Abstract
The structure and function of conserved motifs constituting the apex of Stem I in T-box mRNA leaders are investigated. We point out that this apex shares striking similarities with the L1 stalk (helices 76–78) of the ribosome. A sequence and structure analysis of both elements shows that, similarly to the head of the L1 stalk, the function of the apex of Stem I lies in the docking of tRNA through a stacking interaction with the conserved G19:C56 base pair platform. The inferred structure in the apex of Stem I consists of a module of two T-loops bound together head to tail, a module that is also present in the head of the L1 stalk, but went unnoticed. Supporting the analysis, we show that a highly conserved structure in RNAse P formerly described as the J11/12–J12/11 module, which is precisely known to bind the elbow of tRNA, constitutes a third instance of this T-loop module. A structural analysis explains why six nucleotides constituting the core of this module are highly invariant among all three types of RNA. Our finding that major RNA partners of tRNA bind the elbow with a same RNA structure suggests an explanation for the origin of the tRNA L-shape.
INTRODUCTION
T-box leaders are 5′ regulatory elements found in some bacterial mRNAs (1–3). Their interaction with cellular tRNAs, which determines whether the messenger will be fully transcribed and/or translated, has been shown to involve two specific regions of the tRNA: the anticodon and the 3′ acceptor arm (2,4). The anticodon interacts with an internal loop (called Specifier) present at the bottom of Stem I, the first major stem–loop of the leader. The acceptor arm may interact with a downstream antiterminator stem–loop if it is not aminoacylated (4). Although other interactions are thought to occur based on experimental assays, the nature of these interactions remains unclear (2,3). Moreover, several conserved motifs on the T-box leader, in particular an AG-box and a GNUG-box on the apex of Stem I (5), have no known function yet (2,3).
Based on a structural analysis in which the apex of Stem I is compared with modules of the ribosome and the RNAse P, we show that the AG- and GNUG- motifs are parts of a single functional structure consisting of a head-to-tail double T-loop module, the role of which is to bind the elbow of tRNAs through a platform–platform stacking interaction. Consistent with this finding, a molecular model shows that the structural organization of Stem I allows the simultaneous binding of both the anticodon and the elbow of a tRNA. The resulting improved model of tRNA–T-box interaction explains the effects of mutations in Stem I and tRNA observed in mRNA in vivo and in vitro expression systems.
Taken together, our results reveal that three major classes of RNA (the ribosome, RNAse P and T-box leaders) use a same structural motif to bind the elbow of tRNA, a motif that was thus far only described in RNAse P. This finding highlights a fundamental reason for the presence of the G19:C56 platform, which is a signature of the tRNA L-shape.
MATERIALS AND METHODS
T-box conservation analysis
The T-box Stem I alignment was assembled as follows. We sought T-boxes in the 5′ upstream sequences of bacterial genes retrieved from NCBI bacterial genomes using the RNAMotif program (6) and a T-box descriptor (Supplementary Text S2) comprising the basis of Stem I and the rho-independent terminator at the 3′ end of the T-box. No sequence constraint was applied to the apical loop. RNAMotif hits were filtered to retain only those located upstream of aminoacyl-tRNA synthetase genes, which provided a collection of 131 T-boxes. The Muscle program (7) was initially used to refine the alignment; however, it generated disorders within the helical part of the alignment. Starting from the RNAMotif output, a manual alignment of the apex region of Stem I was performed instead, ensuring that it would fit the secondary structure model and locations of the AGNNA and UGNNA motifs (Figure 3A). The final alignment is available as Supplementary Data S1. Sequence conservation at the apex of Stem I was displayed using Weblogo (8).
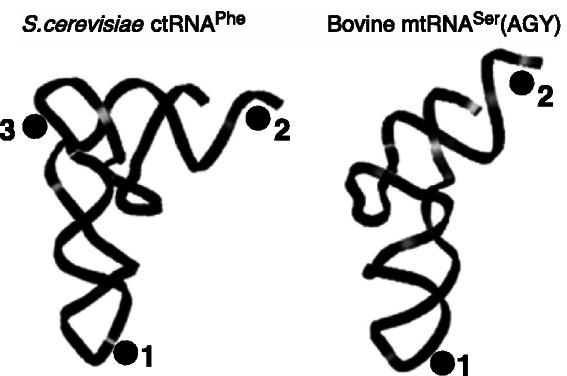
Figure 3.
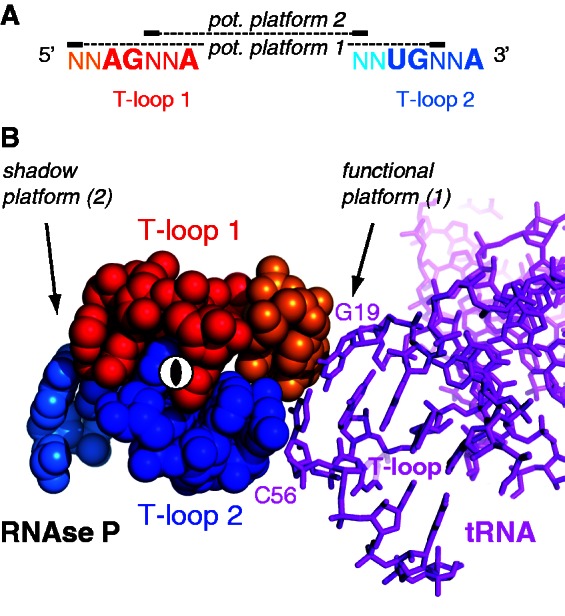
Platform locations in the sequence pattern and 3D structure of the head-to-tail double T-loop module. (A) Sequence pattern of all known head-to-tail double T-loop module. The two potential (pot.) platforms are constituted by a base pair between the penultimate residue preceding one T-loop and the central residue of the other T-loop. Only one platform is functional in all known structures. (B) Platform–platform interaction between the functional platform of the head-to-tail double T-loop module of T. maritima RNAse P and the G19:C56 platform of the elbow of tRNAPhe [pdb 3Q1Q (12)]. The two T-loop motifs are related by a 2-fold rotation axis (shown at the center). The cyan residue of the shadow platform is included within P12 stem–loop (Figure 1B). The color code is identical to that of Figure 1.
Modeling of ProI Stem I
ProI Stem I was chosen for modeling among other T-box leaders because a structure of the U,CGC C-loop motif within Stem Ia (Figure 4A) was available (residues 2659–2663 and 2705–2707 of Deinococcus radiodurans 23S rRNA, pdb 1NJP). The structure of the L1 stalk of Thermus thermophilus 23S rRNA (pdb 1VSA) was used as a starting material because its apex possesses a topology similar to that of Stem I (Figure 1A and D). Bases were substituted and structural changes implemented using the Assemble software (15) to accommodate the ProI Stem I sequence while approximately fitting a tRNA. Other structural motifs used in the construction are as follows: Thermotoga maritima RNAse P (pdb 3Q1Q: head-to-tail double T-loop module) and Bacillus subtilis tyrS T-box leader (pdb 2KZL: structure of the k-turn and Specifier loop domains). The resulting model of Stem I is shown in Figure 5B. Because no tRNApro structure was available, a tRNAPhe (pdb 1EHZ) is shown instead in Figure 5B and in the Supplementary pdb Files (Model of Stem I: Supplementary Data S2; tRNAPhe pdb 1EHZ: Supplementary Data S3). A final optimization of the contacts with this non-cognate tRNA was not realized, and Stem I was left in a slightly ‘open’ conformation.
Figure 4.
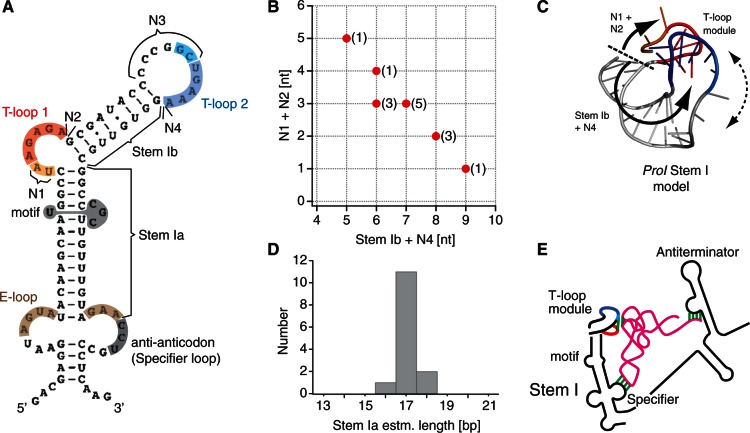
Analysis of Stem I structures in T-box leaders. (A) Definition of structural parameters determined in 14 experimentally assessed T-box leaders (Supplementary Table S1). The secondary structure shown is from ProI Stem I, adapted from (22) (Stem Ia = 17, including a non-canonical motif and an E-loop; Stem Ib = 8; N1 = 2; N2 = 0; N3 = 6; N4 = 0). (B) Correlation between (Stem Ib + N4) and (N1 + N2). Note that N4 and N2 almost do not contribute to the correlation (Supplementary Table S1). (C) Apex of B. subtilis ProI Stem I model (‘Materials and Methods’ section), from top. The two curved arrows represent the main axes of the structural contributions of the parameters of the correlation in (B). The dotted double arrow indicates a possible adjustment of the orientation of the platform as a function of the parameters of the correlation in (B). (D) Distribution of the estimated length of Stem Ia. See Supplementary Table S1 for the method of estimation. (E) Improved model of the interaction between a tRNA and a T-box leader, with the head-to-tail double T-loop module constituting a wedge for the elbow of the tRNA. The color code is identical to that of Figure 1.
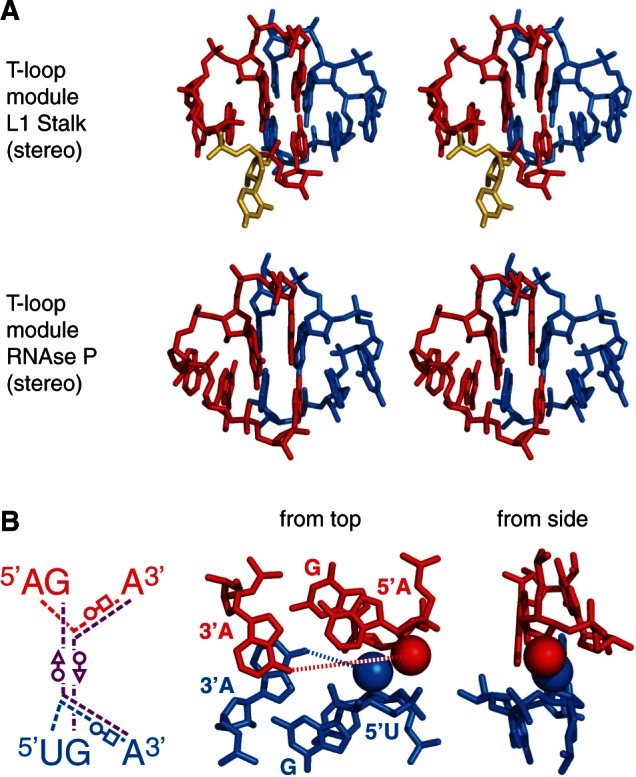
Figure 1.
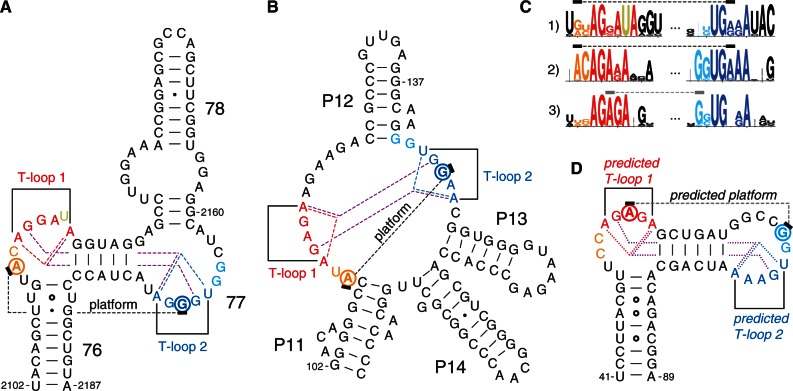
Secondary structures of RNA elements displaying a crystallographically established (A and B) or a predicted (D) head-to-tail double T-loop module. T-loops are shown in red (first T-loop along the strand, T-loop 1) and blue (T-loop 2). The two residues preceding a T-loop are shown in orange (T-loop 1) and cyan (T-loop 2). The dotted lines show the (established of predicted) base pairs defining the core of the T-loop module (detailed in Figure 2B) and the functional platform (residues outlined with a circle). (A) B. subtilis L1 stalk (23S rRNA). The established structure and numbering are from the crystal structure of T. thermophilus 23S rRNA (pdb 1VSA) (9). (B) T. maritima RNAse P: region of the head-to-tail double T-loop module (P11–P14) (10,11). The tertiary interactions are from the crystal structure pdb 3Q1Q (12). (C) Sequence conservation encompassing the two T-loops of the structures in A, B and D: 1) L1 stalk (23S rRNA), 2) J11/12–J12/11 module of RNAse P and 3) apex of Stem I in T-box leaders. Sequence logos (highest value = 2 bits) were produced using the Weblogo interface (8) while using the following sequence alignments: 23S rRNA: primary alignments of 592 sequences from three phylogenetic domains, from the Comparative RNA Web site (13); bacterial RNAse P, class A: 340 sequences from the Ribonuclease P database (14); and T-box Stem I: alignment generated from 131 T-box sequences (‘Materials and Methods’ section). (D) B. subtilis tyrS T-box leader: apex of Stem I. Secondary structure (solid lines only) and base numbering according to (2).
Figure 5.
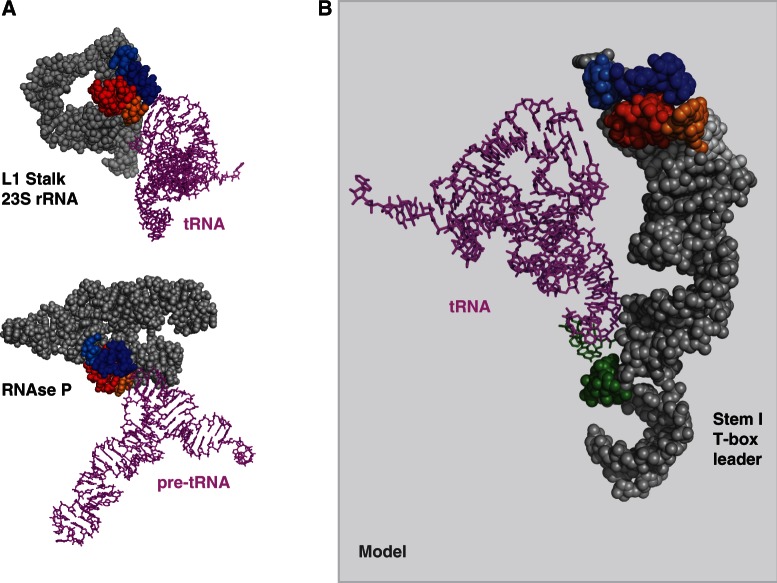
Crystallographically established (A) and model (B) of tRNA–RNA binding in which a stacking interaction between the G19:C56 base pair platform defining the elbow of a tRNA and a platform of a head-to-tail double T-loop module is involved. (A) L1 stalk–tRNA (pdb 1VSA) and RNAse P–pre-tRNA (pdb 3Q1Q) interactions. Only a fragment of RNAse P is shown. (B) Model of B. subtilis ProI Stem I–tRNA interaction (see ‘Materials and Methods’ section and Supplementary Data S2 and S3). The secondary structure of Stem I is shown in Figure 4A. A tRNAPhe (pdb 1EHZ) is shown instead of a tRNAPro (for which no structure is available). A final optimization of the contacts with this non-cognate tRNA was not realized, and Stem I was left in a slightly ‘open’ conformation. The color code is identical to that of Figure 1.
RESULTS
The L1 stalk suggests the organization of the apex of Stem I in T-box leaders
A comparison of the secondary structures of the apex of Stem I of the well-studied B. subtilis TyrS T-box leader (2) with bacterial 23S rRNA reveals a striking homology between this apex and the L1 stalk (Figure 1A and D): the bulge loop containing the AG motif and the apical loop containing the GNUG motif are organized similarly to helices 76 and 77, respectively. An examination of a high-resolution crystal structure of the L1 stalk of T. thermophilus 16) (no such crystal structure is available for B. subtilis) shows that the stem of helix 77 causes the bulge loop between residues 2162 and 2173 to fold back and form a complex interaction with the bulge loop of helix 76 (residues 2111–2119). While helix 78 has no equivalent in Stem I, the crystal structure shows that this helix does not interfere with the topology of the loop–loop interaction, implying that it may also characterize the apex of Stem I. A detailed examination of the loop–loop interaction shows that residues 2167–2171 (helix 77) form a T-loop (T-loop 2 in Figure 1A), which binds the loop of helix 76 in the opposite direction. This second bulge loop also includes a well-characterized T-loop (T-loop 1 in Figure 1A), from residues 2114 to 2119. An extra residue (almost always a U, shown in gold in Figures 1A, C and 2A) is present in between the fourth and the fifth nucleotide. It is involved in an outer interaction with helix 78, and virtually does not alter the characteristic shape of the T-loop (Figure 2A, top). The structure generated by the kissing interaction between these two T-loops consists of a module we call the head-to-tail double T-loop module (Supplementary Figure S1). The apical residue (2169) of T-loop 2 forms a platform with the penultimate residue (2112) preceding T-loop 1, on which the conserved G19:C56 base pair constituting the elbow of tRNA can stack, an interaction that is involved in the removal of deacylated tRNA form the ribosome (17,18). This suggests that the apex of Stem I in T-box leaders is involved in the binding of the elbow of tRNAs as well. The issue is examined further below.
Figure 2.
Core structure of the head-to-tail double T-loop module. (A) Stereo view of the head-to-tail double T-loop module of 23S rRNA L1 stalk [pdb 3U4M (16)] and RNAse P [pdb 3Q1Q (12)]. (B) Core of the head-to-tail double T-loop module, involving the highly conserved AG..A and UG..A residues of the two T-loops (Figure 1C). The two base triple (Leontis/Westhof nomenclature), stacking on each other, are found in both 23S rRNA L1 stalk and RNAse P crystal structures. The structure on the right (pdb 3Q1Q) outlines the oxygen atoms of the hydroxyl groups of both A5′ and U5′ closing residues (shown as van der Waals spheres). The distance between this oxygen atom and A3′ N6 of the corresponding T-loop (dotted lines) is 8.78 Å (L1 stalk) and 9.25 Å (RNAse P) for the A5′–A3′ closing residues, and 6.13 Å (L1 stalk) and 5.46 Å (RNAse P) for the U5′–A3′ closing residues. The color code is identical to that of Figure 1.
Sequence conservation analysis (Figure 1C) reveals that helix 77 and the apical loop of Stem I share a similar conservation pattern comprising T-loop 2, suggesting that this structural motif may also be present on the apical loop of Stem I. Another striking conservation analogy concerns the residues forming T-loop 1 on helix 76 and the AG motif on the bulge loop of Stem I. This motif, however, does not include an extra uridine residue, which makes it a typical motif found in T-loops (19). In addition, the topology of the strands connecting the two potential T-loops on the apex of Stem I is similar to that of the L1 stalk (Figure 1A and D). This suggests that this apex may also be characterized by the presence of a head-to-tail double T-loop module.
A head-to-tail double T-loop module is also present in RNAse P
Because the structural analogy between the L1 stalk and the apex of Stem I in T-box leaders was not complete, we sought additional clues in other RNA structures. We realized that a structure described as the J11/12–J12/11 module in RNAse P (10,11) matches the description of the head-to-tail double T-loop module of the L1 stalk (Figure 2A). Although the strands connecting the two T-loops in the RNAse P module are folded differently from those of Stem I (Figure 1), the conservation pattern of these T-loops shares even more similarities with the pattern found on the apex of Stem I (Figure 1C). In particular, this module is constituted by two canonical T-loops, without extra residue. Just as in the L1 stalk, this structure generates a base pair platform enabling the docking of the elbow of tRNA (12), reinforcing the assumption that the apex of Stem I has this function. A comparison of all three patterns of conservation (Figure 1C) reveals a highly invariant motif of the kind AGNNA … UGNNA encompassing the two T-loops (Figures 2B and 3A). While each of the five residues of any T-loop does show some preferences for certain bases, T-loops are usually best characterized by their structure (19). This conservation, therefore, suggests the presence of higher structural constraints connected with the formation of the module itself. A high-resolution crystal structure of the L1 stalk of T. thermophilus (16) and a crystal structure of the RNAse P of T. maritima (12) enables a detailed characterization of the core of the head-to-tail double T-loop module. The structure of this core, identical in both molecules, precisely involves all six residues with the highest overall conservation (Figure 1C). It is constituted by two base triples stacking on each other (shown by dashed lines in Figure 1A, B and D; detailed interactions are shown in Figure 2B, left). One base triple consists of a trans W.-C./Hoogsteen base pair between the closing residues of a T-loop (A:A or U:A), while the A3′ closing residue makes an additional trans W.-C/sugar edge base pair with the conserved G in second position of the other T-loop. The other base triple is generated symmetrically, both T-loops being related by a 2-fold rotation axis. In this way, each A3′ closing residue of a T-loop is filling the characteristic gap (19,20,21) of the other T-loop (Figure 3B and Supplementary Figure S1). A structural explanation for why the two T-loops have a different 5′ closing nucleotide (5′U or 5′A) lies in a potential clash between their 2′ hydroxyl groups due to the strong interpenetration of the two loops. This clash is resolved if one closing base pair (A:A) is wider than the other one (U:A) (Figure 2B, right). Because of the symmetry, each side of the head-to-tail double T-loop module potentially displays a platform for tRNA docking, which is constituted by a base pair between the central nucleotide of one T-loop and the penultimate residue (5′–3′) preceding the first nucleotide of the other T-loop (Figure 3A). In both RNAse P and L1 stalk, only platform 1 is functional, leaving platform 2 as a ‘shadow’ platform (Figure 3B).
Structural constraints exerted on the six residues forming the core of all known head-to-tail double T-loop modules may explain why they are highly conserved in both RNAse P and L1 stalk. The presence of this peculiar conservation pattern on the apex of Stem I in T-box leaders is therefore in itself a strong indication of the presence of the module on this RNA.
The apex of Stem I in T-box leader is docking the elbow of tRNA
The above structural comparison of the apex of Stem I of T-box leaders with the L1 stalk and RNAse P suggests that it bears a head-to-tail double T-loop module constituting a wedge for the elbow of tRNA. This interaction, which would add to the two established ones involving the anticodon and the 3′ acceptor arm (2,3), is supported by several results. The formation of the T-loop module itself on the apex is possible only if some structural requirements are fulfilled. An analysis of the secondary structure of experimentally assessed T-box leaders (Figure 4A and Supplementary Table S1) reveals that a distance conservation rule is narrowly observed, which is highlighted by a correlation (Figure 4B): when the strands connecting T-loop 1 to the base of Stem Ib (N1 and N2) are long, the shortest path leading to the 3′ side of T-loop 2 (Stem Ib + N4) is short, and vice versa. Thus, the longest identified Stem Ib brings T-loop 2 back onto the top of T-loop 1 owing to the curvature of the double helix, enabling the formation of the module almost without any adjustment (N1 = 1; N2 = 0). This geometrical feature can be best visualized on our molecular model (Figure 4C). A reanalysis of published MgCl2 structure probing gels also supports the presence of the module (Supplementary Figure S2A): both predicted T-loops are resistant to Mg2+, whereas nucleotides predicted to be single stranded are overall more prone to cleavage, with the notable exception of the two residues preceding T-loop 2 (included in N3). The penultimate residue before T-loop 2 is part of potential platform 2 (Figure 3A), which is thus likely to be the functional platform of Stem I because a well-structured platform is expected to be more resistant to Mg2+ cleavage. Furthermore, a comparison between N1 and N3 residues clearly designates platform 2 as the functional one. Gel structure probing is thus consistent with the presence of the module, and provides an important clue about the actual functional platform. Note that tRNA binding does not reduce the cleavage propensity of the predicted functional platform, which is already robust against Mg2+ cleavage without tRNA (Supplementary Figure S2A, right). Another feature that may help discriminate the functional platform in Stem I is the base composition of known functional and shadow platforms. A comparison of the two platforms does not show any clear conservation signal that would be typical of the functional platform in both L1 stalk and RNAse P (Figure 1C). However, a clear distinction is revealed in L1 stalk by establishing a statistics of the purine/pyrimidine (R/Y) composition of the residues constituting the two platforms, a result that can be expressed in terms of normalized information (Table 1): both residues of the functional platform (platform 1) are almost exclusively purines (information = 0.08), whereas this preference is clearly not as strong in the shadow platform (information = 0.82). Remarkably, the results are inverted in Stem I (T-box leader), for which it is platform 2 that has the lowest information (all platforms are R:R). In conjunction with the fact that some Stem I cannot form any potential platform 1 (N1 < 2), this result thus also points out platform 2 as the functional platform in T-box leaders (structural constraints show that the occurrence of either possibility is highly unlikely, see below). The unclear situation of RNAse P (both informations are at ∼0.2) may be due to a different structural context around this T-loop module. In particular, a G residue of the shadow platform is involved in the first base pair of stem P12 (Figure 1B), which might explain why it is conserved.
Table 1.
Purine/pyrimidine (R/Y) composition of the two potential platforms in head-to-tail double T-loop modules of RNAse P, L1 stalk (23S rRNA) and T-box leaders
| Base pair | RNAse P |
L1 stalk (23S rRNA) |
Stem I (T-box) |
|||
|---|---|---|---|---|---|---|
| Pot. platform 1 | Pot. platform 2 | Pot. platform 1 | Pot. platform 2 | Pot. platform 1 | Pot. platform 2 | |
| R:R | 304 | 313 | 578 | 296 | 48 | 131 |
| R:Y | 32 | 27 | 10 | 178 | 6 | 0 |
| Y:R | 0 | 0 | 3 | 84 | 18 | 0 |
| Y:Y | 0 | 0 | 0 | 30 | 52 | 0 |
| Total | 336 | 340 | 591 | 588 | 124 | 131 |
| Information | 0.23a | 0.20 | 0.08a | 0.82 | 0.84 | 0.00b |
The numbers of each potential (pot.) platform (see Figure 3A for the definition) were established from the same alignments used in the T-loop conservation analysis (Figure 1C). The total numbers of the two platforms in each structure are not identical owing to the occasional unavailability of some residues (involved in other interactions) or some alignment issues. The information (I) associated with each platform is calculated as I = −Σ pi log4 pi, where pi = Total/no. platform i (platform = RR, RY, YR or YY).
Detailed results are shown on Supplementary Table S2.
aKnown functional platform (platform 1) of RNAse P and L1 stalk.
bPredicted functional platform (platform 2) of Stem I in T-box leaders.
A constraint implied by the above results concerns the distance between the T-loop module and the anti-anticodon, which must match the (highly conserved) distance between the elbow and the anticodon of a tRNA. Figure 4D shows that the length of Stem Ia, which was estimated from a uniform set of rules to include motifs of unknown structure (Supplementary Table S1), is narrowly centered around an average value of 17 ± 1 bp, which is of the order of the expected distance.
All published results of biochemical assays we are aware of are also in full agreement with our structural interpretation. In vitro and in vivo antitermination assay experiments in which tRNA modifications were investigated show that, in contrast to other parts of the tRNA, the length of the anticodon stem is critical: while a single base pair insertion results in a similar activity, larger insertions have a dramatic effect on antitermination efficiency (23), as predicted by our analysis. Also in agreement with our model, similar effects are observed when the G19:C56 base pair constituting the elbow of the tRNA is altered (24,25), or when critical residues forming the T-loop module of Stem I are substituted, which would prevent the platform–platform interaction from occurring (Supplementary Text S1). Taken together with our findings, these published results show that the wedge is critical for antitermination (Figure 4E).
To assess the plausibility of the simultaneous binding of a tRNA elbow and anticodon to Stem I, a molecular model of B. subtilis ProI Stem I (Figure 4A for the secondary structure) was built from an existing structure of T. thermophilus L1 stalk (pdb 1VSA), which provided the general topology. The head-to-tail double T-loop module was from T. maritima RNAse P (pdb 3Q1Q), and could fit the structure with only minor structural adjustments. Structural changes were implemented and the interactions optimized using the Assemble software (15) to accommodate the ProI Stem I sequence while approximately fitting a tRNA.
The model (Figures 4C and 5B; Supplementary Data S2 and S3) was obtained with only smooth adjustments, with the exception of a kink at the junction between Stem Ia and Stem Ib (Supplementary Figure S2B, left), which was necessary to bend the T-loop module toward the anti-anticodon so that it can accommodate a tRNA. In vitro experiments with a GlyQS T-box leader do suggest the presence of a kink stabilized by Mg2+ at this specific position (U50 in GlyQS T-box leader numbering), as it is exceptionally prone to Mg2+ cleavage, in a concentration-dependent manner (Supplementary Figure S2B, right). Following a similar MgCl2 dependence, tRNA–GlyQS T-box leader binding experiments and in vitro transcription read-through assays (24) show that high concentrations of MgCl2 are necessary to observe significant tRNA binding and transcription antitermination, respectively. With the only assumption that the relation between the kink and the Mg2+ experiments equally apply to GlyQS and ProI Stem I, our model provides an explanation for the Mg2+ binding dependence.
The model clearly shows that platform 2 is the functional platform docking the elbow of tRNA in T-box leaders, confirming a prediction based on information analysis (Table 1) and Mg2+ structure probing (Supplementary Figure S2). Remarkably, the structure of Stem I can resolve two concomitant constraints: the distance between the anti-anticodon and the platform, and the orientation of the platform itself, which depends on the length of Stem Ia. We noticed that non-canonical motifs are almost always present in Stem Ia (Supplementary Table S1). In conjunction with local adjustments of the orientation of the T-loop module (as determined by the parameters of Figure 4B), these may contribute to correcting the phase of the helix so that the anti-anticodon and the platform are aligned. For instance, ProI Stem Ia has a C-loop motif (Figure 4A), which is known to increase the helical twist between the two Watson–Crick base pairs flanking the motif, and at the same time provide some local flexibility (26). Overall, it appears that Stem I is a highly constrained structure in which delicate adjustments position the docking platform with respect to the anti-anticodon.
In summary, our results consistently indicate that the apex of Stem I in T-box leaders displays a head-to-tail double T-loop module with a platform for tRNA docking (Figures 4E and 5B; see also Supplementary Text S2).
DISCUSSION
Our study reveals that a recurrent structural motif, the head-to-tail double T-loop module, is used in three classes of RNA to bind the elbow of tRNAs: the L1 stalk of the ribosome, RNAse P and Stem I of T-box leaders (Figure 5). Although no structure is available yet that could further demonstrate the presence of the module on the apex of Stem I, we believe that our present analysis makes the case strong enough to claim that the module is indeed present on this apex. Thus far, this special T-loop arrangement was only noticed in RNAse P, where it is known as the J11/12–J12/11 module (10,11), while the module of the L1 stalk is described for the first time in the present article based on existing crystal structures.
The widespread distribution of a module with such a peculiar function suggests that the stabilization of the elbow is a general requirement for proper tRNA handling by the interacting RNA. This interaction, which is at the same time unspecific and universal to most tRNAs, appears to complement those involving the anticodon and the 3′ acceptor arm (Figure 6).
Figure 6.
Universal contact regions of a canonical tRNA (left) and a mitochondrial tRNA (right). 1 = anticodon (contact through base pairing); 2 = acceptor arm (contact through base pairing); 3 = platform (contact through stacking). Adapted from (27) [see also (28)].
It may seem surprising that a large structural motif has emerged repeatedly in evolution for the purpose of binding the tRNA elbow. Two properties of the head-to-tail double T-loop module may explain why it is most suitable for tRNA docking. First, the core of the structure is a very dense packing made up of highly conserved residues that are almost exclusively purines (Figure 2B). This certainly contributes to generate a stable platform, most suitable for forming a strong stacking interaction with the G19:C56 mirror platform of the tRNA, also known for its stability (27). Second, the head-to-tail double T-loop module is versatile because two platforms (related by a 2-fold rotation axis) are potentially available. Both L1 stalk and RNAse P selected platform 1 as the functional platform, whereas, according to our analysis, Stem I selected platform 2.
Because a T-loop reaches a high stability only when an outer residue intercalates into the characteristic gap present between the fourth and the fifth nucleotide (21), the remarkable reciprocal gap complementation occurring between the two T-loops of the head-to-tail double T-loop module, clearly visible in Figure 3B, is worth noticing. This complementation does not occur in a related double T-loop arrangement formerly described as the nested double T-loop (29), which also has a platform for trans stacking (Supplementary Figure S1). These two types of T-loop arrangements suggest that, although T-loops are still involved in several other kinds of structures, they do have optimal properties to generate platforms for trans stacking. In the case of T-box leaders, a rationale for the presence of a head-to-tail double T-loop module is that the docking platform can be oriented around the axis defined by Stem Ia by an adjustment of the two correlated variables of Figure 4B. This may enable an alignment of the platform with the anti-anticodon so that Stem I can fit a tRNA. Interestingly, the kink introduced in the ProI Stem Ia model contributes to optimally position Stem Ib at approximately 90° from Stem Ia (Supplementary Figure S2B).
The universality of the docking interaction described here may account for the origin of the elbow (and thus the L-shape) of tRNA. It has been suggested that the primary function of the L-shape is to enable the 3′ ends of A- and P-site-bound tRNAs to meet during peptide bond formation (30). A bend on the tRNA is indeed required to compensate for the angle between the anticodon stems of these tRNAs, which is generated by a kink on the mRNA between the A and P sites (31). However, other shapes besides the L-shape could work just as well for that purpose, such as the simpler ‘boomerang-like’ shape found in mitochondrial tRNAs that are lacking the D-loop–T-loop kissing interaction, for instance 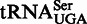 (28). Many mitochondrial tRNAs are indeed characterized by the absence or a reduced size of the D-loop and/or T-loop (32), which implies that they do not have a platform for docking (Figure 6). Consistent with this fact, both L1 stalk and RNAse P of mitochondria lack the RNA segments generating the head-to-tail double T-loop module (helices 76–77 and J11/12–J12/11, respectively) (33–36), and therefore do not have a platform for tRNA docking. Furthermore, T-box leaders are also unknown in mitochondria. The reduced complexity of the mitochondrial system may reflect that of the primitive ribosome, predicted to lack protuberances such as the L1 stalk (37).
(28). Many mitochondrial tRNAs are indeed characterized by the absence or a reduced size of the D-loop and/or T-loop (32), which implies that they do not have a platform for docking (Figure 6). Consistent with this fact, both L1 stalk and RNAse P of mitochondria lack the RNA segments generating the head-to-tail double T-loop module (helices 76–77 and J11/12–J12/11, respectively) (33–36), and therefore do not have a platform for tRNA docking. Furthermore, T-box leaders are also unknown in mitochondria. The reduced complexity of the mitochondrial system may reflect that of the primitive ribosome, predicted to lack protuberances such as the L1 stalk (37).
Along this line, we propose that the elbow (and the associated platform) is likely a late addition to the structure of tRNA. It has long been suggested that the original tRNA was a short molecule bearing only the two functions necessary for decoding genetic information: an anticodon stem–loop and a 3′ acceptor arm (38–41). On the evolutionary time scale, an elbow with a platform could pop up in the form of a D- and T-loop kissing interaction on an existing curved tRNA structure. This additional contact region, optimally positioned at 90° in between the anticodon and the 3′ acceptor arm (Figure 6), would be retained because it provides a great potential in terms of optimizing the kinetics (and therefore the information processing) of any tRNA–RNA interaction. The present study suggests that the docking platform provided by a unique structural motif, the head-to-tail double T-loop module, would favor this evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1 and 2, Supplementary Text 1 and 2 and Supplementary Datasets 1–3.
FUNDING
Funding for open access charge: Université Paris-Sud.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 2.Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol. Mol. Biol. Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol. 1994;176:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rollins SM, Grundy FM, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 1997;25:411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 6.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif—an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 10.Krasilnikov AS, Yang X, Pan T, Mondragón A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 11.Krasilnikov AS, Yang X, Pan T, Mondragón A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 12.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JW. The Ribonuclease P Database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jossinet F, Ludwig TE, Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tishchenko S, Gabdulkhakov A, Nevskaya N, Sarskikh A, Kostareva O, Nikonova E, Sycheva A, Moshkovskii S, Garber M, Nikonov S. High-resolution crystal structure of the isolated ribosomal L1 stalk. Acta Crystallogr. D Biol. Crystallogr. 2012;68(Pt. 8):1051–1057. doi: 10.1107/S0907444912020136. [DOI] [PubMed] [Google Scholar]
- 17.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 18.Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl Acad. Sci. USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krasilnikov AS, Mondragón A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–1119. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang Z, Jaeger L, Shea JE. Probing the structural hierarchy and energy landscape of an RNA T-loop hairpin. Nucleic Acids Res. 2007;35:6995–7002. doi: 10.1093/nar/gkm719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brill J, Hoffmann T, Putzer H, Bremer E. T-box-mediated control of the anabolic proline biosynthetic genes of Bacillus subtilis. Microbiology. 2011;157:977–987. doi: 10.1099/mic.0.047357-0. [DOI] [PubMed] [Google Scholar]
- 23.Yousef MR, Grundy FJ, Henkin TM. tRNA requirements for glyQS antitermination: a new twist on tRNA. RNA. 2003;9:1148–1156. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 25.Grundy FJ, Collins JA, Rollins SM, Henkin TM. tRNA determinants for transcription of the Bacillus subtilis tyrS gene. RNA. 2000;6:1131–1141. doi: 10.1017/s1355838200992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frazer-Abel AA, Hagerman PJ. Core flexibility of a truncated metazoan mitochondrial tRNA. Nucleic Acids Res. 2008;36:5472–5481. doi: 10.1093/nar/gkn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinberg S, Gautheret D, Cedregren R. Fitting the structurally diverse animal mitochondrial tRNAsSer to common three-dimensional constraints. J. Mol. Biol. 1994;236:982–989. doi: 10.1016/0022-2836(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 29.Jaeger L, Verzemnieks EJ, Geary C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2007;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marqués V, Nierhaus KH. tRNA: structure and function. In: Nierhaus KH, Wilson D, editors. Protein Synthesis and Ribosome Structure: Translating the Genome. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim; 2004. p. 149. [Google Scholar]
- 31.Selmer M, Dunham CM, Murphy FV, IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 32.Helm M, Brulé H, Friede D, Giegé R, Pütz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 34.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc. Natl Acad. Sci. USA. 2009;106:9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klimov PB, Laceyknowles L. Repeated parallel evolution of minimal rRNAs revealed from detailed comparative analysis. J. Hered. 2011;103:283–293. doi: 10.1093/jhered/esr005. [DOI] [PubMed] [Google Scholar]
- 36.Rossmanith W. Of P and Z: mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta. 2012;1819:1017–1026. doi: 10.1016/j.bbagrm.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bokov K, Steinberg SV. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009;457:977–980. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- 38.Kuhn H, Waser J. Molecular self-organization and the origin of life. Angew. Chem. Int. Ed. Engl. 1981;20:500–520. [Google Scholar]
- 39.Di Giulio M. On the origin of protein synthesis: a speculative model based on hairpin RNA structures. J. Theor. Biol. 1994;171:303–308. doi: 10.1006/jtbi.1994.1233. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann J. Physico-chemical constraints connected with the coding properties of the genetic system. J. Theor. Biol. 2000;202:129–144. doi: 10.1006/jtbi.1999.1045. [DOI] [PubMed] [Google Scholar]
- 41.Lehmann J, Cibils M, Libchaber A. Emergence of a code in the polymerization of amino acids along RNA templates. PLoS One. 2009;4:e5773. doi: 10.1371/journal.pone.0005773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.