Abstract
The stress-responding protein, GADD45α, plays important roles in cell cycle checkpoint, DNA repair and apoptosis. In our recent study, we demonstrate that GADD45α undergoes a dynamic ubiquitination and degradation in vivo, which process can be blocked by the cytotoxic reagent, arsenite, resulting in GADD45α accumulation to activate JNKs cell death pathway, thereby revealing a novel mechanism for the cellular GADD45α functional regulation. But the factors involved in GADD45α stability modulations are unidentified. Here, we demonstrated that MDM2 was an E3 ubiquitin ligase for GADD45α. One of MDM2-binding partner, ribosomal protein S7, interacted with and stabilized GADD45α through preventing the ubiquitination and degradation of GADD45α mediated by MDM2. This novel function of S7 is unrelated to p53 but seems to depend on S7/MDM2 interaction, for the S7 mutant lacking MDM2-binding ability lost its function to stabilize GADD45α. Further investigations indicated that arsenite treatment enhanced S7–MDM2 interaction, resulting in attenuation of MDM2-dependent GADD45α ubiquitination and degradation, thereby leading to GADD45α-dependent cell death pathway activation. Silencing S7 expression suppressed GADD45α-dependent cytotoxicity induced by arsenite. Our findings thus identify a novel function of S7 in control of GADD45α stabilization under both basal and stress conditions and its significance in mediating arsenite-induced cellular stress.
INTRODUCTION
GADD (growth arrest and DNA-damage inducible) 45α is a multi-functional protein implicated in a variety of cellular stress responses (1,2). Upregulation of GADD45α expression has been shown to link to the G2/M checkpoint of the cell cycle by inhibiting CDC2 kinase activity (3). In addition, GADD45α also functions in modulating genomic DNA demethylation and DNA-damage repair responses, and therefore contributing to the genome stability (4,5). Furthermore, induction of GADD45α represents an important cellular mechanism to trigger the apoptosis in responding to various harmful stresses (6–8).
The induction of GADD45α expression can be regulated at multiple steps, including the transcriptional, post-transcriptional, translational and post-translational events, and the mechanisms involved in these processes are also complicated (2,4,7,9–11). In our previously study, we demonstrated that GADD45α is a liable protein and undergoes constitutive ubiquitination and proteasome-dependent degradation in the resting cells. Arsenite, a toxic chemical agent with anticancer usage, has effects to interrupt endogenous GADD45α ubiquitination and degradation, leading to GADD45α protein accumulation and cellular apoptosis (7,12). These findings provide a new model for GADD45α functional regulation through the modulation of its protein stability in vivo. However, the factors and mechanisms involved in the cellular GADD45α protein stability regulation are still poorly understood yet.
The oncoprotein MDM2 (mouse counterpart of HDM2 in human) is a ring-finger-containing E3 ubiquitin ligase, which is well known as a feedback regulator of the tumor suppressor p53. It binds to and mediates the ubiquitination and degradation of p53, thereby contributing greatly to the cellular p53 functional control. Various damaging stresses can trigger the interruption of constitutive p53–MDM2 interaction, leading to p53 accumulation and fully functional activation to mediate diverse cellular biological effects (13). A lot of regulatory factors have been demonstrated to act on MDM2 and modulate p53 stability and transactivity under various stress conditions (14). The latest players in MDM2–p53 pathway regulation are a subset of ribosomal proteins (RPs) with well-established extraribosomal functions, including the large subunit RPs, L5 (15), L11 (16–18), L23 (19,20), L26 (21,22), and the small subunit RPs, S7 (23,24) and S3 (25). The binding of these RPs to MDM2 have been reported with effects of blocking the E3 ubiquitin ligase activity of MDM2, thereby contributing to p53 upregulation and functional activation under the ribosomal biogenesis stress conditions (26–28). Furthermore, each of the ribosomal protein binds to overlapping yet distinct domains within the central region of MDM2. Therefore, the interdependence interaction of the different ribosomal proteins to MDM2 might be critical for regulating the E3 ubiquitin ligase activity of MDM2 by similar but not identical mechanism and indicates the potential and specific roles of these RPs in sensing different types of stress signals (15,17,19,23,26–28).
In this study, we show that GADD45α, MDM2 and a small subunit of RPs, S7, interact with each other in human cells. Interaction with MDM2 serves as a mechanism for GADD45α protein undergoing the dynamic ubiquitination and degradation process in vivo; although by associating with MDM2, S7 has effect to interrupt MDM2-mediated GADD45α degradation and thereby contributing to the GADD45α protein stabilization in cells. We further demonstrate that modulation of the S7–MDM2 interaction is the key mechanism for arsenite to kill cancer cells through activating the GADD45α-dependent apoptotic pathway. Therefore, our results illustrate a novel mechanism for the cellular GADD45α protein stability regulation and identify a novel function for S7 in mediating arsenite-induced apoptotic response.
MATERIALS AND METHODS
Plasmids and reagents
The HA–GADD45α expression plasmid was described in our previous study (7). The following expression plasmids were constructed by regular polymerase chain reaction (PCR) or reverse transcriptase (RT)–PCR to obtain the DNA fragments: FLAG–GADD45α, FLAG–S7, HA–S7, GFP–S7, FLAG–S7–ΔMDM2 (S7 mutant deleting amino acids 59–134 responsible for MDM2 interaction). The details of the primers used for the construction are available on request. The HA–MDM2 and Myc–Ub expression plasmids were kindly provided by Dr Tao Zhou (National Center of Biomedical Analysis, China). Human MDM2 shRNA and S7 shRNA were constructed by using the GeneSuppressor System (Imgenex). The target sequences were also available on request. Human p53 shRNA and the antibodies against myc, ubiquitin, GFP, p-JNK, JNK and β-actin were purchased from Cell Signaling Technology (Beverly, MA, USA). The antibodies against GADD45α, S7, HA, MDM2 and GAPDH were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-FLAG antibody, MG132, cyclohexamide (CHX) and arsenite were purchased from Sigma (St. Louis, MO, USA).
Cell culture and transfection
HepG2, H1299 and 293T cells were maintained in Dulbecco’s modified Eagle’s medium supplement with 10% FBS, 1% penicillin/streptomycin, and 2 mM l-glutamine. The transfections were performed with the Lipofectamine 2000 reagent (Life Technologies Inc., Rockville, MD, USA) according to the manufacturer’s instructions.
RT–PCR and real-time PCR
Total RNA was extracted with TRizol reagent (Life Technologies Inc., Rockville, MD, USA), and cDNAs were synthesized with the ThermoScript™ RT–PCR system (Life Technologies Inc., Rockville, MD, USA) or real-time PCR detection system (Agilent Biotechnologies, Santa Clara, CA, USA) as recommended by the manufactures. To detect the induction of gadd45α transcription, the following oligonucleotides were synthesized and used as specific primers to amplify the human gadd45α cDNA: 5′-atgactttggaggaattctcg-3′ (forward) and 5′-ccgttcagggagattaatc-3′ (reverse). The results were analyzed using the comparative threshold cycle method with GAPDH as an internal control.
Immunoprecipitation and western-blot assay
Cells transfected with various combination of the expression plasmids were incubated for at least 36 h before harvest or exposure to the chemical reagents. Cellular protein preparation, immunoprecipitation (IP) and immunoblot assays were performed as described previously (29,30).
In vitro ubiquitination assay
In vitro ubiquitination assay of GADD45α was performed with the in vitro ubiquitination system (Enzo Biotechnology, New York, NY, USA) as recommended by the manufacture. Briefly, the reactions were conducted at 37°C for 40 min in a 30 -µl volume containing 1× reaction buffer, 20 ng of E1, 125 ng of E2 mixture, 100–400 ng of MDM2 (Sigma), 600 ng of ubiquitin and 250 ng of purified GADD45α protein (Santa Cruz Biotechnology). After terminating the reactions with 5× sodium dodecyl sulfate (SDS) loading buffer, the reaction products were fractionated by SDS–polyacrylamide gel electrophoresis and detected by immunoblotting with the anti-GADD45α antibody.
In vivo ubiquitination assay
HepG2 cells were transfected with Myc–Ub and FLAG–GADD45α expression plasmids with or without combination of the constructs encoding MDM2, S7 or MDM2 and S7 shRNA. The transfected cells were incubated for 30 h and then exposed to MG132 (10 µM) treatment for 4 h before harvesting. Cell lysate was immunoprecipitated with anti-FLAG antibody, and the ubiquitination of GADD45α was detected with anti-GADD45α antibody.
Yeast two-hybrid assay
The full-length human gadd45α cDNA was inserted into pGBKT7 (Clontech Laboratories) to construct the bait plasmid. For screening the potential GADD45α-binding proteins, the bait construct was transformed into the yeast cells pre-transfected with a mammary embryonic liver cDNA library (Clontech Laboratories) following the manufacture’s instruction.
GST-fusion protein association assay
The gadd45α cDNA was inserted into the pGEX-T vector to construct the GST–GADD45α fusion protein expression plasmid. The fusion protein was prepared from BL21 strains by the regular protocols. Next, GST fusion protein bound to the agarose beads was mixed with the total lysate from 293T cells, followed by incubation with gentle rotation. Beads-bounded proteins were eluted with 2× SDS sample buffer and analyzed by immunoblot assay.
Cell apoptosis assay
Cell death incidence on arsenite exposure was determined by flow cytometric analysis after propidium iodide staining of the nuclei as described previously (7,12).
RESULTS
GADD45α is a liable protein, whose ubiquitination and degradation are inhibited under arsenite exposure
We have previously disclosed that GADD45α protein undergoes a constitutive degradation via the ubiquitin–proteasome pathway in the mouse embryonic fibroblasts (MEFs), which process can be blocked by arsenite treatment, and the accumulation of GADD45α and its dependent JNKs cell death pathway activation contribute largely to arsenite-induced apoptotic response in the MEFs (7). More recently, we further demonstrated that induction of GADD45α/JNKs cascade activation also has key roles in signaling arsenite-induced apoptotic effect in HepG2 human hepatoma cells as that observed in the mouse cells (12). These findings prompt us to consider that protein stabilization may constitute an important functional regulation of GADD45α in mammalian cells.
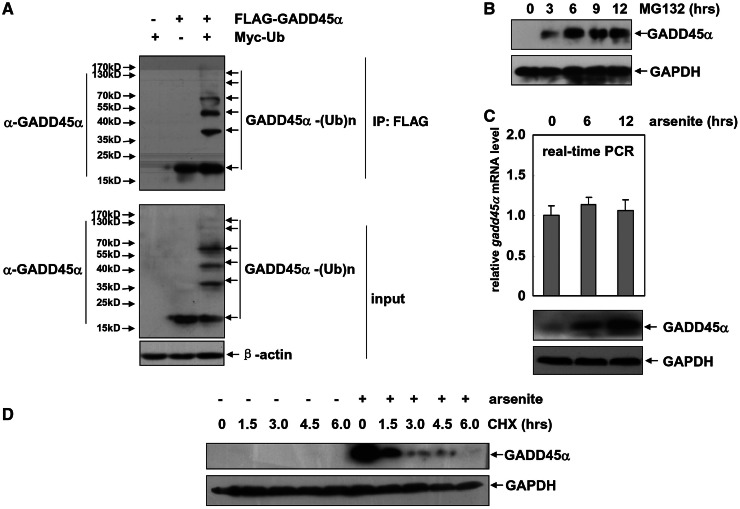
To determine whether a constitutive ubiquitination and degradation of GADD45α proteins also happens in human cells, FLAG–GADD45α and Myc–Ub expression plasmids were co-transfected into the HepG2 cells. We found that a high-molecular weight GADD45α smear characteristic of poly-ubiquitinated GADD45α was obviously presented in the co-transfected cells but not in controlled cells (Figure 1A). Furthermore, treatment with the proteasome inhibitor MG132 was able to significantly accumulate the endogenous GADD45α protein, which is barely detectable before the treatment, in HepG2 cells (Figure 1B). After arsenite exposure, a significant upregulation of GADD45α protein levels was detected in HepG2 cells (Figure1C). However, real-time PCR assays showed that gadd45α mRNA was constitutively expressed in the resting HepG2 cells, and its levels did not get obvious changes after the arsenite treatment during the indicated time (Figure1C). Meanwhile, when HepG2 cells were pre-treated with arsenite and then subjected to the exposure of protein synthesis inhibitor (CHX) after arsenite withdrawal, the pre-accumulated GADD45α could undergo a rapid degradation in a time-dependent manner (Figure 1D). Together, we thus conclude that GADD45α proteins are also constitutively subjected to ubiquitin–proteasome-dependent degradation and blocking this process also constitutes a response of HepG2 cells to the arsenite stress.
Figure 1.
GADD45α is a liable protein, and its proteasome-dependent degradation is blocked by arsenite in the HepG2 cells. (A) HepG2 cells were transfected with combination of the expression plasmids encoding FLAG–GADD45α or Myc–Ub as indicated. Cell extracts were immunoprecipitated with anti-FLAG antibody, and the ubiquitination of GADD45α was detected with anti-GADD45α antibody. (B) HepG2 cells were treated with MG132 (10 µM) for the indicated time, and then the levels of GADD45α were detected. (C) HepG2 cells were treated with arsenite (20 µM) for the indicated time, and then the induction of gadd45α mRNA and protein levels was detected by real-time PCR or western-blot assays, respectively. (D) HepG2 cells were left untreated or treated with arsenite (20 µM) for 12 h and then subjected to CHX (20 µM) exposure at the indicated time after arsenite withdrawal. The degradation of GADD45α was detected by western-blot assay.
Ribosomal protein S7 interacts with GADD45α
To find factors involved in regulating GADD45α protein stability, the full-length GADD45α was used as the bait to screen a human embryonic liver cDNA library through the yeast two-hybrid approach. Several positive clones were found to contain the same cDNA sequences encoding the ribosomal protein S7.
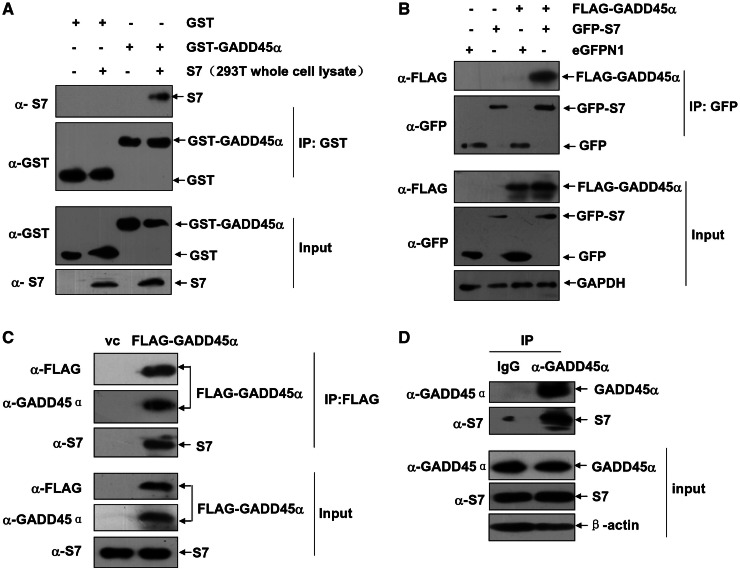
To confirm the putative GADD45α–S7 interaction, the Escherichia coli prepared and purified GST–GADD45α proteins were mixed with the 293T cellular extracts. Then we found that the GST–GADD45α, but not the controlled GST proteins, was able to pull down the cellular expressed S7 (Figure 2A). Co-expressions of the FLAG–GADD45α with GFP–S7 in 293T cells also confirmed the specific interaction between these two ectopic expressed proteins in vivo as judged by the IP assays (Figure 2B). The following IP assays also showed that the ectopic-expressed FLAG–GADD45α was able to form complex with endogenous S7 in the HepG2 cells (Figure 2C). Next, we wanted to address whether an endogenous GADD45α–S7 interaction occurs in human cells. Therefore, the arsenite-treated HepG2 cells were used as model for the extremely low GADD45α expression levels in this cell line under the physiological growth condition (Figure 1B). As indicated in Figure 2D, the accumulated GADD45α induced by the arsenite could bind to S7 endogenously in HepG2 cells. Taken together, we conclude that S7 is a novel GADD45α-binding protein, and such an interaction between GADD45α and S7 could be present in human cells under both basal and the arsenite-stress conditions.
Figure 2.
S7 interacts with GADD45α. (A) The purified GST–GADD45α or GST proteins and 293T whole-cell lysate with high levels of S7 expression were incubated together, and then GST-pull-down assay was performed to examine the in vitro interaction between GADD45α and S7. (B) 293T cells were either transfected with the expression plasmid encoding FLAG–GADD45α or in combination with GFP–S7 construct or eFGPN1 vector. Cell lysate was immunoprecipitated with anti-GFP antibody, and then the immunoprecipitants were probed with anti-GFP and anti-FLAG antibodies. (C) HepG2 cells were transfected with FLAG–GADD45α construct or its control vector. Cell lysate was immunoprecipitated with anti-FLAG antibody, and the immunoprecipitants were probed with anti-FLAG, anti-GADD45α and anti-S7 antibodies. (D) HepG2 cells were treated with arsenite (20 µM) for 12 h, and then cell lysate was immunoprecipitated with anti-GADD45α antibody or rabbit IgG. The immunoprecipitants were probed with anti-GADD45α and anti-S7 antibodies.
S7 enhances GADD45α stability by suppressing GADD45α ubiquitination and degradation
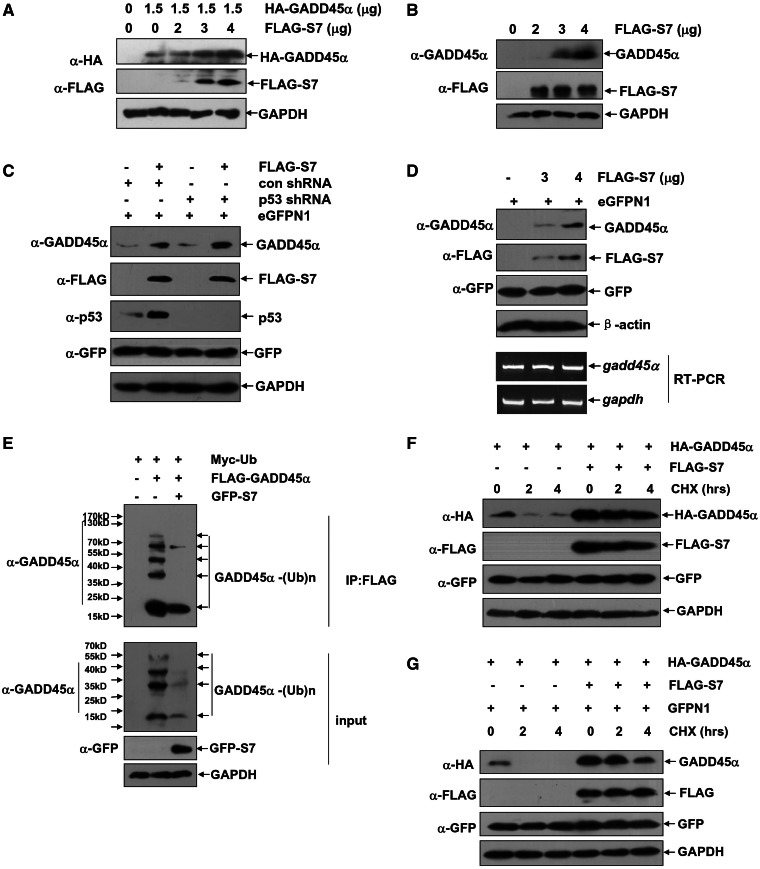
To test whether S7 has any roles in regulating GADD45α stability, a single dose of HA–GADD45α and the increasing amount of FLAG–S7 were co-transfected into HepG2 cells, and then a dose-dependent effect of S7 overexpression on enhancing the original GADD45α levels was found (Figure 3A). Consistently, ectopic overexpression of S7 also caused a significant accumulation of endogenous GADD45α proteins in HepG2 cells (Figure 3B). These data thus suggest that S7 has a positive role in regulating the cellular GADD45α expression.
Figure 3.
S7 upregulates GADD45α protein stability by attenuating GADD45α ubiquitination and degradation under physiological conditions. (A) HepG2 cells were left untreated or transfected with a single dose of expression plasmid encoding HA–GADD45α (1.5 µg) with or without combination of increasing amount of FLAG–S7 construct (2, 3 and 4 µg). The expression levels of the exogenous GADD45α were detected with anti-HA antibody. (B) HepG2 cells were transfected with increasing amount of FLAG–S7 construct (2, 3 and 4 µg). The expression levels of the endogenous GADD45α were detected with anti-GADD45α antibody. (C) HepG2 cells were transfected with p53 shRNA or its control vector with or without combination of FLAG–S7 construct (3 µg). The expression levels of the endogenous GADD45α and p53 were detected. The cells were also co-transfected with eGFPN1 vector (100 ng) and the expression level of GFP served as the control for both transfection efficiency and global translation in HepG2 cells. (D) H1299 cells were transfected with increasing amount of FLAG–S7 construct (3 and 4 µg). The expression levels of the endogenous GADD45α and the transcription of gadd45α mRNA were detected by western-blot and RT–PCR assay, respectively. (E) HepG2 cells were transfected with Myc–Ub and FLAG–GADD45α constructs with or without combination of the expression plasmid encoding GFP–S7. The cell lysate was immunoprecipitated with anti-FLAG antibody, and then the ubiquitination of GADD45α was detected with anti-GADD45α antibody. (F and G) HepG2 (F) or H1299 (G) cells were transfected with the expression plasmid encoding HA–GADD45α (1 µg) with or without combination of FLAG–S7 construct (3 µg). Then the cells were subjected to CHX (20 µM) exposure at the indicated time, and the degradation of GADD45α was detected with anti-HA antibody.
S7 was previously shown with effect on upregulating expression levels of p53, an established transcriptional activator for the gadd45α gene (1,2,9,23,24). Indeed, an increase of p53 protein levels could be found in the ectopic S7-overexpressed HepG2 cells as compared with the controlled cells (Figure 3C). However, blocking the inducible p53 upregulation by a p53-specific shRNA co-transfection did not remove the effect of the overexpressed S7 on GADD45α accumulation in these cells (Figure 3C). Moreover, ectopic overexpression of S7 in the well-known p53-deficient human non–small-cell carcinoma H1299 cells also caused the similar response of GADD45α accumulation (Figure 3D). We, therefore, prefer to interpret that the major role for S7 in enhancing GADD45α expression is unrelated to its putative function involving in p53 regulation. Further supporting that non-transcriptional mechanisms are involved in the S7 overexpression-dependent GADD45α accumulation, we showed that gadd45α mRNA levels remained roughly unchanged in H1299 cells with and without ectopic S7 overexpression (Figure 3D). Based on the aforementioned data, we anticipate that the positive effect of S7 on GADD45α expression may result from the modulation of the process of GADD45α ubiquitination and degradation in cells. To address this possibility, plasmids encoding FLAG–GADD45α and Myc–Ub were co-transfected into the HepG2 cells with or without a combination of GFP–S7 overexpression. As shown in Figure 3E, the signs for the ubiquitin-conjugated GADD45α were almost undetectable in the presence of S7 overexpression. In addition, the degradation dynamics assay showed that half-life for the ectopic expressed GADD45α was significantly elongated in the S7 overexpressing HepG2 cells as compared with that in the controlled cells (Figure 3F). The same response was also readily observed in the H1299 cells (Figure 3G). Therefore, we propose that S7 functions as a GADD45α stabilizer, and its overexpression has the effect of suppressing GADD45α ubiquitination and degradation, thereby increasing cellular GADD45α protein expression levels.
MDM2 mediates GADD45α ubiquitination and degradation
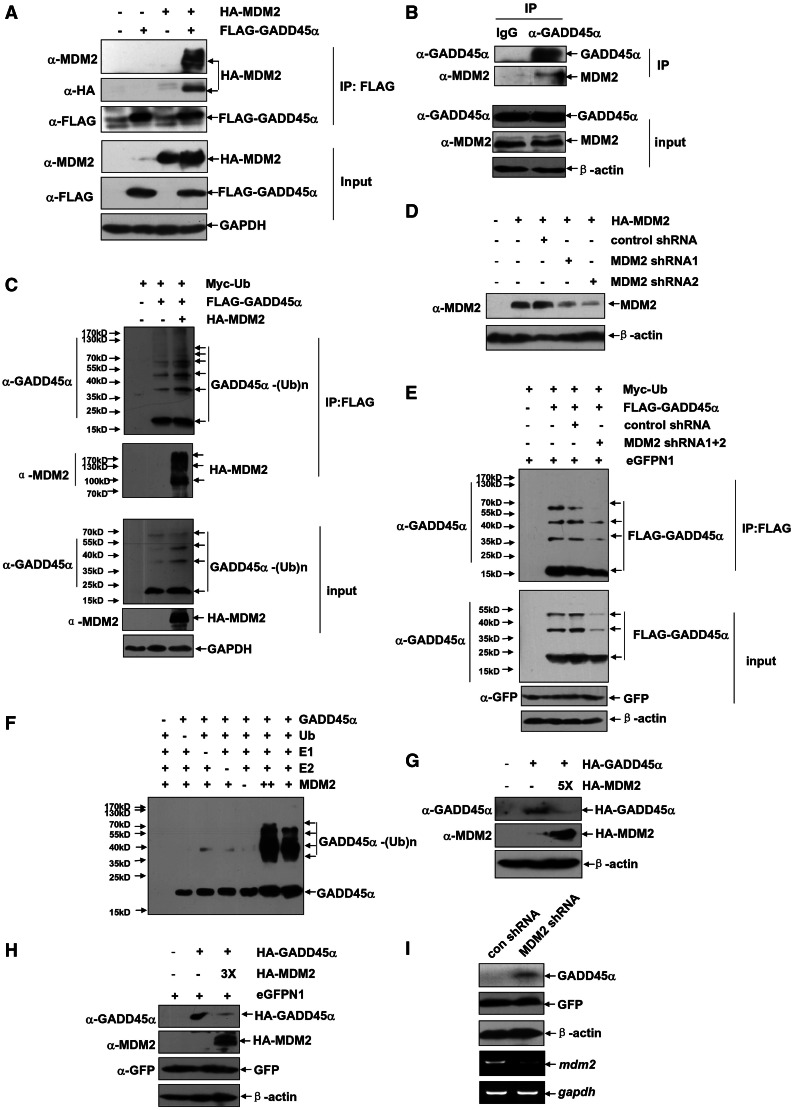
MDM2 has been known as the E3 ubiquitin ligase for p53 and some other proteins as well. Previous studies showed that S7 can bind to and suppress MDM2’s activity on mediating p53 ubiquitination, thereby enhancing p53 protein levels in cells (23,24). It is, therefore, reasonable to ask whether S7 also stabilizes GADD45α via the same mechanism. To test this possibility, we first determined whether GADD45α could be a substrate for MDM2. 293T cells were co-transfected with HA–MDM2 and FLAG–GADD45α and then IP was performed with anti-FLAG antibody. Data in Figure 4A showed that HA–MDM2 can be detected in the anti-FLAG immunoprecipitants. Endogenous MDM2–GADD45α association was also detectable with the arsenite-treated HepG2 cells as model (Figure 4B). These data thus indicate that MDM2 has the potential to interact with GADD45α in vivo.
Figure 4.
MDM2 functions as an E3 ubiquitin ligase for GADD45α. (A) 293T cells were either transfected with the expression plasmid encoding FLAG–GADD45α or in combination with HA–MDM2 construct. The cells were subjected to MG132 (10 µM) treatment for 4 h before harvesting. Then cell lysate was immunoprecipitated with anti-FLAG antibody, and the immunoprecipitants were probed with anti-FLAG, anti-HA and anti-MDM2 antibodies. (B) HepG2 cells were treated with arsenite (20 µM) for 12 h, and then cell lysate was immunoprecipitated with anti-GADD45α antibody or rabbit IgG. The immunoprecipitants were probed with anti-GADD45α and anti-MDM2 antibodies. (C) HepG2 cells were transfected with Myc–Ub and FLAG–GADD45α constructs with or without combination of the expression plasmid encoding HA–MDM2. Cell lysate was immunoprecipitated with anti-FLAG antibody, and then the ubiquitination of GADD45α was detected with anti-GADD45α antibody. (D) HepG2 cells were transfected with the HA–MDM2 construct with or without combination of control shRNA or MDM2 shRNAs. Then the efficiency of MDM2 shRNAs was determined by western-blot assay. (E) HepG2 cells were transfected with Myc–Ub and FLAG–GADD45α constructs with or without combination of control shRNA or MDM2 shRNAs mixture. Cell lysate was immunoprecipitated with anti-FLAG antibody, and then the ubiquitination of GADD45α was detected with anti-GADD45α antibody. (F) The purified GADD45α protein (250 ng) was incubated in the in vitro ubiquitination reaction buffer with E1 (20 ng), E2 mixture (125 ng), MDM2 (100 or 400 ng) and ubiquitin (600 ng) for 40 min at 37°C. Then the ubiquitination of GADD45α was detected by immunoblotting with the anti-GADD45α antibody. (G) HepG2 cells were transfected with HA–GADD45α construct (0.25 µg) with or without combination of overdosed HA–MDM2 expression plasmid (1.25 µg). Then the expression levels of GADD45α were detected. (H) H1299 cells were transfected with HA–GADD45α construct (0.5 µg) with or without combination of overdosed HA–MDM2 expression plasmid (1.5 µg). Then the expression levels of GADD45α were detected. (I) H1299 cells were left untreated or transfected with MDM2 shRNA. Then the expression levels of endogenous GADD45α and mdm2 mRNA were detected.
We further provided evidences that co-expression with HA–MDM2 could enhance GADD45α poly-ubiquitination in HepG2 cells (Figure 4C). On the contrary, knockdown endogenous MDM2 expression by its specific shRNA decreased GADD45α ubiquitination levels significantly (Figure 4D and E). Moreover, MDM2 directly mediated the ubiquitination of the purified GADD45α protein according to the results obtained from the in vitro ubiquitination assay (Figure 4F). These data thus indicated that MDM2 is an E3 ubiquitin ligase for GADD45α.
To further clarify the critical role of MDM2 involving in GADD45α protein stability regulation in human cells, we next showed that co-expression with overdosed HA–MDM2 significantly reduced the ectopic GADD45α levels in both HepG2 and H1299 cells (Figure 4G and H). In contract, MDM2 depletion by specific shRNA transfection remarkably increased endogenous GADD45α protein expression in H1299 cells (Figure 4I). Because of p53 deficiency in H1299 cells, we believe that the effects of MDM2 on cellular p53 and GADD45α protein stability regulations are two separate processes.
S7 stabilizes GADD45α by binding to and inhibiting MDM2-mediated GADD45α ubiquitination and degradation
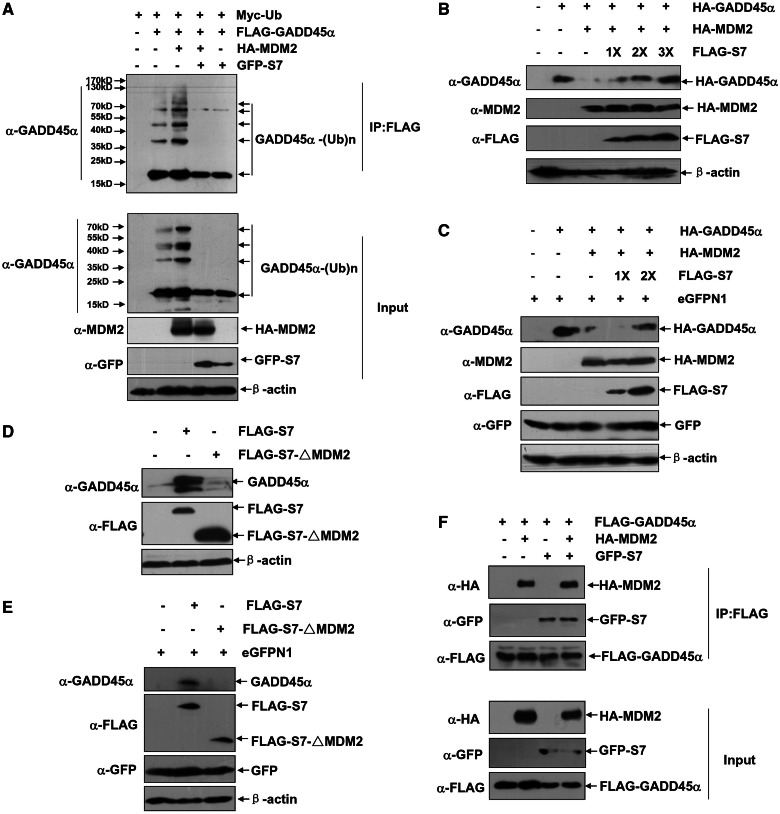
It has been showed in the previous study that S7 binds to and prevents MDM2 from accessing to p53, thereby blocking the ubiquitination and degradation of p53 in cells (23). We wonder whether this model is also applied to the action of S7 on stabilizing GADD45α proteins in vivo. To this end, Myc–Ub, FLAG–GADD45α, HA–MDM2 and GFP–S7 were co-expressed in HepG2 cells. As indicated in the Figure 5A, the effect of MDM2 on mediating GADD45α poly-ubiquitination was significantly suppressed by S7 co-expression. Moreover, with the increased amounts of S7 overexpression, it also displayed a dose-dependent effect on rescuing MDM2-mediated GADD45α degradation (Figure 5B). The same results were also obtained with the H1299 cells as model (Figure 5C), further supporting the effect of S7 on GADD45α upregulation independent of p53. However, deletion of the MDM2-binding site rendered the mutant S7–ΔMDM2 (23) losing the function of accumulating GADD45α in both HepG2 and H1299 cells under the same experimental conditions (Figure 5D and E). These results thus indicate that, like its manner in the MDM2–p53 regulation, a direct binding is also necessary for S7 to inhibit MDM2-mediated GADD45α ubiquitination and degradation in vivo. However, unlike its setting in regulating MDM2–p53 interaction (23), S7 did not interrupt the MDM2–GADD45α association in vivo (Figure 5F). As shown in this figure, the protein amounts for HA–MDM2 in the FLAG–GADD45α immunoprecipitates from cells with and without the GFP–S7 overexpression remained unchanged, although the MDM2-dependent GADD45α ubiquitination was significantly decreased in the GFP–S7 expressing cells (Figure 5A). The mechanism for how S7 repressing MDM2-mediated GADD45α ubiquitination and degradation without interrupting MDM2/GADD45α interaction is currently unknown yet.
Figure 5.
S7 attenuates MDM2-mediated GADD45α ubiquitination and degradation. (A) HepG2 cells were transfected with Myc–Ub expression plasmid in combination of FLAG–GADD45α, HA–MDM2 or GFP–S7 constructs as indicated. Then the ubiquitination of GADD45α was detected with anti-GADD45α antibody. (B) HepG2 cells were left untreated or transfected with HA–GADD45α construct (0.25 µg) with or without combination of HA–MDM2 expression plasmid (1.25 µg) and increasing amount of FLAG–S7 construct (1.25, 2.5 and 3.75 µg, respectively). Then the levels of GADD45α were detected. (C) H1299 cells were left untreated or transfected with HA–GADD45α construct (0.5 µg) with or without combination of HA–MDM2 expression plasmid (1.5 µg) and increasing amount of FLAG–S7 construct (1.5 and 3.0 µg, respectively). Then the levels of GADD45α were detected. (D and E) HepG2 (D) or H1299 (E) cells were transfected with equal amount of FLAG–S7 or FLAG–S7–ΔMDM2 constructs (3 µg), and then the levels of the endogenous GADD45α were detected. (F) 293 T cells were transfected with combination of FLAG–GADD45α, HA–MDM2 or GFP–S7 constructs as indicated. The cells were subjected to MG132 (10 µM) treatment for 4 h before harvesting. Then cell lysate was immunoprecipitated with anti-FLAG antibody, and the immunoprecipitants were probed with anti-FLAG, anti-GFP or anti-MDM2 antibodies.
Arsenite exposure enhances S7–MDM2 interaction, which subsequently downregulates GADD45α ubiquitination
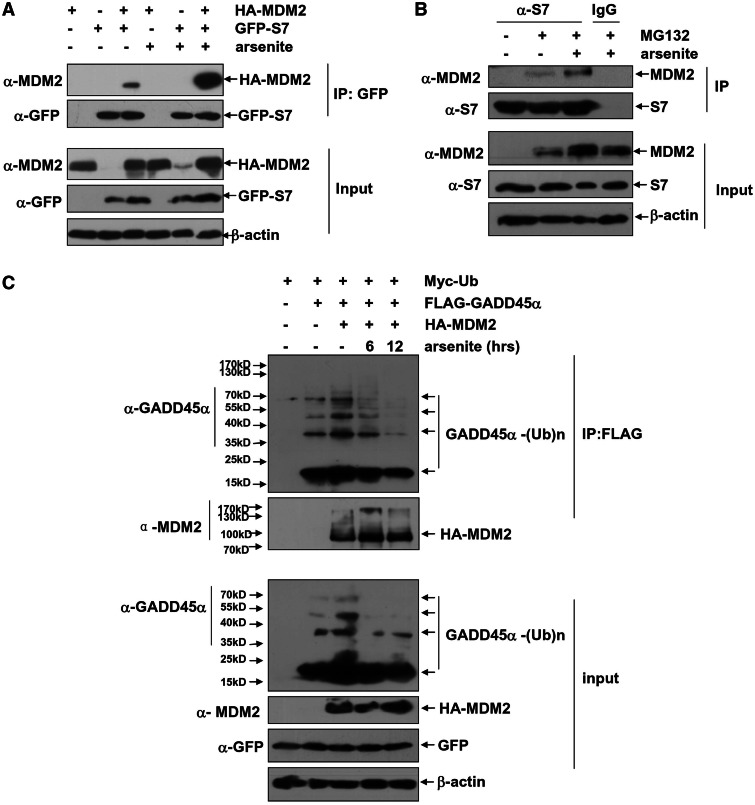
As we have demonstrated that arsenite stimulation has effect on accumulating GADD45α by blocking its ubiquitination and degradation in both human and mouse cells, then we asked whether the role of S7 for suppressing MDM2-mediated GADD45α degradation accounts for the mechanism in this process. By co-expressing HA–MDM2 and GFP–S7 in HepG2 cells, we found that the MDM2–S7 interaction was drastically enhanced after arsenite treatment, evidenced by the amounts of HA–MDM2 in the GFP–S7 immunoprecipitates significantly increased in the sample from arsenite-treated cells (Figure 6A). Consistently, an increased endogenous MDM2–S7 interaction was also detectable in the arsenite-treated HepG2 cells (Figure 6B). Notably, an increase of endogenous MDM2 protein levels was also observed in HepG2 cells after arsenite exposure, whereas the protein levels for endogenous S7 remained roughly unchanged under the same conditions (Figure 6B). Moreover, MDM2-mediated GADD45α ubiquitination was remarkably inhibited after arsenite exposure (Figure 6C). Based on these data, we proposed that associating and blocking MDM2 to mediate GADD45α ubiquitination and degradation by S7 is a mechanism for arsenite-induced GADD45α accumulation in cells.
Figure 6.
Arsenite exposure enhances S7–MDM2 interaction, which subsequently abrogates GADD45α ubiquitination. (A) HepG2 cells were either transfected with HA–MDM2 or in combination with GFP–S7. Then the cells were left untreated or exposed to arsenite (20 µM) for 8 h. Cell lysate was immunoprecipitated with anti-GFP antibody, and the immunoprecipitants were probed with anti-GFP and anti-MDM2 antibodies. (B) HepG2 cells were left untreated or pre-treated with MG132 (10 µM) for 6 h followed by exposure to arsenite (20 µM) for 8 h. Cell lysate was immunoprecipitated with anti-S7 antibody or the mouse IgG, and then the immunoprecipitants were probed with anti-S7 and anti-MDM2 antibodies. (C) HepG2 cells were transfected with Myc–Ub expression plasmid with or without combination of FLAG–GADD45α or HA–MDM2 constructs. Then the transfected cells were left untreated or exposed to arsenite (20 µM) for the time indicated. Cell lysate was immunoprecipitated with anti-FLAG antibody, and then the ubiquitination of GADD45α was detected with anti-GADD45α antibody.
S7 mediates arsenite-induced cellular apoptosis through the GADD45α-JNKs pathway
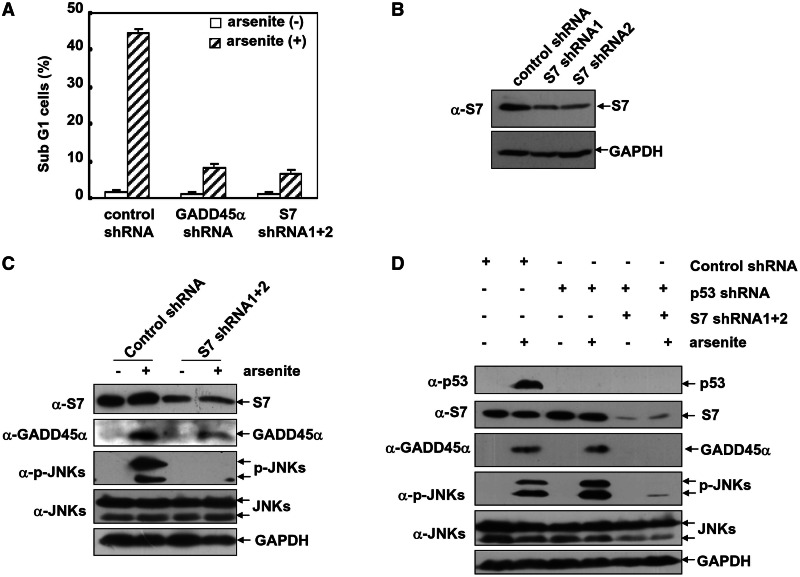
We finally addressed the significances for the S7-dependent GADD45α regulation in cellular stress responses. According to our previous reports, GADD45α–JNKs cascade functions an apoptotic signaling pathway in mice cells to the arsenite stress response (7). Consistently, stimulation with arsenite (20 µM) triggers a robust apoptotic response in HepG2 cells (Figure 7A), associated with induction of GADD45α accumulation and JNKs activation (Figure 7C). However, pre-transfection with a pair of S7 shRNAs, which individually displayed a modest (Figure 7B) but changed to a robust S7 knockdown effect in combination (Figure 7C), greatly reduced the inducible GADD45α accumulation and JNKs activation in HepG2 cells by the subsequent arsenite treatment (Figure 7C). Also like the case of GADD45α shRNA transfection, introduction of S7 shRNAs also significantly attenuated arsenite-induced apoptosis in HepG2 cells by the same extent (Figure 7A). Together, these data indicate that S7 is critical for mediating GADD45α-dependent cell death response under arsenite exposure.
Figure 7.
Knockdown S7 levels impair GADD45α-dependent cell apoptotic pathway activation in the arsenite-treated HepG2 cells. (A) HepG2 cells were transfected with the control shRNA, p53 shRNA or combination of S7 shRNA1 and 2. The cell death incidence was detected by flow cytometric assay 24 h after arsenite exposure. And the cells accumulated in the sub G1 phase were presented. (B) HepG2 cells were transfected with either the control shRNA or the individual S7 shRNA1 or S7 shRNA 2. Then the efficiency of each shRNA was determined with anti-S7 antibody. (C) HepG2 cells were transfected with either the control shRNA or S7 shRNA mixture. Then the cells were left untreated or exposed to arsenite (20 µM) for 12 h. The induction of GADD45α expression and JNKs activation was detected. (D) HepG2 cells were transfected with either the control shRNA or p53 shRNA with or without S7 shRNA mixture. Then the cells were treated as described in C. The induction of GADD45α and p53 expression, as well as JNKs activation, was detected.
As arsenite treatment also triggered a p53 induction in these cells (Figure 7D), the p53-specific shRNA was transfected into HepG2 cells before S7 shRNA introduction. As shown in Figure 7D, silencing p53 expression did not affect arsenite-induced GADD45α accumulation and JNKs activation, and co-transfection with p53 shRNA failed to affect the effect of S7 shRNAs on suppressing the inducible GADD45α accumulation and JNKs activation in the arsenite-treated HepG2 cells (Figure 7D). Thus, we prefer to exclude that p53 induction would be a predominant mechanism for the S7-dependent activation of the GADD45α–JNKs cell death pathway under the cellular arsenite stress.
DISCUSSION
GADD45α induction has been functionally linked to multiple cellular damaging responses and with implications in the cell fate determination under these conditions (1,2). Although transcriptional controls are generally regarded as the major mechanism for the cellular GADD45α regulations, other post-transcriptional mechanisms have also been implicated (1,2,7,9–11). We previously demonstrate that arsenite exposure induces GADD45α expression via blocking its constitutive ubiqitination and degradation, thereby revealing a novel model of the post-translational modification for GADD45α and highlighting the importance of the protein stability modulations on this protein in the maintenance of cellular homeostasis (7). Here, we extended the study to explore the cellular mechanisms on GADD45α protein stability regulations. We identified the MDM2 as an E3 ligase for mediating GADD45α ubiquitination and degradation and S7 as the key factor for preventing GADD45α from MDM2-dependent degradation pathway, thereby throwing new lights on the mechanisms of the cellular GADD45α stability regulations and the corresponding significance in cell fate determination under certain damaging stress conditions, such as the arsenite.
S7 belongs to a member of small subunit of the ribosome proteins but is recently shown with novel function of modulating the MDM2-dependent p53 ubiquitination and degradation in mammalian cells (23,24). In that case, S7 enters for p53 functional regulation via binding to MDM2, with the outcome of interrupting MDM2 access to p53 and thereby freeing p53 from MDM2-mediated ubiquitination and degradation pathway. In this study, we got evidences to show GADD45α served as a substrate of MDM2 in vitro and was subjected to a constitutive ubiquitination and degradation process mediated by MDM2 in vivo, and that the effect of S7 on suppressing GADD45α ubiquitination and degradation was dependent on its MDM2-binding capacity, as mutation disrupting this interaction rendered S7 losing its function to accumulate GADD45α. These data thus can support our notion that S7 enhances GADD45α protein stability by binding to repress the MDM2 action on mediating GADD45α ubiquitination in vivo. However, as the gadd45α has been established as a p53-targeted gene, one might also expect that the observation of S7 on GADD45α protein upregulation is derived from the outcome of its potential effect on p53 functional activation. But, as the effect for S7 on modulating GADD45α expression remained intact in p53-deficient or p53 shRNA-silencing cells, we excluded the possible involvement of p53 in S7-dependent GADD45α accumulation. On the other hand, the S7 association seemed to not interrupt MDM2–GADD45α interaction in vivo, which markedly contrasted to the model proposed for the action of S7 on the MDM2–p53 modulation (23). How can S7 bind to repress MDM2-mediated GADD45α ubiquitination without affecting MDM2–GADD45α association is waiting further elucidated. We prefer to interpret this as distinctive situation, for S7 can bind to MDM2 and GADD45α simultaneously, thereby representing a novel independent function for S7.
The significance for S7 on GADD45α protein stabilization is also reflected by its key role in turning on the GADD45α–JNKs apoptotic signaling pathway in cellular arsenite response. Notably, although the importance for the presence of S7 in the arsenite-induced GADD45α accumulation and JNKs activation is true, no significant changes in the S7 levels were detected during the course of cellular arsenite response. Given that the role of S7 in this cellular reaction is to bind to interrupt MDM2 on mediating GADD45α ubiquitination, it raises an interesting question of why the same levels of S7 do not inhibit this process under the physiological condition. We consider that cells may possess additional mechanisms on controlling the endogenous S7–MDM2 interaction so that make it an inducible event under certain cellular growth conditions. Indeed, the endogenous MDM2 protein level is extremely low in resting cells and is remarkably upregulated associating with the obvious presence of the S7–MDM2 interaction after arsenite stimulation. This observation may suggest that certain signaling events are elicited to trigger the endogenous S7–MDM2 interaction under this stress conditions. In fact, we have previously demonstrated that the arsenite-induced GADD45α–JNKs pathway activation is distinctively dependent on a particular role of the β subunit of the IκB kinase (IKK) complex, IKKβ (7). Whether IKKβ has function to confer special modifications on S7 or MDM2, or both, during the cellular arsenite response is worth of investigating. As arsenite has been used for the clinical treatment of some human cancers, further elucidating the molecular events involved in the regulation of the S7–MDM2–GADD45α signaling axle is of medical important.
FUNDING
Funding for open access charge: National Key Research and Development Program on Fundamental Sciences [973 Project, 2011CB503803]; National Natural Science Foundation of China [30970594, 31171342 and 31270797]; Beijing Natural Science Foundation [5102035 to L.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Moskalev AA, Smit-McBride Z, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Tacutu R, Fraifeld VE. Gadd45 proteins: Relevance to aging, longevity and age-related pathologies. Ageing Res. Rev. 2012;11:51–66. doi: 10.1016/j.arr.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao M, NIng G, Chuanshu H, Lun S. Diverse roles of GADD45a in stres signaling. Curr. Protein Pep. Sci. 2009;10:388–394. doi: 10.2174/138920309788922216. [DOI] [PubMed] [Google Scholar]
- 3.Wang XW, Zhan Q, Coursen JD, Khan MA, Kontny HU, Yu L, Hollander MC, Oonnor PM, Fornace AJ, Harris CC. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl Acad. Sci. USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 5.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA Demethylation in Zebrafish Involves the Coupling of a Deaminase, a Glycosylase, and Gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, et al. Induction of GADD45 and JNK/SAPK-Dependent Apoptosis following Inducible Expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 7.Song L, Li J, Zhang D, Liu ZG, Ye J, Zhan Q, Shen HM, Whiteman M, Huang C. IKKβ programs to turn on the GADD45α–MKK4–JNK apoptotic cascade specifically via p50 NF-κB in arsenite response. J. Cell Biol. 2006;175:607–617. doi: 10.1083/jcb.200602149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 9.Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, Gariboldi M, Myers TG, Weinstein JN, Pommier Y, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol. Cell. Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Q, Bhatia D, Zhang Y, Meighan T, Castranova V, Shi X, Chen F. Incorporation of an internal ribosome entry site-dependent mechanism in arsenic-induced GADD45a expression. Cancer Res. 2007;67:6146–6154. doi: 10.1158/0008-5472.CAN-07-0867. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Bhatia D, Xia H, Castranova V, Shi X, Chen F. Nucleolin links to arsenic-induced stabilization of GADD45a mRNA. Nucleic Acids Res. 2006;34:485–495. doi: 10.1093/nar/gkj459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao M, Dong W, Hu M, Yu M, Guo L, Qian L, Guo N, Song L. GADD45α mediates arsenite-induced cell apoptotic effect in human hepatoma cells via JNKs/AP-1-dependent pathway. J. Cell. Biochem. 2010;109:1264–1273. doi: 10.1002/jcb.22509. [DOI] [PubMed] [Google Scholar]
- 13.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2009;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 14.Manfredi JJ. The Mdm2–p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580–1589. doi: 10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai MS, Lu H. Inhibition of MDM2-mediated p53 Ubiquitination and Degradation by Ribosomal Protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 16.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, Jin C, Chen H, Zhang L, Yang X, et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 2010;38:6544–6554. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair. 2009;8:1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 27.Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget. 2011;2:234–238. doi: 10.18632/oncotarget.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong W, Li Y, Gao M, Hu M, Li X, Mai S, Guo N, Yuan S, Song L. IKKa contributes to UVB-induced VEGF expression by regulating AP-1 transactivation. Nucleic Acids Res. 2012;40:2940–2955. doi: 10.1093/nar/gkr1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song L, Gao M, Dong W, Hu M, Li J, Shi X, Hao Y, Li Y, Huang C. p85[alpha] mediates p53 K370 acetylation by p300 and regulates its promoter-specific transactivity in the cellular UVB response. Oncogene. 2011;30:1360–1371. doi: 10.1038/onc.2010.506. [DOI] [PMC free article] [PubMed] [Google Scholar]