Abstract
The DNase I accessibility and chromatin organization of genes within the nucleus do correlate to their transcriptional activity. Here, we show that both serum starvation and overexpression of Tip5, a key regulator of ribosomal RNA gene (rDNA) repression, dictate DNase I accessibility, facilitate the association of rDNA with the nuclear matrix and thus regulate large-scale rDNA chromatin organization. Tip5 contains four AT-hooks and a TAM (Tip5/ARBP/MBD) domain, which were proposed to bind matrix-attachment regions (MARs) of the genome. Remarkably, the TAM domain of Tip5 functions as nucleolar localization and nuclear matrix targeting module, whereas AT-hooks do not mediate association with the nuclear matrix, but they are required for nucleolar targeting. These findings suggest a dual role for Tip5’s AT-hooks and TAM domain, targeting the nucleolus and anchoring to the nuclear matrix, and suggest a function for Tip5 in the regulation of higher-order rDNA chromatin structure.
INTRODUCTION
Mammalian cells synthesize the 47S precursor for ribosomal RNA (rRNA) from multicopy genes. During recent years, several chromatin-dependent regulators of rRNA transcription were discovered, which take part in the balancing of this highly energy-demanding metabolic activity of the cell [reviewed in (1)]. Compared with promoter-specific actions of these chromatin regulators, little is known about their role in large-scale spatial organization and distribution of actively transcribed versus inactive rRNA gene copies in the nucleus. The synthesis of 47S pre-rRNA from active rDNA takes place at the fibrillar center/dense fibrillar component (FC/DFC border) of the mammalian nucleolus, whereas inactive rDNA is localized within the FC or outside of nucleoli [for a recent review see (2)]. It has been demonstrated earlier that changes in the ribosome synthesis activity result in alterations of nucleolar architecture when cells are treated with different inhibitors of ribosome biogenesis or serum starved (3–5). Part of the morphological alterations in nucleolar structure may be correlated to rDNA chromatin movements, which accompany changes in the transcriptional activity of rRNA genes.
In addition to the visual inspection of nuclear morphology, nuclear matrix isolation enables a simple biochemical characterization of large-scale chromatin organization. The nuclear matrix was originally defined as a component of nuclei that resists extensive DNase I digestion and salt extraction (6). It contains mainly intermediate filament proteins like lamins, heterogeneous nuclear ribonucleoprotein particles, specific non-histone chromatin proteins and associated DNA, which represents the matrix-attachment regions (MARs) of the genome. MARs, which are supposed to anchor chromatin loops to the nuclear matrix constitutively or transiently, have been implicated in the regulation of gene expression and replication [for a review see (7)]. Importantly, specific enrichment of rDNA in nuclear matrix preparations has been demonstrated by using biochemical (8) and cell biology methods (9).
Previous studies on rDNA chromatin regulation revealed the role of the nucleolar remodeling complex (NoRC) in nucleosome positioning, transcriptional repression, epigenetic silencing and replication timing (10–14). NoRC consists of two subunits, the ATPase subunit Snf2h and the large, regulatory subunit Tip5 (15). More recently, the association of these two proteins with the transcriptional co-repressor CtBP (C-terminal–binding protein) was also reported, and a non-nucleolar chromatin regulatory function of this tripartite complex has been described (16). The role of Tip5 in the inactivation of rRNA transcription has been demonstrated to involve cooperation with proteins, such as TTF-I, HDACs and Dnmts (12,14,17). Tip5 not only has numerous protein interacting domains but also has several predicted AT-hooks and the TAM domain. AT-hooks are small peptide motifs, which mediate binding to the minor groove and thereby alter the architecture of DNA (18–20). The TAM domain shows sequence homology to the methyl-CpG–binding domain (MBD) found in transcriptional repressor proteins that selectively bind methylated DNA (18,21). However, the TAM domain of Tip5 has been shown to bind to DNA irrespective of its DNA methylation status (15) and also associates with the structured rDNA promoter RNA (22). As the TAM domain and AT-hooks are predicted MAR binders (18), we hypothesized that Tip5 could mediate the anchoring of rDNA to the nuclear matrix and, thus, separate silenced rDNA repeats from active ones. To elucidate the contribution of transcriptional repression, and particularly that of Tip5, to the control of large-scale organization of rDNA chromatin, the association of rDNA with the nuclear matrix was analyzed after serum starvation and overexpression of Tip5. In subsequent experiments, the DNA-binding activities of single AT-hook domains of the Tip5 protein were characterized in vitro, and the role of AT-hooks and the TAM domain in sub-nuclear localization and nuclear matrix association of Tip5 was investigated in vivo.
MATERIALS AND METHODS
Cell culture
IMR90 human lung embryonic fibroblasts, HeLa human cervix carcinoma cells and HEK293 human embryonic kidney cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum, 100 U/ml of penicillin and 100 mg/ml of streptomycin and incubated at 37°C in 5% CO2. IMR90 cells were cultured for 1 week in serum-free medium in serum starvation experiments. HeLa and HEK293 cells were used in transfection experiments, and DNA was delivered with the FuGENE HD (Roche) transfection reagent according to the manufacturer’s instructions. Tip5 sequences were derived from mouse Tip5 cDNA (GenBank Acc. No AJ309544).
Immunofluorescence
Cells were rinsed twice in phosphate-buffered saline (PBS) at 37°C. Fixation was performed in 4% paraformaldehyde in PBS for 10 min at room temperature. During the last minute, few drops of 0.5% Triton X-100 in PBS were added to the cells. Afterwards cells were washed three times in PBS with 0.01% Triton X-100 for 3 min at room temperature, followed by 5 min in PBS with 0.5% Triton X-100. Finally, cells were washed with PBS-T (PBS with 0.1% Tween20) twice for 5 min at room temperature and stored at 4°C. All the subsequent steps were carried out at 37°C. Cells were washed in PBS-T three times for 5 min before antibody staining. Afterwards they were incubated with the primary antibody in 4% bovine serum albumin (BSA) in PBS-T for 1 h in a humidified chamber. Cells were then again washed in PBS-T three times for 3 min and incubated with the suitable secondary antibody in 4% BSA in PBS-T for 1 h. After washing with PBS-T twice for 5 min, the DNA was counterstained with 50 ng/ml of DAPI in PBS-T for 5 min. Slides were rinsed again in PBS-T and mounted in Vectashield mounting medium (Vector). For detection of Tip5, cells were fixed in methanol and acetone as previously described (15) and then processed as described earlier in the text. Images were taken on a Zeiss Axiovert 200 inverted fluorescence microscope using the Axio Vision software.
Nuclear matrix isolation
The isolation of nuclear matrix was carried out essentially as described (6,23,24). A total of 2.5–5 × 106 cells were washed in 1 ml of PBS, pH 7.4, (5 min, 1000g centrifugation), and cells were extracted in 200 µl of cytoskeleton buffer (10 mM PIPES, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, supplemented with Protease Inhibitor Cocktail (Roche), 1 mM TCEP and 0.5% Triton X-100). After 5 min incubation at 4°C, soluble cytoplasmic proteins were separated by centrifugation at 5000g for 3 min (supernatant = ‘CP’ cytoplasmic fraction). Chromatin was solubilized by DNA digestion with 400 U of RNase-free DNase I (Roche) in 110 µl of CSK buffer plus protease inhibitors for 90 min at 37°C with shaking at 300 g. In particular experiments, RNase A at 0.3 mg/ml final concentration was also added at this step. Then 50 µl of ammonium sulfate was added from a 1 M stock solution in CSK buffer to a final concentration of 0.25 M, and, after 5 min incubation at 4°C on rotating wheel, samples were pelleted again by centrifugation at 5000 g for 3 min (supernatant = ‘CHR’ chromatin fraction). The pellet was further extracted with 100 µl of 2 M NaCl in CSK buffer for 10 min at 4°C on rotating wheel and then centrifuged at 5000g for 3 min. This treatment removes all accessible DNA and histones from the nucleus (supernatant = ‘2M’ 2 M salt wash fraction, which was diluted 1:2 with water before SDS–PAGE). The remaining pellet was solubilized in 200 µl of 8 M urea buffer (10 mM Tris–HCl, 100 mM NaH2PO4 × H2O, 8 M urea; pH 8.0) and was considered the nuclear matrix-containing fraction (‘NM’). Hundred microliters of each fraction was dialyzed against 1× TE buffer and subsequently digested with RNaseA (37°C, 1 h) and proteinase K (50°C o/n), precipitated, dissolved in 50 µl of ddH2O and controlled on 1% agarose gel (10 µl), and the ‘NM’ fraction was subjected to qPCR analysis. In all, 25 µl of 5× Lämmli buffer was added to the remaining 100 µl of the fractions, and 20 µl each was subjected to western blot analysis or Coomassie staining after boiling the probes at 95°C, 10 min and separating on denaturing SDS–polyacrylamide gels.
Nuclear matrix preparations in situ
In situ preparations of nuclear matrix for immunofluorescence analysis was done essentially as described (25). For protease inhibition, the complete EDTA-free tablets (Roche) were used. Chromatin digestion was performed using 500 U/ml of recombinant DNase I, RNase-free (Roche) at 30°C for 30 min. After matrix preparation, cells were fixed in 4% paraformaldehyde in PBS at room temperature for 15 min. Immunofluorescence detection was carried out as described earlier in the text.
Quantitative PCR
To measure the quantities of rDNA fragments in the nuclear matrix fractions from different experiments, approximately equal amounts of the isolated nuclear matrix-associated DNA were used as templates from control versus treated cells. The interferon β (IFNβ) promoter MAR, which is not bound by Tip5, served as normalization control, and the amount of rDNA promoter, 28S and intergenic spacer (IGS) MAR sequences were quantified in the isolated DNA fractions. The oligonucleotides were as follows: IFNβF 5′-TGC TCT CCT GTT GTG CTT CTC CAC, IFNβR 5′-ATA GAT GGT CAA TGC GGC GTC C, Hr42857F 5′-ATG GTG GCG TTT TTG GGG AC, Hr42964R 5′-CGA AAG ATA TAC CTC CCC CG, Hr9661F 5′-CGA ATG ATT AGA GGT CTT GGG GC, Hr9860R 5′-TGG GGT CTG ATG AGC GTC GG, Hr36189F 5′-TCG CCG ACT CTC TCT TGA CTT G, Hr36399R 5′-TGG AGC ACA GTG ACA CAA CTA TGG. The numbers of human rDNA oligos (Hr) indicate the position relative to the transcription start site (+1) in the repeat unit (GenBank Acc. No. U13369).
Antibodies
To detect proteins on western blots or by immunofluorescence rabbit polyclonal α-Upstream Binding Transcription Factor, RNA Polymerase I (UBF) (sc-9131, Santa Cruz Biotechnology), α-lamin A/C (sc-20681, Santa Cruz Biotechnology), α-RPA194 (sc-28714, Santa Cruz Biotechnology), α-Tip5 (15), α-Snf2h, α-RPA116 (26), α-Brg1, α-Mi-2 (both were a kind gift of A. Brehm) and mouse monoclonal α-fibrillarin (38F3, Novus Biologicals) and α-tubulin (ab7291, Abcam) antibodies were used at appropriate dilutions.
Expression and purification of recombinant proteins
GST–AT-hook constructs were prepared by inserting oligonucleotides containing the coding region for the peptides listed in Figure 4A and flanking BamHI/SalI restriction sites into pGEX4T3. The fusion proteins were expressed in the Escherichia coli Rosetta strain as follows: the transformed bacteria were induced at OD600 = 0.6 by adding IPTG at 0.5 mM final concentration and incubated for 3–4 h at 37°C. Cells were then harvested and affinity purified on Glutathione Sepharose 4B beads (Pharmacia) in batch according to the manufacturer’s protocol. Protein concentration of the elution fractions was estimated by using the Bradford assay (Bio-Rad).
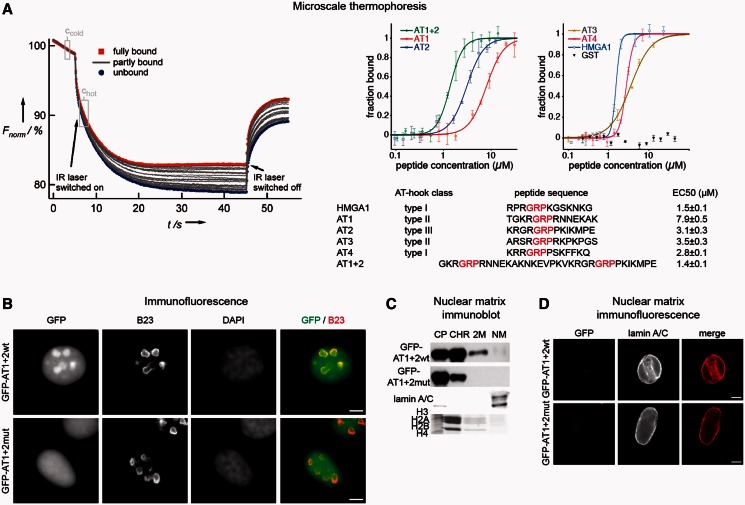
Figure 4.
Analyses of Tip5’s AT-hook domains in microscale thermophoresis, immunofluorescence and nuclear matrix isolation experiments. (A) Microscale thermophoresis experiments. The scheme of a measurement is shown on the left side. The curves represent individual reactions, in which the DNA concentration was kept constant and the AT-hook concentration varied. Thermophoresis was induced by heating the reaction with an infrared (IR) laser. Fluorescence intensity changes are plotted as a function of time. Ccold and Chot indicate the time points where fluorescence measurements were carried out. The curves representing full binding and no binding are labeled in red and blue, respectively, whereas the gray curves represent partial binding. For a detailed description of the assay please refer to Baaske et al. (27). The results of the measurements are shown on the right side. The indicated amounts of the different Tip5 AT-hook peptides were analyzed for DNA binding at constant, 50 nM concentration of a 29 bp, Cy5-labeled rDNA IGS oligonucleotide. The amount of bound DNA was plotted against the peptide concentration, and the concentrations where half of the oligonucleotide is bound, the EC50 values, were determined. The data points of the plot represent mean ± standard deviation values of four measurements. The list of the analyzed AT-hooks is shown below the diagrams. GRP core motifs of the peptides are indicated in red. The classification of AT-hooks according to Aravind and Landsman (18) is shown. The numerical EC50 values for each peptide are shown on the right side. (B) Immunofluorescence experiments show the sub-nuclear localization of the wild-type and mutated versions of the first two AT-hooks. The GFP-tagged proteins were stained with a rabbit polyclonal GFP antibody, the nucleoli with a mouse monoclonal B23 antibody and the nuclear DNA with DAPI. Bars indicate 5 µm. (C) Immunoblot and Coomassie gel pictures of nuclear matrix preparations. HEK293 cells were transfected with the indicated GFP-Tip5 protein-encoding plasmid DNA for 72 h, and nuclear matrices were prepared. CP, CHR, 2 M and NM indicate cytoplasmic, soluble chromatin, high-salt wash and nuclear matrix fractions, respectively. Core histones and lamin A/C served as controls for the CHR/2 M and NM fractions, respectively. (D) The double AT-hook domain of Tip5 does not mediate nuclear matrix association. HeLa cells were transfected with the indicated GFP-Tip5 protein-encoding plasmid DNA for 48 h; nuclear matrices were prepared in situ and analyzed by immunofluorescence. The names of the peptides are shown on the left. Lamin A/C detection served as control for nuclear matrix preparations. Bars indicate 5 µm.
Gel retardation assays
To analyze the DNA-binding activity of recombinant AT-hooks, gel retardation assays were performed as follows: 34 bp, double-stranded synthetic oligonucleotides (sense strand sequences: rDNA En: 5′-TGG ATC TTT TTT TTT TTT TTT CTT TTT TCC TCC A, rDNA IGS: 5′-AAG TCG TGC CTA AAA TAA ATA TTT TTC TGG CCA A, IFNβ: 5′-GGA GAA GTG AAA GTG GGA AAT TCC TCT GAA TAG A, GC ctrl: 5′-CAC GCG CTC GCG CAC GCG CTC GCG CAC GCG CTC) were incubated with recombinant AT-hook proteins in 180 mM NaCl, 1 mM MgCl2, 0.01% (w/v) BSA, 10 mM Tris, pH 8.1, and 5% glycerol. Ten-microliter reactions were incubated for 10 min on ice, and protein–DNA complexes were separated on 6% native polyacrylamide gels in 0.4× TBE buffer at 4°C applying the constant voltage of 100 V. The gels were stained with SybrSafe, and DNA was visualized on a transilluminator.
Microscale thermophoresis
All microscale thermophoresis measurements were performed at room temperature using Monolith NT™ Hydrophobic Capillaries and the Monolith NT.015T device by NanoTemper technologies with the laser being on for 40 s resulting in a temperature increase of 6 K (27). Binding reactions took place in a solution containing 100 mM NaCl, 50 mM HEPES, pH 7.4, 0.05% (v/v) NP-40 and 5 mM MgCl2 in final concentration, as well as one type of double-stranded, fluorescently labeled oligonucleotide as template (rDNA En: 5′-TCT TTT TTT TTT TTT TTC TTT TTT CCT CC, Cy5-labeled; GC ctrl: 5′-CAC GCG CTC GCG CAC GCG CTC GCG CAC GCG CTC, Cy3-labeled) with a constant concentration of 50 nM. For each binding analysis, a titration series was prepared with varying concentrations of the respective peptide as indicated in the plots. All curves were plotted with the KaleidaGraph software, and the thermophoresis signals were fitted with the Hill equation corrected for the minimum (Min) and maximum (Max) values of the binding curve: Y(c) = Min + (Max− Min)/(1 + EC50/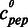 )n, with Y (c) being the thermophoresis signal,
)n, with Y (c) being the thermophoresis signal, 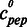 the variable concentration of the respective peptide, EC50 the concentration where half of the oligonucleotide is bound and n being the Hill-coefficient. Thermophoresis signals were normalized to fraction bound (X) by X = [Y(c)− Min]/(Max− Min), error bars (std) were normalized by stdnorm = std(c)/(Max− Min), and both were plotted and fitted as described earlier in the text. KD values were not determined, as the gel retardation experiments showed that multiple protein–DNA complexes appear at higher peptide concentrations (Figure 3B).
the variable concentration of the respective peptide, EC50 the concentration where half of the oligonucleotide is bound and n being the Hill-coefficient. Thermophoresis signals were normalized to fraction bound (X) by X = [Y(c)− Min]/(Max− Min), error bars (std) were normalized by stdnorm = std(c)/(Max− Min), and both were plotted and fitted as described earlier in the text. KD values were not determined, as the gel retardation experiments showed that multiple protein–DNA complexes appear at higher peptide concentrations (Figure 3B).
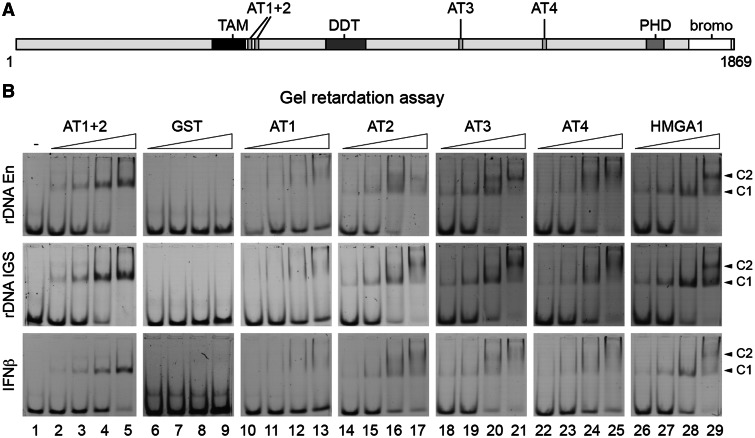
Figure 3.
DNA-binding features of Tip5’s AT-hook domains. (A) Domain structure of Tip5, the large subunit of the NoRC complex. TAM domain; ATx, AT-hook; DDT, domain in different transcription and chromosome remodeling factors; PHD, plant homeodomain zinc-finger; bromo, bromodomain. The numbers below the scheme indicate amino acids. (B) Gel retardation assays. Lane 1 shows in the upper panel an rDNA enhancer (rDNA En) probe, in the middle panel an rDNA intergenic spacer (rDNA IGS) probe and in the lower panel an IFNβ promoter (IFNβ) probe alone. Increasing amounts (18, 36, 72 and 144 pmol) of the different Tip5 AT-hook peptides AT1 + 2 (lanes 2–5), the negative control GST (lanes 6–9), AT1 (lanes 10–13), AT2 (lanes 14–17), AT3 (lanes 18–21), AT4 (lanes 22–25) and the positive control second AT-hook of HMGA1 (‘HMGA1’, lanes 26–29) were analyzed for DNA binding on 1.25 pmol DNA. Different protein–DNA complexes are indicated with arrows and labeled with C1 and C2.
RESULTS
Serum starvation induces global changes in nucleolar architecture and enrichment of rDNA in the nuclear matrix
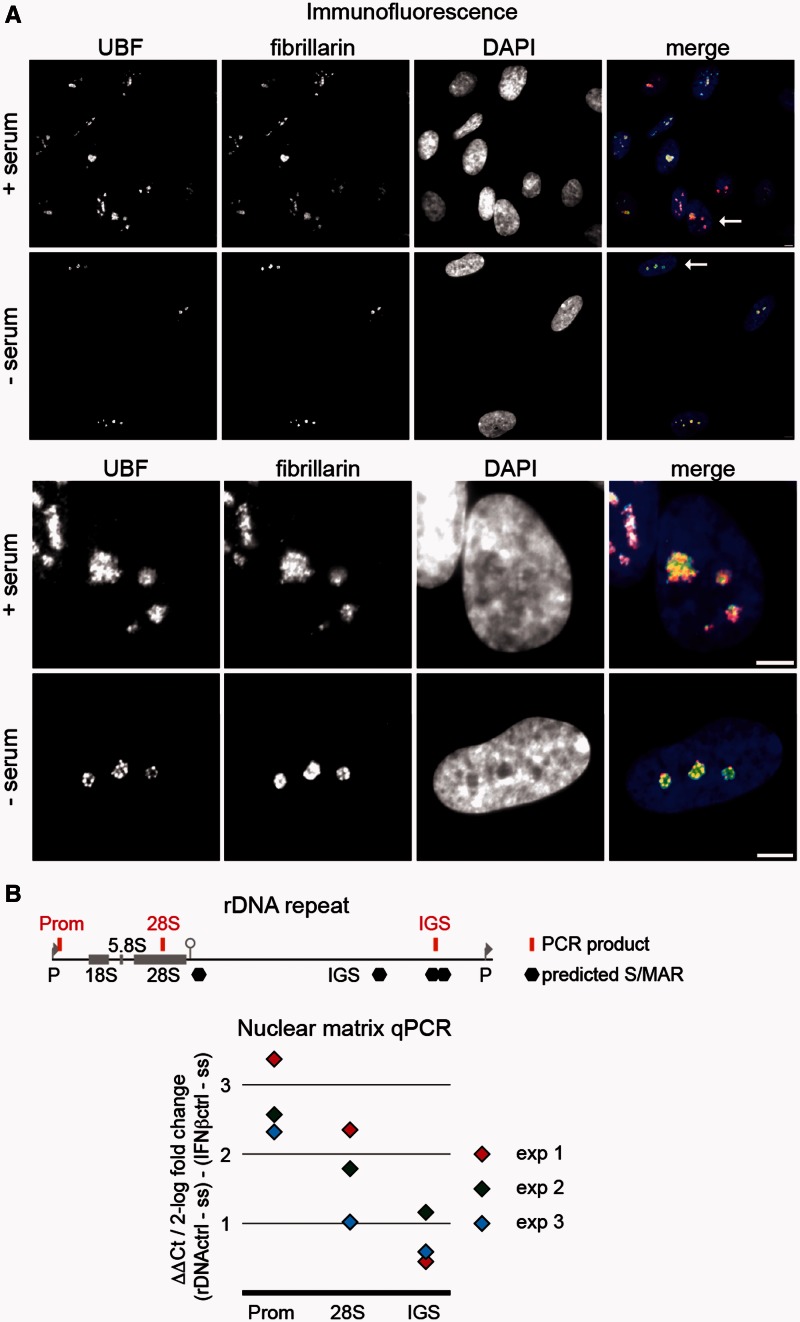
To monitor changes in nucleolar structure, which correlate to repression of rRNA synthesis, immunofluorescence experiments were performed and the distributions of UBF, fibrillarin and Pol I were compared in serum-starved and normally proliferating IMR90 human embryonic lung fibroblasts (Figure 1A and Supplementary Figure S1). Serum starvation led to reduction of nucleolar size and focal compactions of UBF and Pol I signals within the nucleolus. Based on these results and similar observations in previous reports (28–30), we assumed that the spatial organization of rDNA chromatin in the nucleolus is changed after repression of rRNA synthesis. To test this hypothesis, the relative amounts of various rDNA fragments in isolated nuclear matrix fractions of control and serum-starved cells were quantified and compared with the level of the IFNβ promoter, which is a bona fide MAR (31), stably associated with the nuclear matrix, and contains a well-characterized binding site for the AT-hook protein HMGA1 (19). We assumed that alterations in the relative amount of selected rDNA regions compared with this specific MAR reflect the changes in the association of rDNA with the nuclear matrix. First, the putative MARs of the human rDNA were determined in silico by using a formerly developed web tool (32). Predicted MARs localize to the IGS of rDNA as shown in Figure 1B. Real-time qPCR reactions were established to quantify the amount of one selected rDNA IGS sequence (IGS) that is localized between two predicted neighboring MAR, as well as two additional rDNA regions, which are not predicted MARs (Figure 1B). One of these sites, the rDNA promoter (Prom), is a binding site of Tip5. Tip5 possesses four AT-hooks and a TAM domain and, therefore, potentially targets its binding sites to the nuclear matrix. Another sequence was selected from the rDNA coding region (28S) where no Tip5 binding occurs (12). Thus, our experimental system allows the monitoring of MAR- and Tip5-dependent and -independent associations of rDNA sequences with the nuclear matrix. Similar amounts of purified nuclear matrix template DNA were analyzed from normally growing and serum-starved cells in quantitative real-time PCR reactions. Threshold cycle (Ct) differences between serum-starved and control cells were determined at each of the three different regions of the human rDNA and normalized to the Ct differences of the IFNβ promoter (Figure 1B). The results of three biological replicates are shown and demonstrate that the three tested rDNA regions are accumulating in the nuclear matrix on serum starvation. They are sequestered to different extent: the IGS sequence is enriched ∼1.5- to 2-fold in the matrix fraction compared with the IFNβ promoter, the coding region 2- to 5-fold, whereas the promoter region is enriched 5- to 10-fold.
Figure 1.
Serum starvation induces global changes in nucleolar architecture and enrichment of rDNA in the nuclear matrix. (A) Immunofluorescence detection of the nucleolar proteins UBF and fibrillarin in control and serum starved IMR90 human embryonic fibroblasts. The nuclei were counterstained with DAPI. The lower panel shows the enlargements of the selected cells that are indicated with an arrow on the merged image of the upper panel. Bars indicate 5 µm. (B) Real-time qPCR analysis of serum starvation-induced changes in the nuclear matrix association of rDNA. The scheme of the rDNA repeat is shown at the top, 18S, 5.8S, 28S and IGS mark the coding regions and the intergenic spacer of the human rDNA (GenBank Acc. No. U13369). Brackets and abbreviations indicate the location of rDNA regions analyzed in qPCR reactions (Prom, promoter; 28S, rRNA coding region). Predicted MARs are labeled with hexagons. Similar amounts of purified nuclear matrix template DNA were subjected real-time qPCR from normally growing and serum-starved cells. Ct differences between serum-starved and control cells were determined at the three different human rDNA regions and normalized to the Ct differences of the IFNβ promoter. The results of three biological replicate experiments are shown in different colors.
Tip5 is associated with the nuclear matrix and targets the rDNA to the nuclear matrix
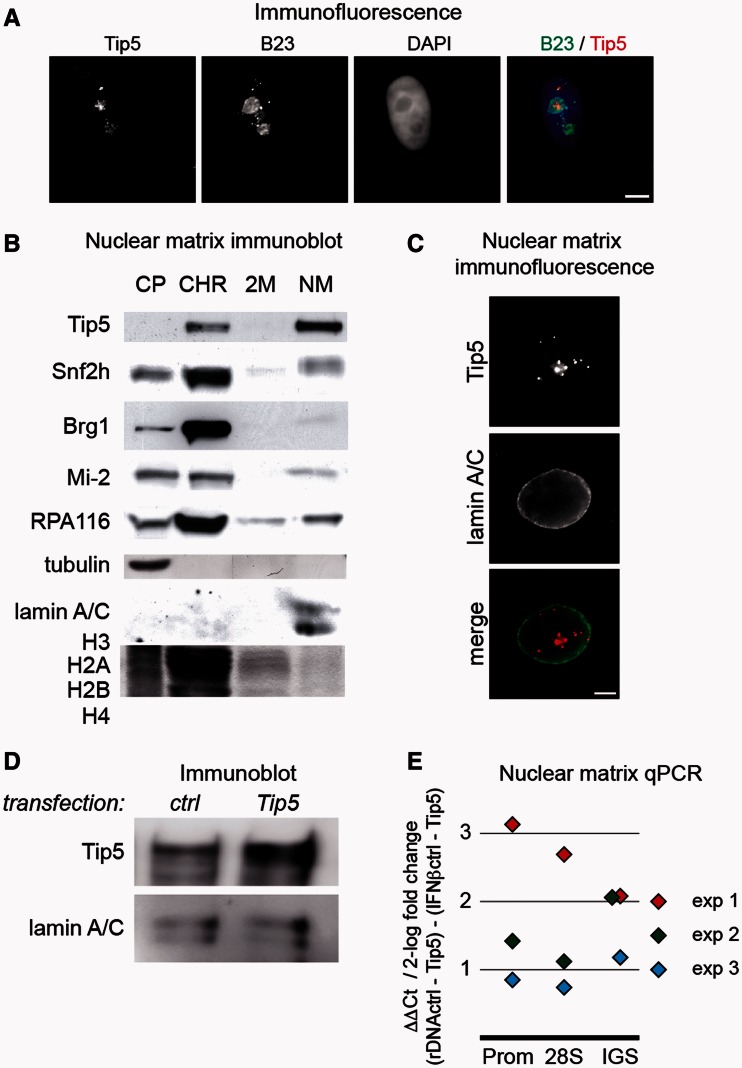
Because the NoRC subunit Tip5 contains several predicted MAR-binding domains (18), we tested its potential to target rDNA to the nuclear matrix. First, the sub-cellular localization of Tip5 was investigated by immunofluorescence. The result showed that Tip5 predominantly, but not exclusively, localizes to nucleoli, which were marked by B23 immunostaining (Figure 2A). Next, we monitored the distribution of Tip5 in the fractions of nuclear matrix preparations. Whole-cell extracts of HEK293 human embryonic kidney cells were fractionated, and the resulting cytoplasmic, chromatin, high-salt wash and nuclear matrix fractions were analyzed by immunoblotting (Figure 2B). The localization of lamin A/C in the matrix fraction, α-tubulin in the cytoplasmic fraction and large and small amounts of histone proteins in the chromatin fraction and wash fraction, respectively, served as controls for the nuclear matrix preparations. The results showed that two pools of Tip5 co-exist in the cell. These pools were found in the chromatin and nuclear matrix fractions, where the majority of the protein is located. In contrast, other chromatin remodeling complex subunits, i.e. Brg1, Snf2h and Mi-2, appeared preferentially in the chromatin fraction. Moreover, the distribution of Pol I in the different fractions demonstrated that not all nucleolar transcription factors are concentrated in the nuclear matrix. As the RNA-binding activity of Tip5 was recently reported (22), we also performed the nuclear matrix assay in the presence of RNaseA to test for RNA-dependent binding. Our results show that the matrix-bound fraction of Tip5 is not sensitive to digestion with RNaseA, but chromatin-bound Tip5 requires RNA for its stable chromatin association (Supplementary Figure S2). In addition to cell fractionation, the association of Tip5 with the nuclear matrix was investigated by immunofluorescence experiments in HeLa cervix carcinoma cells (Figure 2C). Similar to lamin A/C, Tip5 was clearly detectable in situ in the nuclear matrix after extensive DNase I digestion and chromatin extraction. To test whether there is a serum starvation-dependent change in the association of Tip5 with the nuclear matrix, immunoblot experiments were performed. The results illustrate that there is no detectable loss of Tip5 in the nuclear matrix, the vast majority of the protein remains in this fraction (Supplementary Figure S3).
Figure 2.
Tip5 is associated with the nuclear matrix and targets the rDNA to the nuclear matrix. (A) Immunofluorescence experiments show the predominantly nucleolar localization of endogenous Tip5 in HeLa cells. Tip5 was stained with a rabbit polyclonal antibody, the nucleoli with a mouse monoclonal B23 antibody and the nuclear DNA with DAPI. Bars indicate 5 µm. (B) Tip5 is associated with the nuclear matrix. Western blot and Coomassie gel pictures of nuclear matrix preparations from human HEK293 cells. CP, CHR, 2 M and NM indicate cytoplasmic, soluble chromatin, high-salt wash and nuclear matrix fractions, respectively. Tubulin, core histones and lamin A/C served as controls for the CP, CHR/2 M and NM fractions, respectively. Immunoblot detections of Tip5, Snf2h, Brg1, Mi-2 and RPA116, the second largest subunit of RNA polymerase I, are shown. (C) Immunofluorescence detection of the association of Tip5 with the nuclear matrix. Nuclear matrix of HeLa cells was prepared in situ and analyzed by immunofluorescence. Lamin A/C served as control. The bar indicates 5 µm. (D) Immunoblot detection of Tip5 overexpression. HEK293 cells were transfected with full-length, Flag-tagged Tip5 for 72 h, and cellular extracts were prepared and analyzed by immunoblotting. Lamin A/C served to normalize for differences in loading. (E) Real-time qPCR analysis of Tip5 overexpression-induced changes in the nuclear matrix association of rDNA. For details about the experimental set-up see Figure 1B and the text. The results of three biological replicates are shown.
The fact that Tip5 contains a number of DNA-binding domains that potentially bind to MAR sequences, and that the majority of the protein is present in the nuclear matrix fraction suggested that Tip5 could be involved in the nuclear matrix targeting of rDNA. To test this hypothesis, we measured the relative amounts of rDNA in the nuclear matrix fraction of Tip5- and mock-transfected HEK293 cells as described earlier in the text. The immunoblot results show that Tip5 was strongly overexpressed 72 h post-transfection (Figure 2D). DNA quantification revealed that all three regions of the rDNA repeat were enriched in the nuclear matrix fraction, thus indicating that Tip5 targets rDNA to the nuclear matrix. The amount of IGS, coding region and promoter sequences increased ∼2- to 8-fold in the matrix fraction compared with the IFNβ MAR control. There was only a minor difference between the matrix association levels of different rDNA regions within the individual biological replicate experiments (Figure 2E).
DNA-binding features of potential MAR-binding domains of Tip5
Tip5, the large subunit of NoRC, contains a tandem PHD-bromodomain, which is involved in protein–protein interactions, furthermore a variety of nucleic acid-binding domains, e.g. AT-hooks and the TAM domain (Figure 3A), which were proposed to bind MARs (18). To begin with the functional characterization of Tip5’s potential MAR-binding domains, DNA-binding assays were performed. The DNA-binding properties of the TAM domain have been analyzed in our previous study (15); however, the four AT-hooks remained to be investigated. Thus, the four individual AT-hooks and the combination of the first two AT-hooks of Tip5 were expressed and purified as GST-tagged recombinant proteins and subjected to gel retardation assays. The well-characterized second AT-hook of the HMGA1 protein (19) served as a control in the DNA-binding assays (abbreviated as HMGA1 later in the text). Two AT-rich sites from the rDNA IGS were selected in addition to the previously characterized HMGA1-binding site of the IFNβ promoter, and the DNA-binding properties of the purified AT-hooks were tested. The gel retardation experiments showed that (i) all AT-hook domains are bona fide DNA-binding elements; (ii) the individual AT-hooks bound the different sequences with similar affinity; (iii) more than one AT-hook molecule bound to one DNA molecule in the case of the individual AT-hooks, as indicated by the supershifts; (iv) there was only one protein–DNA complex in the case of the larger, double AT-hook AT1 + 2 protein. This suggests that in the double AT-hook construct, both AT-hooks contact the short, 34-bp DNA fragments, not leaving space for an additional AT-hook to bind this DNA (Figure 3B). To monitor the sequence preference of the AT-hook binding, gel retardation assays were performed in parallel using AT- and GC-rich sequences. The results showed that the GC-rich template was less efficiently bound under identical experimental conditions (Supplementary Figure S4A).
The double AT-hook acts as a nucleolar targeting domain
To continue with the characterization of Tip5’s potential MAR-binding domains and to determine the MAR binder with highest affinity, AT-hook–DNA interactions were quantified in microscale thermophoresis experiments. This novel method enables the measurement of molecular interactions in solution based on the monitoring of molecular movement in a thermal gradient (27). We have measured thermally induced kinetics of a fluorescently labeled AT-rich site from the rDNA incubated with a serial dilution of the different AT-hooks. The analysis of the normalized thermophoresis curves provided us with the equilibrium constant concentration values for each AT-hook, when 50% of the DNA was bound by the protein (EC50). The binding constants exhibit clear differences between the individual AT-hooks, displaying slightly weaker affinities than the HMGA1 control. The EC50 value of the double AT-hook AT1 + 2 is higher than the EC50 of the individual AT-hooks, suggesting that both domains contact DNA simultaneously and reveal a binding affinity similar to HMGA1 (Figure 4A). To examine the sequence preference of AT-hook binding in a quantitative manner, the Cy5-labeled AT-rich rDNA sequence was mixed with equimolar amounts of a Cy3-labeled GC-rich DNA fragment, and the EC50 values were determined for AT2 and AT1 + 2 in a competitive-binding assay. The results reinforced the observations of the gel retardation experiments in that the AT-rich sequence was bound with higher affinity (Supplementary Figure S4B).
After identifying the double AT-hook as the strongest putative MAR binder, the nuclear matrix association of this protein domain was investigated in transient transfection experiments (Figure 4B–D). A wild-type and a mutant version of the double AT-hook domain was fused to GFP resulting in the GFP–AT1 + 2 wt and GFP–AT1 + 2mut constructs. In the latter one, the RGR core motifs of both AT-hooks were mutated to the DGD tripeptide that was previously shown to loose DNA-binding activity (33). First, the sub-cellular localization was analyzed in immunofluorescence experiments, which showed that GFP–AT1 + 2 wt predominantly localizes to nucleoli. In contrast, GFP–AT1 + 2mut was evenly distributed in the nucleus (Figure 4B). The results clearly demonstrate that the first two AT-hooks serve as nucleolar targeting module. Surprisingly, nuclear matrix analyses of cellular fractions (Figure 4C) and fixed cells (Figure 4D) showed that despite the in vitro MAR-binding activity and nucleolar targeting, the double AT-hook domain is not sufficient to mediate association with the nuclear matrix.
The TAM domain mediates nuclear matrix association and nucleolar targeting
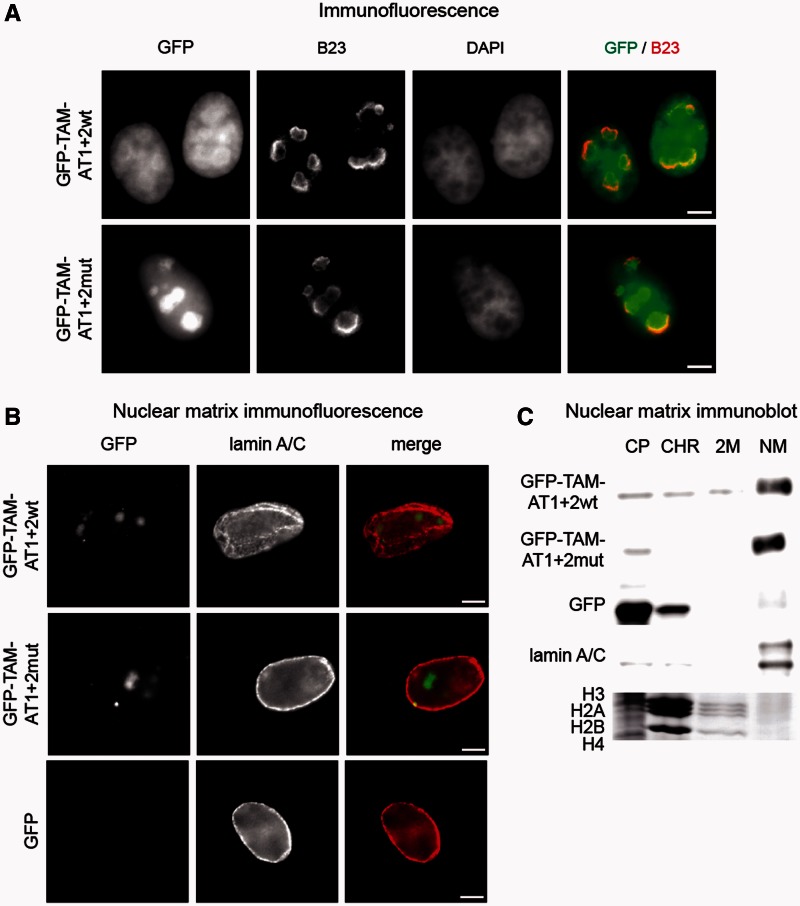
To test whether the other putative MAR-binding domain, the TAM domain, is required to mediate association with the nuclear matrix, the GFP–AT1 + 2 wt and GFP–AT1 + 2mut constructs were extended with this domain resulting in the GFP–TAM–AT1 + 2 wt and GFP-TAM-AT1 + 2mut constructs. Again, the sub-cellular localization of these two GFP-tagged Tip5 proteins was analyzed first. Immunofluorescence experiments showed slightly different sub-nuclear distributions of the proteins. GFP–TAM–AT1 + 2mut was predominantly localized in nucleoli, suggesting that the TAM domain is sufficient for nucleolar targeting, similar to the first two AT-hooks. In contrast, the GFP–TAM–AT1 + 2 wt protein was rather enriched in perinucleolar chromatin and entered only to lesser extent the nucleolus, with higher protein levels detected in the nucleoplasma (Figure 5A). Interestingly, nuclear matrix analyses of fixed cells (Figure 5B) and cellular fractions (Figure 5C) revealed that the TAM domain is necessary and sufficient to mediate the association of the GFP–TAM–AT proteins with the nuclear matrix.
Figure 5.
The TAM domain mediates nuclear matrix association and nucleolar targeting. (A) Immunofluorescence experiments show the sub-nuclear localization of the TAM domain fused to either wild-type or mutated versions of the first two AT-hooks. The GFP-tagged proteins were stained with a rabbit polyclonal GFP antibody, the nucleoli with a mouse monoclonal B23 antibody and the nuclear DNA with DAPI. Bars indicate 5 µm. (B) Immunofluorescence detection of the association of Tip5 proteins with the nuclear matrix. HeLa cells were transfected with GFP or the indicated GFP–Tip5 protein-encoding plasmid DNA for 48 h; nuclear matrices were prepared in situ and analyzed by immunofluorescence. The names of the peptides are shown on the left. Lamin A/C served as control for nuclear matrix preparations. Bars indicate 5 µm. (C) Immunoblot and Coomassie gel pictures of nuclear matrix preparations. HEK293 cells were transfected with GFP or the indicated GFP–Tip5 protein-encoding plasmid DNA for 72 h, and nuclear matrices were prepared. CP, CHR, 2 M and NM indicate cytoplasmic, soluble chromatin, high-salt wash and nuclear matrix fractions, respectively. Core histones and lamin A/C served as controls for the CHR/2 M and NM fractions, respectively.
Even though the nucleolar matrix targeting domain could be assigned to the TAM domain, the sequestering of the rDNA to the nuclear matrix fraction requires a functional Tip5 molecule. DNA quantification after expressing similar levels of the double AT and TAM-AT hook constructs did not exhibit an increase of rDNA in the matrix, as shown for the Tip5 protein, showing that the TAM domain is required for nucleolar matrix targeting; however, additional DNA-binding domains in Tip5 are required for rDNA binding (Supplementary Figure S5).
DISCUSSION
Targeting rRNA genes to the nuclear matrix
rRNA gene repression either by serum starvation or Tip5 overexpression results in a significant enrichment of rDNA in the nuclear matrix. The chromatin remodeling complex NoRC is a key factor required for repression of the gene by repositioning the promoter bound nucleosome and initiates heterochromatin formation by its interaction with HDACs and Dnmts (10,12,14). Here, we show that NoRC regulates higher-order rDNA chromatin organization, in that it is part of the nuclear matrix and induces the recruitment of rDNA to the matrix. Our results suggest that in addition to its well-defined role in regulating local chromatin structures at the rDNA promoter, NoRC is involved also in large-scale chromatin domain organization of the rDNA locus.
The association of mammalian rDNA with the nuclear matrix was shown earlier by several independent laboratories. Genome-scale biochemical (8) and cell biology experiments (9) unambiguously demonstrated the specific enrichment of rDNA in the nuclear matrix. However, neither the transcriptional activity of the nuclear matrix-associated rDNA nor the sequences within the rDNA repeat unit, which mediate the association with the nuclear matrix, were identified in these studies. Regarding these questions, seemingly contradictory models were proposed: Keppel suggested that the entire rDNA repeat unit is associated with the nuclear matrix (34), whereas others found that the coding sequence itself (35) or non-transcribed regions flanking the 47S rRNA coding sequence are predominantly enriched in the nuclear or nucleolar matrix (36–38). With regard to the transcriptional activity of nuclear matrix-associated DNA, it was suggested on one side that active rDNA is associated with the nuclear matrix (34,36), and on the other side that the nuclear matrix contains transcriptionally inactive rDNA (38), which could also represent sequences that are being replicated (39). These discrepancies can be explained largely by differently used terminology and differences in the experimental procedures. The nuclear matrix (6), nuclear scaffold (40) and nuclear skeleton (41) are operational definitions, which are based on biochemical fractionation approaches. The experimental procedures include different endonuclease digestions followed by high-salt or low-salt extractions, or the fractionation is carried out at physiological salt concentration. Remarkably, the concentration of DNase I and the incubation time of the endonuclease digestion vary frequently between the protocols of different laboratories, which may affect the observed association of the rDNA with the nuclear matrix as shown in an initial study (8). The addition of nucleolus isolation steps to the nuclear matrix isolation procedure in particular studies (37,38) further complicates the comparability of the published data about the nuclear matrix association of rDNA. Here, we prepared the nuclear matrix by applying extensive DNase I digestion and high-salt extractions essentially as described in former publications (6,23,24) and named the last insoluble fraction as nuclear matrix, according to the nomenclature of the initial publication (6). It is important to note here that this nuclear matrix does not represent an identifiable sub-nuclear structure (42,43). However, its protein content largely overlaps with that of the nucleoskeleton, a well-defined, intermediate filament-based protein network of the nucleus (44). Moreover, the DNA content of the nuclear matrix represents a fraction of the genome, which is resistant to extensive DNase I digestion, and specific sequences that are enriched in this fraction possess gene regulatory functions (7,45). As active, open chromatin structures are highly accessible to nucleases (46) and active rDNA is largely nucleosome-depleted (47), we suppose that predominantly inactive rDNA repeats are associated with the nuclear matrix. Our results suggest that the entire rDNA repeat can be associated with the nuclear matrix. The relatively moderate effects on the rDNA IGS MAR indicate that this region was probably associated with the nuclear matrix already before the serum starvation; thus, it could represent a nucleation site for the association.
Tip5 is a nuclear matrix-associated protein and targets rDNA to the nuclear matrix
In addition to DNase I inaccessible genomic regions, the nuclear matrix consists of various proteins and RNA molecules. In this study, we demonstrated that the large proportion of the protein resides in the nuclear matrix fraction, and thus identified Tip5 as a nuclear matrix-associated protein. Next, the role of RNA in mediating the association of Tip5 with chromatin was investigated in the nuclear matrix assay. The finding that chromatin-associated Tip5 was sensitive to RNaseA treatment suggests co-existence of two functionally different Tip5 populations in the cell. It is tempting to speculate whether the binding of Tip5 to this mobile chromatin fraction is mediated by the regulatory pRNA, which is transcribed from the rDNA promoter (22), and/or by other RNA species. Tip5, the large, regulatory subunit of the NoRC complex, is a key regulator of rDNA repression (1). Our data on Tip5-dependent nuclear matrix targeting of rDNA indicate that besides its other functions, Tip5 also regulates the DNase I accessibility of rDNA in the nucleus, i.e. nucleolar topology. To our surprise, not only the IGS MAR, but also the Tip5-binding site at the promoter, further a 28S rRNA coding region, where no Tip5 binding occurs, were enriched in the nuclear matrix fraction after overexpression of Tip5. This suggests that in addition to a possible direct nuclear matrix targeting, NoRC-mediated silencing also augments the association of rDNA with the nuclear matrix. We propose a model in which Tip5 plays a key role in recruiting the rDNA to the nuclear matrix and NoRC-mediated heterochromatin formation and chromatin compaction leads to limited DNase I accessibility and the accumulation of large rDNA chromatin domains in the nuclear matrix. Taken together, our results provide insights into the activity-dependent large-scale organization of nucleolar rDNA chromatin and reveal a novel function of Tip5 in this process.
A role for TAM and AT-hook domains in nucleolar targeting and association of Tip5 with the nuclear matrix
Tip5 contains the TAM domain and four minor groove binder AT-hooks, which are supposed to bind MARs and mediate nuclear matrix association (18). To identify Tip5’s protein domain, which shows the highest affinity to a MAR and could thus mediate association with the nuclear matrix, the DNA-binding features of the AT-hooks were investigated in gel retardation and microscale thermophoresis experiments. It was already shown that the TAM domain binds considerably less efficiently to DNA than the AT-hooks (15). Similar DNA-binding affinities were detected for three AT-hooks, whereas one of them bound less efficiently to all three DNA fragments tested. In summary, the comparison of experimentally observed DNA-binding activities of the AT-hooks showed the following order: AT1 < AT2 ≈ AT3 ≈ AT4 < HMGA1 in contrast to the expected AT1 ≈ AT3 < AT2 < AT4 ≈ HMGA1, which is based on the classification described previously (18). Quantification of the DNA-binding efficiencies also revealed that the combination of the first two AT-hooks bound most efficiently to DNA. Thus, this double AT-hook domain along with its mutant was tested for nuclear matrix-binding activity. To our surprise, the result was negative and, therefore, this domain and its mutant were extended with the TAM domain and tested again for nuclear matrix-binding activity. The results revealed that the TAM domain is a nuclear matrix targeting domain, which is in agreement with its proposed role (18). In addition, both the TAM domain and the double AT-hook domain of Tip5 were identified as nucleolar targeting sequences. Finally, the targeting of rDNA to the nuclear matrix by these Tip5 domains was investigated, where we could not detect significant changes in the matrix association of rDNA on overexpression of the different proteins. This result indicates that additional parts of Tip5 are required for the specific enrichment of rDNA in the nuclear matrix. We speculate that overexpression of these domains could result in genome-wide MAR binding, which prevents detectable rDNA-specific targeting effects. In contrast, overexpression of the full-length Tip5 clearly showed such an effect. In summary, our findings suggest a dual role for Tip5’s double AT-hook and TAM domain, targeting the nucleolus and anchoring to the nuclear matrix, and a function for Tip5 in regulating large-scale rDNA chromatin organization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Deutsche Forschungsgemeinschaft [SFB960]; Bayerische Genomforschungsnetzwerk [BayGene] (to G.L.); Elite Network of Bavaria (to K.Z.). Funding for open access charge: German Research Foundation (DFG) within the funding programme Open Access Publishing.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Alexander Brehm and Ingrid Grummt for antibodies against Mi-2, Brg1 and RPA116 and Katharina Filarsky and Philipp Baaske for technical help.
REFERENCES
- 1.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell. Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Verdun D. In: The Nucleolus. Olson MOJ, editor. New York, NY: Springer New York; 2011. pp. 3–28. [Google Scholar]
- 3.Chan PK, Aldrich M, Busch H. Alterations in immunolocalization of the phosphoprotein B23 in HeLa cells during serum starvation. Exp. Cell Res. 1985;161:101–110. doi: 10.1016/0014-4827(85)90494-x. [DOI] [PubMed] [Google Scholar]
- 4.Granick D. Nucleolar necklaces in chick embryo fibroblast cells. II. Microscope observations of the effect of adenosine analogues on nucleolar necklace formation. J. Cell Biol. 1975;65:418–427. doi: 10.1083/jcb.65.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haaf T, Ward DC. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 1996;224:163–173. doi: 10.1006/excr.1996.0124. [DOI] [PubMed] [Google Scholar]
- 6.Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 7.Ottaviani D, Lever E, Takousis P, Sheer D. Anchoring the genome. Genome Biol. 2008;9:201. doi: 10.1186/gb-2008-9-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardoll DM, Vogelstein B. Sequence analysis of nuclear matrix associated DNA from rat liver. Exp. Cell. Res. 1980;128:466–470. doi: 10.1016/0014-4827(80)90083-x. [DOI] [PubMed] [Google Scholar]
- 9.Craig JM, Boyle S, Perry P, Bickmore WA. Scaffold attachments within the human genome. J. Cell Sci. 1997;110(Pt. 21):2673–2682. doi: 10.1242/jcs.110.21.2673. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Längst G, Grummt I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006;25:5735–5741. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Santoro R, Koberna K, Grummt I. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 2005;24:120–127. doi: 10.1038/sj.emboj.7600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 13.Strohner R, Németh A, Nightingale KP, Grummt I, Becker PB, Längst G. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol. Cell. Biol. 2004;24:1791–1798. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I. NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emelyanov AV, Vershilova E, Ignatyeva MA, Pokrovsky DK, Lu X, Konev AY, Fyodorov DV. Identification and characterization of ToRC, a novel ISWI-containing ATP-dependent chromatin assembly complex. Genes Dev. 2012;26:603–614. doi: 10.1101/gad.180604.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Németh A, Strohner R, Grummt I, Längst G. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 2004;32:4091–4099. doi: 10.1093/nar/gkh732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 20.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 21.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 22.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 23.He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J. Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes JC, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vecerova J, Koberna K, Malinsky J, Soutoglou E, Sullivan T, Stewart CL, Raska I, Misteli T. Formation of nuclear splicing factor compartments is independent of lamins A/C. Mol. Biol. Cell. 2004;15:4904–4910. doi: 10.1091/mbc.E04-07-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seither P, Iben S, Grummt I. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J. Mol. Biol. 1998;275:43–53. doi: 10.1006/jmbi.1997.1434. [DOI] [PubMed] [Google Scholar]
- 27.Baaske P, Wienken CJ, Reineck P, Duhr S, Braun D. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed. Engl. 2010;49:2238–2241. doi: 10.1002/anie.200903998. [DOI] [PubMed] [Google Scholar]
- 28.O’Mahony DJ, Xie WQ, Smith SD, Singer HA, Rothblum LI. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J. Biol. Chem. 1992;267:35–38. [PubMed] [Google Scholar]
- 29.Yamamoto K, Yamamoto M, Hanada KI, Nogi Y, Matsuyama T, Muramatsu M. Multiple protein-protein interactions by RNA polymerase I-associated factor PAF49 and role of PAF49 in rRNA transcription. Mol.Cell. Biol. 2004;24:6338–6349. doi: 10.1128/MCB.24.14.6338-6349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seither P, Zatsepina O, Hoffmann M, Grummt I. Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma. 1997;106:216–225. doi: 10.1007/s004120050242. [DOI] [PubMed] [Google Scholar]
- 31.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 32.Kramer JA, Singh GB, Krawetz SA. Computer-assisted search for sites of nuclear matrix attachment. Genomics. 1996;33:305–308. doi: 10.1006/geno.1996.0198. [DOI] [PubMed] [Google Scholar]
- 33.Bourachot B, Yaniv M, Muchardt C. The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol. Cell. Biol. 1999;19:3931–3939. doi: 10.1128/mcb.19.6.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keppel F. Transcribed human ribosomal RNA genes are attached to the nuclear matrix. J. Mol. Biol. 1986;187:15–21. doi: 10.1016/0022-2836(86)90402-x. [DOI] [PubMed] [Google Scholar]
- 35.Schwarzacher HG, Mosgoeller W. Ribosome biogenesis in man: current views on nucleolar structures and function. Cytogenet. Cell Genet. 2000;91:243–252. doi: 10.1159/000056853. [DOI] [PubMed] [Google Scholar]
- 36.Smith HC, Rothblum LI. Ribosomal DNA sequences attached to the nuclear matrix. Biochem. Genet. 1987;25:863–879. doi: 10.1007/BF00502606. [DOI] [PubMed] [Google Scholar]
- 37.Stephanova E, Stancheva R, Avramova Z. Binding of sequences from the 5′- and 3′-nontranscribed spacers of the rat rDNA locus to the nucleolar matrix. Chromosoma. 1993;102:287–295. doi: 10.1007/BF00352403. [DOI] [PubMed] [Google Scholar]
- 38.Bolla RI, Braaten DC, Shiomi Y, Hebert MB, Schlessinger D. Localization of specific rDNA spacer sequences to the mouse L-cell nucleolar matrix. Mol. Cell. Biol. 1985;5:1287–1294. doi: 10.1128/mcb.5.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Little RD, Platt TH, Schildkraut CL. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirkovitch J, Mirault ME, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 41.Jackson DA, Yuan J, Cook PR. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J. Cell Sci. 1988;90(Pt. 3):365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- 42.Hancock R. A new look at the nuclear matrix. Chromosoma. 2000;109:219–225. doi: 10.1007/s004120000077. [DOI] [PubMed] [Google Scholar]
- 43.Pederson T. Half a century of “the nuclear matrix”. Mol. Biol. Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks'. Nat. Rev. Mol. Cell. Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 45.Gavrilov AA, Zukher IS, Philonenko ES, Razin SV, Iarovaia OV. Mapping of the nuclear matrix-bound chromatin hubs by a new M3C experimental procedure. Nucleic Acids Res. 2010;38:8051–8060. doi: 10.1093/nar/gkq712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves R. Transcriptionally active chromatin. Biochim. Biophys. Acta. 1984;782:343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- 47.Hamperl S, Wittner M, Babl V, Perez-Fernandez J, Tschochner H, Griesenbeck J. Chromatin states at ribosomal DNA loci. Biochim. Biophys. Acta. 2013;1829:405–417. doi: 10.1016/j.bbagrm.2012.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.