Abstract
The Schizosaccharomyces pombe genome is one of the smallest among the free-living eukaryotes. We further reduced the S. pombe gene number by large-scale gene deletion to identify a minimal gene set required for growth under laboratory conditions. The genome-reduced strain has four deletion regions: 168.4 kb in the left arm of chromosome I, 155.4 kb in the right arm of chromosome I, 211.7 kb in the left arm of chromosome II and 121.6 kb in the right arm of chromosome II. The deletions corresponded to a loss of 223 genes of the original ∼5100. The quadruple-deletion strain, with a total deletion size of 657.3 kb, showed a decreased ability to uptake glucose and some amino acids in comparison with the parental strain. The strain also showed increased gene expression of the mating pheromone M-factor precursor and the nicotinamide adenine dinucleotide phosphate -specific glutamate dehydrogenase. There was also a 2.7-fold increase in the concentration of cellular adenosine triphosphate, and levels of the heterologous proteins, enhanced green fluorescent protein and secreted human growth hormone were increased by 1.7- and 1.8-fold, respectively. The transcriptome data from this study have been submitted to the Gene Expression Omnibus (GEO: http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE38620 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vjkxjewuywgcovc&acc=GSE38620).
INTRODUCTION
Microorganisms have various genes that are expressed for adaptation to a particular condition. These genes are thought to be unnecessary under optimized conditions, including nutrient sources, temperature and aeration. A minimal gene set required for cellular viability gives important clues about evolutionary origins. In addition, investigation of a minimal gene set can be used to construct minimal genome factories for some industrial purposes. In the development of microbial production systems, computer modeling and simulation of the cellular metabolic systems have been used to optimize metabolic networks (1). However, a complete understanding of a predicted metabolic system is often difficult in microorganisms, as complex intracellular metabolic pathways often interfere with desired results. Recently, reduction of genome size has been examined in some microorganisms to simplify intracellular metabolic pathways while maintaining growth efficiency. In the production of a minimum genome factory, it has been proposed that effective use of intracellular energy can be achieved by the elimination of unnecessary genes (2). Comparative genomics support this hypothesis; it has been speculated that genome reduction can be a selective process favoring bacterial adaptation to a low-nutrient environment for effective energy use (3). Several genome-reduced microorganisms were reported to have beneficial properties. In Escherichia coli, benefits included improvement of electroporation efficiency, accurate propagation of recombinant genes and unstable plasmids (4) and an increase in l-threonine production (5). In Bacillus subtilis, increases in production and secretion levels of heterologous enzymes were noted (6,7). Saccharomyces cerevisiae is the only eukaryotic organism for which a genome-reduced strain has been reported (8). The size of this deleted genome was 531.5 kb [4.4% of the 12.16 Mb total genome size archived in the Saccharomyces genome database (http://www.yeastgenome.org/cache/genomeSnapshot.html)] and contained 247 Open Reading Frames (ORFs). Genome reduction in eukaryotes is difficult, owing to more complicated cell systems and a greater number of essential genes compared with prokaryotes. The proportion of essential genes of the total number of genes has been estimated for these microorganisms: S. cerevisiae 18.7% (9), E. coli 7.0% (10) and B. subtilis 6.6% (11).
The Schizosaccharomyces pombe genome project was completed in 2002 (12). The estimated whole genome size is 13.8 Mb and is distributed on three chromosomes. In all, 12.57 Mb of the genome, excluding rDNA, has been sequenced (http://www.pombase.org/status/statistics). However, the published S. pombe genomic sequence initially included four gaps and three undetermined regions. A gap sequence in the left arm of chromosome II has since been filled (13), but the other regions remain undefined. In S. pombe, a single-systematic genome-wide gene deletion collection (14) and a pilot study deleting 100 genes (15) have been reported. S. pombe essential genes constitute 26.1% of total genes (1260/4836), which is a higher percentage than in other model organisms described earlier in the text. The S. pombe total gene number (recently reported in PomBase as ∼5100 genes) is one of the smallest among free-living eukaryotes. Genetic manipulation of S. pombe has been established, and S. pombe is a useful tool for genetic research. We, therefore, selected S. pombe as a suitable model free-living eukaryote to investigate a minimal gene set required for cell growth. The S. pombe genome was reduced by deletion of terminal regions of chromosomes I and II, and a 657.3 kb (5.2% of 12.57 Mb) deletion strain was generated. Here, we report cellular function profiling of the genome-reduced S. pombe, including growth efficiency, nutrient uptake efficiency, gene expression and metabolite production. We also report on the production of heterologous proteins in the genome-reduced S. pombe.
MATERIALS AND METHODS
Strains and culture conditions
S. pombe strains used in this study are shown in Table 1. S. pombe strains were grown on yeast extract with supplements (YES) medium, containing 0.5% Bacto yeast extract (BD, Sparks, MD, USA), 3% d-glucose (Wako, Osaka, Japan) and SP supplements (50 mg/l l-leucine, l-histidine hydrochloride monohydrate, l-lysine monohydrate, uracil and adenine sulfate); yeast extract peptone dextrose (YPD) medium, containing 1% Bacto yeast extract, 2% Bacto peptone (BD, Sparks, MD, USA) and 2% d-glucose; minimal medium with agar (MMA) plates supplemented with 250 mg/l l-leucine (Wako) and/or 100 mg/l uracil (Wako) and/or 500 mg/l 5-fluoroorotic acid monohydrate (5-FOA, Wako) as necessary for auxotroph selection; and Edinburgh minimal media (EMM, MP Biomedicals, Solon, OH, USA) for production of ethanol, enhanced green fluorescent protein (EGFP) and adenosine triphosphate (ATP).
Table 1.
Genome-reduced strains and heterologous protein production strains
| Strain | Genotype | Source | NBRP IDa |
|---|---|---|---|
| ARC010 | h− leu1-32 ura4-D18 | JY743 | |
| ARC001 | h− leu1-32 | ATCC38399 | |
| ARC032 | h− | JY1 | |
| IGF719 | h− leu1-32 ura4-D18 int::SPAC1F8.07cb ALT-D1797 | This study | |
| IGF718 | h− leu1-32 ura4-D18 int::SPAC1F8.07cb ALT-D1684 | This study | FY21635 |
| IGF524 | h− leu1-32 ura4-D18 ART-D1554 | This study | FY21636 |
| IGF525 | h− leu1-32 ura4-D18 BLT-D2117 | This study | FY21637 |
| IGF526 | h− leu1-32 ura4-D18 BRT-D1216 | This study | FY21638 |
| IGF740 | h− leu1-32 ura4-D18 int::SPAC1F8.07cb ALT-D1684 ART-D1554 BLT-D2117 BRT-D1216 | This study | FY21649 |
| IGF725 | h− leu1-32 ura4 int::SPAC1F8.07cc ALT-D1797 | This study | |
| IGF724 | h− leu1-32 ura4 int::SPAC1F8.07cc ALT-D1684 | This study | FY21650 |
| IGF628 | h− leu1-32 ART-D1554 | This study | FY21651 |
| IGF629 | h− leu1-32 BLT-D2117 | This study | FY21652 |
| IGF630 | h− leu1-32 BRT-D1216 | This study | FY21653 |
| IGF742 | h− leu1-32 ura4 int::SPAC1F8.07cc ALT-D1684 ART-D1554 BLT-D2117 BRT-D1216 | This study | FY21664 |
| IGF816 | h− leu1 int::SPBC1683.05d ura4-D18 BLT-D2117 | This study | |
| IGF817 | h− leu1 int::SPBC1683.06ce ura4-D18 BLT-D2117 | This study | |
| ASP3880 | h− leu1 int::EGFPf | This study | |
| ASP3881 | h− leu1 int::EGFPf ura4 int::SPAC1F8.07cc ALT-D1684 | This study | |
| ASP3882 | h− leu1 int::EGFPf ART-D1554 | This study | |
| ASP3883 | h− leu1 int::EGFPf BLT-D2117 | This study | |
| ASP3884 | h− leu1 int::EGFPf BRT-D1216 | This study | |
| ASP3894 | h− leu1 int::EGFPf ura4 int::SPAC1F8.07cc ALT-D1684 ART-D1554 BLT-D2117 BRT-D1216 | This study | |
| ASP3341 | h− leu1 int::hGHf | This study | |
| ASP4187 | h− leu1 int::hGHf ura4 int::SPAC1F8.07cc ALT-D1684 ART-D1554 BLT-D2117 BRT-D1216 | This study | |
| ASP3895 | h− leu1 int::hTFf | This study | |
| ASP3909 | h− leu1 int::hTFf ura4 int::SPAC1F8.07cc ALT-D1684 ART-D1554 BLT-D2117 BRT-D1216 | This study |
aThe NBRP ID indicates identification data in the Yeast Genetic Resource Center (YGRC, Osaka, Japan, http://yeast.lab.nig.ac.jp/nig/) as part of the National Bio Resource Project (NBRP). The genome-reduced strains constructed in this study were deposited with the YGRC.
bSPAC1F8.07c with endogenous promoter and terminator was integrated next to the ura4-D18 region.
cSPAC1F8.07c with endogenous promoter and terminator was integrated next to ura4 without ura4 disruption.
dSPBC1683.05 or eSPBC1683.06c with endogenous promoter and terminator, respectively, were integrated next to leu1 without leu1 disruption.
fHeterologous protein with hCMV promoter and hLPI terminator was integrated next to leu1 without leu1 disruption.
Construction of Latour fragments for S. pombe genome reduction
S. pombe genome-reduced strains were constructed using the Latour system as previously described (16). Modification fragments for targeting a deletion region, named a Latour fragment, were amplified from genomic DNA of S. pombe ARC032 using several primers (shown in Supplementary Table S1). The Latour fragments consisted of two homologous regions for integration into a targeted chromosomal locus, the ura4+ gene selection marker and an ∼200 bp direct repeat sequence that flanks the desired deletion region and that drives loop-out deletion (Supplementary Figure S2). The fragments were amplified using KOD Plus (Toyobo, Osaka, Japan), with an initial denaturation of 94°C for 2 min and 30 cycles of 10 s at 98°C, 30 s at 55°C and 1 min/kb at 68°C. Fragments harboring ∼20 bp of identical DNA sequence were fused by step-down PCR performed as follows: an initial denaturation of 94°C for 2 min, followed by 20 cycles of denaturation of 98°C for 10 s and extension of 68°C for 1 min/kb, annealing temperatures of 65°C, 63°C, 61°C, 59°C and 57°C for 30 s each for two cycles and 55°C for 30 s for 10 cycles.
Transformation
S. pombe strains were cultured in YES medium at 32°C for 48 h to stationary phase as a pre-culture. Fifteen microliters of pre-culture medium was inoculated into 5 ml of YES medium and grown to logarithmic phase (5 × 107 cell/ml) at 32°C for ∼16 h. Competent cells were prepared by the lithium acetate-based transformation method described previously (17). The transformed cells were selected on MMA plates, including leucine and/or uracil. Transformants were purified by single-colony isolation from the auxotrophic selection medium.
The 5-FOA treatment of S. pombe
The 5-FOA is metabolized in S. pombe by orotidine-5′-phosphate (OMP) decarboxylase, encoded by the SPCC330.05c/ura4 gene, to the toxic compound, 5-fluorouracil, causing death. A strain that has no working OMP decarboxylase shows 5-FOA resistance. In the Latour system, a 5-FOA-resistant strain indicates deletion of the targeted region harboring ura4 using loop-out homologous recombination at direct repeats (Supplementary Figure S2), or an ura4 mutant strain that has deactivated OMP decarboxylase.
S. pombe that had been transformed with the Latour fragment was cultured in YES medium at 32°C for 2 days until the stationary phase. A 100-µl aliquot of culture was spread on MMA medium, including 5-FOA and auxotrophic supplement, and incubated at 32°C until colony formation. The 5-FOA-resistant strains were purified by single-colony isolation.
Confirmation of genome modification
Colony PCR was used to confirm genome modification in S. pombe. Colonies grown on selective medium were picked with a sterile toothpick and transferred to 20 μl of zymolyase solution [2.5 mg/ml zymolyase 20T (Seikagaku biobusiness, Tokyo, Japan), 1.2 M sorbitol and 20 mM phosphate, pH 7.6] for single-colony isolation. The zymolyase solutions containing a selected colony were incubated at 37°C for ∼1 h. A 0.5-μl aliquot of the zymolyase solution containing a selected colony was transferred into 20 μl of KOD dash (Toyobo) PCR reaction mixture. The conditions for colony PCR consisted of an initial denaturation of 94°C for 5 min and 30 cycles of 94°C for 30 s, 50°C for 1 min and 74°C for 1 min/2 kb.
Measurement of maximum specific growth rate (μmax) and final cell density
S. pombe was cultured in 5 ml of YES medium at 32°C for 48 h as a seed culture. One hundred microliters of seed culture was inoculated into 5 ml of fresh YES medium and cultured at 32°C for 24 h as a pre-culture. A 5-μl aliquot of pre-culture medium was inoculated into 5 ml of fresh YES medium in an L-shaped test tube. Cells were cultured at 32°C with measurement of optical density at 660 nm (OD660) every 5 min using a bio-photorecorder TN-1506 and TVS062CA (Advantec Toyo, Tokyo, Japan). The μmax values were calculated from growth rates between OD660 = 0.05–2.0 using the following equation: μ = 2.303 (log10OD2–log10OD1)/(t2–t1). OD1 and OD2 are optical densities of cells at time t1 and t2, respectively. Among ∼150 time points, all combinations of two time points, t1 and t2, were used to calculate μ-values using Excel 2007 software (Microsoft, Redmond, WA, USA), and the maximum μ-value was taken as μmax.
The 4′,6-diamidino-2-phenylindole, dihydrochloride staining of S. pombe cells
S. pombe was cultured in YES medium at 32°C, and 1 ml of culture was harvested at the logarithmic phase (OD660 = 3.0). Collected cells were washed in 1 ml of ice-cold 1× phosphate-buffered saline and resuspended in a suitable volume of 1× phosphate-buffered saline. The cell solution was mixed with an equal volume of 50 μg/ml of 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Dojindo, Kumamoto, Japan) solution. The cells were observed and photographed with an Olympus BX50 microscope equipped with a BX-FLA fluorescence attachment (Olympus, Tokyo, Japan).
Measurement of glucose and ethanol concentrations in culture
S. pombe was cultured in 5 ml of EMM medium at 32°C after seeding with a pre-culture, as described for the measurement of the μmax rate. Cultures were collected during the logarithmic growth phase. Glucose and ethanol concentrations were measured in culture supernatants using a BF-5 enzyme electrode glucose or ethanol sensor (Oji scientific instruments, Hyogo, Japan), respectively, according to the manufacturer’s instructions.
Transcriptome analysis
Total RNA was isolated from S. pombe cells using an RNeasy mini kit (Qiagen, Germantown, MD, USA). Antisense RNA (aRNA) was synthesized from 1 μg of total RNA and labeled with Cy3 and Cy5 using an Amino Allyl Message Amp II aRNA kit (Ambion, Austin, TX, USA). The mixture of Cy3- and Cy5-labeled aRNA and hybridization buffer was hybridized on a 3D-Gene S. pombe 6 K Yeast Oligo Chip (Toray, Tokyo, Japan) for 16 h at 37°C. The hybridized array was scanned with a GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA), and fluorescence ratios were determined using GenePix software.
Metabolome analysis
S. pombe strains were cultured in EMM medium at 32°C to OD660 = 5.0 in the logarithmic phase. Approximately 1 × 109 cells were harvested with a suction-filtering system using a 0.4 μm pore size filter and washed twice with 10 ml of milli-Q water. The filter was then immersed in 2 ml of methanol containing 5 μM Internal Standard Solution 1 (provided by Human Metabolome Technologies) and sonicated for 30 s to re-suspend the cells into the methanol. All further steps, including the cell lysis treatment and capillary electrophoresis–time-of-flight mass spectrometer (CE–TOFMS) analysis, were performed by Human Metabolome Technologies (Tsuruoka, Japan) (18–20).
Measurement of ATP concentration
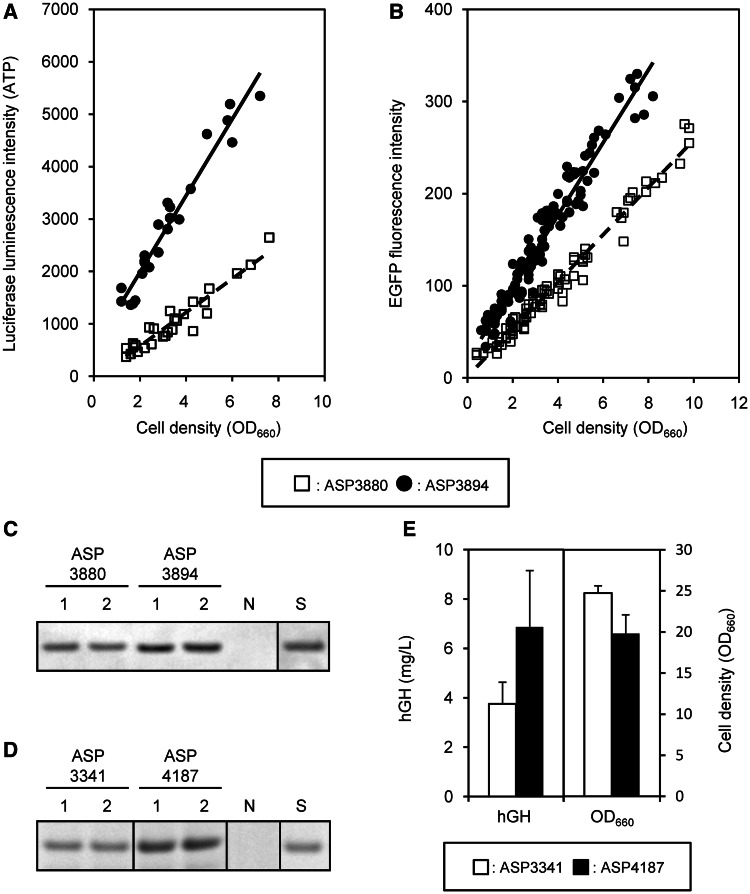
Intracellular ATP was measured using the BacTiter-Glo Microbial Cell Viability Assay Kit (Promega, Madison, WI, USA), which is a luciferase-based luminescence emission system. S. pombe strains grown in EMM medium were harvested every 2 h, and 100 μl of culture was added to 100 μl of BacTiter-Glo Regent. Luminescence was measured every minute for 15 min. The maximum luminescence value of each strain was used for comparisons of ATP concentration, as shown in Figure 7A.
Figure 7.
ATP concentration and heterologous protein production in the quadruple-deletion strain. (A) Time-dependent ATP concentrations were measured by luciferase analysis in six independent EMM cultures, and they were plotted to the intersection graph of luciferase intensity with cell density OD660. (B) Time-dependent EGFP production levels were measured by EGFP fluorescence analysis in ∼20 independent EMM cultures, and they were plotted to the intersection graph of EGFP fluorescence with cell density OD660. (C) EGFP production level in logarithmic phase (OD660 = 5.0) in two independent YPD cultures was measured by western blot and densitometric analysis. S: 2 ng of EGFP standard, N: ARC032 as negative control. (D) Secreted hGH in two independent YPD culture supernatant after 4 days was detected by SDS–PAGE analysis. S: 0.4 μg of hGH standard, N: ARC032 as negative control. (E) Average hGH concentration and cell density OD660 of six independent YPD (pH 6.0 buffering) cultures are shown. The hGH concentration was determined by densitometric analysis of hGH signals on a SDS–PAGE gel against hGH standard protein. The P-value is 0.028 for hGH concentration.
Measurement of EGFP fluorescence and concentration
EGFP-producing S. pombe was cultured at 32°C in EMM medium, and 200 μl of culture was harvested into a 96-well microtiter plate. EGFP fluorescence was measured at an excitation wavelength of 490 nm and emission wavelength of 530 nm using an MTP-810 Lab (Corona Electric, Hitachinaka, Japan) microplate reader. Background fluorescence was measured in S. pombe ARC032, which has no EGFP gene expression.
EGFP protein was measured by western blot analysis. S. pombe culture was grown to ∼OD660 = 5.0 at 32°C in YPD medium, and the cell pellet was lysed with CelLytic Y solution (Sigma, St. Louis, MO, USA). Cell lysis solution equivalent to 5 μl of culture before centrifugation was used for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis. For western blot analysis, mouse anti-GFP monoclonal antibody Ab-2 (Thermo Fisher Scientific, Fremont, CA, USA) and alkaline-phosphatase–labeled goat anti-mouse IgG (H + L) (Kirkegaard & Perry Laboratories, Gaithersburg, MD, USA) were used as first and second antibodies, respectively.
Measurement of secreted human growth hormone
Human growth hormone (hGH)-producing S. pombe was grown for 4 days at 32°C in YPD medium (pH 6.0) buffered with 0.3 M 2-morpholinoethanesulfonic acid monohydrate (MES, Dojindo, Kumamoto, Japan) to reduce acidic protease activity in the culture supernatant. One hundred microliters of culture supernatant was mixed with 10 μL of cold trichloro-acetic acid (TCA, Wako, Osaka, Japan) and chilled on ice for 1 h. The chilled TCA solution was centrifuged at 14 000 g for 20 min. The precipitate was washed twice with cold ethanol to remove TCA. Dried precipitate was dissolved in 10 μl of dye solution and analyzed by SDS–PAGE. Secreted hGH concentration was estimated by densitometric analysis against hGH standard protein (AspenBio Pharma Inc., CO, USA).
RESULTS
Examination of regions tolerant of large-scale deletion at the termini of chromosomes I and II
To reduce the S. pombe genome, we explored large deletable regions containing non-essential genes. By comparing systematic deletion information from S. cerevisiae (9), we inferred that large regions at the termini of both the left and right arms of S. pombe chromosomes I and II contained only non-essential genes. To examine the most-terminal essential genes required for viability in a minimal medium, the Latour system was used for partial deletion analyses (16). Deletion analysis revealed that the four deficient strains, SPAC13G6.05c/trs33 on the left arm of chromosome I (ALT), SPAC29B12.07/sec16 on the right arm of chromosome I (ART), SPBC1198.04c/zas1 on the left arm of chromosome II (BLT) and SPBC1289.12/usp109 on the right arm of chromosome II (BRT), did not grow on MMA medium. We determined that trs33, sec16, zas1 and usp109 are the last essential genes at the termini of chromosomes I and II (Supplementary Figure S1) and are required for viability in minimal medium. We, therefore, inferred that the terminal regions located on the telomeric sides of the four essential genes could be deleted.
In the S. pombe genome database, PomBase (http://www.pombase.org/), SPAC1F8.07c, SPBC1348.06c, SPBC359.02/alr2 and SPBC1198.02/dea2 are also designated as essential genes and are located on the telomeric sides of trs33 and zas1, respectively. However, we determined that SPAC1F8.07c, SPBC1348.06c, alr2 and dea2 were deletable. Furthermore, deletion of the genes, except for SPAC1F8.07c, which encodes a putative pyruvate decarboxylase, had no effect on growth rate. Because the SPAC1F8.07c-deficient strain exhibited a highly reduced growth rate (data not shown), SPAC1F8.07c was left in the genome to maintain growth stability. The telomeric regions were also maintained for this reason.
Deletion of large-scale regions from the termini of chromosomes I and II
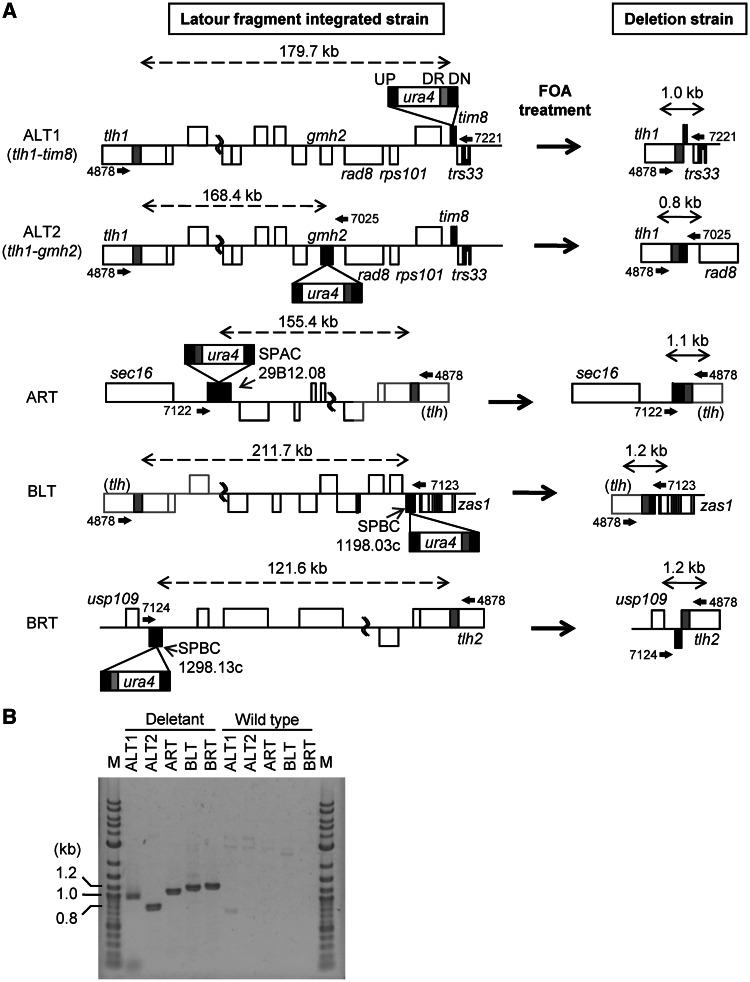
Large-scale deletion was carried out using the Latour system, which requires a direct repeat sequence outside of a target region to drive loop-out deletion (Supplementary Figure S2). ALT, ART and BLT had undetermined regions at the terminal ends of the chromosomes, and the most-terminal genes on the three chromosome ends were uncertain. In contrast, the sequence of the BRT region has been completely determined up to the telomeric repeat. We speculated that the three unsequenced regions were almost identical to the terminal of BRT because the available sequence for the termini of the left and right arms of chromosomes I and II showed a high level of sequence similarity (12,21,22). The highly homologous sequenced regions (>98%) located within the terminal regions are ∼41 (ALT), 20 (ART), 21 (BLT) and 40 kb (BRT) from the SPAC977.06 (ALT), SPAC750.02c (ART), SPBC1348.07 (BLT) and SPBPB2B2.14c (BRT) genes, respectively (Supplementary Figure S3). Therefore, ∼200 bp of direct repeat sequence for use as a Latour fragment was designed from the internal sequence of SPBCPT2R1.08c/tlh2 in BLT. The terminal deletion regions were designed to maintain the intact telomeric regions on the chromosomes in genome-reduced strains. The direct repeat sequence from tlh2 was also used for ART and BLT deletions, and the fragment also included the SPAC212.11/tlh1 sequence on ALT.
The Latour fragments, consisting of a direct repeat and ura4+ selection marker, were inserted into SPAC13G6.04/tim8 (ALT), SPAC29B12.08/clr5 (ART), SPBC1198.03c (BLT) and SPBC1289.13c (BRT) in proximity to the essential genes, by homologous recombination. To delete a target region by homologous recombination, the strains carrying the Latour fragment and ura4+ marker were cultured on 5-FOA selection medium. The 5-FOA-resistant strains were confirmed by PCR, using primers located outside of a targeted deletion region (Figure 1A, Supplementary Table S1 and sequence files). A ∼1-kb fragment was amplified from the chromosome of the 5-FOA-resistant strains containing a successful loop-out deletion and resultant genome reduction (Figure 1B). In contrast, if the target regions were not deleted, a fragment was not amplified because the annealing locations of the primers were >100 kb apart. Large-scale chromosome deletions in the termini of the left and right arms of chromosomes I and II successfully occurred (deletion details are described in the Supplementary Sequence Files). The results suggested that the unsequenced terminal regions of ART and BLT have similar sequences to that of the BRT terminus. The ALT deletion region between tlh1 and tim8 was 179 712 bp, including a predicted 1717-bp gap-filling sequence. The ART deletion region between clr5 and a putative tlh gene was 155 419 bp, including a predicted 6102 kb terminal unsequenced region. The BLT deletion region between a putative tlh gene and SPBC1198.03c was 211 745 bp, including a predicted 17 139 bp terminal unsequenced region and a 17 881-bp gap-filling sequence determined in our previous study (13). The BRT deletion region between SPBC1289.13c and tlh2 was 121 649 bp. SPAC1F8.07c was inserted adjacent to the SPCC330.05c/ura4 locus before ALT deletion to maintain growth stability. The deletion genes were categorized using the S. pombe GO slim gene association file (http://www.pombase.org/browse-curation/fission-yeast-go-slim-terms) in PomBase (Table 2), and the deletion genes are listed in Supplementary Table S2.
Figure 1.
Large-scale deletions of chromosome terminal fragments for genome reduction. (A) Schematic diagrams of large-scale deletions using Latour fragments. The Latour fragment, indicated by the box, consists of four regions: UP, indicated by black box: upstream region of targeted site of recombination for integration of Latour fragment into the chromosome; ura4, indicated by white box: selection marker; DR, indicated by gray box: direct repeat outside of targeted deletion region for homologous recombination by FOA treatment; DN, indicated by black box: downstream region of targeted site of recombination for integration of Latour fragment into the chromosome. Black boxes on each chromosome represent an integration site for the Latour fragment, and the gray box is a direct repeat included in the Latour fragment. Undetermined regions of nucleotide sequence are indicated by gray lines, and they are estimated from the right arm of chromosome II. Arrows above and below the chromosome show primers used to confirm large-scale deletion. Numbers near the arrows give the primer ID. Double-headed dashed arrows show the length of the targeted deletion region, and double-headed solid arrows show the length after the large-scale deletion by FOA treatment. (B) Electrophoresis of DNA to confirm large-scale deletion. The fragments generated after deletion of terminal regions were confirmed by direct colony PCR from deletion strains using primers described in Figure 1A. In the wild-type strain, the distances between primer pairs were >120 kb; therefore, no fragment was detected. M: 2-log DNA ladder (New England Biolabs, Ipswich, MA, USA).
Table 2.
Summary of deletion genes
| Name | Term | ALT | ART | BLT | BRT | Subtotal |
|---|---|---|---|---|---|---|
| Transmembrane transport | GO:0055085 | 8 | 10 | 20 | 3 | 41 |
| Cellular amino acid metabolic process | GO:0006520 | 4 | 5 | 4 | 1 | 14 |
| Generation of precursor metabolites and energy | GO:0006091 | 3 | 3 | 2 | 0 | 8 |
| Protein glycosylation | GO:0006486 | 3 | 0 | 0 | 4 | 7 |
| Carbohydrate metabolic process | GO:0005975 | 0 | 2 | 2 | 2 | 6 |
| Cell adhesion | GO:0007155 | 2 | 1 | 2 | 1 | 6 |
| Regulation of transcription, DNA-dependent | GO:0006355 | 2 | 0 | 4 | 0 | 6 |
| DNA recombination | GO:0006310 | 2 | 0 | 0 | 1 | 3 |
| Vitamin metabolic process | GO:0006766 | 2 | 0 | 0 | 1 | 3 |
| Lipid metabolic process | GO:0006629 | 1 | 0 | 1 | 0 | 2 |
| Meiosis | GO:0007126 | 2 | 0 | 0 | 0 | 2 |
| Nucleobase-containing small molecule metabolic process | GO:0055086 | 0 | 0 | 2 | 0 | 2 |
| Transcription, DNA-dependent | GO:0006351 | 1 | 1 | 0 | 0 | 2 |
| DNA replication | GO:0006260 | 0 | 0 | 0 | 1 | 1 |
| Cellular cell wall organization or biogenesis | GO:0070882 | 0 | 1 | 0 | 0 | 1 |
| Chromosome segregation | GO:0007059 | 1 | 0 | 0 | 0 | 1 |
| Cofactor metabolic process | GO:0051186 | 0 | 0 | 1 | 0 | 1 |
| Conjugation with cellular fusion | GO:0000747 | 0 | 1 | 0 | 0 | 1 |
| Cytoskeleton organization | GO:0007010 | 0 | 0 | 0 | 1 | 1 |
| Nitrogen cycle metabolic process | GO:0071941 | 0 | 0 | 0 | 1 | 1 |
| Protein targeting | GO:0006605 | 0 | 1 | 0 | 0 | 1 |
| Signaling | GO:0023052 | 0 | 1 | 0 | 0 | 1 |
| Other | 23 | 22 | 16 | 13 | 74 | |
| Conserved unknown | 5 | 1 | 2 | 3 | 11 | |
| Sequence orphan | 6 | 1 | 3 | 3 | 13 | |
| Pseudogene | 5 | 0 | 4 | 4 | 13 | |
| Dubious | 0 | 1 | 0 | 0 | 1 | |
| Total deletion genes | 70 | 51 | 63 | 39 | 223 |
The deletion genes were categorized using GO slim annotation associated with PomBase. Genes annotated as ‘conserved unknown’, ‘sequence orphan’, ‘pseudogene’ and ‘dubious’ are categorized separately from GO slim annotation. Non-categorized genes are shown as ‘other’. Supplementary Table S2 shows details.
Modification of the deletion region for recovery of growth rate
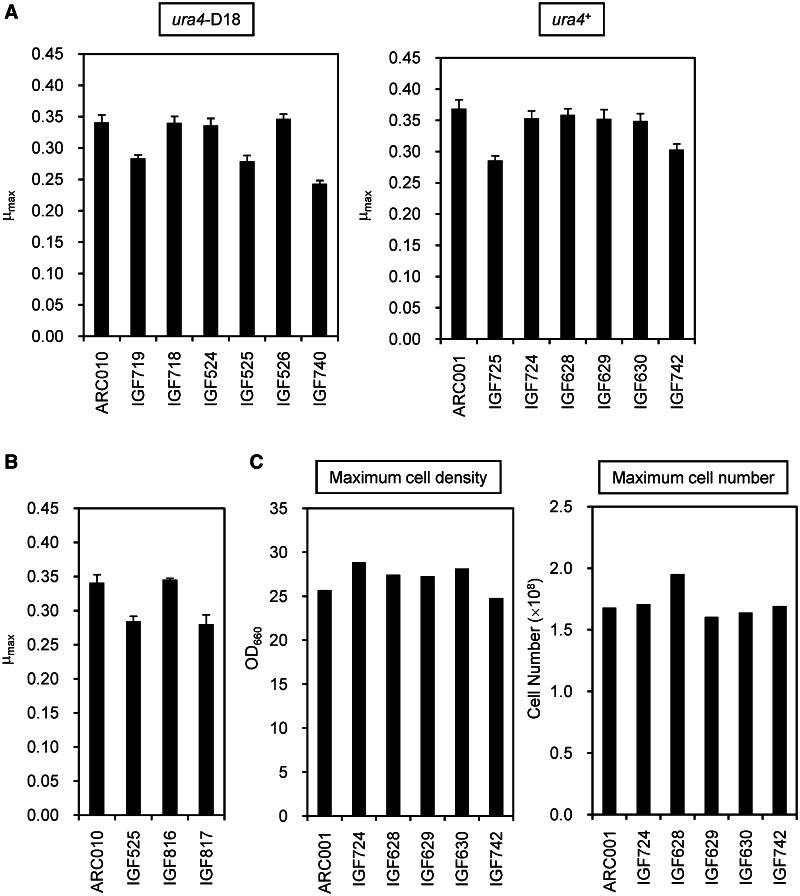
Maintaining a growth rate similar to that of the parental strain is important in the process of genome reduction. The genome-reduced strains with single-terminal region deletions were initially characterized by measuring the maximum specific growth rate (μmax) in YES medium at 32°C. IGF719 and IGF525 showed decreased growth rates (Figure 2A), suggesting that the decrease was associated with deleted genes in ALT and BLT. To identify the gene responsible for recovering the growth rate of IGF719, genes deleted from ALT except for SPAC1F8.07c were re-inserted next to the SPBC1A4.02c/leu1 locus. Reconstructed strains containing the SPAC13G6.01c/rad8 and SPAC13G6.02c/rps101 genes showed a similar growth rate to the parental strain (data not shown). In the ALT deletion region, rad8 and rps101 are located near trs33; hence, the deletion region was shortened to the region between tlh1 and SPAC5H10.13c/gmh2 (Figure 1A). The growth recovery strain, named IGF718, had a deletion size of 168 447 bp and grew at a similar rate to the non-deleted parent strain ARC010 (Figure 2A).
Figure 2.
Growth properties affected by genome reduction. (A) Maximum specific growth rates (μmax) of the genome-reduced strains. The ura4+ strains were generated by reintroducing ura4+ into ura4-D18 strains. (B) Recovery of growth efficiency by reintroduction of a predicted uracil transporter. SPBC1683.05 (IGF816) and SPBC1683.06c (IGF817), including intergenic regions between upstream and downstream genes, were integrated between leu1-32 and top2 using a leu1 selection marker. (C) Maximum cell density and cell number of the genome-reduced strains. The strains were cultured in YES medium. The values of the highest cell density and cell number during culture were defined as the maximum cell density and the maximum cell number, respectively. The cell densities were measured with a 96-well plate reader, and the cell numbers were measured using a hemocytometer.
In S. pombe, ura4+ is frequently used as an auxotrophic marker. However, we have observed that ura4-deficient strains tend to lyse in YPD medium, which contained rich nutrient sources (data not shown). We re-inserted ura4+ at the original locus on chromosome III to maintain a stable growth rate in the genome-reduced strains. As a result, almost all ura4+ re-insertion strains showed an improved growth rate in YES (Figure 2A) and YPD media (data not shown).
IGF629, generated by re-insertion of ura4+ into IGF525, showed improvement of growth rate to the level of the parental strain. We postulated that the BLT deletion region included a gene related to the UMP biosynthesis pathway, because ura4 is an intermediate gene in the UMP de novo pathway. In comparison with S. cerevisiae, SPBC1683.05 is homologous to uridine permease YBL042C/FUI1 (32% amino acid identity) and uracil permease YBR021W/FUR4 (28%), whereas SPBC1683.06c is homologous to uridine nucleosidase YDR400W/URH1 (31%). IGF816, constructed by re-insertion of SPBC1683.05 into IGF525, recovered its growth rate to the level of the parental strain (Figure 2B). From this we inferred that SPBC1683.05 relates to uracil uptake in UMP biosynthesis in S. pombe.
Based on the aforementioned growth analysis results, we used the ura4+ strains carrying the modified ALT deleted region between tlh1 and gmh2 in the following analysis. If necessary, SPBC1683.05 could be re-inserted into the BLT plus ura4 deletion strain in an additional genetic modification step because decreased growth rate in IGF525 caused by SPBC1683.05 deletion is complemented by ura4+ re-insertion.
The order of deletion of the chromosomal terminal regions is key to construction of a quadruple-deletion strain
Construction of triple-deletion strain ΔART BLT BRT was relatively straightforward and resulted in a deletion of 488.8 kb. To construct a quadruple-deletion strain, SPAC1F8.07c was inserted adjacent to the ura4 locus in the ΔART BLT BRT strain to maintain growth stability, and the Latour fragment for ALT deletion was integrated into the ALT region of the triple-deletion strain. An initial PCR screen of 5-FOA-resistant strains revealed that a quadruple-deletion strain was not obtained from >200 candidate colonies. We then attempted several different strategies to construct the quadruple-deletion strain: ART deletion from the ΔALT BLT BRT strain, BLT deletion from the ΔALT ART BRT strain and BRT deletion from the ΔALT ART BLT strain. By changing the order of deletion, a quadruple-deletion strain was successfully constructed. The ratio of ART or BRT deletion from each triple deletion strain was 94 and 70%, respectively. In contrast, the ratio of BLT deletion from the triple deletion strain was 19%. Thus, the order of region deletion was important for constructing a quadruple-deletion strain.
We examined why the deletion efficiency of ALT or BLT from each triple-deletion strain was low. A partial fragment in the ALT deletion region was re-inserted next to the leu1 locus in the ΔART BLT BRT strain. The deletion ratio of ALT from the ΔART BLT BRT strain with the re-inserted partial fragment was then examined. The strain with inserted SPAC977.11 showed an improvement of deletion efficiency to 100%. In the same way, re-insertion of SPAC977.11 led to improvement of the BLT deletion ratio from 19 to 88% in the ΔALT ART BRT strain. The BLT deletion region contains a region identical to SPAC977.11 and SPAC977.12 in the ALT deletion region. However, SPAC977.12 had no effect on the deletion efficiency. SPAC977.11 codes for a CRCB domain protein (PF02537 in the Pfam protein families database), and CrcB in E. coli is a predicted membrane protein and functions in chromosome condensation (23). Although it is not clear how SPAC977.11 functions to influence deletion efficiency, SPAC977.11 is likely to play a role in the deletion efficiency of ALT or BLT from the ΔART BLT BRT and ΔALT ART BRT strains, respectively.
The quadruple-deletion slightly affects growth rate and cell size
We evaluated growth efficiency of the quadruple-deletion strain in YES medium. Single-deletion strains, IGF724, IGF628, IGF629 and IGF630, showed no decrease in μmax. However, the quadruple-deletion strain, IGF742, showed a slight decrease in comparison with the other strains (Figure 2A), although the maximum cell density and the maximum cell number of the quadruple-deletion strains reached the same level as the non-deleted parent strain (Figure 2C).
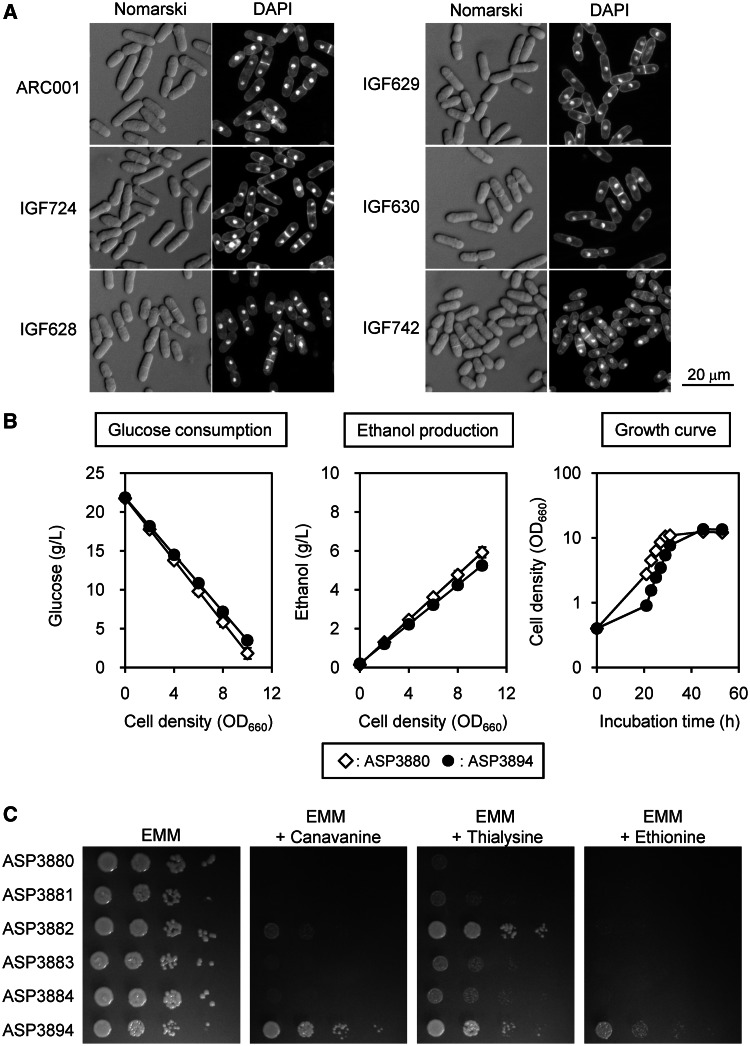
To analyze cell morphology, genome-reduced strains were cultured in YES medium to an OD660 of 3.0, and cells were then observed directly using Nomarski differential interference contrast optics and DAPI fluorescence microscopy. Genome-reduced strains showed few morphological defects and little abnormal nuclear distribution (Figure 3A). The cell size of genome-reduced strains, especially in IGF742, was reduced in comparison with the parental strain (Table 3). Nutrient stress has been reported to result in reduced cell size (24), raising a possibility that IGF742 may have some defect in nutrient uptake.
Figure 3.
Cell morphology, nutrient uptake properties and ethanol production of the genome-reduced strains. (A) Cells were visualized under an Olympus BX50 microscope equipped with an UPlanFL ×40 objective lens and WH 10 × 22 eyepiece. DAPI fluorescence images were collected using a DAPI-specific filter set. (B) Glucose consumption, ethanol production and growth curves of ASP3880 and ASP3894 cultured in EMM medium. The linear graphs of glucose consumption and ethanol production were drawn using average values of five independent cultures, estimated by linear approximations (Supplementary Figure S4 and Supplementary Table S3 show details). (C) Sensitivity to amino acid analogs. Cultures of the genome-reduced strains were serially diluted 10-fold, and 3 µl of dilute solution was spotted on the plates. The plates were incubated at 32°C for 5 days. Canavanine, 60 mg/l; thialysine, 200 mg/l; ethionine, 100 mg/l.
Table 3.
Cell size of genome-reduced strains
| Strain | Deletion region | Cell length (μm) | SD(±) | P-value |
|---|---|---|---|---|
| ARC001 | 14.0 | 0.8 | ||
| IGF724 | ALT | 12.7 | 0.9 | 1.5E-10 |
| IGF628 | ART | 11.8 | 1.1 | 6.2E-19 |
| IGF629 | BLT | 12.8 | 1.1 | 2.5E-07 |
| IGF630 | BRT | 12.0 | 1.2 | 1.2E-15 |
| IGF742 | ALT BRT BLT BRT | 10.9 | 1.1 | 1.0E-27 |
The cell lengths of 50 septated cells were measured using ImageJ and averaged. The P-value of genome-reduced strains was calculated against ARC001.
Glucose uptake efficiency is decreased during late logarithmic phase in the quadruple-deletion strain
To determine glucose uptake efficiency, non-auxotrophic strain ASP3880 and genome-reduced strain ASP3894, derived from ARC001 and IGF742, respectively, by re-insertion of leu1+, were incubated in EMM medium. Although lag phase in ASP3894 tended to be slightly longer than that of ASP3880 in EMM medium, the generation times were similar (Figure 3B). In addition, ASP3894 showed a slight but significant decrease in glucose consumption (P = 0.0080, t-test, n = 5) and ethanol production (P = 0.0046) compared with ASP3880 in the late logarithmic phase (Figure 3B, Supplementary Figure S4 and Supplementary Table S3).
S. pombe has eight hexose transporters. In ASP3894, three hexose transporters, SPAC1F8.01/ght3, SPBC1683.08/ght4 and SPBC1348.14c/ght7, were deleted. Ght3 and Ght4 were previously reported to not transport glucose in S. cerevisiae complementation tests (25). Functional analysis of Ght7 has not yet been reported, and ght7 expression was not detected in the logarithmic phase in transcriptome analysis (data not shown). Therefore, deletion of the three hexose transporters is unlikely to significantly affect glucose uptake rate in ASP3894.
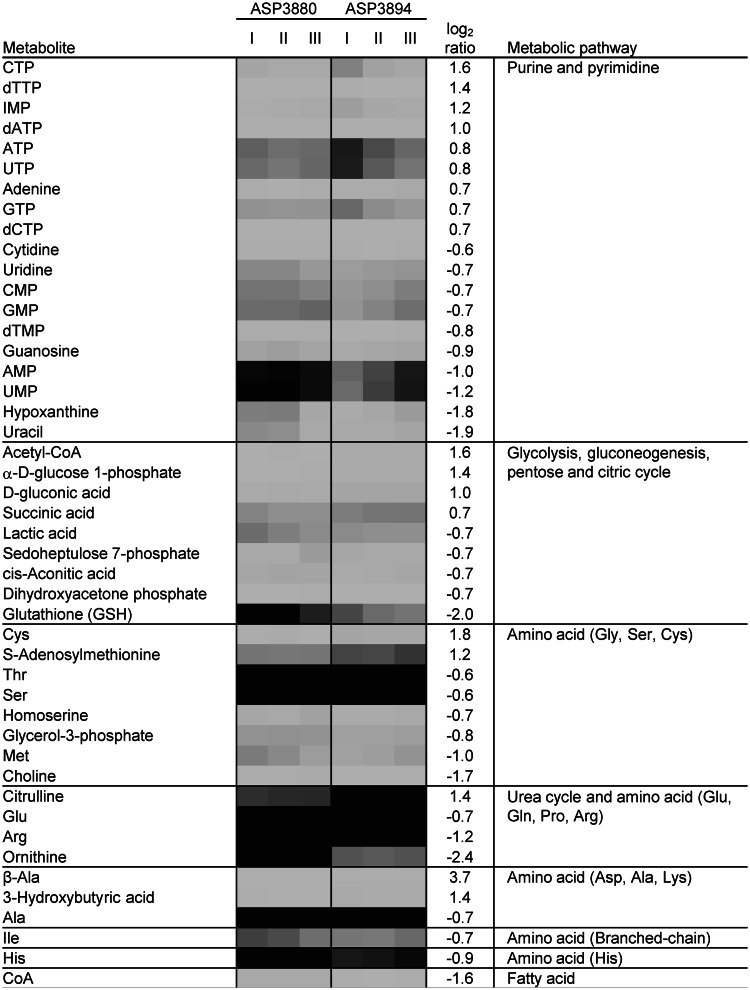
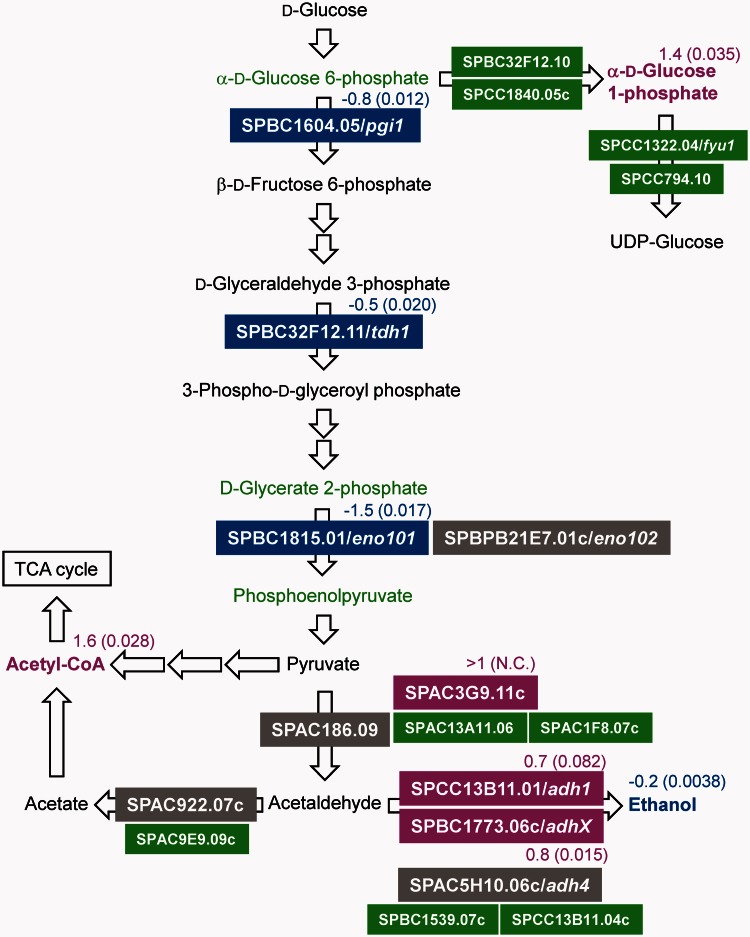
Metabolome analysis (described later in the text) showed that α-d-glucose 1-phosphate was increased in ASP3894 (Figure 6). The slight reduction of glucose uptake efficiency was postulated to be related to accumulation of α-d-glucose 1-phosphate. However, the transcriptome analyses (described later in the text) showed gene expression of the phosphoglucomutases, SPBC32F12.10 and SPCC1840.05c, which convert α-d-glucose 6-phosphate to α-d-glucose 1-phosphate, to be almost equal to that of ASP3880 (Figure 4). UTP-glucose-1-phosphate uridylyltransferase genes, SPCC1322.04/fyu1 and SPCC794.10, which convert α-d-glucose 1-phosphate to UDP-glucose, were also expressed at equal levels. Therefore, the reason for the increased level of α-d-glucose 1-phosphate in ASP3894 is unknown.
Figure 6.
Metabolite profile of the quadruple-deletion strain in logarithmic phase growth. Cells were harvested from three different logarithmic phase cultures (OD660 = 5.0) of each strain. Cellular metabolite mass was estimated by CE–TOFMS analysis, and the quadruple-deletion strain ASP3894 was compared with the parental strain ASP3880. The metabolites for which mass levels had changed in a log2 ratio of more than ±0.5 between ASP3880 and ASP3894 were categorized by each metabolic pathway. The log2 ratio is the average of three independent analyses. The gray shading indicates cellular metabolite concentration. The dark gray indicates high metabolite concentration and the light gray indicates low-metabolite concentration.
Figure 4.
Change of gene expression and metabolite in glycolytic pathway. The gene expression level and metabolite concentration of ASP3894 were compared with ASP3880. These data, except for ethanol, were obtained from transcriptome analysis and metabolome analysis. The ethanol concentrations were measured by an enzyme electrode sensor BF-5 (Figure 3B). Gene name is shown in color filled box. Metabolite name is shown without box. The colors represent increased or decreased levels of gene expression and metabolite: blue, decrease; green, equal; magenta, increase; black, not detected; gray, deletion gene in ASP3894. The log2 ratio (ASP3894/ASP3880) is shown near the gene or metabolite (N.C. = not calculated.). The parenthetical number indicates P-value of three independent transcriptome or metabolome analyses. Non-indicated genes and metabolites in the glycolytic pathway showed no significant change in comparison of ASP3894 with ASP3880.
On the other hand, ethanol production in ASP3894 was slightly reduced (Figure 3B). In the glycolytic pathway, although amounts of α-d-glucose 6-phosphate, d-glycerate 2-phosphate and phosphoenolpyruvate were not significantly changed, gene expression of SPBC1604.05/pgi1, coding for glucose-6-phosphate isomerase, SPBC32F12.11/tdh1, coding for glyceraldehyde-3-phosphate dehydrogenase, and especially SPBC1815.01/eno101, coding for enolase, was decreased in ASP3894 (Figures 4 and 5C). SPAC186.09, coding for a predicted pyruvate decarboxylase, and SPAC5H10.06c/adh4, coding for alcohol dehydrogenase, are in the ethanol fermentation pathway and were deleted in ASP3894. SPAC186.09 was not expressed in ASP3880; therefore, deletion of this gene in ASP3894 may have little influence on ethanol production. Although adh4 was expressed at a low level in ASP3880, SPCC13B11.01/adh1, which is the main alcohol dehydrogenase (26), was still present in ASP3894, and its expression was induced (Figure 4). The deletion of adh4 is also likely to have little influence on ethanol production. The reduction in ethanol seemed to result from a decrease in the carbon source flow in the glycolytic pathway. Therefore, the slight reduction of glucose uptake in ASP3894 may result from decreased efficiency of the glycolytic pathway.
Figure 5.
Gene expression profile of the quadruple-deletion strain in the logarithmic phase. Gene expression levels were determined by transcriptome analysis, and the quadruple-deletion strain ASP3894 was compared with the parental strain ASP3880. Total RNA was extracted from three independent logarithmic phase cultures (OD660 = 5.0) of each strain. The transcriptome analysis was performed three times using each total RNA. Numbers above the strain numbers are serial numbers of each transcriptome analysis. The genes with expression levels that were reproducibly changed were categorized using the GO biological process. (A–D) a gene list of change in log2 ratio more than ±1; (E) more than ±0.5. Gray shading indicates fluorescence intensity of Cy 5 (ASP3880) or Cy 3 (ASP3894). Dark gray and white indicate high- and low-fluorescence intensity, respectively. The log2 ratios are the average of three transcriptome analyses, and N. A. indicates not available, as no fluorescence intensity in either strain. (A) Conjugation with cellular fusion (GO:0000747), (B) carbohydrate metabolic process (GO:0005975), (C) generation of precursor metabolites and energy (GO:0006091), (D) cellular amino acid metabolic process (GO:0006520) and (E) nitrogen cycle metabolic process (GO:0071941).
Amino acid uptake efficiency is decreased in the quadruple-deletion strain
To measure amino acid uptake efficiency, the genome-reduced strains were cultured on EMM agar plates containing toxic amino acid analogs canavanine, thialysine and ethionine. ASP3894 showed resistance to these toxic amino acid analogs (Figure 3C), suggesting that uptake of arginine, lysine and methionine was reduced in ASP3894. Non-auxotrophic strain ASP3882, derived from IGF628, also showed resistance to canavanine and thialysine. We inferred that the main cause for the observed decrease in uptake of arginine and lysine was the deletion of SPAC869.11/cat1, which is a basic amino acid permease located in the ART deletion region (27). Furthermore, ASP3894 showed strong resistance to canavanine and ethionine compared with the single-deletion strains. For ASP3894, 223 genes were deleted, which were 70 genes in ALT, 51 genes in ART, 63 genes in BLT (including five genes in gap-filling sequence) (13) and 39 genes in BRT deletion region. Nine of the 223 deleted genes are categorized as ‘amino acid transmembrane transport (GO:0003333)’ by the Gene Ontology (GO) biological process in PomBase; ART contains such four genes, BLT each contains such three genes and ALT and BRT each contain one such gene. Therefore, it is likely that about one-third of the 27 amino acid transporters in the ‘amino acid transmembrane transport (GO:0003333)’ are deleted in the quadruple-deletion strain. Hence, we speculated that the multiplex deletion of nine amino acid transporters caused a decrease in amino acid uptake, and minor arginine or methionine transporters are located in the right and left arms of chromosomes I and II.
Expression of sexual development genes and genes associated with non-glucose metabolism is induced in the quadruple-deletion strain
To examine the effect of genome reduction on gene expression, transcriptome analysis was carried out. Gene expression in ASP3894 was compared with that in ASP3880, after culture in EMM medium to the logarithmic phase (OD660 = 5.0). Genes with log2 ratio (ASP3894/ASP3880) expression changes of >2-fold (either positive or negative) were mainly those with roles in ‘conjugation with cellular fusion (GO:0000747)’ and ‘carbohydrate metabolic process (GO:0005975)’, as categorized using the S. pombe GO slim annotation associated with PomBase. The genes that showed similar expression changes in three independent transcriptome analyses were used in the categorization.
Expression of M-factor precursors categorized as being involved in ‘conjugation with cellular fusion (GO:0000747)’ increased in ASP3894 (Figure 5A). However, ASP3894 total RNA was extracted in the logarithmic phase, at which time ammonia, used as a nitrogen source, remained in the culture medium. The proliferation rate of ASP3894 was not reduced, and the growth of ASP3894 was independent of the uptake of some amino acids because EMM medium contained no amino acids as a nitrogen source. Therefore, the increases in M-factors were speculated to be independent of the deficiency in amino acid uptake. Depression of Tor2 function, which is a component of TORC1, was reported to mimic the response to nitrogen starvation and initiate sexual development (28). In addition, depression of Tor2 function caused small cell morphology (28–30). We noted that ASP3894 derived from IGF742 was also slightly small. It would be interesting to envisage that activity of Tor2 in ASP3894 was decreased.
In terms of ‘carbohydrate metabolic process (GO:0005975)’, expression of the SPAPB24D3.10c/agl1 gene, coding for extracellular maltose α-glucosidase (31), and the SPAC22A12.11/dak1 gene, coding for dihydroxyacetone kinase in the glycerol assimilation pathway (32), was induced in ASP3894 (Figure 5B). The genes were reported to be induced under glucose limitation. However, the expression of SPBC1198.14c/fbp1 and SPCC191.11/inv1, which are the main glucose-repressed genes, was not induced in ASP3894. SPBC1683.07/mal1 and SPAC977.16c/dak2, which have the same function as agl1 and dak1, were deleted in ASP3894. Both mal1 and dak2 were originally expressed at a low level in ASP3880 (data not shown); therefore, it seems unlikely that the increase in agl1 and dak1 expression was a phenomenon resulting from deletions of mal1 and dak2. At late logarithmic phase, glucose uptake by ASP3894 was slightly reduced (Figure 3B). Therefore, it is likely that the genome-reduction causes a partial diauxic shift in the late logarithmic phase of ASP3894 growth.
Gene expression and metabolite concentrations in the ammonia metabolic cycle are altered in the quadruple-deletion strain
The concentrations of metabolites related to the urea cycle in ASP3894 were altered in comparison with ASP3880. In S. cerevisiae, ammonia as a nitrogen source in EMM medium is used by two metabolic reactions; it is transferred to 2-oxoglutarate for production of glutamate by nicotinamide adenine dinucleotide phosphate (NADP)-specific glutamate dehydrogenase, and it is transferred to glutamate for production of glutamine by glutamine synthase (33). In S. pombe, the NADP-specific glutamate dehydrogenase encoded by SPCC622.12c/gdh1, which is categorized as ‘cellular amino acid metabolic process (GO:0006520)’ in the S. pombe GO slim annotation associated with PomBase, was reported to play an important role in ammonia assimilation (34,35). Mutation of gdh1 resulted in slow growth in liquid minimal medium with ammonium as the sole nitrogen source. Expression of gdh1 in ASP3894 was increased in EMM medium (Figure 5D), but glutamate levels in ASP3894 were decreased (Figure 6). Therefore, glutamate use is likely to be increased in ASP3894. Glutamate is used as an amino group donor in amino acid synthetic pathways. ASP3894 showed no significant increase in total amino acids. Although we have not performed any further analysis, it remains possible that both biosynthesis of amino acids from glutamate and their use are augmented in this strain.
For the urea cycle of ASP3894, citrulline concentration increased, whereas concentration of arginine and ornithine decreased (Figure 6). S. pombe has two arginases, encoded by SPBP26C9.02c/car1 and SPAC3H1.07/aru1. Arginine is hydrolyzed to ornithine and urea by arginase. In ASP3894, aru1 expression was slightly induced (Figure 5E), which may partly explain the decrease of arginine.
ATP concentration is increased in the quadruple-deletion strain
Metabolome analysis using CE–TOFMS showed that changes in metabolite concentration in the logarithmic phase were mainly observed in purine and pyrimidine biosynthesis pathways. Interestingly, the concentration of nucleoside triphosphates, including ATP, which play an important role as energy sources or activators in cell metabolic processes, tended to be increased, in contrast to the decrease in the nucleoside monophosphates in ASP3894 (Figure 6). Variance of ATP concentration among three independent ASP3894 cultures was speculated to be caused by different lag times between sampling and cell fixation because ATP is rapidly used for cellular metabolism. To reduce potential errors, the ATP concentration was also measured immediately after time-course sampling with an ATP bioluminescence assay kit (BacTiter-Glo), which is a luciferase-based luminescence emission system; therefore, the lag time between sampling and measurement was minimized. Increase in ATP concentration in the logarithmic phase was proportional to cell density, and ASP3894 produced approximately 2.7-fold more ATP than ASP3880 (Figure 7A). There are two possibilities for the observed differences in ATP concentration: the increase in ATP may be caused by decreases in the amount consumed; or the ATP synthetic pathway may be activated. A couple of our findings are consistent with the latter interpretation. IMP, which is an intermediate metabolite of the ATP synthetic pathway, was increased in ASP3894. In addition, the level of succinic acid in ASP3894 was increased compared with ASP3880 (Figure 6). The conversion of succinic acid to fumaric acid, which is a part of the electron transport chain generating ATP, may be activated. Glutamate is changed to succinic acid via 2-oxoglutarate or 4-Aminobutanoate (GABA). Increasing the expression of gdh1 may provide additional substrate to facilitate electron transport and increase ATP generation. In any cases, the observed increase in ATP concentration suggested that the intracellular energy level was increased in ASP3894.
Expression of heterologous proteins is increased with reduction in genome size
We evaluated the production of heterologous proteins in the quadruple-deletion strain that exhibited increased ATP concentration. Heterologous protein expression cassettes with a human cytomegalovirus promoter (hCMV) and human lipocortin I terminator (hLPI) were integrated into the ARC001 and IGF742 genomes between leu1 and top2 on chromosome II using a previously described method (36).
First, growth-dependent changes in intracellular EGFP levels were measured in the logarithmic phase of cultures in EMM medium. ASP3894, derived from IGF742, exhibited a 1.7-fold increase in intensity of EGFP fluorescence compared with ASP3880 (Figure 7B). Western blot analysis showed that the EGFP concentration in ASP3894 was ∼1.5-fold that in ASP3880 at OD660 = 5.0 in YPD medium (Figure 7C). We also evaluated the growth-dependent changes in EGFP production levels in the other genome-reduced strains in the logarithmic phase. These strains had single-, double- or triple-region deletions with various combinations of the chromosomal terminals, ALT, ART, BLT and BRT (Supplementary Table S4). ASP3882, a single-chromosomal terminal deletion strain derived from IGF628, showed a 1.3-fold increase in the ratio of EGFP fluorescence compared with ASP3880. The other single-chromosomal terminal deletion strains had similar EGFP fluorescence levels to ASP3880. Double- and triple-chromosomal terminal deletion strains derived from ASP3882 showed an increased ratio of EGFP fluorescence compared with ASP3882, but no strains showed an increase in EGFP fluorescence greater than that of the ASP3894 quadruple-deletion strain (data not shown). These findings are consistent with a view that the production ratio of heterologous protein increased with the size of genome reduction.
Second, efficiency of human growth hormone (hGH) secretion in YPD medium was measured in 4-day cultures. Total secreted proteins containing hGH in 100 μl of culture supernatant were electrophoresed on SDS–polyacrylamide gels after TCA precipitation. ASP4187, derived from IGF742, showed ∼1.8-fold increase in hGH secretion compared with ASP3341, derived from ARC001 (Figure 7D and E). In addition, a similar effect of genome reduction for human transferrin (hTF) secretion was observed (Supplementary Figure S5). These results suggest that genome reduction can be a valuable tool for the construction of heterologous protein production hosts.
DISCUSSION
We succeeded in reducing the S. pombe genome size while maintaining growth rate. In the genome-reduced strain, the genome size and the gene number were reduced by 657.3 kb and 223 genes, respectively. The gene number of the 657.3-kb deletion strain is currently the smallest among eukaryotic model organisms. We determined that the most-terminal essential genes in the left and right arms of chromosomes I and II were trs33 (ALT), sec16 (ART), zas1 (BLT) and usp109 (BRT). However, it has been reported that SPAC1F8.07c (ALT), SPBC1348.06c (BLT), alr2 (BLT) and dea2 (BLT), which are located on the telomeric side of trs33 and zas1, are also essential for growth (14). IGF629, which does not include SPBC1348.06c, alr2 or dea2, showed no decrease in growth rate in comparison with the parental strain. It was unknown whether the phenotype is a particular property of our laboratory strain.
The quadruple-deletion strain showed an increase in the expression of nitrogen starvation-response genes in logarithmic phase cultures. The quadruple-deletion strain was speculated to have decreased TORC1 activity, but the known TORC1 components and TORC1 regulation genes were not included in the 223 deleted genes. Why does the quadruple-deletion strain show a response mimicking nitrogen starvation in the logarithmic phase? We suggest that the nitrogen starvation response in the quadruple-deletion strain might be related to AMP-activated protein kinase (AMPK) activity. Mammalian AMPK acts as a cellular energy sensor that can be activated by metabolic stress, including low-glucose conditions. AMPK switches on catabolic pathways generating ATP and switches off non-essential processes consuming ATP (37). The quadruple-deletion strain showed a 2.7-fold increase in ATP concentration in comparison with the wild-type strain. We, therefore, speculate that activation of AMPK is stimulated by an unknown cue caused by genome reduction. S. pombe has two AMPK catalytic subunit genes, SPCC74.03c/ssp2 and SPAC23H4.02/ppk9 (38). Recently, Ssp2 has been reported to regulate gene expression in glucose catabolite repression through phosphorylation of Scr1 transcription factor (39). However, the details of the enzymatic functions of S. pombe AMPK remain unclear. Mammalian AMPK downregulates TORC1 activity through phosphorylation of TSC2 (40) and a raptor gene of TORC1 (41). It has also been suggested that S. pombe may contain the AMPK–TORC1 pathway, which is similar to that found in mammalian cells (42). S. pombe has two tsc genes and a raptor gene, SPAC22F3.13/tsc1, SPAC630.13c/tsc2 and SPAC57A7.11/mip1, which were identified as homologs of human TSC1, TSC2 and Wat1/Pop3, respectively (28,43,44). Therefore, we postulated that S. pombe AMPK, which was stimulated by an unknown cue caused by genome reduction, downregulated TORC1 activity and led to a mimicking of the nitrogen starvation response in the quadruple-deletion strain.
The production rate of the heterologous protein, EGFP, in the quadruple-deletion strain was increased ∼1.7-fold in comparison with the parental strain during logarithmic phase growth. Based on transcriptome and metabolome analysis results, we suggest a possible reason for the increased EGFP expression in the deletion mutant. An increase in intracellular GTP required for ribosomal activity might lead to increases in translation and EGFP production. Nitrogen use efficiency might be facilitated by increases in gene expression of gdh1, a component of the ammonia assimilation cycle. In addition, an increase in intracellular ATP might lead to activation of amino acid biosynthesis reactions. However, an increase in concentration of intracellular amino acids was not observed, and some intracellular amino acids in the quadruple-deletion strain were decreased. It was unclear whether the decrease in amino acids was caused by decreased biosynthesis or by increased use efficiency for the heterologous protein.
S. pombe has been considered to be an attractive host for the production of heterologous glycoproteins derived from higher animals (45,46). In the quadruple-deletion strain, three α-1,2-galactosyltransferase genes (SPAC5H10.11/gmh1, SPAC5H10.13c/gmh2 and SPBC1289.13c/gmh6) and an acetylglucosaminyltransferase gene (SPAC5H10.12c/otg1) were deleted. In a previous study, the molecular weight of N-linked oligosaccharide in a gmh1 deletion mutant was almost the same as in the wild-type, whereas that in Δgmh2 and Δgmh6 mutants was reduced, and the O-linked oligosaccharide profile in Δgmh2 and Δgmh6 mutants was also different from the wild-type (47). In addition, otg1 did not influence the formation of oligosaccharide (48). Glycosylation may be influenced in the quadruple-deletion strain because gmh2 and gmh6 were deleted. However, SPCC736.04c/gma12, a major α-1,2-galactosyltransferase gene, remains in the quadruple-deletion strain. Therefore, we speculate that deletion of gmh2 and gmh6 does not have a significant influence on oligosaccharide structures.
To construct a genome-reduced strain with a simpler gene set, we aim to further reduce the gene number in S. pombe. A genome-reduced strain is suitable for modeling and simulation of cellular metabolism. Problems with unpredictability have often been caused by the complex intracellular metabolic system, where the elucidation of all cellular metabolic intermediates has not been completed. A simplified cellular metabolism, while maintaining satisfactory growth and requisite functions (e.g. productivity of heterologous protein), will lead to success in the control of intracellular metabolism in accord with metabolic modeling and simulations, and it is beneficial for the production of biological materials.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–5 and Supplementary Sequences.
FUNDING
It is a part of The Development of Basic Technologies for Advanced Production Methods Using Microorganism Functions of New Industrial Science and Technology Frontiers by the Ministry of Economy, Trade and Industry (METI) and is entrusted by the New Energy and Industrial Technology Development Organization (NEDO). Funding for open access charge: Asahi Glass Co., Ltd.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Yuko Giga-Hama who gave support and insightful comments.
REFERENCES
- 1.Medema MH, van Raaphorst R, Takano E, Breitling R. Computational tools for the synthetic design of biochemical pathways. Nat. Rev. Microbiol. 2012;10:191–202. doi: 10.1038/nrmicro2717. [DOI] [PubMed] [Google Scholar]
- 2.Fujio T. Minimum genome factory: innovation in bioprocesses through genome science. Biotechnol. Appl. Biochem. 2007;46:145–146. doi: 10.1042/BA20060117. [DOI] [PubMed] [Google Scholar]
- 3.Moya A, Gil R, Latorre A, Pereto J, Pilar Garcillan-Barcia M, de la Cruz F. Toward minimal bacterial cells: evolution vs. design. FEMS Microbiol. Rev. 2009;33:225–235. doi: 10.1111/j.1574-6976.2008.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Posfai G, Plunkett G, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 5.Mizoguchi H, Sawano Y, Kato J, Mori H. Superpositioning of deletions promotes growth of Escherichia coli with a reduced genome. DNA Res. 2008;15:277–284. doi: 10.1093/dnares/dsn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manabe K, Kageyama Y, Morimoto T, Ozawa T, Sawada K, Endo K, Tohata M, Ara K, Ozaki K, Ogasawara N. Combined effect of improved cell yield and increased specific productivity enhances recombinant enzyme production in genome-reduced Bacillus subtilis strain MGB874. Appl. Environ. Microbiol. 2011;77:8370–8381. doi: 10.1128/AEM.06136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morimoto T, Kadoya R, Endo K, Tohata M, Sawada K, Liu S, Ozawa T, Kodama T, Kakeshita H, Kageyama Y, et al. Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis. DNA Res. 2008;15:73–81. doi: 10.1093/dnares/dsn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami K, Tao E, Ito Y, Sugiyama M, Kaneko Y, Harashima S, Sumiya T, Nakamura A, Nishizawa M. Large scale deletions in the Saccharomyces cerevisiae genome create strains with altered regulation of carbon metabolism. Appl. Microbiol. Biotechnol. 2007;75:589–597. doi: 10.1007/s00253-007-0859-2. [DOI] [PubMed] [Google Scholar]
- 9.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 10.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. February 21 (doi: 10.1038/msb4100050; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki M, Idiris A, Tada A, Kumagai H, Giga-Hama Y, Tohda H. The gap-filling sequence on the left arm of chromosome 2 in fission yeast Schizosaccharomyces pombe. Yeast. 2008;25:673–679. doi: 10.1002/yea.1613. [DOI] [PubMed] [Google Scholar]
- 14.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decottignies A, Sanchez-Perez I, Nurse P. Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 2003;13:399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirashima K, Iwaki T, Takegawa K, Giga-Hama Y, Tohda H. A simple and effective chromosome modification method for large-scale deletion of genome sequences and identification of essential genes in fission yeast. Nucleic Acids Res. 2006;34:e11. doi: 10.1093/nar/gnj011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Ohashi Y, Hirayama A, Ishikawa T, Nakamura S, Shimizu K, Ueno Y, Tomita M, Soga T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. Biosyst. 2008;4:135–147. doi: 10.1039/b714176a. [DOI] [PubMed] [Google Scholar]
- 19.Soga T, Ishikawa T, Igarashi S, Sugawara K, Kakazu Y, Tomita M. Analysis of nucleotides by pressure-assisted capillary electrophoresis-mass spectrometry using silanol mask technique. J. Chromatogr. A. 2007;1159:125–133. doi: 10.1016/j.chroma.2007.05.054. [DOI] [PubMed] [Google Scholar]
- 20.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 21.Chikashige Y, Tsutsumi C, Okamasa K, Yamane M, Nakayama J, Niwa O, Haraguchi T, Hiraoka Y. Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct. Funct. 2007;32:149–161. doi: 10.1247/csf.07036. [DOI] [PubMed] [Google Scholar]
- 22.Hansen KR, Ibarra PT, Thon G. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 2006;34:78–88. doi: 10.1093/nar/gkj415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sand O, Gingras M, Beck N, Hall C, Trun N. Phenotypic characterization of overexpression or deletion of the Escherichia coli crcA, cspE and crcB genes. Microbiology. 2003;149:2107–2117. doi: 10.1099/mic.0.26363-0. [DOI] [PubMed] [Google Scholar]
- 24.Fantes P, Nurse P. Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp. Cell Res. 1977;107:377–386. doi: 10.1016/0014-4827(77)90359-7. [DOI] [PubMed] [Google Scholar]
- 25.Heiland S, Radovanovic N, Hofer M, Winderickx J, Lichtenberg H. Multiple hexose transporters of Schizosaccharomyces pombe. J. Bacteriol. 2000;182:2153–2162. doi: 10.1128/jb.182.8.2153-2162.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakurai M, Tohda H, Kumagai H, Giga-Hama Y. A distinct type of alcohol dehydrogenase, adh4+, complements ethanol fermentation in an adh1-deficient strain of Schizosaccharomyces pombe. FEMS Yeast Res. 2004;4:649–654. doi: 10.1016/j.femsyr.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Aspuria PJ, Tamanoi F. The Tsc/Rheb signaling pathway controls basic amino acid uptake via the Cat1 permease in fission yeast. Mol. Genet. Genomics. 2008;279:441–450. doi: 10.1007/s00438-008-0320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 2007;27:3154–3164. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakashima A, Sato T, Tamanoi F. Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J. Cell Sci. 2010;123:777–786. doi: 10.1242/jcs.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisman R, Roitburg I, Schonbrun M, Harari R, Kupiec M. Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics. 2007;175:1153–1162. doi: 10.1534/genetics.106.064170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen ML, Krook DJ, De Graaf K, van Dijken JP, Pronk JT, de Winde JH. Physiological characterization and fed-batch production of an extracellular maltase of Schizosaccharomyces pombe CBS 356. FEMS Yeast Res. 2006;6:888–901. doi: 10.1111/j.1567-1364.2006.00091.x. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzawa T, Ohashi T, Hosomi A, Tanaka N, Tohda H, Takegawa K. The gld1+ gene encoding glycerol dehydrogenase is required for glycerol metabolism in Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 2010;87:715–727. doi: 10.1007/s00253-010-2586-3. [DOI] [PubMed] [Google Scholar]
- 33.Magasanik B. Ammonia assimilation by Saccharomyces cerevisiae. Eukaryot. Cell. 2003;2:827–829. doi: 10.1128/EC.2.5.827-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perysinakis A, Kinghorn JR, Drainas C. Biochemical and genetical studies of NADP-specific glutamate dehydrogenase in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 1994;26:315–320. doi: 10.1007/BF00310495. [DOI] [PubMed] [Google Scholar]
- 35.Perysinakis A, Kinghorn JR, Drainas C. Glutamine synthetase/glutamate synthase ammonium-assimilating pathway in Schizosaccharomyces pombe. Curr. Microbiol. 1995;30:367–372. doi: 10.1007/BF00369864. [DOI] [PubMed] [Google Scholar]
- 36.Idiris A, Tohda H, Sasaki M, Okada K, Kumagai H, Giga-Hama Y, Takegawa K. Enhanced protein secretion from multiprotease-deficient fission yeast by modification of its vacuolar protein sorting pathway. Appl. Microbiol. Biotechnol. 2009;85:667–677. doi: 10.1007/s00253-009-2151-0. [DOI] [PubMed] [Google Scholar]
- 37.Hardie DG. Minireview: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology. 2003;144:5179–5183. doi: 10.1210/en.2003-0982. [DOI] [PubMed] [Google Scholar]
- 38.Hanyu Y, Imai KK, Kawasaki Y, Nakamura T, Nakaseko Y, Nagao K, Kokubu A, Ebe M, Fujisawa A, Hayashi T, et al. Schizosaccharomyces pombe cell division cycle under limited glucose requires Ssp1 kinase, the putative CaMKK, and Sds23, a PP2A-related phosphatase inhibitor. Genes Cells. 2009;14:539–554. doi: 10.1111/j.1365-2443.2009.01290.x. [DOI] [PubMed] [Google Scholar]
- 39.Matsuzawa T, Fujita Y, Tohda H, Takegawa K. Snf1-like protein kinase Ssp2 regulates glucose derepression in Schizosaccharomyces pombe. Eukaryot. Cell. 2012;11:159–167. doi: 10.1128/EC.05268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 41.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakashima A, Tamanoi F. In: The Enzymes: Structure, Function and Regulation of TOR Complexes From Yeasts to Mammals. Hall MN, Tamanoi F, editors. Vol. 28. Amsterdam: part B. Academic Press/Elsevier; 2010. pp. 167–187. [Google Scholar]
- 43.Alvarez B, Moreno S. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 2006;119:4475–4485. doi: 10.1242/jcs.03241. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto S, Bandyopadhyay A, Kwiatkowski DJ, Maitra U, Matsumoto T. Role of the Tsc1-Tsc2 complex in signaling and transport across the cell membrane in the fission yeast Schizosaccharomyces pombe. Genetics. 2002;161:1053–1063. doi: 10.1093/genetics/161.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giga-Hama Y, Kumagai H. Expression system for foreign genes using the fission yeast Schizosaccharomyces pombe. Biotechnol. Appl. Biochem. 1999;30:235–244. [PubMed] [Google Scholar]
- 46.Takegawa K, Tohda H, Sasaki M, Idiris A, Ohashi T, Mukaiyama H, Giga-Hama Y, Kumagai H. Production of heterologous proteins using the fission-yeast (Schizosaccharomyces pombe) expression system. Biotechnol. Appl. Biochem. 2009;53:227–235. doi: 10.1042/BA20090048. [DOI] [PubMed] [Google Scholar]
- 47.Ohashi T, Nakakita S, Sumiyoshi W, Yamada N, Ikeda Y, Tanaka N, Takegawa K. Structural analysis of alpha1,3-linked galactose-containing oligosaccharides in Schizosaccharomyces pombe mutants harboring single and multiple alpha-galactosyltransferase genes disruptions. Glycobiology. 2011;21:340–351. doi: 10.1093/glycob/cwq167. [DOI] [PubMed] [Google Scholar]
- 48.Ohashi T, Fujiyama K, Takegawa K. Identification of novel alpha1,3-galactosyltransferase and elimination of alpha-galactose-containing glycans by disruption of multiple alpha-galactosyltransferase genes in Schizosaccharomyces pombe. J. Biol. Chem. 2012;287:38866–38875. doi: 10.1074/jbc.M112.347351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.