Abstract
Activation-induced cytidine deaminase (AID) deaminates deoxycytidine (dC) to deoxyuracil (dU) at immunoglobulin loci in B lymphocytes to mediate secondary antibody diversification. Recently, AID has been proposed to also mediate epigenetic reprogramming by demethylating methylated cytidines (mC) possibly through deamination. AID overexpression in zebrafish embryos was shown to promote genome demethylation through G:T lesions, implicating a deamination-dependent mechanism. We and others have previously shown that mC is a poor substrate for human AID. Here, we examined the ability of bony fish AID to deaminate mC. We report that zebrafish AID was unique among all orthologs in that it efficiently deaminates mC. Analysis of domain-swapped and mutant AID revealed that mC specificity is independent of the overall high-catalytic efficiency of zebrafish AID. Structural modeling with or without bound DNA suggests that efficient deamination of mC by zebrafish AID is likely not due to a larger catalytic pocket allowing for better fit of mC, but rather because of subtle differences in the flexibility of its structure.
INTRODUCTION
Activation-induced cytidine deaminase (AID) is a B lymphocyte-specific member of the apolipoprotein B mRNA-editing catalytic component (APOBEC) family of Zn-dependent cytidine deaminases (1,2). AID converts deoxycytidine (dC) to deoxyuridine (dU) on single-stranded DNA (ssDNA), preferentially in trinucleotide WRC (W = A/T, R = A/G) motifs (3–8). In activated B cells where it is primarily expressed, AID induces mutations and double-strand breaks that initiate somatic hypermutation and class switch recombination (9–11). Inherited mutations of AID that incapacitate function result in hyper-IgM immunodeficiency syndrome (10,12). Although its activity is largely restricted to the Ig loci, AID also initiates off-target mutations and double-stranded breaks that can lead to transformations (13–17).
Although the roles of AID in immunity and lymphomagenesis are well established, a more recent body of evidence implicates AID in epigenetic reprogramming through genome demethylation (18,19). First, AID has been reported to be expressed in pluripotent tissues, such as oocytes, primordial germ cells and embryonic stem cells, although at considerably lower levels than in activated B lymphocytes (19,20). Second, primordial germ cells from AID−/− mice have hyermethylated genome (21). Third, in a heterokaryon system of human somatic cell re-differentiation, AID is required for promoter demethylation (22). Fourth, in zebrafish embryos, AID knockdown and overexpression corresponded with promoter hyper and hypomethylation, respectively, and morpholino knockdowns of AID led to developmental abnormalities (23).
The 5-methylctyosine (mC) in CpG motifs constitutes the most common form of genome methylation associated with transcriptional repression (24–30). In terminally differentiated human cells, the majority of CpGs are methylated (31–34). In principle, mC can be demethylated either through DNA replication or enzymatic processes. mC can be hydroxylated to form 5hmC by TET family enzymes, although the extent of their involvement in vivo, as well as the pathways which may act downstream to process 5hmC, is poorly understood (35–42). It is also possible that either mC or 5hmC is deaminated by AID or other deaminases, leading to dT or 5hmU, respectively, that can be substrates of thymine DNA glycosylase (TDG) or uracil glycosylase (18,20–23).
The involvement of AID in mC demethylation may or may not be through direct deamination. AID may act as a scaffolding molecule, recruiting or stabilizing complexes with the ultimate function of demethylation. In support of this possibility, dozens of putative binding partners for AID have already been proposed, some of which are involved in DNA repair and transcription (43). Alternatively, several lines of evidence suggest that AID can directly deaminate mC. First, deamination of mC generates G:T mismatches, which were observed in the zebrafish experimental system (23). Second, MBD4, which recognizes G:T mismatches, was found to act synergistically with AID towards demethylation (23). Third, enzymatically inactive AID harboring a catalytic site point mutation failed to mediate hypomethylation (23).
On the other hand, AID-deficient mice or individuals do not exhibit developmental defects (9,12,18,44), suggesting that the role of AID in genome demethylation may be significantly more subtle and/or redundant than in zebrafish. In agreement with this, we and others showed that mC is a poor substrate for human AID (6,45). In contrast, another study found that human AID efficiently deaminates mC (20), whereas two more recent studies have confirmed our previous finding (46,47). We have previously shown that bony fish AID exhibit distinct biochemical characteristics compared with their human counterpart (48). Here, we investigated the activity of purified bony fish AID on mC. We found that in contrast to its human and other bony fish orthologs, zebrafish AID efficiently deaminates mC. This finding suggests that genome demethylation through direct mC deamination by AID is more likely in zebrafish than humans.
MATERIALS AND METHODS
AID expression and purification
EcoRI fragments encoding the ORF of medaka AID (Ol-AID) and tetradon AID (TnAID) were synthesized based on identified genomic sequences (http://uswest.ensembl.org/index.html) and cloned into pGEX-5X-3 (GE Healthcare, USA) to generate GST-AID expression constructs in the same manner we have described for human AID (Hs-AID), channel catfish AID (Ip-AID) and zebrafish AID (Dr-AID) (4,48). Domain-swapped AID constructs were generated by synthesis of full ORF sequence (Genscript, USA), and point mutants were generated by site-directed mutagenesis. GST-AID was purified as described previously (4). In total, three independent preparations of Hs-AID, Ip-AID, Dr-AID, Tn-AID, Ol-AID, domain-swapped and mutant AIDs purified in parallel were used. SDS–PAGE with bovine serum albumin loading standards was used to equalize the amount of GST-AID used in experiments.
Preparation of substrates
Preparation of partially single-stranded bubble substrates for AID deamination has been described previously (4). Briefly, six bubble substrates were used differing only in the dinucleotide sequence upstream of the target C nucleotide, constituting a WRC or non-WRC motif, with either dC or mC as the target of deamination (Figure 1A). In all, 2.5 pmol of the target strand was 5′-labeled with [γ-32P] dATP using polynucleotide kinase (NEB, USA), followed by purification through mini-Quick spin DNA columns (Roche, Indianapolis, IN, USA) and annealing of 2-fold excess (5 pmol) of the complementary strand.
Figure 1.
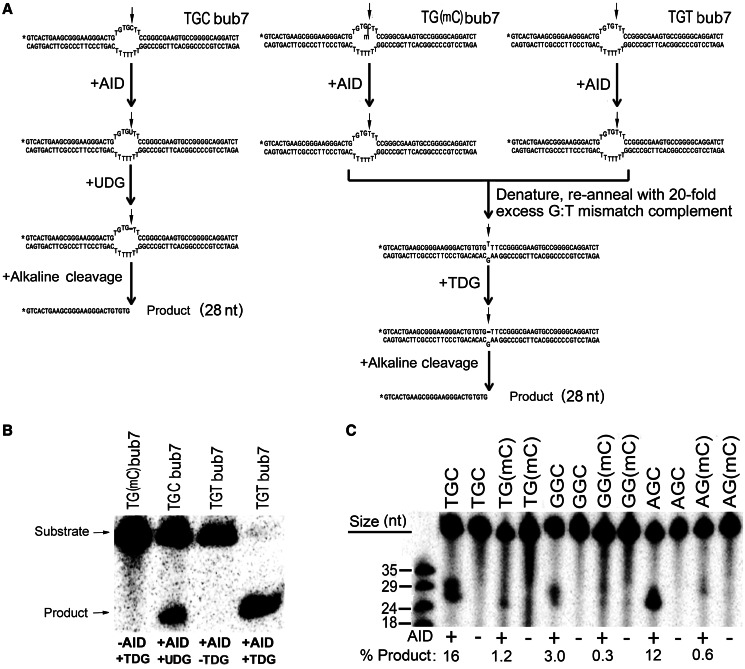
Experimental scheme for measurement of deamination activity on 5-methylcytosine. (A) Typical bubble type substrates used in this study are shown. TGCbub7 denotes a substrate bearing the WRC motif TGC located in a seven-nucleotide-long bubble region. Left shows the scheme for the UDG-based alkaline cleavage assay on the WRC bearing substrate TGCbub7. The middle shows the scheme for the TDG-based alkaline cleavage assay on the WR(mC) bearing substrate TG(mC)bub7. Right shows a control reaction for the TDG-based alkaline cleavage assay to detect the activity of AID on mC. The substrate is TGTbub7, which is the expected product of TG(mC)bub7 after deamination by AID. (B) Control reactions for the efficiency and specificity of TDG. In all, 50 fmol substrate was incubated with AID, TDG or both (as shown in A). This control was included in each subsequent experiment to ensure 100% TDG efficiency. (C) Comparison of Hs-AID activity on dC versus mC located either in a WRC motif (TGC, AGC) or non-WRC motif (GGC). In all, 50 fmol substrate was incubated with AID. A typical alkaline cleavage gel used to measure the generation of deaminated product is shown and percentage of product formation is shown below each lane.
Alkaline cleavage deamination assay
The standard alkaline cleavage assay for measuring the activity of AID on dC has been described previously (7,49). A modified version of the assay was used to measure AID activity on substrates containing mC (Figure 1A). Briefly, 25 fmol substrate was incubated with 0.25 μg of GST-AID in 100 mM phosphate buffer, pH 7.3, in a volume of 10 μl at the optimal temperature for each AID (Hs-AID: 37°C, fish AID: 25°C) for times ranging from 30 min to overnight. AID was deactivated at 85°C for 30 min. For substrates containing dC, the volume was increased to 20 μl by adding 7.8 μl H2O, 2 μl 10× UDG buffer and 0.2 μl (1 U) UDG enzyme (NEB, USA), followed by 30-min incubation at 37°C to excise the uracil and generate an alkali-labile abasic site (Figure 1A, left). For substrates containing mC, the volume was then increased to 20 μl by adding 8 μl of H2O, 1 μl of 1 M KCl and 1 μl of 1 pmol/μl stock (40-fold excess) of a fully complementary strand. Annealing was performed to generate a G:T mismatch double-stranded substrate. Volume was increased to 30 μl by adding 6 μl of H2O, 3 μl of 10× TDG buffer and 1 μl (1 U) TDG enzyme (Trevigen, UK) followed by overnight incubation at 65°C to excise the thymine (Figure 1A, middle and right). NaOH was added to 100 mM, and the sample was heated to 96°C for 8 min to cleave the abasic site and electrophoresed on a 14% denaturing gel. Control reactions are shown in Figure 1B. The gels were exposed to a Kodak Storage Phosphor Screen GP (Bio-Rad) and visualized using a PhosphorImager (Bio-Rad, Hercules, CA, USA) on Quantity One software (Bio-Rad, Hercules, CA, USA).
Data collection and quantitation
Densitometry was performed using ImageLab Analysis Software (Bio-Rad, Hercules, CA, USA). For each experiment, all AID:substrate reaction combinations were carried out in duplicate and loaded on multiple gels. Individual lanes of each gel were quantitated three times, thus obtaining an average value for the lane, representing a single-data point (typically the triplicate measurements of each lane as well as the duplicate reaction lanes within each experiment contained a variability of <5%). The data points are the mean of the values obtained in this manner from all experiments (four to six experiments for each AID:substrate combination, yielding 8–12 values per data point). The data were graphed using GraphPad Prism Software. Error bars represent standard deviations.
Structural models and substrate docking
Structure models of AID were produced by submission to the Swiss protein data bank (http://swissmodel.expasy.org/repository/) using either the crystal or NMR-determined structure of the catalytic domain of the family member APOBEC3G (PDB IDs: 3EIU, 2JYW) (50–52). The template was either chosen as best fit by the database or manually chosen, and the lowest energy state predictions were generated. Generation and analysis of predicted models were performed using Pymol v1.30. Stick and sphere structures of cytosine and 5-methylcytosine were constructed in ChemDraw v12.0 (http://www.cambridgesoft.com/software/ChemDraw/) and exported into Pymol to scale. Manual orientation of docked cytosine and 5-methylcytosine bases was performed based on the requirement for proximity of its atoms to specific residues in the putative catalytic pocket of AID for deamination to proceed. Additionally, docking of 2′-deoxycytidine and 5-methyl-2′-deoxycytidine nucleosides was simulated using Autodock Vina (53), while allowing for flexibility of nucleoside docking into the putative catalytic pocket of AID to examine possible orientations in which the deamination reaction is possible.
RESULTS
Hs-AID is minimally active on mC on an optimal ssDNA structure
We previously reported that mC in CpG motifs was not efficiently deaminated by purified His- or GST-tagged AID in long (∼500 bp) stretches of ssDNA (45). A second study using purified GST-tagged AID found that mC located in the WRC motif AGC on a 27-nt-long ssDNA substrate was deaminated ∼10-fold less efficiently than dC (6). In contrast, another study found a less crass preference for dC over mC estimated at ∼3-fold when considering the WRC motif AGC (20). As AID mutates a WRC motif 6- to 8-fold more efficiently than a non-WRC motif (3–6), the results of the latter study indicate that given varied sequence contexts, mC can be deaminated by AID more efficiently than dC. Two recent studies have re-examined the efficiency of mC deamination by AID (46,47). An antibiotic gene reversion assay in bacteria revealed that AID deaminated mC at least an order of magnitude less efficiently than dC and barely above background mutations levels (46), whereas another study that measured the activity of purified MBP-tagged AID on an ssDNA substrate containing the WRC motif TGC, found AID to be 10- to 16-fold less active on mC as compared with dC (47). We reasoned that some of the differences among studies in the degree of preference for dC over mC may lie in the structure and sequence context of the substrates, which we and others have previously shown to be an important determinant of deamination efficiency by AID (8,49). To this end, we measured the activity of purified GST-AID on a substrate in which the dC or mC target of deamination is located in the middle of a 7-nt-long bubble region (Figure 1A), which we have previously shown to be an optimal structure for deamination by AID, supporting several fold higher levels of activity than fully single-stranded DNA (4,43). The activity of AID on dC is detected through alkaline cleavage of an abasic site generated by the removal of the ensuing uracil (Figure 1A, left). To generate abasic sites after AID activity on mC, we used the enzyme TDG, the primary activity of which is to remove thymine bases at G:T mismatches (54) (Figure 1A, middle). To ensure that this experimental approach accurately detects the deamination of mC by AID, we used a control substrate that contains a TGT motif as would result from the action of AID on a TG(mC) substrate (Figure 1A, right). As shown in Figure 1B, the control substrate was fully cleaved in a TDG-dependent manner. Thus, this method effectively detects the product of mC deamination by AID. To test whether human AID (Hs-AID) can deaminate mC in varying sequence contexts, we measured its activity on two WRC [TGCbub7 versus TG(mC)bub7 and AGCbub7 versus AG(mC)bub7] and one non-WRC [GGCbub7 versus GG(mC)bub7] substrates (Figure 1C). On average, mC was deaminated 22 -, 20- and 16-fold less efficiently than dC in TGC, AGC and GGC motifs, respectively. We conclude that in an optimal ssDNA structure for AID, its activity on mC is significantly diminished as compared with dC.
Zebrafish AID efficiently deaminates methylated cytidines
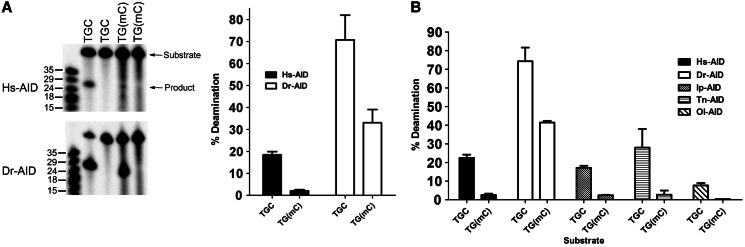
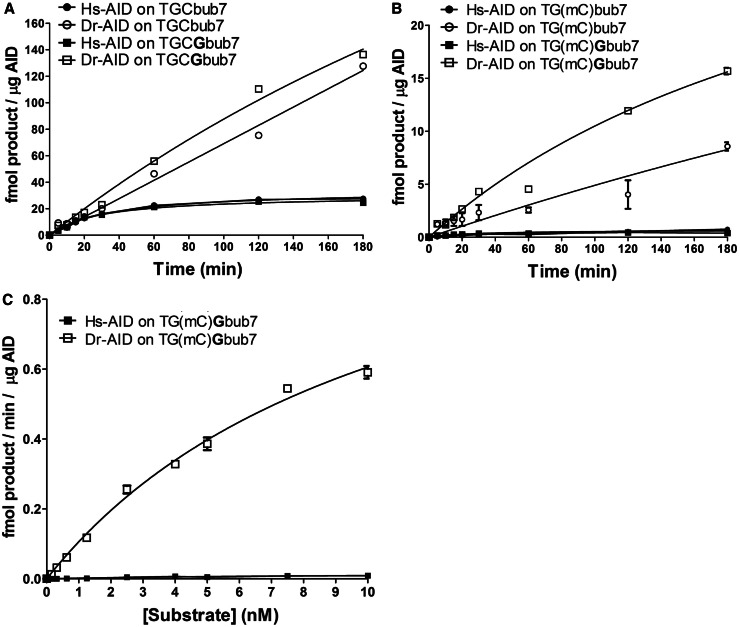
Given that the proposed role of AID in genome demethylation is largely based on data from the zebrafish, we sought to determine whether zebrafish AID (Dr-AID) can deaminate mC. We have previously shown that Dr-AID has a higher catalytic rate than Hs-AID (48). To allow for the generation of sufficient mC deamination by Hs-AID, both Hs- and Dr-AID were incubated overnight with TGC and TG(mC), as TGC is the most preferred WRC motif for both (Figure 2A). On average, Dr-AID generated 3.6-fold more deamination product on TGCbub7 as compared with Hs-AID (68 versus 18%). Like Hs-AID, Dr-AID deaminated mC less efficiently than dC; however, in contrast to 10-fold less deamination by Hs-AID (1.8 versus 18%), Dr-AID generated 2-fold less product on TG(mC)bub7 (35 versus 68%). To assess whether the relative lack of discrimination between dC and mC is a property shared by other bony fish AID, we examined AID from the channel catfish (Ip-AID), Japanese killifish medaka (Ol-AID) and the puffer fish tetradon (Tn-AID). We found that like Hs-AID, these deaminated mC 8- to 19-fold less efficiently than dC (Ip-AID: 17 versus 2.1%, Ol-AID: 7.7 versus 0.4%, Tn-AID: 28 versus 2.7%) (Figure 2B). To compare the initial catalytic rates of Hs- and Dr-AID on mC, we performed enzyme kinetics. We included a second substrate TGCGbub7 that is similar to TGCbub7 but contains a dG residue after the target dC, constituting a CpG motif as may be subject to methylation in vivo. As we previously reported (48), Dr-AID was more active on both TGCbub7 and TGCGbub7 than Hs-AID (Figure 3A). The difference in product formation was apparent after a few minutes of incubation, growing to 4.5-fold at 180 min. As shown in Figure 3B, the activity of Dr-AID on the TG(mC)bub7 was 5-fold higher than that of Hs-AID after a 5-min incubation, with the difference growing to 10-fold at 60 min and 20-fold at 180 min. For TG(mC)Gbub7, the activity of Dr-AID was 7-fold higher than Hs-AID after 5 min, with the difference growing to 12-fold at 60 min and 40-fold at 180 min. Interestingly, the activity of Dr-AID on mC was higher by ∼2-fold when it was in the context of a typical CpG motif in TG(mG)G as compared with TG(mC)T. This finding further suggests that Dr-AID has the enzymatic potential to contribute to genome demethylation in the zebrafish. We then measured the Michaelis–Menten kinetics of mC deamination by Hs- and Dr-AID (Figure 3C). On TG(mC)Gbub7, the Kcat:Km ratio for Dr-AID was estimated to be ∼60-fold higher than that of Hs-AID. We conclude first that the activity of Dr-AID on mC is unique among all bony fish orthologs tested here, second that Dr-AID can deaminate mC more efficiently than AID from other species deaminate dC and third that the initial rates of mC deamination by Dr-AID indicate a unique specificity for mC.
Figure 2.
Dr-AID exhibits high activity on mC. (A) Left shows alkaline cleavage assay gels showing the deamination of TGCbub7 or TG(mC)bub7 by of Hs-AID (top) and Dr-AID (bottom). Right shows the graphed percentage of deaminated product. Hs- and Dr-AID were incubated overnight with 50 fmol substrate at 37°C and 25°C, respectively. (B) Deamination of TGCbub7 and TG(mC)bub7 by AID of three other bony fish (Medaka: Ol-AID, tetradon: Tn-AID and Channel catfish: Ip-AID) compared with the activities of Hs- and Dr-AID. Each AID was incubated with 50 mol substrate overnight at optimal temperature (37°C and 25°C, respectively, for Hs- and fish AIDs).
Figure 3.
Deamination kinetics comparing the activities of Hs-AID and Dr-AID. (A) Comparison of activity kinetics between Hs-AID and Dr-AID on seven-nucleotide bubble substrates bearing dC in a WRC or a WRCG motif. In all, 50 fmol substrate was incubated with AID for various times. Product formation for 1 µg AID is shown as a function of incubation time. (B) Comparison of activity kinetics between Hs-AID and Dr-AID on seven-nucleotide bubble substrates bearing mC in a WRC or a WRCG motif. (C) Deamination kinetics to compare the deamination rates of dC and mC in a WRCG motif by Hs-AID and Dr-AID. Various concentrations of the substrate TG(mC)Gbub7 ranging from 0.1 to 10 nM were incubated with AID. Velocity was calculated as the amount of deaminated product generated by a given amount of AID in a unit of time and plotted against substrate concentration.
Sequence context differentially influences the activity of AID orthologs on mC
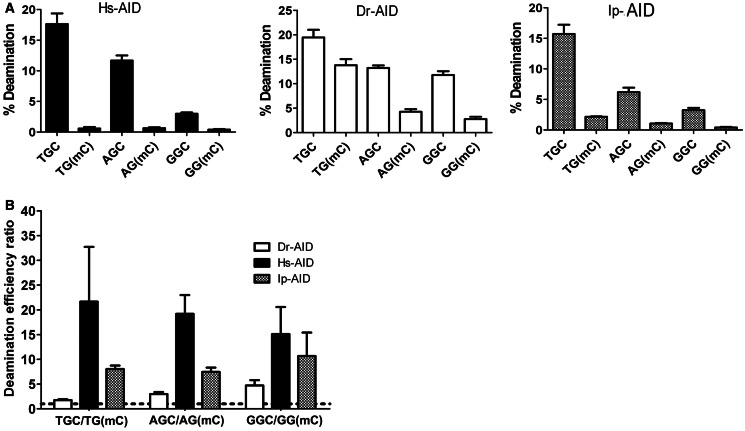
We have previously shown that although purified Dr-AID and Ip-AID possess general WRC specificity similar to Hs-AID, they exhibit subtle alterations in their sequence preference (5,48). We sought to examine whether the relatively high level of Dr-AID activity on mC is a unique feature of the WRC motif TGC. To this end, we measured the activities of Dr-AID, Hs-AID and Ip-AID on WRC non-WRC motifs (Figure 4A). We found that Dr-AID deaminated mC 1.7-, 3- and 4.7-fold less efficiently than dC in TGC, AGC and GGC motifs, respectively, compared with 8-, 7.5- and 11-fold for Ip-AID and 22-, 20- and 16-fold for Hs-AID (Figures 1C and 4B). We conclude first that Dr-AID is more active on mC than Hs-AID or Ip-AID regardless of sequence context, and second that in contrast to Hs-AID and Ip-AID, which disfavor mC compared with dC almost equally regardless of sequence context, Dr-AID discriminates against mC to a lesser degree (1.7- and 3-fold versus 4.7-fold) in a WRC motif.
Figure 4.
The influence of WRC sequence specificity of AID on mC activity. (A) Left, middle and right show the activities of Hs-, Dr- and Ip-AID, respectively, on C and mC, located in two WRC motifs (TGC, AGC) or a non-WRC motif (GGC). AID was incubated with 40 fmol substrate for 1 h. Percentage of deamination product was quantitated and graphed. (B) The ratio of percentage of deamination product for C/mC was calculated for each AID and graphed to show the fold preference for C over mC, in each of the WRC and non-WRC sequence motifs. Dotted line represents a ratio of 1.
The role of putative catalytic pockets of Hs- and Dr-AID in differential discrimination against mC
One possible mechanism for the low activity of Hs-AID on mC may be a size restriction against the methyl moiety imposed by its catalytic site. In support of this, a recent study reported an inverse correlation between the ‘bulk’ of cytidine side chain modifications and deamination efficiency by AID (47). Alternatively, but not mutually exclusively, it is possible that a restriction is imposed by chemical interactions between the methyl side chain and catalytic residues of Hs-AID that impede deamination. To examine these possibilities, we sought to dock either the C or mC base or respective nucleoside versions into the putative catalytic pockets of Hs- and Dr-AID. The structure of AID has not yet been determined. Thus, we used models of the AID catalytic pocket that we previously generated based on the resolved structure of the catalytic region of the family member APOBEC3G (43,48) (Figure 5A).
Figure 5.
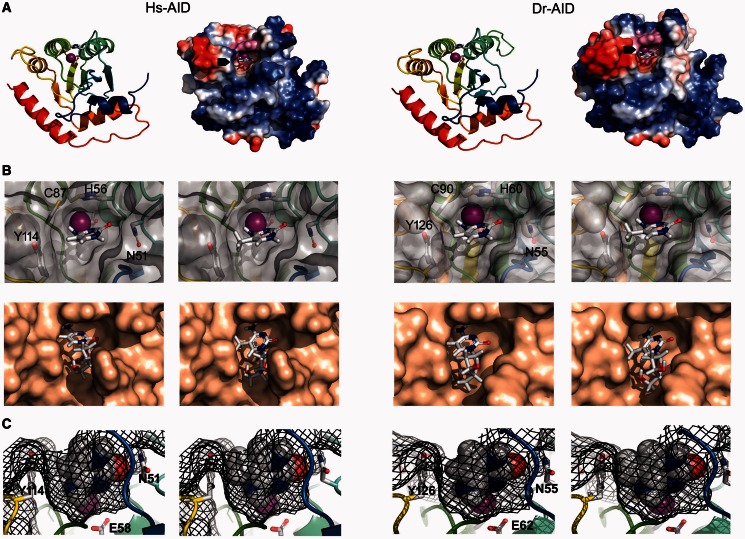
Models of the putative catalytic pockets of Hs- and Dr-AID with docked C or mC. (A) Models of the AID monomer were generated based on the solved structure of the APOBEC3G catalytic region. Left shows a ribbon and surface charge diagram of Hs-AID and right shows the same for Dr-AID. N- to C-terminus progression is shown in color from blue to red. The ribbon diagram shows the putative catalytic (Zn-coordinating) residues are shown in stick (Hs-AID: H56, E58, C87, C90; Dr-AID: H60, E62, C99, C102). The surface topology and charge models show positively and negatively charged residues are blue and red, respectively. The putative catalytic pockets containing the Zn-coordinating residues are shown in magenta, with a docked cytidine base inside the pocket as marked by black arrows. (B) Modeled top down view of the surface topology of the putative catalytic pocket of Hs-AID (left) or Dr-AID (right), with either C or mC bases (top row) using manual docking, or by using Autodock software to dock dC or mC nucleoside versions (bottom row). Zn is shown as a magenta sphere. Residues that surround the catalytic pocket and affect the fit of C or mC are shown. (C) Mesh diagram of the modeled side view of the catalytic pockets of Hs-AID (left), or Dr-AID (right), showing the fit of C (left side) or mC (right side). The mesh diagram illustrates the spaces occupied by the atomic radii of both substrate and AID. Zn is shown as a magenta sphere. Residues that surround the catalytic pocket and affect the fit of C or mC are shown.
To manually dock C or mC into the putative catalytic pockets of Hs- Dr-AID, we took into account the mechanism of deamination by AID. This mechanism can be speculated on with a high degree of confidence, based on that of other cytidine deaminases, the resolved structure of family member APOBEC3G and the model structure of AID itself (2,50,51,55–57). A Zn ion required for nucleophilic attack during deamination is clinched by the triad of Zn-coordinating residues (in Hs-AID: H56, C87, C90, and Dr-AID: H60, C99, C102). The carboxylate ion of a glutamic acid (E58 and E62 in Hs- and Dr-AID, respectively) coordinates a water molecule, and through proton shuttling, it converts it into a reactive hydroxide ion. Hydrolytic deamination of C occurs through a nucleophilic attack by ZnOH on the C4 position of the pyrimidine ring to which the exocyclic amino group is bonded. After proton donation, the deprotonated side chain of E58 stabilizes the N-H group of the intermediate through a hydrogen bond. Based on this reaction mechanism, we used three criteria for situating C and mC in the putative catalytic pocket: first, C4 is juxtaposed to the Zn ion. Second, the cyclic N3 and the hydrogen donor of the E58 side chain are proximal allowing for proton donation in the first step of the deamination reaction. Third, the exocyclic carbonyl group must be proximal to N51 in Hs-AID and N55 in Dr-AID. This hydrogen bond seems to be vital for deamination catalysis: this asparagine is proximal to the catalytic site and 100% conserved among orthologs. Furthermore, mutation of its equivalent residue (N244A) in APOBEC3G abolished deamination, and the equivalent residue in Staphylococcus aureus tRNA adenosine deaminase (TadA) has been shown to stabilize the substrate by hydrogen bonding (50,58). Using these criteria, we positioned C and mC into the putative catalytic pockets of Hs- and Dr-AID (Figure 5B, top). To validate our manual dockings, we also used auto-docking software, allowing for fit of the larger nucleoside versions dC or mC. We observed a range of rotations of the substrate molecule in which deamination was plausible, but the lowest energy state confirmed our positioning of the substrate within the putative catalytic pockets (Figure 5B, bottom). To assess fit, we modeled spaces occupied by atomic radii (Figure 5C). The dimensions of the catalytic pockets of Hs- and Dr-AID appear comparable (8.3 × 7.2 × 6.8 versus 8.3 × 6.7 × 6.8 Å). The dimensions of C and mC are 7.8 × 8.3 × 4.2 and 8.8 × 8.3 × 4.2 Å, respectively, suggesting that C readily fits into the catalytic pocket, whereas the fit of mC is taut (Figure 5C). Other than its size thus restricting fit, we did not observe a chemical interaction between the methyl moiety and residues constituting the ‘floor’ or ‘walls’ of the putative catalytic pockets that may hinder deamination catalysis. With the caveat that actual structures are not known, we conclude that the fit of C and mC into the modeled catalytic pockets of Hs- and Dr-AID explains the preference for dC over mC but not the lower degree of discrimination between C and mC specific to Dr-AID.
The role of catalytic efficiency in differential activity of Dr-AID on mC
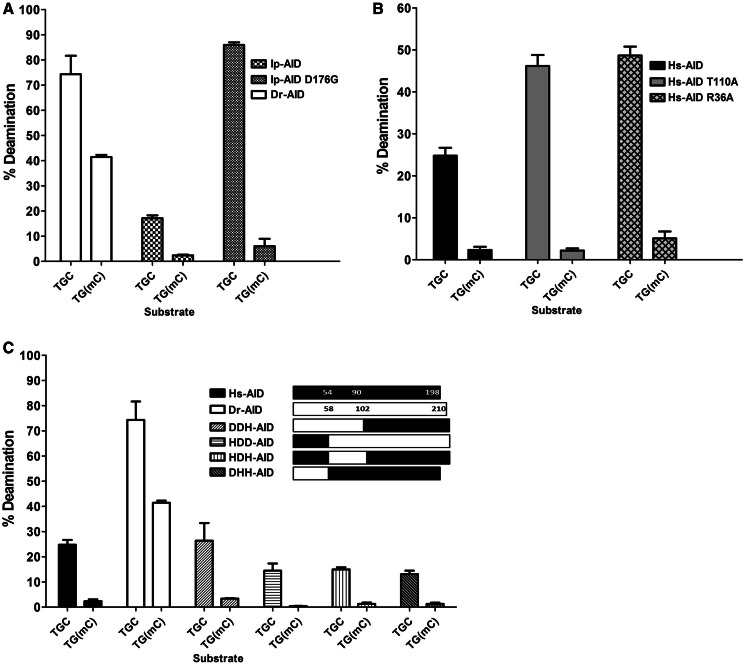
We previously showed that Dr-AID has a higher catalytic efficiency on dC than Hs-AID (48). We sought to examine whether efficient deamination of mC by Dr-AID is a general function of its more robust activity. We reasoned that discrimination between substrates with subtly different chemical features may be less apparent in a more highly active AID enzyme, and that the size restriction of mC may be overcome by higher overall higher turnover of substrate providing more likelihood of fit into the catalytic pocket for a given unit of time. In support of this notion, Dr-AID is also less discriminating between non-WRC and WRC motifs, exhibiting ∼2-fold less activity on the non-WRC motif GGC as compared with the WRC motif TGC, in contrast to a ∼6 fold for Hs-AID (Figure 4) (48). We previously reported that a single mutation (D176G) in the C-terminal domain of Ip-AID results in an enzyme nearly identical to Dr-AID with respect both to catalytic rate and ssDNA-binding affinity (Kcat and Kd) (48). We found that despite its identical activity profile to Dr-AID on dC, Ip-AID D176G does not have appreciable activity on mC (Figure 6A). In addition, we generated two point mutants of Hs-AID (R36A and T110A) with several fold increased Kcat (data not shown) and compared their activities on dC and mC. Despite increased activity on dC, neither produced a disproportionate increase of deamination at mC (Figure 6B). Thus, mutations in AID that we have previously shown to increase its catalytic activity on dC did not result in a gain of activity on mC. We conclude that the activity of Dr-AID on mC is a biochemical property that is independent of its high-catalytic efficiency on dC.
Figure 6.
The activity of AID mutants and domain-swapped AIDs on mC. (A) Comparison of the deamination activities of Dr-AID, Ip-AID and Ip-AID (D176G) on TGCbub7 and TGC(mC)bub7. (B) Comparison of the deamination activities of Hs-AID, Hs-AID (R36A) and Hs-AID (T110A) on TGCbub7 and TG(mC)bub7. (C) Comparison of the deamination activities of Hs-AID, Dr-AID and hybrid enzymes with domains swapped between Hs- and Dr-AID on TGCbub7 and TGC(mC)bub7. AID was incubated with 50 fmol substrate for 3 h, and percentage of deamination product was graphed.
Based on the position of the catalytic residues and homology to other deaminases, AID can be divided into N-, catalytic and C-terminal domains (Supplementary Figure S1; defined by arrows). To determine whether an individual domain of Dr-AID can transfer its high activity on mC, we constructed four domain-swapped enzymes between Hs-AID and Dr-AID. We found that none of these hybrid enzymes was more active on mC than Hs-AID (Figure 6C). From these results and given the collection of regions swapped in the hybrid enzymes, we conclude that this property of Dr-AID cannot be attributed to a portion of AID in the manner in which we apportioned the primary structure and is rather an attribute that likely involves multiple residues that are discontinuous in the primary structure.
DISCUSSION
Methylation of promoter region cytidines at C5 is a marker of silenced genes, in normal differentiation, as well as in oncogenesis (24–34,59–64). Several recent lines of evidence implicate AID in genome demethylation (18,19): first, overexpression, knockdown or absolute deficiencies in zebrafish and mice correlate with changes in levels of methylation in the genome and/or in introduced plasmids (22,23). Second, AID has been reported to be expressed at low levels in germ cells where promoter demethylation is a major contributor to pluripotency (20). Third, in a zebrafish embryogenesis model, AID knockdowns exhibited gross abnormalities in neuronal development, concomitant with hypermethylation at loci encoding for transcription factors involved in neurogenesis. Evidence for the involvement of a G:T mismatch intermediate expected from mC deamination, as well as ablation of the hypomethylation activity of catalytically dead AID mutants, suggests that genome demethylation in zebrafish can proceed through AID-mediated deamination of mC (23).
In contrast to zebrafish, humans and mice deficient in AID bear no developmental abnormalities, suggesting that the role of AID in genome demethylation is either negligible or redundant. This is not surprising, first given that genome demethylation can proceed through a multitude of enzymatic and chemical processes, and second that other deaminases, such as APOBEC-1, APOBEC-2 and APOBEC-3 A, have been suggested to play a role (20,40,46,65). Although the role of AID in genome demethylation in humans seems redundant, it brought forth the question of whether AID is capable of deaminating cytidines carrying a methyl group at the C5 position. To date, four reports have shown that Hs-AID deaminates mC with ∼10- to 100-fold lower efficiency as dC (6,45–47). In contrast, one report showed that Hs-AID can deaminate mC at a comparable efficiency as dC, given a comparison of dC and mC located in different sequence contexts (20). It is formally possible that differences in the observed degree of AID’s preference for dC over mC are due to differing experimental parameters, such as sources of AID expression, purification methods and ssDNA structures, or the local sequences in which dC or mC were located. However, the reports that show AID deaminates mC at a very low efficiency have used a host of purification and assay methods, including prokaryotic and eukaryotic expression, and evaluating the activity of AID on mC in bacterial genomes, in vitro using purified His-, GST- or MBP-tagged AID on fully or partially single-stranded substrates and in various sequence contexts (6,45–47). Here, we report that even in an optimal ssDNA structure and regardless of whether the mC is positioned in a WRC or non-WRC sequence motif, it is deaminated poorly by Hs-AID. Taking together that mC is a poor substrate for Hs-AID, that AID expression is largely restricted to activated B cells where it targets the Ig locus >90% of time (43,66,67), and that AID expression levels in germ cells are low, it appears unlikely that AID would be a significant contributor to genome-wide demethylation in humans. It is more likely that AID may play a minor and redundant role, the exact nature of which is yet to be determined.
We have previously shown that AID of bony fish have different biochemical properties compared with human AID (48). Thus, we hypothesized that the more prominent role of AID in genome demethylation in zebrafish may be indicative of its enzymatic capacity to act on mC. We show that this is indeed the case, and that this biochemical property seems to be unique among AID of bony fish tested here. The activity of Dr-AID on mC can be explained neither by an overall more accommodating modeled catalytic site nor by its more robust enzymatic rate as compared with Hs-AID. In the absence of an experimentally determined structure, we used modeling to gain insight into the mechanism of high-Dr-AID activity on mC. In modeling the structure of Hs-AID, we noted that Y114 was the closest residue to the methyl group of mC docked in the catalytic pocket (Supplementary Figure S2A). We note that Y114 is conserved in AID of all species (Supplementary Figure S1, unpublished observations M. Larijani). Two recent studies have reported that in contrast to APOBEC3G, APOBEC3A is able to efficiently deaminate mC (46,65). Like the case in our study, this difference in mC activity could also not be attributed to an overt structural difference between the two enzymes. Based on modeling the APOBEC3A structure, it was postulated that its equivalent residue to the Y114 of Hs-AID (Y130) was positioned with its side chain further away from the catalytic pocket as compared with Hs-AID (46). We noted in our models that this residue is located at the start of a loop that encircles the putative catalytic pocket (Supplementary Figure S2A and B). Engraftment of this loop from APOBEC3A to Hs-AID transferred the ability to efficiently deaminate mC, suggesting that the composition of the loop supports a positioning of this tyrosine residue that is more conducive to docking of mC (46). However, unlike the case of APOBEC3A, we noted that this tyrosine in Dr-AID (Y126) was positioned similarly to Y114 in Hs-AID and seemed equally likely to cause steric hindrance with the fit of mC (Supplementary Figure S2B). A comparison of AID models based on the NMR- versus X-ray–determined structure of the APOBEC3G catalytic domain revealed that the majority of residues held similar conformations; however, this tyrosine was rotated by ∼180° and was shifted away from the catalytic pocket in Hs-AID and Dr-AID, when modeled on the NMR template (Supplementary Figure S2C). We conclude that this tyrosine and the loop harboring it are likely to undergo a high degree of protein breathing, explaining their different positioning in solution and crystal forms. In addition, we noted that this loop is longer in Dr-AID than Hs-AID by one negatively charged residue (E130) that can allow for further flexibility (Supplementary Figure S1). These observations suggest that because of differences in the charge make up and length of the loop harboring Y126 in Dr-AID, this tyrosine may occupy a lower energy state in the ‘away’ conformation, as compared with Hs-AID (Supplementary Figure S2B and C), thus allowing for better fit of mC more often. For conclusive insights into the molecular basis of the difference in mC activity between Dr-AID and Hs-AID, we look forward to the resolution of their structures.
Whatever the structural basis of higher activity of Dr-AID on mC may be, our results provide a mechanistic explanation for the prominent role for AID in genome demethylation in zebrafish. It is possible that in humans, APOBEC3A functions as a major contributor to genome demethylation, but in zebrafish that lacks APOBEC3A, this role is delegated to AID. On the other hand, mice and other bony fish examined here have neither APOBEC3A nor an AID capable of deaminating mC. This suggests strongly that even though some APOBEC family deaminases are enzymatically capable of genome demethylation, this function is mainly carried out by other pathways, and that zebrafish may be an exception. As AID is present in the earliest jawed vertebrates (e.g. sharks), it is thought to be the ancestral member of the APOBEC family (68,69). On the most recent end of the APOBEC family evolutionary spectrum is the APOBEC3 conglomerate, having expanded from one to seven members in primates (2). In this light, our finding that AID from zebrafish but not three other bony fish species efficiently deaminates mC, combined with the finding that APOBEC3A but not APOBEC3G is able to efficiently deaminate mC (46,65) presents a difficulty in assigning a linear evolutionary association to activity on mC. It seems more likely that the ability to deaminate mC is a somewhat secondary byproduct of overall differences in the internal folding of the enzyme structure among APOBEC family members. It will be of interest to determine whether this enzymatic feature of AID corresponds with a differential role in genome demethylation among species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Canadian Institutes of Health Research CIHR [MOP111132 to M.L.]; Beatrice Hunter Cancer Research Institute (to J.J.K.); Libyan-North American scholarship program (to M.S.); CIHR New Investigator award (to M.L.). Funding for open access charge: CIHR, Canada.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 2.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 4.Larijani M, Petrov AP, Kolenchenko O, Berru M, Krylov SN, Martin A. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol. Cell. Biol. 2007;27:20–30. doi: 10.1128/MCB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larijani M, Frieder D, Basit W, Martin A. The mutation spectrum of purified AID is similar to the mutability index in Ramos cells and in ung(−/−)msh2(−/−) mice. Immunogenetics. 2005;56:840–845. doi: 10.1007/s00251-004-0748-0. [DOI] [PubMed] [Google Scholar]
- 6.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 10.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 12.Durandy A, Peron S, Fischer A. Hyper-IgM syndromes. Curr. Opin. Rheumatol. 2006;18:369–376. doi: 10.1097/01.bor.0000231905.12172.b5. [DOI] [PubMed] [Google Scholar]
- 13.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 14.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, III, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 17.Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim YM, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 2010;24:2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teperek-Tkacz M, Pasque V, Gentsch G, Ferguson-Smith AC. Epigenetic reprogramming: is deamination key to active DNA demethylation? Reproduction. 142:621–632. doi: 10.1530/REP-11-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 21.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev. Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song F, Mahmood S, Ghosh S, Liang P, Smiraglia DJ, Nagase H, Held WA. Tissue specific differentially methylated regions (TDMR): Changes in DNA methylation during development. Genomics. 2009;93:130–139. doi: 10.1016/j.ygeno.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Yang MC, Yang TP. Evidence that silencing of the HPRT promoter by DNA methylation is mediated by critical CpG sites. J. Biol. Chem. 2001;276:320–328. doi: 10.1074/jbc.M007096200. [DOI] [PubMed] [Google Scholar]
- 28.Kroft TL, Jethanandani P, McLean DJ, Goldberg E. Methylation of CpG dinucleotides alters binding and silences testis-specific transcription directed by the mouse lactate dehydrogenase C promoter. Biol. Reprod. 2001;65:1522–1527. doi: 10.1095/biolreprod65.5.1522. [DOI] [PubMed] [Google Scholar]
- 29.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Pomerantz JH, Sen G, Palermo AT, Blau HM. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc. Natl Acad. Sci. USA. 2007;104:4395–4400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doerfler W. In pursuit of the first recognized epigenetic signal–DNA methylation: a 1976 to 2008 synopsis. Epigenetics. 2008;3:125–133. doi: 10.4161/epi.3.3.6249. [DOI] [PubMed] [Google Scholar]
- 34.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc. Natl Acad. Sci. USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, Scholer H, Walter J. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larijani M, Martin A. The biochemistry of activation-induced deaminase and its physiological functions. Semin. Immunol. 2012;24:255–263. doi: 10.1016/j.smim.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Durandy A, Revy P, Fischer A. Hyper-immunoglobulin-M syndromes caused by an intrinsic B cell defect. Curr. Opin. Allergy Clin. Immunol. 2003;3:421–425. doi: 10.1097/00130832-200312000-00002. [DOI] [PubMed] [Google Scholar]
- 45.Larijani M, Frieder D, Sonbuchner TM, Bransteitter R, Goodman MF, Bouhassira EE, Scharff MD, Martin A. Methylation protects cytidines from AID-mediated deamination. Mol. Immunol. 2005;42:599–604. doi: 10.1016/j.molimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 2012;40:9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dancyger AM, King JJ, Quinlan MJ, Fifield H, Tucker S, Saunders HL, Berru M, Magor BG, Martin A, Larijani M. Differences in the enzymatic efficiency of human and bony fish AID are mediated by a single residue in the C terminus modulating single-stranded DNA binding. FASEB J. 2012;26:1517–1525. doi: 10.1096/fj.11-198135. [DOI] [PubMed] [Google Scholar]
- 49.Larijani M, Martin A. Single-stranded DNA structure and positional context of the target cytidine determine the enzymatic efficiency of AID. Mol. Cell. Biol. 2007;27:8038–8048. doi: 10.1128/MCB.01046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, Kobayashi N, Yokoyama S, Takaku H, Katahira M. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair. 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 56.Mejlhede N, Neuhard J. The role of zinc in Bacillus subtilis cytidine deaminase. Biochemistry. 2000;39:7984–7989. doi: 10.1021/bi000542t. [DOI] [PubMed] [Google Scholar]
- 57.Ko TP, Lin JJ, Hu CY, Hsu YH, Wang AH, Liaw SH. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- 58.Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- 59.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiota K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet Genome Res. 2004;105:325–334. doi: 10.1159/000078205. [DOI] [PubMed] [Google Scholar]
- 62.Sato S, Yagi S, Arai Y, Hirabayashi K, Hattori N, Iwatani M, Okita K, Ohgane J, Tanaka S, Wakayama T, et al. Genome-wide DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) residing in mouse pluripotent stem cells. Genes Cells. 2010;15:607–618. doi: 10.1111/j.1365-2443.2010.01404.x. [DOI] [PubMed] [Google Scholar]
- 63.van Vlodrop IJ, Niessen HE, Derks S, Baldewijns MM, van Criekinge W, Herman JG, van Engeland M. Analysis of promoter CpG island hypermethylation in cancer: location, location, location! Clin. Cancer Res. 2011;17:4225–4231. doi: 10.1158/1078-0432.CCR-10-3394. [DOI] [PubMed] [Google Scholar]
- 64.Das PM, Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 65.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parsa JY, Basit W, Wang CL, Gommerman JL, Carlyle JR, Martin A. AID mutates a non-immunoglobulin transgene independent of chromosomal position. Mol. Immunol. 2007;44:567–575. doi: 10.1016/j.molimm.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 67.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat. Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 69.Barreto VM, Magor BG. Activation-induced cytidine deaminase structure and functions: a species comparative view. Dev. Comp. Immunol. 2011;35:991–1007. doi: 10.1016/j.dci.2011.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.