant similarity also exist in other lower eukaryotes, including the pathogenic yeasts like Candida albicans (Supplementary Figure S5). The Bmt2 homologues are likely performing the same function in these organisms. However, the lack of any rRNA modification details from these organisms prevents us from validating this notion. Interestingly, both SPOUT and RFM (Rossmann-like fold methyltransferases) MTases have been discovered to catalyse the m1A modification (58). However, in yeast, only RFM MTases have been discovered to be involved in m1A methylation. A recent crystal structure analysis of RsmC, a bacterial RFM methyltransferase that catalyses the methylation of m2G1207 of 16S rRNA in E. coli, has provided interesting insights into the mechanism of RNA methyltransferases in catalysing the base modification (59). Intriguingly, the N- and C-terminal domains of the RsmC protein form a canonical Rossmann-like fold methyltransferase fold with slight variations. These two structurally related N- and C-terminal domains of RsmC are probably the result of a gene duplication event followed by domain subfunctionalization. The N-terminal of RsmC provides the substrate specificity, whereas the C-terminal binds to S-adenosyl methionine and catalyses the modification reaction. Therefore, it is tempting to speculate that the base modification reaction requires two Rossmann-like methyltransferase fold domains, one for the substrate binding and other for the SAM binding and catalysis. Interestingly, most of the base methyltransferase with just one RFM fold have been discovered to function either as a homo dimer or as a heterodimer. This has also been recently shown for Bud23 that needs an assistance of another methyltransferase adaptor protein Trm112, for the base modification (60). Intriguingly, apart from sharing sequence homology at the C-terminal Rossmann fold domain, the Rrp8 and Bmt2 also share a significant homology at N-terminal domain (Supplementary Figure S6). The N- and C-terminal domains of Rrp8 have already been predicted to have a coiled coil conformation (39). The Bmt2 is also predicted to carry the coiled coil structural motif at the C-terminal (data not shown). Interestingly, the coiled coil conformation has been suggested to facilitate the oligomerization or dimerization of the proteins. It is tempting to speculate that the coiled coil domains of Rrp8 and Bmt2 promote the dimerization of these proteins, which would also suggest that, these two proteins function as a homodimer for the modification reaction. Moreover, the exchange of D116A in the N-terminal of Bmt2 completely abolished the catalytic function of protein. It is tempting to speculate that this amino acid residue is important for either substrate binding or for protein–protein interaction. Nevertheless, a detailed structural analysis of these proteins is required to address this perception.
In summary, we showed that Bmt2 is a nucleolar Rossmann-like fold methyltransferase responsible for the m1A2142 methylation of 25S rRNA in S. cerevisiae. We provide evidences that the lack of modification provides hydrogen peroxide and anisomycin sensitivity along with mild defect in subunit joining. The identification of Bmt2 and the respective rDNA mutations reported in here will enable future structural analyses of ribosomes lacking these modifications and provide the bases for the future understanding of the precise role of these modifications in ribosome function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–6 and Supplementary Reference [61].
FUNDING
Funding for open access charge: DFG: Deutsche Forschungsgemeinschaft (En134-9) and Excellence Cluster: Macromolecular Complexes.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
S.S. would like to thank DAAD for the award of PhD scholarship. The authors thank Roman Martin for helping with primer extension and Dr Britta Meyer for helping with confocal microscopy. They also thank Jun Yang for preparing the schematic illustration of mung bean nuclease protection assay and for careful reading of the manuscript. They acknowledge Dr Christian Peifer for his fruitful and critical discussions.
REFERENCES
- 1.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yi C, Pan T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grosjean H. Fine-Tuning of RNA Functions by Modification and Editing. 2005;Vol. 12 Topics in Current Genetics, Springer. [Google Scholar]
- 4.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways—2012 update. Nucleic Acids Res. 2013;41(Database issue):D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker M, Muller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012;40:11648–11658. doi: 10.1093/nar/gks956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2005;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young BD, Weiss DI, Zurita-Lopez CI, Webb KJ, Clarke SG, McBride AE. Identification of methylated proteins in the yeast small ribosomal subunit: a role for SPOUT methyltransferases in protein arginine methylation. Biochemistry. 2012;51:5091–5104. doi: 10.1021/bi300186g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 14.Lilley DM. The ribosome functions as a ribozyme. Chembiochem. 2001;2:31–35. doi: 10.1002/1439-7633(20010105)2:1<31::AID-CBIC31>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 15.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 17.White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of Pre-40S subunits. Mol. Cell. Biol. 2008;28:3151–3161. doi: 10.1128/MCB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, et al. The bowen-conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of 1191 in yeast 18S rRNA. Nucleic Acids Res. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafontaine D, Delcour J, Glasser AL, Desgrès J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3'-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 20.Klootwijk J, Planta RJ. Analysis of the methylation sites in yeast ribosomal RNA. Eur. J. Biochem. 1973;39:325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- 21.Brand RC, Klootwijk J, Steenbergen TJM, Kok AJ, Planta RJ. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 2008;75:311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- 22.Veldman GM, Klootwijk J, de Regt VC, Planta RJ, Branlant C, Krol A, Ebel JP. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakin A, Lane BG, Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry. 1994;33:13475–13483. doi: 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- 24.Peifer C, Sharma S, Watzinger P. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 2013;41:1151–1163. doi: 10.1093/nar/gks1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postma L, Lehrach H, Ralser M. Surviving in the cold: yeast mutants with extended hibernating lifespan are oxidant sensitive. Aging. 2009;1:957–960. doi: 10.18632/aging.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 27.Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 28.Veniamin S, Sawatzky LG, Banting GS, Glerum DM. Characterization of the peroxide sensitivity of COX-deficient yeast strains reveals unexpected relationships between COX assembly proteins. Free Radic. Biol. Med. 2011;51:1589–1600. doi: 10.1016/j.freeradbiomed.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 29.McEntee CM, Hudson AP. Preparation of RNA from unspheroplasted yeast cells (Saccharomyces cerevisiae) Anal. Biochem. 1989;176:303–306. doi: 10.1016/0003-2697(89)90313-8. [DOI] [PubMed] [Google Scholar]
- 30.Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 31.Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toh SM, Xiong L, Bae T, Mankin AS. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2007;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley LA, Sternberg M. Protein structure prediction on the Web: a case study using the Phyre server [abstract] Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 35.Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 36.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue):W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 38.Bennett-Lovsey RM, Herbert AD, Sternberg MJE, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 39.Wlodarski T, Kutner J, Towpik J, Knizewski L, Rychlewski L, Kudlicki A, Rowicka M, Dziembowski A, Ginalski K. Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome. PLoS ONE. 2011;6:e23168. doi: 10.1371/journal.pone.0023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Micura R, Pils W, Höbartner C, Grubmayr K, Ebert MO, Jaun B. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res. 2001;29:3997–4005. doi: 10.1093/nar/29.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helm M, Giegé R, Florentz C. A Watson−Crick base-pair-disrupting methyl group (m 1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999;38:13338–13346. doi: 10.1021/bi991061g. [DOI] [PubMed] [Google Scholar]
- 42.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 43.White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell. Biol. 2008;28:3151–3161. doi: 10.1128/MCB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 45.Kitahara K, Kajiura A, Sato NS, Suzuki T. Functional genetic selection of Helix 66 in Escherichia coli 23S rRNA identified the eukaryotic-binding sequence for ribosomal protein L2. Nucleic Acids Res. 2007;35:4018–4029. doi: 10.1093/nar/gkm356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8:e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meskauskas A, Russ JR, Dinman JD. Structure/function analysis of yeast ribosomal protein L2. Nucleic Acids Res. 2008;36:1826–1835. doi: 10.1093/nar/gkn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang X-H, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF. Structural and functional roles of the N1- and N3-protons of at tRNA's position 39. Nucleic Acids Res. 1999;27:3543–3549. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishitani R, Yokoyama S, Nureki O. Structure, dynamics, and function of RNA modification enzymes. Curr. Opin. Struct. Biol. 2008;18:330–339. doi: 10.1016/j.sbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA. 2010;16:1317–1327. doi: 10.1261/rna.2057810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan CTY, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juhnke H, Charizanis C, Latifi F, Krems B, Entian KD. The essential protein fap7 is involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 2000;35:936–948. doi: 10.1046/j.1365-2958.2000.01768.x. [DOI] [PubMed] [Google Scholar]
- 54.Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S Pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poole AM, Kobayashi T, Ganley ARD. A positive role for yeast extrachromosomal rDNA circles? Extrachromosomal ribosomal DNA circle accumulation during the retrograde response may suppress mitochondrial cheats in yeast through the action of TAR1. Bioessays. 2012;34:725–729. doi: 10.1002/bies.201200037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu M, Novotny GW, Douthwaite S. Methylation of 23S rRNA nucleotide G745 is a secondary function of the RlmAI methyltransferase. RNA. 2004;10:1713–1720. doi: 10.1261/rna.7820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ero R, Peil L, Liiv A, Remme J. Identification of pseudouridine methyltransferase in Escherichia coli. RNA. 2008;14:2223–2233. doi: 10.1261/rna.1186608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motorin Y, Helm M. RNA nucleotide methylation. WIREs RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 59.Demirci H, Gregory ST, Dahlberg AE, Jogl G. Crystal structure of the Thermus thermophilus 16 S rRNA methyltransferase RsmC in complex with cofactor and substrate guanosine. J. Biol. Chem. 2008;283:26548–26556. doi: 10.1074/jbc.M804005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DLJ, Heurgue-Hamard V. Trm112 Is Required for Bud23-Mediated Methylation of the 18S rRNA at Position G1575. Mol. Cell. Biol. 2012;32:2254–2267. doi: 10.1128/MCB.06623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chernoff YO, Vincent A, Liebman SW. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.