Abstract
Sequencing of RNAs (RNA-Seq) has revolutionized the field of transcriptomics, but the reads obtained often contain errors. Read error correction can have a large impact on our ability to accurately assemble transcripts. This is especially true for de novo transcriptome analysis, where a reference genome is not available. Current read error correction methods, developed for DNA sequence data, cannot handle the overlapping effects of non-uniform abundance, polymorphisms and alternative splicing. Here we present SEquencing Error CorrEction in Rna-seq data (SEECER), a hidden Markov Model (HMM)–based method, which is the first to successfully address these problems. SEECER efficiently learns hundreds of thousands of HMMs and uses these to correct sequencing errors. Using human RNA-Seq data, we show that SEECER greatly improves on previous methods in terms of quality of read alignment to the genome and assembly accuracy. To illustrate the usefulness of SEECER for de novo transcriptome studies, we generated new RNA-Seq data to study the development of the sea cucumber Parastichopus parvimensis. Our corrected assembled transcripts shed new light on two important stages in sea cucumber development. Comparison of the assembled transcripts to known transcripts in other species has also revealed novel transcripts that are unique to sea cucumber, some of which we have experimentally validated.
INTRODUCTION
Transcriptome analysis has been revolutionized by next-generation sequencing technologies (1). The sequencing of polyadenylated RNAs (RNA-Seq) is rapidly becoming standard practice in the research community owing to its ability to accurately measure RNA levels (2,3), detect alternative splicing (4) and RNA editing (5), determine allele (6) and isoform-specific expression (7,8) and perform de novo transcriptome assembly (9–11).
Although RNA-Seq experiments are often more accurate than their microarray predecessors (2,7), they still exhibit a high error rate. These errors can have a large impact on the downstream bioinformatics analysis and lead to wrong conclusions regarding the set of transcribed mRNAs. One class of errors concerns biases in the abundance of read sequences due to RNA priming preferences (12,13), fragment size selection (14,15) and GC-content (16). Sequencing errors, which are a result of mistakes in base calling of the sequencer (mismatch), or the insertion or deletion of a base (indel), are another important source of errors for which no general solution for RNA-Seq is currently available. For example, error rates of up to 3.8% were observed when using Illumina’s GenomeAnalyzer (17).
A common approach to sequencing error removal is read trimming of bad-quality bases from the read end to improve downstream analysis (4,18). Such an approach reduces the absolute amount of errors in the data but can also lead to significant loss of data, which affects our ability to identify lowly expressed transcripts.
A number of approaches were primarily proposed for the correction of DNA sequencing data (19). These methods use suffix trees (20,21), k-mer indices (22,23) and multiple alignments (24). While successful, as we show in ‘Results’ section, these approaches are not always suited for RNA-Seq data. Unlike genome sequencing, which often results in uniform coverage, transcripts exhibit non-uniform expression levels. The only error correction method that we are aware of that explicitly targets non-uniform coverage data is Hammer (25). Unfortunately, Hammer cannot be used to correct reads, as it only outputs corrected k-mers of much shorter length. Even after contacting the authors of Hammer and using their implementation, we could not use it with standard methods for read alignment or assembly, and we are not aware of other articles that had. Finally, all the above methods often fail at the border of alternatively spliced exons, which may lead to false-positive corrections.
Other sequencing error correction methods have been designed for tag-based sequencing or microRNA sequencing where the read spans the complete tag or transcript region under investigation (26–28). These methods, including SEED (28), are based on clustering similar read sequences, but do not consider partially overlapping read sequences, alternative splicing and the correction of indel errors.
Here we present the first general method for SEquencing Error CorrEction in Rna-seq data (SEECER) that specifically addresses the shortcomings of previous approaches. SEECER is based on a probabilistic framework using hidden Markov models (HMMs). SEECER can handle different coverage levels of transcripts, joins partially overlapping reads into contigs to improve error correction, avoids the association of reads at exon borders of alternative splicing events and supports the correction of mismatch and indel errors. Because SEECER does not rely on a reference genome, it is applicable to de novo RNA-Seq. We tested SEECER using diverse human RNA-Seq datasets and show that the error correction greatly improves performance of the downstream assembly and that it significantly outperforms previous approaches. We next used SEECER to correct RNA-Seq data for the de novo transcriptome assembly of the sea cucumber. The ability to accurately analyze de novo RNA-Seq data allowed us to identify both conserved and novel transcripts and provided important insights into sea cucumber development.
MATERIALS AND METHODS
Overview of SEECER
Error correction of a read is done by trying to determine its context (overlapping reads from the same transcript) and using these to identify and correct errors. SEECER builds a set of contigs from reads, where each contig is theoretically a subsequence of a transcript. Ideally, we would like each contig to be exactly one transcript. However, in several cases, transcripts may share common subsequences owing to sequence repeats or alternative splicings. In such cases, each contig in our model represents an unbranched subsequence of some transcript.
We use a profile HMM to represent contigs. Such models are appropriate for handling the various types of read errors we anticipate (including substitutions and insertion/deletion). Owing to several restrictions imposed by the read data, even though we may need to handle a large number of contigs, learning these HMMs can be done efficiently (linearly in the size of the reads assigned to the contig).
Contig HMM
Profile HMM is a HMM that was originally developed to model protein families to allow multiple sequence alignment with gaps in the protein sequences (see Supplement). Here, we extend profile HMMs to model the sequencing of reads from a contig. We thus call this a contig HMM. Each contig HMM includes a consensus sequence based on the set of reads assigned to this contig. The consensus is constructed from the most probable output nucleotides of the match states. Using this consensus sequence we can make correction to the reads assigned to this contig HMM.
The core functionality of SEECER is constructing the contig HMM from sequencing reads. We now outline the details of each step in the following sections.
Pool of reads
We maintain a global pool  (Figure 1, step 0) of reads during the execution of our method. SEECER creates many threads, each independently builds a separate contig HMM. For each such HMM, we start with a random read as the seed and iteratively extend it using overlapping reads. See supplement for discussion on how to avoid collision between two HMMs.
(Figure 1, step 0) of reads during the execution of our method. SEECER creates many threads, each independently builds a separate contig HMM. For each such HMM, we start with a random read as the seed and iteratively extend it using overlapping reads. See supplement for discussion on how to avoid collision between two HMMs.
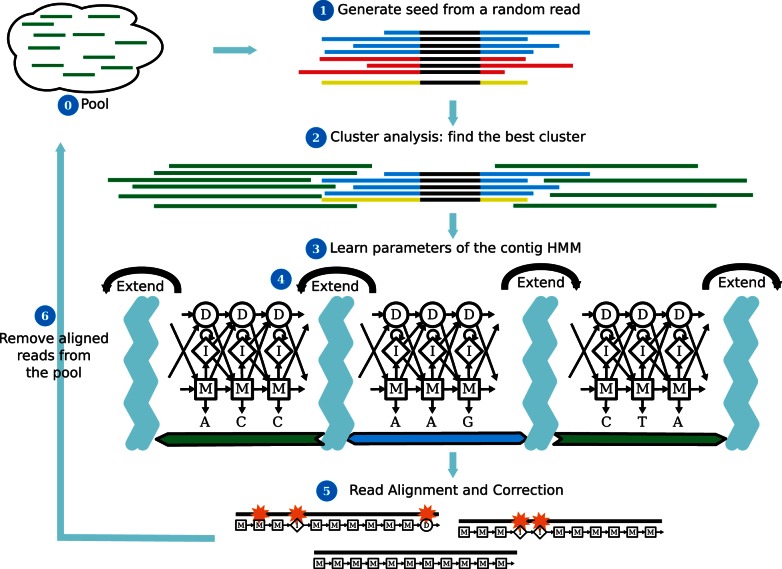
Figure 1.
An overview of SEECER. Step 1: We select a random read that has not yet been assigned to any contig HMM. Next, we extract all reads with at least k consecutive nucleotides that overlap with the selected read. Step 2: We cluster all reads and then select the most coherent subset as the initial set of the contig HMM. Step 3: We learn an initial HMM using the alignment specified by the k-mer matches of selected reads. Step 4: We use the consensus sequence defined by the contig HMM to extract additional reads from our unassigned set. These additional reads are used to extend the HMM in both directions. Step 5: When no more reads can be found to extend the HMM, we determine for each of the reads that were used to construct the HMM the likelihood of being generated by this contig HMM. For those with a likelihood above a certain threshold, we use the HMM consensus to correct errors. Step 6: We remove the reads that are assigned or corrected from the unassigned pool. See ‘Materials and Methods’ section for complete details.
Selecting an initial set of reads for a contig HMM
Using the seed read, we obtain an initial set of reads to use for constructing the HMM contig (Figure 1, step 1). We build a k-mer hash dictionary, where the keys are k-mers and the values are the indices of the reads and the position of the k-mers within them. This hash table could be large, and hence we discard k-mers appearing in less than c reads (here we use c = 3). Counting of k-mers is efficiently done using Jellyfish (29), a parallel k-mer counter. After counting, only k-mers that appear at least c times are stored in a hash table that also records the positions of the k-mer within a read, and as a result, we keep memory requirements as small as possible. Read sequences are saved in the ReadStore from the SeqAn library (30).
SEECER starts the contig construction by selecting (without replacement) a random read (or seed) s from the pool  of reads. We use the dictionary to retrieve a set
of reads. We use the dictionary to retrieve a set 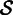 of reads
of reads 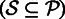 such that each read in
such that each read in 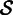 shares at least one k-mer with the seed s. At the same time, we record the locations of the shared k-mers among the reads to construct a multiple-sequence alignment
shares at least one k-mer with the seed s. At the same time, we record the locations of the shared k-mers among the reads to construct a multiple-sequence alignment 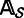 . For each column i (
. For each column i (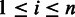 ) of
) of 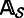 , let Ti be the nucleotide that is the most frequent in that column. Let
, let Ti be the nucleotide that is the most frequent in that column. Let 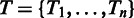 be set of such nucleotides from all columns. Using our current alignment we define
be set of such nucleotides from all columns. Using our current alignment we define 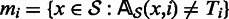 , that is, mi is the set of reads that have a mismatch with Ti. For each read x, we also define
, that is, mi is the set of reads that have a mismatch with Ti. For each read x, we also define 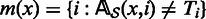 . In other words, m(x) is the set of columns for which x has a mismatch with T.
. In other words, m(x) is the set of columns for which x has a mismatch with T.
Cluster analysis of reads initially retrieved by k-mer overlaps
Because it is only based on k-mer matches, our initial set 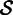 is most likely from a mixture of different transcripts. This situation arises from genomic repeats and alternative splicing. To build a homogenous contig, we use cluster analysis to identify the largest subset
is most likely from a mixture of different transcripts. This situation arises from genomic repeats and alternative splicing. To build a homogenous contig, we use cluster analysis to identify the largest subset 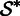 of
of 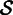 , which satisfies a quality measure.
, which satisfies a quality measure.
To identify the largest subset, the main challenge is in distinguishing genuine errors from other intrinsic difference such as repeats. Note that real biological differences should be supported by a set of reads with similar mismatches to the consensus. This means that we could identify a set of reads associated with intrinsic differences by looking at the intersections of mis. Based on this intuition, we use a clustering algorithm (spectral clustering (31) and a spectral relaxation of k-means (32)) to find clusters of columns in M. These clusters are used to identify coherent subsets of reads, which we then use as the initial set for learning a contig HMM. See supplement for details on how to extract a biologically similar subset using this clustering method and on how to efficiently implement the clustering step.
Learning the parameters of the contig HMM
SEECER has two learning options (Figure 1, step 3). In the first one, we implemented online expectation maximization (EM) algorithm (33) in which we restricted the alignment to have at most v indels to speed up the Forward–Backward algorithm. In the second one, we estimate the parameters based on the alignment of reads using k-mer positions. The first option is much slower than the second because we have to run Forward–Backward algorithm until the EM converges. The second option is faster because we only need to do one pass over all reads. Our experiments show that the second option is good enough for correction and keeps the runtime tractable because often the set of reads is consistent and the amount of errors is low, therefore yielding a good read alignment.
Consensus extension using Entropy
We discard positions in the contig HMM with high entropy of the emission probabilities in the match states. Entropy is a probabilistic statistic, which captures the uncertainty in the discrete distribution of emissions. Positions with high entropy (default max entropy = 0.6) indicate that the initial alignment estimation is not reliable because the set of reads is not consistent. For example, at splitting positions in alternative splicing events, reads from different isoforms may be retrieved, which will lead to high entropy. By discarding these ambiguities, we improve the contig quality and reduce false-positive corrections.
Contig extension
Before contig extension (Figure 1, step 4) all parameters learned for the HMM thus far are fixed. We iteratively extend the contig HMM by repeatedly retrieving more reads sharing k-mers with the new consensus using the dictionary. Each additional read is partially aligned to the HMM, and read bases that are not overlapping the HMM are used to learn the newly extended columns of the HMM, repeating cluster analysis and entropy computation. This iterative process stops when we cannot retrieve any new reads or extend the consensus further.
Probabilistic assignment and correction of reads
After the construction of the contig HMM, each read that was used in the construction is aligned to the HMM using Viterbi’s algorithm. Reads whose log-likelihood of being generated by the contig HMM exceeds a threshold of −1 are considered ‘assigned’ to that HMM. We also restrict the number of corrections for a single read to five to avoid making false-positive corrections. Finally, assigned reads are removed from the pool of reads (Figure 1, step 6).
Handling of ambiguous bases and poly-A tails
We remove ambiguous bases (Ns) from the read sequences before running SEECER by randomly substituting an N with one of the nucleotides (A,T,G,C). However, if there are regions with many Ns in a read, we discard the whole read unless these regions occur at the end, in which case, we truncate and keep the read if the new truncated length is at least half of the original. Reads that have >70% of their bases all As or all Ts are also discarded, as they likely originate from sequenced poly-A tails.
Human datasets and comparison with other methods
Three human paired-end RNA-seq datasets were downloaded for the comparisons: 55 M reads of length 45 bp (ID SRX011546, http://www.ncbi.nlm.nih.gov/sra/) (6), 64 M reads of length 76 bp (34) were downloaded from the GEO database (35) (Accession: GSM759888) and 145 M reads of length 101 bp from the ENCODE consortium (http://genome.ucsc.edu/cgi-bin/hgFileUi?g=wgEncodeCshlLongRnaSeq). The spliced alignment of reads was performed using TopHat version 1.3.3 and Bowtie version 0.12.5 (36). Number of aligned reads is reported for uniquely mapped reads as described in (3).
Quake version 0.3 (22) was run as suggested in the manual for RNA-Seq data, the k-mer size was set to 18 and the automatic cutoff mode was disabled, instead all k-mers with count 1 were classified as erroneous. The other programs were run as follows: Coral version 1.4 (24) with the -illumina option, HiTEC 64 bit version 1.0.2 (21) with options 57000000 4, and Echo version 1.12 (23) with options –ncpu 8 -nh 1024 -b 2000000.
To measure the accuracy of the read error correction methods, we used Tophat to align original and corrected reads to the human reference sequence. Using the reference sequence as ground truth, we used the following definitions (37): a false positive was a base that was changed (corrected) although it was correct in the original read. A true positive was a base that was corrected to the nucleotide in the reference. A false negative was a base that was not corrected even though it is wrong, while a true negative was a base that was left uncorrected and aligned with the reference. The gain metric was computed as explained in (37).
De novo RNA-Seq assembly
We used Oases (version 0.2.5) for the de novo RNA-Seq assembly for the human and sea cucumber datasets. Similar to (11), we conducted a merged assembly for k = 21, … , 35 using default parameters. SEED (version 1.5.1) was run with default parameters, and the resulting cluster sequences were used as input to Oases as described in (28). The evaluation of the human assemblies was conducted by aligning assembled transfrags to the human genome with Blat version 34 (38) and comparing with Ensembl 65 transcript annotation to derive 80% and full-length covered transcripts, as previously described (11). The evaluation metrics were computed using custom scripts.
Sea cucumber sequencing and validation
Gravid Parastichopus parvimensis adults were spawned by heat shock and embryos grown in artificial sea water at 15°C. Total RNA was extracted from 2-day-old gastrula and 6-day-old larvae using the Total Mammalian RNA Miniprep kit (Sigma). RNA was sent to the Wistar Institute for library preparation with Illumina adaptors and 72-bp paired-end sequencing was performed on a Solexa Genome Analyzer II. First strand cDNA synthesis was performed with the iScript Select cDNA Synthesis Kit (BioRad).
From the top 100 expressed transfrags that were expressed in both time points, 14 were randomly selected, seven with a match to either RefSeq or Swissprot and seven without a match. For the validation, PCR primers were designed with Primer3Plus (39) to amplify ∼300–500 bp products corresponding to the 14 selected transfrags (primer sequences are provided in Supplementary Table S11). The PCR was performed using GoTaq (Promega) standard protocols on RNA samples from the first time point.
Sea cucumber transcriptome analysis
For peptide searches, we used Blastx (40) with an E-value cutoff of 10−5 to avoid spurious alignments in Swissprot (41) and the Sea Urchin known proteome (SPU_peptide.fasta at http://www.spbase.org/SpBase/download/). Similarly, for the search in Refseq (42), we used Blastn with the same cutoff. The expression of all assembled transfrags was quantified using RNA-seq by expectation maximization (RSEM) with default parameters (43) after read alignment of the reads to the transfrags with Bowtie (44). The Gene Ontology (GO) annotation for the known and predicted Sea Urchin proteome was downloaded from SpBase (annotation.build6.tar at http://www.spbase.org/SpBase/download/). GO enrichment analysis was done using FuncAssociate 2.0 (45) with a multiple-testing corrected P-value cutoff of 0.05.
Computational infrastructures
SEECER and other error correction methods were run with an 8 core Intel Xeon CPU with 2.40 GHz and 128 GB RAM. The de novo assembly with Oases was run on a 48 core AMD Opteron machine with 265 GB RAM.
The running time of SEECER is discussed in Supplements (Supplementary Table S5).
Data access
The new RNA-Seq datasets for the transcriptome of P. parvimensis were deposited on NCBI Sequence Read Archive (SRA) under the Accession SRA052605.
RESULTS
SEECER: A HMM-based RNA-Seq error correction method
Figure 1 presents a high-level overview of SEECER’s read error correction. The overall goal is to model each contig with a HMM allowing us to model substitutions, insertions and deletions. We start by selecting a random read from the set of reads that have not yet been assigned to any HMM contig. Next, we extract (using a fast hashing of k-mers method) all reads that overlap with the selected read in at least k nucleotides. Because the subset of overlapping reads can be derived from alternatively spliced or repeated segments, we next perform clustering of these reads selecting the most coherent subset (see ‘Materials and Methods’ section) for forming the initial set of our HMM contig. Using this set, we learn an initial HMM using the alignment specified by the k-mer matches. This learning step can either directly rely on the multiple alignment of reads or use standard HMM learning (Expectation Maximization) but with a limited number of indels to keep the run time of the Forward–Backward algorithm linear (see ‘Materials and Methods’ section). Next, we use the consensus sequence defined by the HMM to extract more reads from our unassigned set by looking for those that overlap the current consensus in k or more nucleotides. These additional reads likely overlap the edges of the HMM (because those overlapping the center have been previously retrieved) and so they can be used to extend the HMM in both directions in a similar manner to the method used to construct the initial HMM. This process (learning HMM, retrieving new overlapping reads, etc.) repeats until no more reads overlap the current HMM or the entropy at the edges of the HMM exceeds a predefined threshold (see ‘Materials and Methods’ section).
When the algorithm terminates for a HMM, we determine for each of the reads that were used to construct the HMM how likely it is that they have been generated by this contig HMM. For those reads where this likelihood is above a certain threshold, we use the HMM consensus to correct errors in places where the read sequence disagrees with the HMM. We use several filtering steps to avoid false-positive corrections including testing for the number of similar errors at the same position, the entropy of a position in the HMM and the number of corrections made to a single read. See ‘Materials and Methods’ section for complete details.
Robustness and comparison with other methods
We first tested SEECER on human data to compare it with other approaches that are widely used for other sequencing data (primarily DNA sequencing as mentioned above). Unlike de novo RNA-Seq data, when analyzing human data, we can use a reference genome to determine the accuracy of the resulting corrections and assembly. An established metric to measure the success of error correction after read alignment is the gain metric (19), which is defined as the ratio of newly created versus correctly removed errors (see ‘Materials and Methods’ section).
Before testing SEECER on the human data, we used a subset of ∼34 million reads to assess the influence of the two main parameters for SEECER, the length of k for the initial hashing phase and the value for the maximum entropy at a position. Our experiments show that k = 17 works best for this subset (Supplementary Figures S1 and S2) with stable results for similar values. The maximum entropy was set to 0.6, as lower entropy values resulted in fewer corrections because fewer contigs could be constructed (Supplementary Figure S3).
We next have used these parameters to compare SEECER with four other methods for correcting the reads by initially testing their ability to improve the unique alignment of reads to the human genome after correction (see ‘Materials and Methods’ section). We used three diverse datasets to compare SEECER with the k-mer–based methods Quake (22) and ECHO (23), Coral (24), which relies on multiple alignments of reads for correction, as well as with HiTEC (21), which builds a suffix tree and automatically estimates parameters for correction.
The first dataset we used was derived from human T-cell RNA sequencing resulting in 55 million paired-end reads of length 45 bp (6). In Table 1, we list important statistics regarding the success of the error correction methods. Using SEECER, the number of aligned reads increased by 8.4% when compared with the uncorrected reads, much higher than Quake (3.6%), Coral (4.5%) and ECHO (1.3%). Unlike the other methods, error correction with HiTEC did not result in a higher number of reads mapped. Similarly, the number of reads that align without mismatch errors to the reference sequence using SEECER increased by 50%, which was by far the biggest improvement for all methods tested (Supplementary Figure S4). None of the error correction methods uses paired-end information, and therefore the number of properly aligned read pairs can serve as a good indicator for the accuracy of the error correction. Again, SEECER error corrected reads showed the highest improvement with 15% more pairs properly aligned. The gain metric shows the normalized difference between true-positive and false-positive corrections (Supplementary Table S1) and, again, SEECER outperforms the other methods. In addition, we investigated the error bias in terms of read positions and forward/reverse read strands. Figure 2 presents the distribution of mismatches following TopHat alignments relative to the read positions before and after error correction by SEECER. As can be seen, the previously reported bias that higher error rates are found at read ends for Illumina data (17), is observed in our data as well. However, after SEECER error correction, much of this bias is removed, and the corrected reads have a more uniform distribution of mismatches along the read positions. See Supplementary Figures S6–S9 for details on other types of corrections made by SEECER.
Table 1.
Evaluation using a RNA-Seq dataset of 55 M paired-end 45-bp reads of human T cells
| Method | Original | SEECER | Quake | SEED | Coral | HiTEC | Echo |
|---|---|---|---|---|---|---|---|
| Aligned reads (M) | 31.2 | 33.8 (+8.4%) | 32.3(+3.6%) | – | 32.6 (+4.5%) | 31.2 (+0.0%) | 31.6 (+1.3%) |
| Proper read pairs (M) | 22.1 | 25.5 (+15.1%) | 23.4 (+5.8%) | – | 24.0 (+8.7%) | 22.1 (−0.0%) | 22.7 (+2.5%) |
| Zero error reads (M) | 18.3 | 27.3 (+49.6%) | 22 (+20.4%) | – | 23.9 (+30.7%) | 18.3 (0.1%) | 19.6 (+7.2%) |
| Gain | – | 0.56 | 0.25 | – | 0.38 | 0.00 | 0.024 |
| Assembly full length | 1749 | 2120 (+21%) | 1979 (+13%) | 1358 (−22%) | 2092 (+19.6%) | 1713 (−2.7%) | 1916 (+9.6%) |
| Assembly 80% length | 13 852 | 14 833 (+7%) | 14 267 (+3%) | 9686 (−30%) | 14 643 (+5.7%) | 13450 (−2.9%) | 14 273 (+3.0%) |
| Memory (GB) | – | 27 | 32 | – | 34.3 | 49 | 72 |
| Time (hours) | – | 12.25 | 7.25 | – | 2.42 | 6.33 | 13.7 |
Percentages in brackets denote performance compared with original data.
‘–’ means not applicable.
The evaluation is based on Ensembl 65 annotation.
Figure 2.
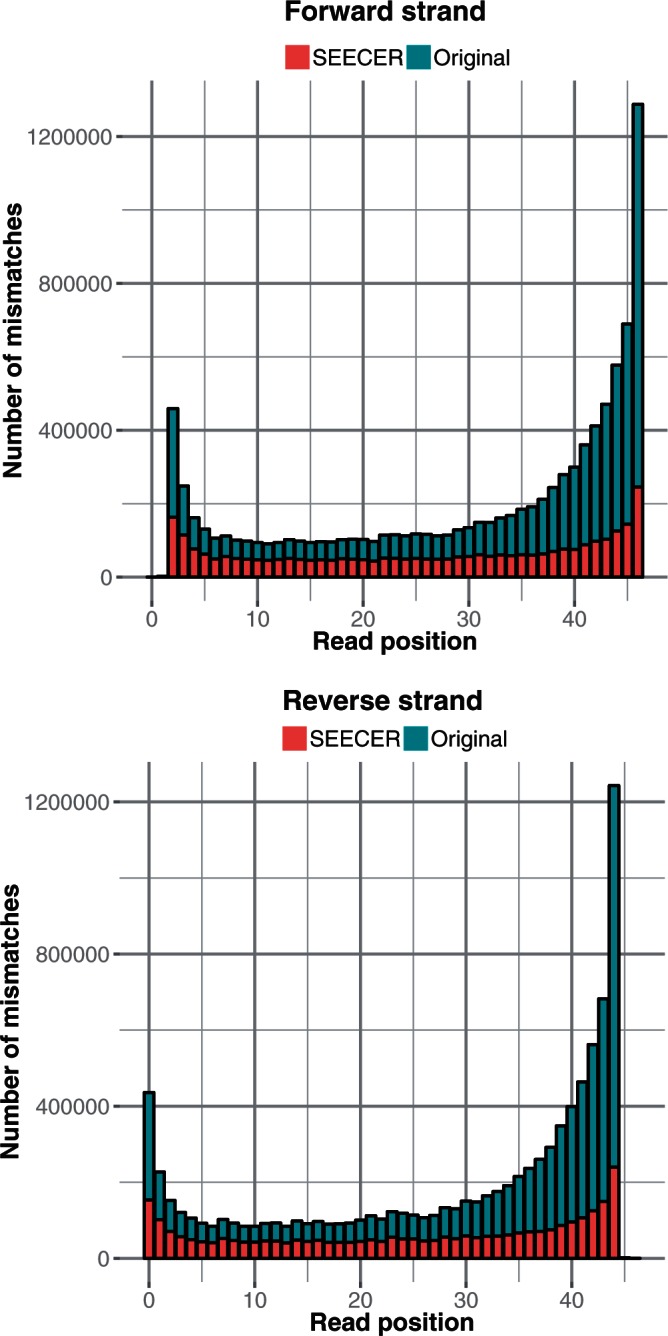
The distribution of mismatches to the reference of pair-mapped reads (using TopHat alignment) of the 55 M paired-end 45 bp reads of human T cells dataset: only reads that are aligned both before and after error correction are shown.
To further test the influence of error correction on downstream analysis, we investigated the ability to identify homozygous single-nucleotide polymorphisms (SNPs) before and after error correction. This analysis demonstrates the usefulness of error correction for such downstream SNP studies and, in particular, shows that using SEECER corrected reads leads to the identification of the highest number of SNPs. See Supplementary Table S6 and Supplementary Figure S10 for details.
While the ability to align individual reads is important, another important goal of de novo RNA-Seq experiments is transcriptome assembly. To test the impact of error correction on downstream assembly, we used the Oases de novo assembler (11). In addition to the read-based error correction methods we compared with above, we have compared with SEED read clustering and subsequent Oases assembly as previously suggested (28). In Table 1, the results for the human T-cell data are shown. An important metric for assembly comparisons is the number of full-length assembled transcripts. Compared with the original reads, after SEECER error correction, 21% more transcripts are reconstructed to full length. SEECER also leads to a 46% increase of detected alternative isoforms (Supplementary Table S4). An example of how Oases benefits from the SEECER error correction is shown in Figure 3 for the gene EIF3CL (see also Supplementary Results). Quake, Echo and Coral led to a lower improvement of assembled full-length transcripts with 13, 9.6 and 19.6%, respectively, whereas SEED and HiTEC resulted in a reduction of full-length reconstructed transcripts of −22 and −2.7%, respectively. The clustering approach used by SEED discards some of the data, which leads to loss of lowly to mid-level expressed transcripts (Supplementary Figure S5). The correction of the human dataset with SEECER took ∼12.25 h, whereas the assembly with Oases took 19 h.
Figure 3.
An illustrating example how Oases benefits from SEECER error correction. Top: Tophat read alignments in the EIF3CL gene for exons 9–13 before (first track) and after (second track) SEECER correction with human data. Colored dots highlight positions with deviations to the reference sequence in the gray read alignments. Bottom: Summary view of the whole region displaying the longest transfrag assembled. Oases assembled the transcript ENST00000380876 (EIF3CL) to 95% of its length with SEECER corrected data (red transfrag), whereas it was only assembled to 45% of its length when using the original data (blue transfrag).
Additional comparisons using larger datasets with longer reads
To test the scalability of SEECER when using datasets with more reads and longer read length, we further tested SEECER on two additional human datatsets: a HeLa cell line dataset of 64 M reads of length 76 bp (GEO Accession: GSM759888) (34) and 145 M reads of length 101 bp from the ENCODE consortium (see ‘Materials and Methods’ section). Owing to the time requirements of the assembly step, we have only focused here on the top three performing methods in our original analysis (SEECER, Quake and Coral). SEECER scales well, and for both datasets, it achieves the best performance for the number of aligned reads, read pairs, full-length assembly and gain (Tables 2 and 3). Additional information about the number of true-positive and false-positive corrections can be found in Supplementary Tables S2 and S3. While SEECER memory requirements scaled more or less linearly with the size of the dataset, Coral’s requirements did not scale in a similar manner. Specifically, we could not run Coral on the largest dataset (Table 3) because its memory requirements were beyond the available memory on the machine we used to test all methods.
Table 2.
Evaluation using a RNA-Seq dataset of 64 M paired-end 76-bp reads of HeLa cell lines
| Method | Original | SEECER | Quake | Coral |
|---|---|---|---|---|
| Aligned reads (M) | 28.9 | 30.9 (+6.9%) | 30.6 (+5.9%) | 29.5 (+2.1%) |
| Proper pairs (M) | 19.4 | 21.4 (+10.4%) | 20.8 (+7.2%) | 20.0 (+2.8%) |
| Zero error reads (M) | 13.7 | 16.9 (+23.4%) | 15.5 (+12.7%) | 14.9 (+8.7%) |
| Gain | – | 0.21 | 0.11 | 0.07 |
| Assembly full | 4067 | 4422 (+8.7%) | 4113 (+1.1%) | 4378 (+7.65%) |
| Assembly 80% | 25 647 | 26 507 (+3.4%) | 25 644 (−0.0%) | 26 414 (+2.99%) |
| Memory (GB) | – | 52 | 32 | 37.3 |
| Time (hours) | – | 20.33 | 1 | 3.5 |
Percentages in brackets denote performance compared with original data.
‘–’ means not applicable.
The evaluation is based on Ensembl 65 annotation.
Table 3.
Evaluation using a RNA-Seq dataset of 145 M paired-end 101-bp reads from the long RNA-seq of IMR90 cell lines from ENCODE Consortium
| Method | Original | SEECER | Quake | Coral |
|---|---|---|---|---|
| Aligned reads (M) | 119.0 | 123.1 (+3.47%) | 121.9 (+2.46%) | – |
| Proper pairs (M) | 81.1 | 85.4 (+5.4%) | 83.5 (+2.9%) | – |
| Zero error reads (M) | 76.2 | 105.3 (+38.2%) | 92.4 (+21.3%) | – |
| Gain | – | 0.58 | 0.32 | – |
| Assembly full | 13 148 | 18 851 (+43.4%) | 14 968 (+13.84%) | – |
| Assembly 80% | 61 522 | 61 178 (−0.6%) | 62 231 (+1.2%) | – |
| Memory (GB) | – | 113 | 60 | >130 |
| Time (hours) | – | 40.25 | 3 | – |
Percentages in brackets denote performance compared with original data.
‘–’ means not applicable.
The evaluation is based on Ensembl 65 annotation.
Assembly of error corrected RNA-Seq sea cucumber data
The sea urchin Strongylocentrotus purpuratus is a model system for understanding the genetic mechanisms of embryonic development, e.g. (46). Other species of echinoderms, including the Californian warty sea cucumber P. parvimensis (Figure 4A), are being developed as comparative developmental model systems, e.g. (47). This work, however, is limited by the absence of a sequenced genome for the sea cucumber. It is thus critical for comparative studies that methods are developed to inexpensively obtain highly accurate transcriptomes for organism for which no sequenced genome exists.
Figure 4.
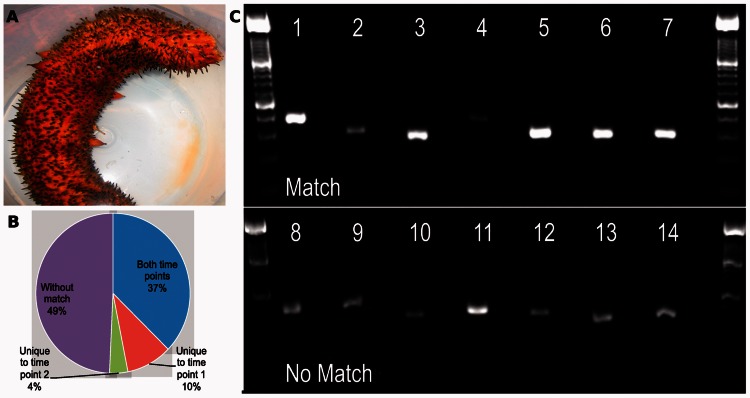
De novo assembly of sea cucumber data. (A) A living sea cucumber Parastichopus parvimensis. (B) Distribution of BlastX matches of sea cucumber transfrags to known sea urchin peptides. The percentages represent the subset of sea urchin peptides that we have significantly matched to at least one transfrag in time point 1 and/or time point 2 and those that were not matched to any transfrag. (C) Ethidium bromide–stained image of PCR products amplified from sea cucumber cDNA. Primer pairs were designed against 14 assembled transfrags, seven of which matched to known peptides of RNAs (top row), and seven other that had no match in the database (bottom row). Standard ladders of 100-bp size are in the first and last lanes. Each lane is followed by the appropriate no template control to demonstrate that amplification was not due to non-specific contamination.
To test how SEECER can help in this direction, we have produced two new datasets for the transcriptome of P. parvimensis. These datasets allow us to determine the expressed mRNAs at the embryonic gastrula (time point 1) and feeding larval (time point 2) stages, which provides insights into the development of this species. Illumina paired-end 72-nt sequencing was conducted and resulted in 88 641 446 and 85 575 446 reads for time points 1 and 2, respectively. We have next used SEECER to correct errors in these datasets, resulting in 28 655 078 and 25 546 050 corrections for 19 465 515 and 17 305 905 reads, respectively. Each corrected read set was then used to produce a de novo RNA-Seq assembly. Error correction took ∼4.7 and ∼4.6 h, whereas de novo assembly took ∼11.3 and ∼13 h for time points 1 and 2. In all, 850 056 transcript fragments (transfrags) were assembled for the embryonic stages (time 1) and 682 913 transfrags for the larval (time 2) stage using Oases (see ‘Materials and Methods’ section).
The only other echinoderm with a sequenced genome is the sea urchin S. purpuratus, which last shared a common ancestor with sea cucumbers ∼350 million years ago (48). Thus, we initially analyzed the similarity between the transfrags we obtained and sea urchin proteins. For the embryonic and larva stages 261 405 and 189 101 transfrags mapped to fragments of 13 330 and 11 793 distinct known peptides in sea urchin (minimum length 50 amino acids for each match). Although we only sequenced RNAs from two developmental stages, thereby not sampling much of the long developmental process and many adult tissues of these organisms, the assembled transfrags from both time points nonetheless matched to >50% of known sea urchin peptides. This suggests both that we have achieved a high-sequence coverage in the assembly, and that many of the sea cucumber genes are already being expressed during early development. In addition, the fact that 14% of these matches were restricted to only one of the two time points suggests that we are able to detect stage-specific developmentally regulated genes, an important requirement for developmental studies (see Figure 4B). To illustrate the usefulness of de novo sequencing, we next performed a GO enrichment analysis for sea urchins’ peptides matched to both time points, and those matched only to time point 1 or time point 2. The results are presented in Supplementary Tables S7–S9.
Time point 1 embryos are undergoing active development including cell movements involved with gastrulation. Larval stages (time point 2) meanwhile are actively swimming and feeding in the water column. As can be seen in the GO analysis, many differences in expression between these stages are of mRNAs that encode for proteins involved in energy metabolism, which is likely due to a switch in how sessile non-feeding embryos and motile feeding larvae use energy resources. We also find an enrichment of expression of genes involved in RNA splicing and translation control in time point 1 (embryos), which may be related to the active transcriptional processing requirements of early embryogenesis. This analysis thus provides an entry point into understanding these important processes.
Although 62–65% of transfrags matched known sea urchin peptides, 297 173 and 255 672 sea cucumber transfrags for time points 1 and 2 did not significantly match any sea urchin peptide (see ‘Materials and Methods’ section). We computed the expression levels of the assembled transfrags and investigated the top 100 expressed transfrags that we could not match to sea urchin peptides from both time points in more detail (see ‘Materials and Methods’ section). In the top 100, 28 and 9 transfrags matched to the RefSeq and Swissprot databases, respectively. Still, we were unable to match 64 transfrags expressed in both time points to any known entry in these databases. To further test the accuracy of our correction and assembly and whether the non-matched transfrags are indeed novel expressed RNAs, we have performed additional follow-up experiments. We selected 14 transfrags that were highly expressed in both time points and performed RT-PCR analysis on these to confirm that the predicted products could be amplified from sea cucumber–derived embryonic cDNA (Figure 4C and Supplementary Table S11). Of the 14, seven were derived from transfrags that matched known peptides and the other seven were derived from transfrags with no match to any of the databases we looked at. As can be seen in Figure 4C, all 14 transfrags were successfully validated, indicating that these are indeed expressed mRNAs and lending support to our correction and assembly procedure.
DISCUSSION
We have developed and tested SEECER, a new method based on profile HMMs to perform error correction in RNA-Seq data. Our method does not require a reference genome. We first learn a contig HMM using a subset of reads and use the HMM to correct errors in reads that are likely associated with the HMM. Our method can handle non-uniform coverage and alternative splicing, both key challenges when performing RNA-Seq. We tested SEECER using complex human RNA-Seq data and have shown that it outperforms several other error correction methods that have been used for RNA-Seq data, in some cases leading to a large improvement in our ability to correctly identify full-length transcripts. We next applied it to perform de novo transcriptome correction and assembly of sea cucumber expression data, providing new insights regarding the development of this species and identifying novel transcripts that cannot be matched to proteins in other species. We note that although a recent report of a 454 sequencing analysis of mixed embryo, larval and adult tissues provides some coverage of an unrelated species, the Japanese sea cucumber Apostichopus japonicas (49), to the best of our knowledge, this is the first published transcriptome of P. parvimensis.
Our analysis of the sea cucumber data indicates that we were able to obtain good transcriptome coverage. The expressed genes from the two developmental stages matched 50% of the protein-coding regions of sea urchin. In addition, de novo correction and assembly was able to accurately detect taxon-specific transcripts. This is critical for comparative development studies, which, in the absence of a genome sequence, often rely on gene discovery from homology searches in related model species. Full appreciation of the role of species-specific genes is essential to understand the developmental origins of animal diversity.
Although one of the main motivations for developing SEECER are applications of de novo RNA-Seq, the human data are useful because alignments allow us to explore the accuracy of the methods, and it is thus a common practice for testing sequencing error correction approaches (19). However, we would like to point out that the classification into false and true positives/negatives is based on the human reference sequence, which may miss haplotype allels. Thus, the false-positive rates reported in the tables may be slightly higher than the real false-positive rates. Nevertheless, we doubt that this approach favors any of the methods because none of them use the reference sequence for performing corrections.
The genome read error correction methods Quake and Coral were able to correct many reads but resulted in a large number of false negatives, as indicated by their lower rates of aligned reads and the drop in the gain statistic compared with SEECER. Coral was the closest to SEECER in terms of the resulting number of full-length assembled transcripts for two of the three datasets. However, Coral seems to suffer from lack of scalability, which may be problematic as dataset size increase. Indeed, its memory requirements for the largest dataset we analyzed were larger than the capacity of our machine cluster.
Our experiments have shown that read clustering leads to a loss of assembled full-length transcripts, especially for low-to-mid level expressed transcripts, because parts of the data are discarded. Owing to non-uniform expression levels in RNA-Seq data, error correction sensitivity critically depends on a method’s ability to detect errors. The performance drop for HiTEC and ECHO, compared with the other methods tested, may be explained by their uniform coverage assumption leading to missing higher frequency errors in highly expressed genes. In contrast, Quake and Coral do not have these strong assumptions and perform much better. However, unlike SEECER, they do not use a probabilistic HMM model and read clustering. These steps allowed SEECER to outperform all other methods in the number of alignable reads, full-length assemblies and false-negative rate with only linear increase in memory requirements for larger datasets.
While we have focused here on the improvement to RNA assembly following error correction, it has been shown that de novo assemblies allow reliable detection of genes that are differentially expressed between two conditions (50). Thus, by improving the resulting assembly, SEECER is likely to improve downstream differential expression analyses as well.
There are many directions to improve SEECER further by using base call quality scores to improve performance on lowly expressed transcripts or using the paired-end information to improve construction of contigs. Currently, SEECER was designed to work without an available reference sequence (de novo RNA-Seq), but an available reference sequence could help with correction of repetitive regions and lowly expressed transcripts.
Finally, while we have primarily developed SEECER for RNA-Seq data, it may also prove useful for single-cell and single-molecule sequencing. In other studies, including metagenomics and ribosome profiling experiments, researchers encounter sequencing data where the coverage is non-uniform and as such SEECER, which does not assume uniformity, can improve the analysis of these data as well.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–11, Supplementary Methods, Supplementary Results, Supplementary Figures 1–11 and Supplementary References [51–55].
FUNDING
National Institutes of Health (NIH) [1RO1 GM085022] and National Science Foundation (NSF) [DBI-0965316 award to Z.B.J.]. Funding for open access charge: NIH [1RO1 GM085022 to Z.B.J.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Animals were collected by Patrick Leahy and Peter Halmay or Marinus Inc under permit number SC-11478 from California Department of Fish and Game to V.F.H. This research was funded by NSF grant IOS-0844948 and the Winters Foundation to V.F.H.
REFERENCES
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marioni J, Mason C, Mane S, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science (New York, N.Y.) 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 4.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 5.Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 6.Heap GA, Yang JHM, Downes K, Healy BC, Hunt KA, Bockett N, Franke L, Dubois PC, Mein CA, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum. Mol. Genet. 2010;19:122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard H, Schulz MH, Sultan M, Nürnberger A, Schrinner S, Balzereit D, Dagand E, Rasche A, Lehrach H, Vingron M, et al. Prediction of alternative isoforms from exon expression levels in RNA-Seq experiments. Nucleic Acids Res. 2010;38:e112. doi: 10.1093/nar/gkq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. De novo assembly and analysis of RNA-seq data. Nat. Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 10.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics (Oxford, England) 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Jiang H, Wong WH. Modeling non-uniformity in short-read rates in RNA-Seq data. Genome Biol. 2010;11:R50. doi: 10.1186/gb-2010-11-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol. Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-Content Normalization for RNA-Seq data. BMC Bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smeds L, Künstner A. ConDeTri–a content dependent read trimmer for Illumina data. PloS One. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Chockalingam SP, Aluru S. A survey of error-correction methods for next-generation sequencing. Brief. Bioinformatics. 2013;14:56–66. doi: 10.1093/bib/bbs015. [DOI] [PubMed] [Google Scholar]
- 20.Schröder J, Schröder H, Puglisi SJ, Sinha R, Schmidt B. SHREC: a short-read error correction method. Bioinformatics (Oxford, England) 2009;25:2157–2163. doi: 10.1093/bioinformatics/btp379. [DOI] [PubMed] [Google Scholar]
- 21.Ilie L, Fazayeli F, Ilie S. HiTEC: accurate error correction in high-throughput sequencing data. Bioinformatics (Oxford, England) 2011;27:295–302. doi: 10.1093/bioinformatics/btq653. [DOI] [PubMed] [Google Scholar]
- 22.Kelley DR, Schatz MC, Salzberg SL. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11:R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao W-C, Chan AH, Song YS. ECHO: a reference-free short-read error correction algorithm. Genome Res. 2011;21:1181–1192. doi: 10.1101/gr.111351.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmela L, Schröder J. Correcting errors in short reads by multiple alignments. Bioinformatics (Oxford, England) 2011;27:1455–1461. doi: 10.1093/bioinformatics/btr170. [DOI] [PubMed] [Google Scholar]
- 25.Medvedev P, Scott E, Kakaradov B, Pevzner P. Error correction of high-throughput sequencing datasets with non-uniform coverage. Bioinformatics (Oxford, England) 2011;27:i137–i141. doi: 10.1093/bioinformatics/btr208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu W, Hashimoto S-I, Morishita S. Efficient frequency-based de novo short-read clustering for error trimming in next-generation sequencing. Genome Res. 2009;19:1309–1315. doi: 10.1101/gr.089151.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wijaya E, Frith MC, Suzuki Y, Horton P. Recount: expectation maximization based error correction tool for next generation sequencing data. Genome Inform. 2009;23:189–201. [PubMed] [Google Scholar]
- 28.Bao E, Jiang T, Kaloshian I, Girke T. SEED: efficient clustering of next-generation sequences. Bioinformatics (Oxford, England) 2011;27:2502–2509. doi: 10.1093/bioinformatics/btr447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics (Oxford, England) 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Döring A, Weese D, Rausch T, Reinert K. SeqAn an efficient, generic C++ library for sequence analysis. BMC Bioinformatics. 2008;9:11. doi: 10.1186/1471-2105-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng AY, Jordan MI, Weiss Y. On spectral clustering: analysis and an algorithm. Adv. Neural Inf. Process. Syst. 2002;2:849–856. [Google Scholar]
- 32.Zha H, Ding C, Gu M, He X, Simon H. Spectral relaxation for k-means clustering. Adv. Neural Inf. Process. Syst. 2001;14:1057–1064. [Google Scholar]
- 33.Liang P, Klein D. Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics. 2009. Online EM for unsupervised models; pp. 611–619. [Google Scholar]
- 34.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Dorman KS, Aluru S. Reptile: representative tiling for short read error correction. Bioinformatics (Oxford, England) 2010;26:2526–2533. doi: 10.1093/bioinformatics/btq468. [DOI] [PubMed] [Google Scholar]
- 38.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Research. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galperin MY, Fernández-Suárez XM. The 2012 nucleic acids research database issue and the online molecular biology database collection. Nucleic Acids Res. 2012;40:D1–D8. doi: 10.1093/nar/gkr1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics (Oxford, England) 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C-H, Minokawa T, Amore G, Hinman V, et al. A genomic regulatory network for development. Science (New York, N.Y.) 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 47.Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl. Acad. Sci. USA. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wada H, Satoh N. Phylogenetic relationships among extant classes of echinoderms, as inferred from sequences of 18S rDNA, coincide with relationships deduced from the fossil record. J. Mol. Evol. 1994;38:41–49. doi: 10.1007/BF00175494. [DOI] [PubMed] [Google Scholar]
- 49.Du H, Bao Z, Hou R, Wang S, Su H, Yan J, Tian M, Li Y, Wei W, Lu W, et al. Transcriptome sequencing and characterization for the sea cucumber Apostichopus japonicus (Selenka, 1867) PloS One. 2012;7:e33311. doi: 10.1371/journal.pone.0033311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nookaew I, Papini M, Pornputtpong N, Scalcinati G, Fagerberg L, Uhln M, Nielsen J. A comprehensive comparison of rna-seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10084–10097. doi: 10.1093/nar/gks804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eddy SR. Profile hidden Markov models. Bioinformatics (Oxford, England) 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 52.Von Luxburg U. A tutorial on spectral clustering. Stat. Comput. 2007;17:395–416. [Google Scholar]
- 53.Saccone SF, Quan J, Mehta G, Bolze R, Thomas P, Deelman E, Tischfield JA, Rice JP. New tools and methods for direct programmatic access to the dbsnp relational database. Nucleic Acids Res. 2011;39:D901–D907. doi: 10.1093/nar/gkq1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emde A-K, Schulz MH, Weese D, Sun R, Vingron M, Kalscheuer VM, Haas SA, Reinert K. Detecting genomic indel variants with exact breakpoints in single- and paired-end sequencing data using splazers. Bioinformatics. 2012;28:619–627. doi: 10.1093/bioinformatics/bts019. [DOI] [PubMed] [Google Scholar]
- 55.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.