Abstract
Choriocarcinomas are embryonal tumours with loss of imprinting and hypermethylation at the insulin-like growth factor 2 (IGF2)-H19 locus. The DNA methyltransferase inhibitor, 5-Aza-2′deoxycytidine (5-AzaCdR) is an approved epigenetic cancer therapy. However, it is not known to what extent 5-AzaCdR influences other epigenetic marks. In this study, we set out to determine whether 5-AzaCdR treatment can reprogram the epigenomic organization of the IGF2-H19 locus in a choriocarcinoma cancer cell line (JEG3). We found that localized DNA demethylation at the H19 imprinting control region (ICR) induced by 5-AzaCdR, reduced IGF2, increased H19 expression, increased CTCF and cohesin recruitment and changed histone modifications. Furthermore chromatin accessibility was increased locus-wide and chromatin looping topography was altered such that a CTCF site downstream of the H19 enhancers switched its association with the CTCF site upstream of the IGF2 promoters to associate with the ICR. We identified a stable chromatin looping domain, which forms independently of DNA methylation. This domain contains the IGF2 gene and is marked by a histone H3 lysine 27 trimethylation block between CTCF site upstream of the IGF2 promoters and the Centrally Conserved Domain upstream of the ICR. Together, these data provide new insights into the responsiveness of chromatin topography to DNA methylation changes.
INTRODUCTION
Genome-wide correlations indicate that DNA methylation, histone modifications and chromatin structure cooperatively regulate gene expression (1). Malignant cells have perturbed epigenetic programmes that result in changes in DNA methylation, histone modifications and possibly chromatin looping conformation to disrupt normal gene regulation (2). DNA methyltransferases and histone deacetylases were the first epigenetic modifiers to be targeted for therapeutic intervention in cancer (3). Decitabine or 5-aza-2′deoxycytidine (5-AzaCdR) is an analogue of deoxycytidine that irreversibly binds to DMNTs to inhibit the maintenance of cytosine methylation during DNA replication (3). 5-AzaCdR has been applied in clinical use for myelodisplastic syndrome (3). It has been observed that 5-AzaCdR does not affect all loci uniformly, and that some genomic regions are more responsive to treatment than others (4). Detailed exploration of DNA methylation changes in specific genomic regions in response to 5-AzaCdR, and their impact on epigenomic organization is required to understand the mechanisms underlying the clinical activity of this drug and to identify predictive markers for responsiveness. At a basic research level, it is still unknown to what extent localized DNA methylation changes can reprogram the structural spatiotemporal compartmentalization of epigenetic marks at a given locus.
Epigenetic regulation of gene expression occurs over long genomic distances, and this is in part due to the chromatin looping structure that underlies the epigenomic organisation of the nucleus. Looping structures may be dynamic with variable interactions occurring tissue specifically, but within a stable framework of invariable loops (5). These invariable loops may be the ‘topological associated domains’ described by Nora et al. (6) or ‘topological domains’ observed by Dixon et al. (7) that were enriched for CTCF at the boundaries. CTCF is a methylation sensitive transcription factor that functions in transcriptional activation, repression and insulation/boundaries [reviewed (8)] and has an architectural role in structuring chromatin loops (5,9). Genome-wide chromatin immunoprecipitation (ChIP) analyses indicate that CTCF-binding sites occur on average every 40–70 kb across the genome (10–13) and could theoretically divide the genome into looping domains with CTCF boundaries at the base of these loops. CTCF recruits cohesin complexes to many of its binding sites (13–16), which stabilizes loop formation, possibly by securing chromatin in cis, analogous to the holding of sister chromatids together (17–20). In addition to CTCF, the polycomb repressive complex 2, known to have a role in maintaining methylation of histone H3 lysine 27 (H3K27me3) (21,22), and several DNA-binding transcription factors have been reported to mediate chromatin loops (22–24).
DNA methylation, chromatin looping conformation and gene expression are well studied in the context of the imprinted insulin-like growth factor 2 (IGF2) and the non-coding RNA gene, H19 locus. The IGF2-H19 locus is regulated epigenetically at a genomic region upstream of the H19 gene known as the imprinting control region (ICR) (Figure 1A). CTCF binds to the ICR on the unmethylated maternal allele to form a chromatin boundary (25–28) and structure the chromatin loops (17–20) that maintain the silencing of the IGF2 gene. Loss of imprinting at this locus is implicated in the aetiology of Beckwith–Wiedemann syndrome (BWS) and Silver–Russell syndrome (SRS) as well as several cancers (29–31). Aberrant chromatin looping has been associated with DNA methylation changes at IGF2-H19 in BWS and SRS (17) and in cancer cell lines (32).
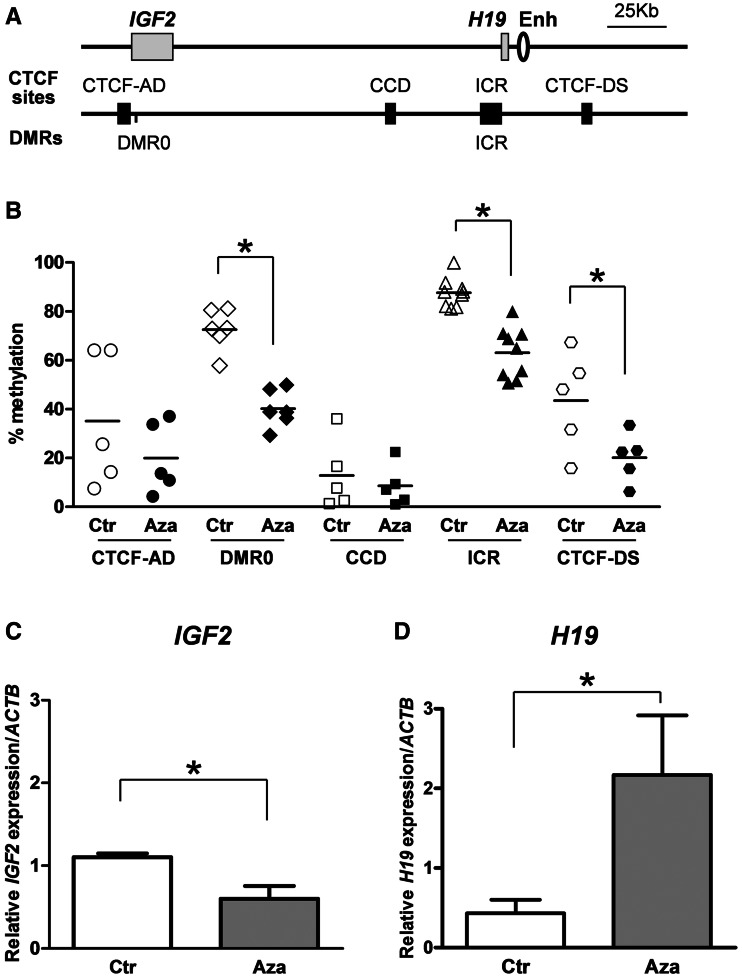
Figure 1.
5-AzaCdR treatment changes DNA methylation and gene expression at the IGF2-H19 locus in JEG3, a choriocarcinoma cell line. (A) Schematic representation of regions analysed for DNA methylation at the IGF2-H19 locus. IGF2 and H19 genes (grey rectangles), downstream enhancer (oval), the differentially methylated regions, DMRs [DMR0 and (the H19) ICR] and CTCF-binding sites (CTCF-AD: CTCT-binding site adjacent to IGF2 DMR0, CCD: the Centrally Conserved Domain, ICR: the H19 imprinting control region and CTCF-DS: CTCF-binding site downstream of H19 enhancers; black rectangles) are indicated. (B) DNA methylation levels at the DMRs and CTCF-binding sites determined by the PSQ. DMR0 and the ICR show hypermethylation and a 50% reduction after 5-AzaCdR treatment. Each point indicates the average methylation score for a single CpG in more than three biological replicates. Horizontal bars indicate the average methylation score for the CpGs in each PSQ assay. (C) and (D) IGF2 and H19 expression changes. Data in B, C and D represent more than three biological replicate 5-AzCdR experiments. Asterisk indicates the significant differences (P < 0.05).
The IGF2-H19 locus is an ideal locus to study the plasticity of chromatin looping conformation on 5-AzaCdR-mediated DNA demethylation and the overall impact upon gene expression and histone methylation. Several adult cancers have a loss of IGF2 imprinting, but IGF2 expression is generally low in cell lines derived from these cancers. Germ-cell tumours such as choriocarcinomas have global disruption of genomic imprinting with hypomethylation at maternally imprinted genes and hypermethylation at paternally imprinted genes (33). The human choriocarcinoma cell line (JEG3) presents many of the biological and biochemical features of human syncytiotrophoblasts and, similar to germ-cell tumours, has loss of imprinting with high levels of IGF2 expression and hypermethylation at the ICR. These cells are known to respond to 5-AzaCdR treatment by reducing DNA methylation at the IGF2-H1919 locus (34), making them suitable for analyses of demethylation-induced epigenetic restructuring of the IGF2-H19 locus.
We herein report that localized changes in DNA methylation at the ICR after 5-AzaCdR treatment leads to a cascade of epigenetic changes, which correlate with concomitant changes in IGF2 and H19 expression. These changes include the recruitment of CTCF and cohesin to the ICR, an overall locus-wide increase in micrococcal nuclease (MNase) accessibility, altered histone modifications and the reshaping of chromatin looping conformation. We show the IGF2 gene is present within a stable looping domain marked by H3K27me3 that is resistant to DNA methylation changes at the wider locus. This stable looping domain is adjacent to a dynamic, DNA methylation-sensitive looping domain. The dynamic looping domain changes its structure by alternating the association of the CTCF site downstream of H19 enhancers (CTCF-DS) between the ICR and a CTCF site upstream of the IGF2 promoters (CTCF-AD). Interactions between enhancers and promoters can occur even when the promoters and enhancers are on different looping domains. Overall, this snapshot of chromatin modelling provides insights into the responsiveness of chromatin looping to changes in DNA methylation and CTCF binding.
MATERIALS AND METHODS
Cell culture
The use of the JEG3 cell line was selected for this study, as it is heterozygous for single nucleotide polymorphisms at IGF2 (rs680) and H19 (rs3741219) and has loss of imprinting such that both parental alleles express IGF2.
JEG3 was cultured in D-MEM supplemented with 10% (v/v) fetal bovine serum and 5% (v/v) non-essential amino acid at 37°C and 5% CO2. For 5-Azacytidine treatment, 5-AzaCdR (A3656, Sigma) was added to the medium at 1 uM for 48 h as previously described (34). For control s each 5-AzaCdR experiment included a parallel cell culture without 5-AzaCdR.
DNA methylation and gene expression analysis of the IGF2-H19 locus
The DNA methylation was assayed by pyrosequencing (PSQ) with the bisulfite-converted DNA as described previously (15) using PSQ™ HS Systems and Pyro Q-C-G™ Software, (Qiagen). Allele-specific methylation at the H19 ICR (rs2071094) was determined by cloning and bisulphite sequencing.
IGF2 and H19 expression analysis was performed by RT-qPCR. Allele-specific expression was determined by sequencing with qPCR products for rs680 at IGF2 and rs3741219 at H19.
Primer sequences and polymerase chain reaction (PCR) conditions are listed in Supplementary Tables S2 and S3.
ChIP
ChIP was performed as described previously (35) using 50 μg of chromatin [quantified by Qubit Fluorometer (Life Technologies)] and 5 μg of antibody. Enrichment at specific genomic regions across the IGF2-H19 locus was determined by qPCR. PCR data were corrected for DNA amount, and enrichment was normalized against input. For ChIPs analysing histone modifications, data were further normalized against total histone H3 enrichment. To compare the enrichment between the control and 5-AzaCdR-treated samples, the data were normalized against a negative control region; FOXA1 for CTCF and RAD21 ChIP, GAPDH for SUZ12 ChIP, and for histone modification ChIPs, we used a previously described negative control region designated ‘Gene desert’ (36). More than three biological replicates were performed for each ChIP (each biological replicate was an individually prepared 5-AzaCdR treated and matched non-treated cell sample processed for ChIP).
A detailed figure showing the location of all primers used in this study is in Supplementary Figure S1. Most PCR amplicons were designed to incorporate CTCF consensus binding sites identified by Kim et al. (12). Primer sequences and PCR conditions are listed in Supplementary Table S4, and the antibodies are used in Supplementary Table S5.
Quantitative chromatin conformation capture
Quantitative chromatin conformation capture (3C) was as described previously (20). Quantitative interaction frequencies were determined using a standard curve generated from a library of chimeric PCR fragments for each interaction. The 3C profiles between biological replicates (n > 3) were normalized to the self-ligation controls as previously described (20). As high association frequencies were detected between the Centrally Conserved Domain (CCD) and other CTCF-binding sites, the association frequencies are depicted as log10 scale (Figure 3).
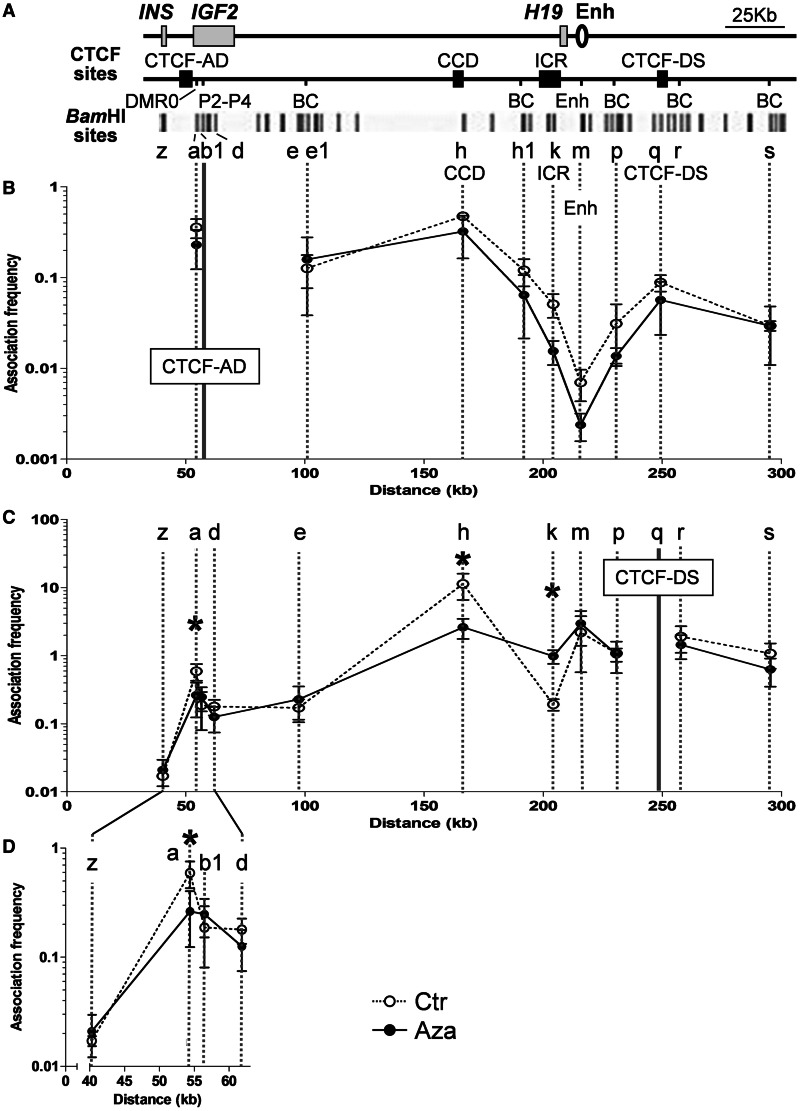
Figure 3.
Changes in chromatin conformation at the IGF2-H19 locus after 5-AzaCdR treatment. (A) Graphic of the IGF2-H19 locus as in Figures 1 and 2 with additional BC regions. BC here indicates the intervening regions between CTCF sites. The bottom track shows positions of BamHI restriction site used in 3C analysis. Vertical dotted lines below the restriction sites, which extend into the graphs in B and C are aligned to the respective restriction site analysed. Solid vertical lines at restriction sites b1 (B) and q (C) indicate 3C anchor sites at CTCF-AD and CTCF-DS, respectively. (B) 3C analysis showing associations with CTCF-AD. Dotted lines with open circles are control profiles of untreated JEG3 cells. Solid lines with filled circles are 3C profiles after 5-AzaCdR treatment. Strong association peaks are seen with the CCD and CTCF-AD, and the association with the CCD persists after 5-AzaCdR treatment. CTCF-AD does not interact with the ICR. (C) 3C associations with CTCF-DS. Strong association peaks are seen with the CCD and CTCF-AD in control. These associations are significantly reduced after 5-AzaCdR treatment, and a new association between CTCF-DS and the ICR is found, as the DNA methylation level decreases at the ICR (Figure 1B). (D) Enlarged graph of CTCF-DS associations with CTCF-AD between restriction sites z and d. The enlarged graph shows the significant reduction in the interactions between CTCF-DS and CTCF-AD more clearly.
Primer sequences for quantitative 3C are listed in Supplementary Table S6.
MNase digestion assay
MNase digestion of chromatin using EpiQ Chromatin Analysis Kit according to the manufacturer’s protocol (Bio-Rad). Briefly, 25 000 cells were harvested and divided into two 100 µL aliquots and resuspended in EpiQ chromatin buffer. To the first aliquot [digestion (D)], 2 µL of EpiQ nuclease was added. Nuclease was not added to the second aliquot as the undigested control (U). Both D and U samples were incubated at 37°C for 1 h, and thereafter the DNA was extracted, and nuclease sensitivity was assessed by qPCR (with primers as in Supplementary Table S4). The data were analysed using EpiQ Chromatin Kit Data Analysis Tool (www.bio-rad.com/epiq) and normalized against the RHO (Rhodopsin) gene in the undigested sample as a negative reference. The GAPDH region was a positive reference gene region for an open chromatin (>95% accessibility).
Statistical analysis
The independent two-sample t-test was carried out for the statistical analysis in all ChIP experiments except assessing CTCF enrichment at the CCD. This was assessed by the paired two-sample t-test. For the DNA methylation study, the statistical significance between 5-AzaCdR-treated and non-treated samples was assessed by paired two-sample t-test for each CpG at a region tested. P < 0.05 was considered significant.
RESULTS
5-AzaCdR treatment reduces DNA methylation with concomitant IGF2-H19 expression changes in JEG3 cells
IGF2 and H19 expression and DNA methylation levels were measured in JEG3 cells before and after 48 h of treatment with 5-AzaCdR. The JEG3 cell line was selected for this study, as it shows loss of imprinting with both the maternal and paternal alleles expressing IGF2. Specifically, the H19 ICR and the IGF2 DMR0 are hypermethylated (Figure 1B, Supplementary Figures S2B and D and S3A) with strong IGF2 expression from both alleles and reciprocally low levels of H19 expression (Figure 1C and D and Supplementary Table S1). This hypermethylation profile at IGF2-H19 in JEG3 is similar to what we previously found in peripheral blood DNA from BWS patients that have biallelic IGF2 expression (30). After 5-AzaCdR treatment, DNA methylation was reduced at the ICR from 87.6 (+/−5.9%) to 63.0% (+/−10.4%) and at the IGF2 DMR0 from 72.5 (+/−8.5%) to 40.2% (+/−7.7%) in JEG3 cells (Figure 1B, Supplementary Figure S2B and D). A small reduction in DNA methylation levels was observed at CTCF sites flanking the locus (Figure 1B, Supplementary Figure S2A and E). Single nucleotide polymorphism analyses in bisulphite cloning indicated that both alleles undergo demethylation at the ICR (Supplementary Figure S3). IGF2 expression decreased with a reciprocal increase in H19 transcription (Figure 1C and D). In accordance with the DNA methylation changes, IGF2 and H19 were still expressed from both alleles (Supplementary Table S1). Thus, in the JEG3 cell line, 5-AzaCdR treatment reduced DNA methylation and changed IGF2 and H19 expression levels, but did not distinguish between alleles.
Changes in CTCF, cohesin and SUZ12-binding profiles at the IGF2-H19 locus in response to reduced DNA methylation
We next examined the CTCF and cohesion-binding profiles before and after 5-AzaCdR. CTCF and cohesin normally bind at the unmethylated ICR and several other sites at the locus (17,20); these sites are referred to as CTCF-AD (CTCT-binding site adjacent to IGF2 DMR0, 5 kb upstream of the IGF2 DMR0), the CCD (37) (40 kb upstream of the ICR) and CTCF-DS (CTCF-binding site downstream of H19 enhancers, 30 kb downstream of the H19 enhancers) (Figures 1A and 2A and Supplementary Figure S1). As expected, CTCF binding was initially undetectable at the hypermethylated ICR and was recruited to the demethylated ICR after 5-AzaCdR treatment. Unexpectedly, CTCF binding also increased significantly at CTCF-AD and the CCD within the wider locus (Figure 2B). Modest DNA methylation changes after 5-AzaCdR treatment were observed at these sites. However, these changes seem too small to account for the marked increase in CTCF binding, especially at the CCD (Figure 1B, Supplementary Figure S2A and C).
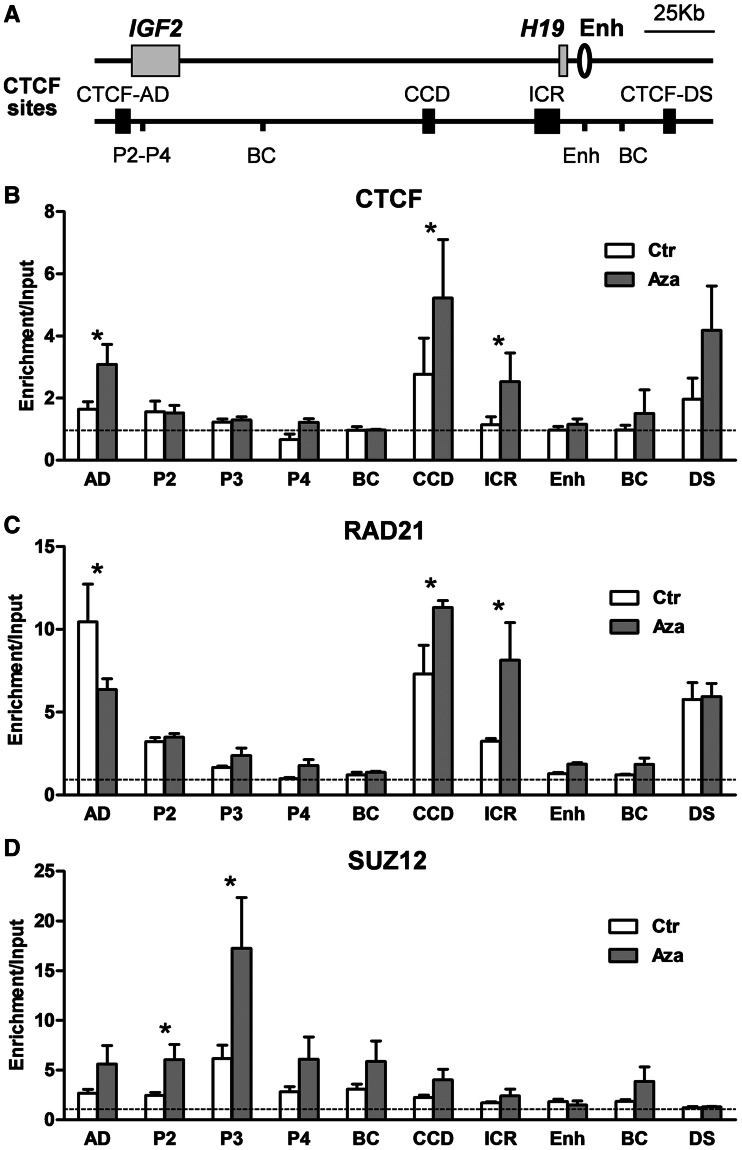
Figure 2.
CTCF and cohesin binding increases at CTCF-binding sites at the IGF2-H19 locus after 5-AzaCdR treatment. (A) Graphic showing a region of IGF2-H19 locus analysed by ChIP. IGF2, H19 genes, H19 downstream enhancer and CTCF-binding sites are annotated as in Figure 1. P2–P4: IGF2 promoters, BC: the non-CTCF-binding sites as intergenic background control regions. (B) ChIP analysis of CTCF, (C) the cohesin subunit, RAD21 and (D) the polycomb repressive complex 2 subunit, SUZ12. The horizontal dotted line indicates the level of enrichment for the negative control region. CTCF and cohesin, but not SUZ12, are enriched at the ICR after 5-AzaCdR treatment. Asterisk indicates the significant difference (P < 0.05) in more than three biological replicate experiments.
ChIP analyses with an antibody to the RAD21 subunit of cohesin confirmed that cohesin is present at CTCF binding sites but also showed enrichment at the hypermethylated ICR and IGF2 P2 promoter in untreated cells. At the ICR, this may be due to a higher sensitivity of the RAD21 antibody in detecting cohesin at a small proportion of unmethylated ICR sequences. The detection of RAD21 at the P2 promoter may similarly be due to the sensitivity of the antibodies used. However, we cannot exclude the possibility that cohesin may be recruited independently of CTCF at this site. Importantly, after 5-AzaCdR treatment, increased enrichment of cohesin was detected at the ICR as well as to the CCD. Cohesin occupancy did not change at CTCF-DS site and was decreased at CTCF-AD site (Figure 2C). These observations confirm that DNA demethylation enables the recruitment of CTCF and cohesin to the ICR and further suggest that localized demethylation has long ranging effects on distant CTCF and cohesin-binding sites.
SUZ12 was previously reported to interact with CTCF and to bind to the ICR and IGF2 promoters (38,39). Here, SUZ12 enrichment was found at the IGF2 promoter P3 in untreated cells. This enrichment significantly increased after 5-AzaCdR treatment (Figure 2D). However, we did not observe a significant SUZ12 enrichment at the ICR (Figure 2D). Therefore, SUZ12 recruitment to the IGF2 promoters is unlikely to be directly due to a CTCF-SUZ12 interaction at the ICR as suggested (38,39).
Chromatin looping conformation changes occur concomitantly with CTCF and cohesin recruitment to the ICR after reduction in DNA methylation
As CTCF and cohesin are recruited to the ICR after 5-AzaCdR treatment, it is likely that the change in DNA methylation also changes the chromatin looping conformation. We have previously reported that with the 3C technique, the strongest proximity associations in the human IGF2 locus are detected between CTCF-binding sites (20), and that hypermethylation at the ICR precludes it from interacting with other CTCF sites at the locus (20). Given the DNA methylation profile of JEG3 cells (Figure 1B and Supplementary Figure S2), we predicted that the looping profiles in this cell line would be similar to paternal allele specific loops as found previously in BWS patients (17). However, cancer cell lines have been shown to have aberrant looping profiles at this locus (32). We therefore analysed 3C interactions between CTCF sites using CTCF-AD and CTCF-DS as anchors in JEG3 cells before and after 5-AzaCdR treatment.
In untreated cells, we found that the ICR had no interaction with CTCF-AD (Figure 3B) or with CTCF-DS (Figure 3C), which was already predicted due to the lack of CTCF binding at the methylated ICR. We saw association peaks between CTCF-AD and CTCF-DS (Figure 3B–D), which is consistent with our previously defined paternal allele looping profile in BWS patients with hypermethylation at the ICR (17,20). The CCD formed the strongest association peaks with CTCF-AD and CTCF-DS (Figure 3B and C). We previously observed biallelic interactions between the CCD and the other CTCF sites in normal cell lines and in patients with SRS or BWS (17,20). Overall, the CTCF-mediated chromatin profile of JEG3 cells was similar to that observed in lymphoblastoid cell lines from BWS patients with hypermethylation at the ICR.
After 5-AzaCdR treatment, the association between CTCF-DS and the CCD was significantly reduced (Figure 3C). This is presumably because of the newly created interaction between the ICR and CTCF-DS (Figure 3C, Figure 6B and C). A significant reduction was also seen in the interaction between CTCF-DS and CTCF-AD (Figure 3C and D). This switch in CTCF-DS association with CTCF-AD to an association with the ICR is consistent with the acquired CTCF binding at the ICR. More importantly, the switching of CTCF-DS associations changes the promoter–enhancer association to favour the reduction of IGF2 and upregulation of H19 expression, which we observe after demethylation of the ICR.
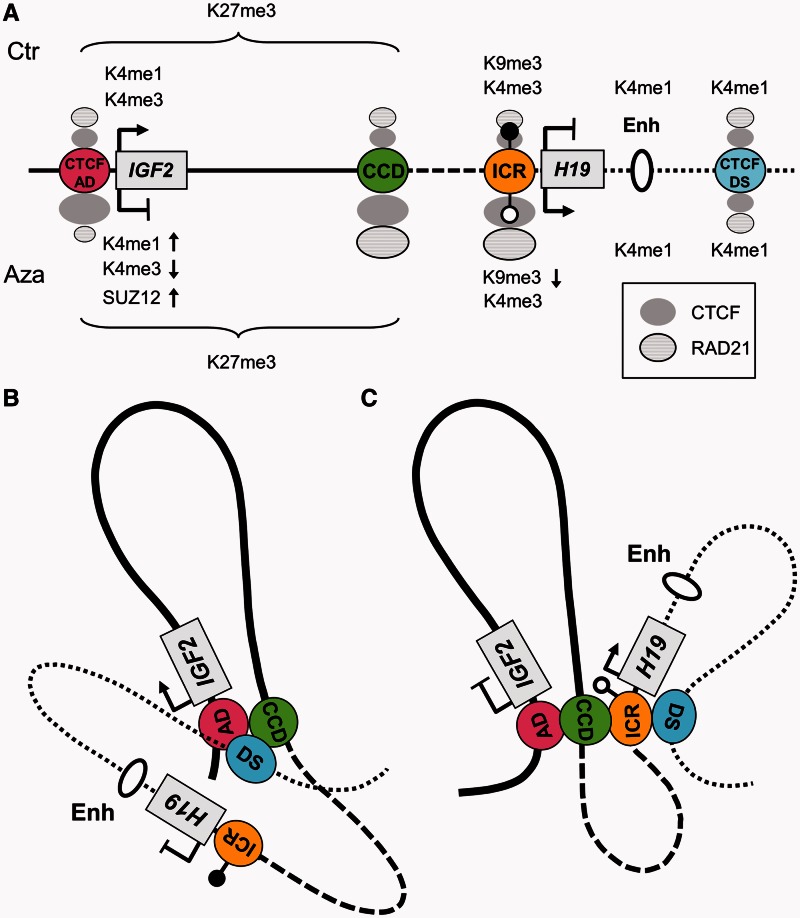
Figure 6.
Schematic model of chromatin conformation before and after DNA demethylation at the ICR. (A) Graphic summarising the changes in gene expression, histone modifications, CTCF, cohesin and SUZ12 binding after 5-AzaCdR treatment. Symbols above the IGF2-H19 graphic represent the untreated control state (Ctr), whereas symbols below represent the changes (arrows up and down) after 5-AzaCdR treatment (Aza). The DNA methylation at the ICR is indicated as black (methylated) or white (unmethylated) lollipops. The CTCF-binding sites (CTCF-AD, the CCD, the ICR and CTCF-DS) are indicated as coloured circles in the IGF2-H19 graphic. Changes in CTCF and RAD21 binding is shown by size differences of the filled grey circles (CTCF) and striped circles (RAD21). The IGF2-H19 graphic is marked with a solid horizontal line between CTCF-AD and the CCD, a dashed line between the CCD and the ICR and a dotted line between the ICR and CTCF-DS. (B) Schematic model of the chromatin conformation in the untreated JEG3 cells. CTCF-DS (blue), which also has H3K4me1 enrichment (H3k4me1/2 is an enhancer mark), interacts with CTCF-AD (red) and the CCD (green). Owing to the CTCF-AD and CTCF-DS interaction, the H19 enhancers (oval) are now located in close proximity to promoters of IGF2. The ICR (orange) does not associate with any of CTCF sites owing to its hypermethylated state. The loop formed between CTCF-AD and the CCD (solid line) is marked by an H3K27me3 BLOC. (C) Schematic of the changes in chromatin conformation after 5-AzaCdR-mediated DNA demethylation at the ICR. Increased CTCF and cohesin binding at the ICR (orange) enables an association between the ICR and CTCF-DS (blue), which forms a new chromatin loop (dotted line) containing H19 and the enhancers. H19 is active within this loop. The interaction between CTCF-AD and the CCD is unchanged (solid line), and H3K27me3 enrichment is unchanged in this stable chromatin loop. The reduced interaction of CTCF-DS with CTCF-AD and the CCD is also accompanied by an increase in SUZ12 binding at IGF2 promoters, a reduction in H3K4me3 at the IGF2 promoters, and reduced IGF2 expression.
DNA demethylation at the ICR did not significantly change the associations between the CCD and CTCF-AD, suggesting that this is a stable loop (Figure 3B). We have previously found this association to be biallelic in normal fibroblast, epithelial and lymphoblast as well as BWS and SRS cell lines (17,20). It is therefore likely that this is a ubiquitous looping domain, which is present in all tissues and on both parental alleles while downstream of the CCD, the ICR and CTCF-DS sites form allele-specific looping domains, which can mobilize the enhancers.
Histone H3 lysine 4 monomethylation is enriched at the enhancers and promoters
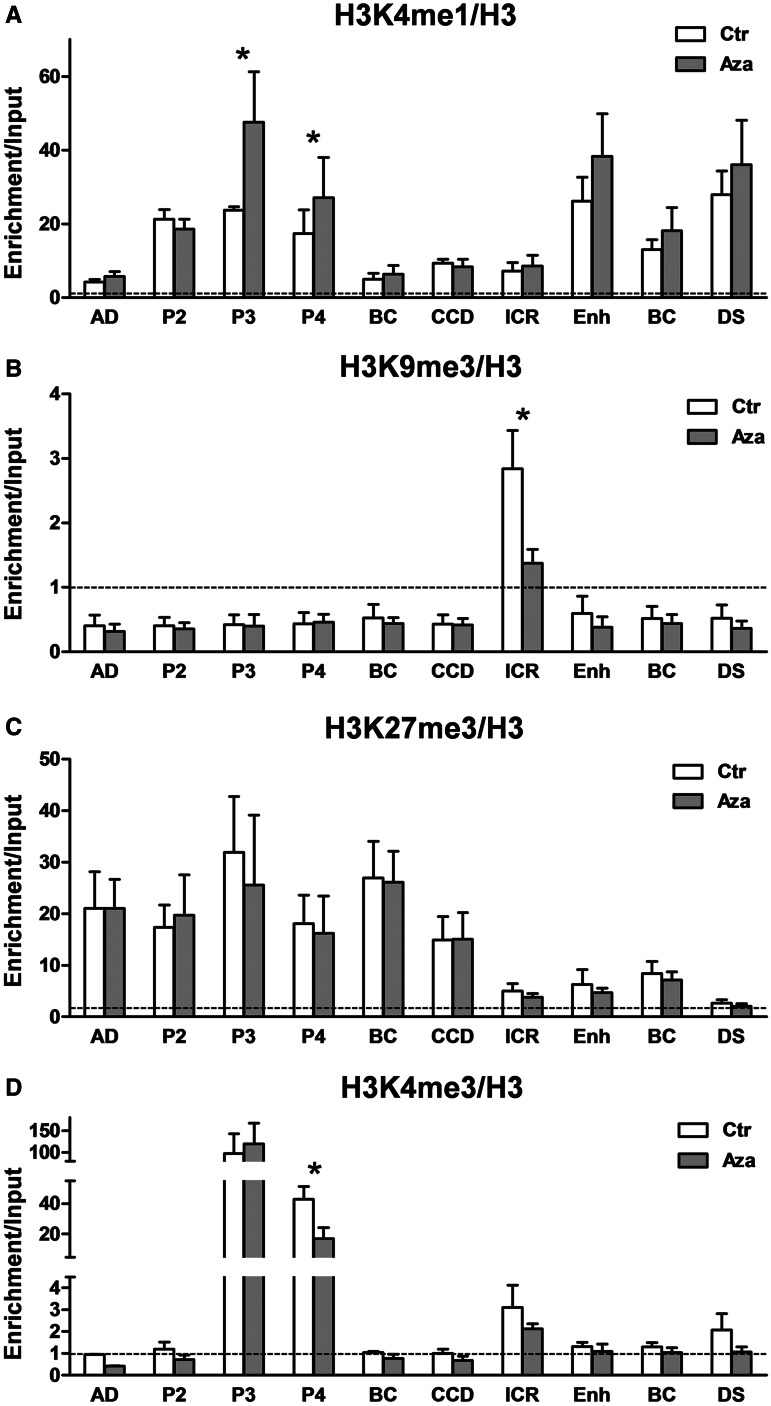
The CCD and CTCF-DS have previously been reported to have enhancer activity in mice (37,40); therefore, we examined whether the CTCF sites at the locus are enriched for histone H3 lysine 4 monomethylation, which is known to mark multiple classes of enhancer elements and some promoters (41–43). We found that H3K4me1 is enriched at the entire locus of IGF2-H19, and prominent enrichment can be seen at the IGF2 promoters and the region between the enhancers downstream of H19 and CTCF-DS (Figure 4A). CTCF-DS was highly enriched for H3K4me1, suggesting that this site may also be an enhancer (Figure 4A).
Figure 4.
Histone modification changes at the IGF2-H19 locus after 5-AzaCdR treatment. (A) Histone H3 lysine 4 monomethylation is enriched at the enhancers, IGF2 P3 and P4 promoters and CTCF-DS. H3K4me1 significantly increases at the P3 and P4 promoters after 5-AzaCdR treatment. (B) Histone H3 lysine 9 trimethylation (H3K9me3) is enriched only at the ICR and significantly decreases after 5-AzaCdR treatment. (C) Histone H3 lysine 27 trimethylation (H3K27me3) is highly enriched within IGF2 3C-loop flanked by CTCF-AD and the CCD. The mark persists in the loop after the cells are treated with 5-AzaCdR. (D) Histone H3 lysine 4 trimethylation (H3K4me3) is strongly enriched at the IGF2 P3 and P4 promoters. After 5-AzaCdR treatment, when IGF2 expression is downregulated, P4 has reduced H3K4me3. Some enrichment of H3K4me3 above background levels is present at the ICR in the control cells, despite the high DNA methylation state and suppressed expression of H19. This does not change after 5-AzaCdR treatment. ChIP profiles of histone modification were normalized to the total histone H3, and a negative control. The horizontal dotted line indicates the level of enrichment for the negative control region. Error bars represent Standard Deviations (SD) of more than three biological replicates, and the statistical significance (asterisk) is considered as P < 0.05.
CTCF-DS corresponds to a conserved enhancer element identified by comparative genomic sequencing and which was verified in mice for its ability to tissue-specifically enhance Igf2-H19 transcription (40). Enhancers downstream of H19 are shared by IGF2, and thus we would expect little or no change of H3K4me1 enrichment with a switch from IGF2 to H19 expression. Indeed, after treatment with 5-AzaCdR, the distribution and levels of H3K4me1 at the enhancer and also at CTCF-DS did not change (Figure 4A). The chromatin looping associations described above indicate that CTCF-DS switches between IGF2 and H19 looping domains by interacting either with CTCF-AD or with the ICR to mediate promoter–enhancer interactions (Figure 3B–D). If CTCF-DS is also an enhancer, it may activate IGF2 in untreated cells directly through its interaction with CTCF-AD in addition to mobilizing the canonical enhancers to the IGF2 promoters. Similarly, in 5-AzaCdR-treated cells, the CTCF-DS could activate H19 via its interaction with the ICR in addition to bringing the enhancers to H19 promoter.
The CCD has been described as an enhancer that can be accessed biallelically by the Igf2 promoters in normal mouse brain (chorioplexus) to bypass the ICR-mediated silencing (37). However, the CCD was not enriched for H3K4me1 in JEG3 (Figure 4A), which would argue against it being an active enhancer in these cells. It therefore seems that in addition to its reported enhancer role, the CCD has an architectural role as a boundary to keep the ‘IGF2-loop’ and ‘H19-loop’ segregated in tissues where the IGF2 and H19 promoters share enhancers (Figure 6B and C). Our results do not preclude the CCD from having a tissue specific enhancer function as well as a boundary function.
DNA demethylation at the ICR is not associated with changes in histone H3 lysine methylation at the ICR
Imprinted ICRs have been reported to have a tri-mark histone signature, trimethylation of histone H3 lysine 4, 9 and 20 (H3K4me3, H3K9me3 and H3K20me3), whereas a bivalent histone mark, H3K27me3-H3K4me3 is associated with promoters of imprinted genes subject to developmental regulation (44). Treatment with 5-AzaCdR is known to affect histone acetylation and methylation possibly as a consequence of gene activation associated with DNA demethylation (45,46). Therefore, we examined histone methylation profiles for H3K4me3, H3K9me3 and H3K27me3 at the ICR.
An enrichment of all three modifications was observed at the ICR in untreated cells (Figure 4B, C and D). After DNA demethylation at ICR by 5-AzaCdR, H3K27me3 and H3K4me3 were still present, whereas H3K9me3 was significantly reduced (Figure 4B–D). Although a reduction in H3K9me3 was expected to occur together with DNA demethylation at ICR, it is surprising to find H3K4me3 already on the predominantly DNA hypermethylated ICR and for it to be unchanged after 5-AzaCdR treatment. One explanation may be that the promoter region of the H19, which is very near to the ICR, may be poised for transcription and be bivalently marked with H3K4me3 and H3K27me3. If this is the case, then upregulation of the poised H19 promoter occurs without changing the bivalent mark.
Downregulation of IGF2 gene expression is accompanied by promoter specific reduction of histone H3K4me3
Further analysis of H3K4me3 across the whole locus showed that in untreated cells H3K4me3 was initially substantially enriched at the active IGF2 promoters (P3 and P4) in keeping with the strong IGF2 expression levels (Figure 4D). A reduction in H3K4me3 at IGF2 promoters would be expected with suppressed IGF2 expression after 5-AzaCdR. This mark was significantly reduced from IGF2 P4, although it did not disappear, and we did not see any change at the P3 (Figure 4D). H3K27me3 was also present at the IGF2 promoters (Figure 4C). The presence of both H3K4me3 and H3K27me3 at the IGF2 promoters suggests that these are also bivalent promoters. If so, then a decrease in H3K4me3 at these promoters resolves the bivalent mark to leave an H3K27me3 mark and a reduction of IGF2 expression. It is possible that the recruitment of SUZ12 to the IGF2 promoters (Figure 2D) maintains the H3K27me3 mark.
H3K27me3 spreads within the IGF2 looping domain bound by two CTCF-binding sites
Unlike H3K9me3 and H3K4me3, which accumulated at distinct regions (the ICR or the IGF2 promoters), we found H3K27me3 was enriched across the whole locus (Figure 4C). Higher levels of H3K27me3 were found spread across an IGF2-looping domain flanked by CTCF-AD and the CCD. Most of the IGF2-looping domain consists of the intergenic region, and H3K27me3 within this region could correspond to a transcriptionally inactive Broad Local Enrichment (BLOC) (47) of H3K27me3. The local enrichment for H3K27me3 within the IGF2-looping domain was >20-fold higher than the negative control region and higher than at the ICR and the H19-looping domain containing the enhancers and CTCF-DS. The H19-looping domain was up to 10-fold more enriched for H3K27me3 than the negative control region. There were no significant changes to the H3K27me3 profile after 5-AzaCdR treatment (Figure 4C), suggesting that the maintenance and turnover of this mark is not responsive to changes in DNA methylation at the ICR. It is noteworthy that the looping association between CTCF-AD and the CCD is still stable after 5-AzaCdR treatment (Figure 3B), which may explain why the H3K27me3 BLOC remains constant.
DNA demethylation of the ICR results in locus-wide nucleosomal remodelling
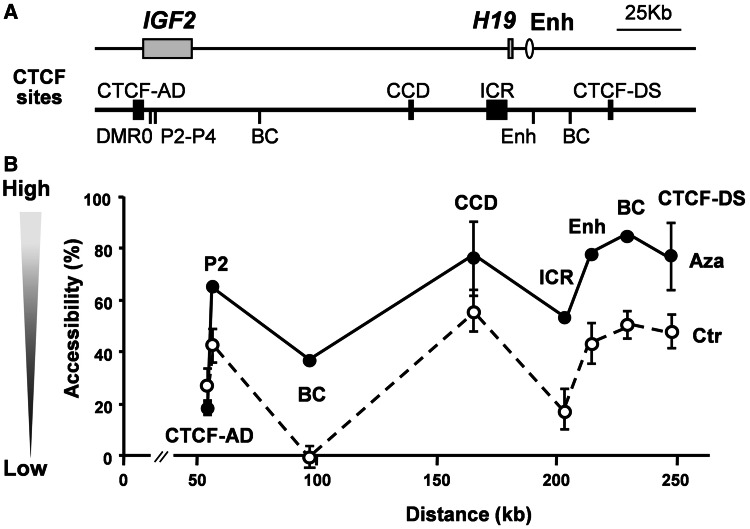
It has recently been reported that nucleosome occupancy changes when gene promoters become demethylated with 5-AzaCdR treatment (48). We therefore measured MNase accessibility across the region using EpiQ Chromatin Analysis Kit. Before 5-AzaCdR treatment, the whole locus showed low accessibility to nuclease digestion. After 5-AzaCdR treatment, the entire locus opened up and nuclease accessibility increased at all regions tested except the CTCF-AD (Figure 5B). This result suggests DNA demethylation at the ICR and possibly recruitment of CTCF not only change the nucleosome density at the ICR but also has an effect on the wider locus. Decreased nucleosome density at the CCD may account for the increased CTCF and cohesin binding observed after 5-AzaCdR treatment (Figure 2B and C).
Figure 5.
Changes in MNase accessibility after 5-AzacCdR treatment. (A) Graphic of the locus as in Figures 1 and 2, showing regions screened for the sensitivity to MNase digestion. (B) Accessibility of the locus to nuclease digestion. In untreated control samples (open circles and a dotted line), the MNase accessibility is low. After 5-AzaCdR treatment (filled circles and a solid line), it is increased locus-wide, except at CTCF-AD.
DISCUSSION
Although 5-AzaCdR is in clinical use as an epigenetic therapy targeting DNA methyltransferases, it is not known to what extent the reduction of DNA methylation impacts on other epigenetic features of a given locus. In this study, we have taken advantage of the choriocarcinoma cell line, JEG3, which is hypermethylated at the IGF2-H19 locus to study the plasticity of epigenomic organization on 5-AzaCdR-mediated DNA demethylation. Our results show that 5-AzaCdR treatment can induce DNA demethylation at specific regions (the ICR and DMR0) and that these localized DNA methylation changes occur together with locus-wide changes in CTCF-cohesin binding, nucleosome accessibility, chromatin looping conformation and gene expression. The extensive epigenetic profiling undertaken in this study provides further novel insights into the roles of CTCF and cohesin in higher order chromatin architecture at the IGF2-H19 locus, with broader significance to transcriptional regulation.
Profiling of chromatin looping interactions at the human IGF2-H19 locus has previously indicated that interactions between CTCF-binding sites separate the IGF2- and H19-looping domains (17,20). In the present study, we have demonstrated the impact of localized DNA demethylation on chromatin organization of the wider locus to refine the model as summarized in Figure 6. First, DNA demethylation at the ICR did not significantly change the associations between the CCD and CTCF-AD, suggesting that this is a stable loop (Figure 3B). Moreover, this association is present in several different cell lines (17,20) and thus likely to be a ubiquitous looping domain that is independent of DNA methylation. The H3K27me3 is particularly high and widespread within this domain in JEG3 cells and also does not change after 5-AzaCdR treatment. SUZ12 is recruited to the IGF2 promoters on demethylation of the ICR and may have a role in silencing IGF2 and/or stabilizing H3K27me3 within the domain.
Second, in contrast to the ubiquitous stable association of CTCF-AD with the CCD, CTCF-DS interactions with CTCF-AD (when IGF2 is active and the ICR is methylated) or with the ICR (when H19 is active and the ICR is unmethylated) form dynamic looping associations. In normal cell lines, these looping associations are allele specific (17). We propose that CTCF-DS associations with other CTCF sites facilitate enhancer–promoter interactions because the enhancer sequences located near to CTCF-DS have to follow where CTCF-DS interacts. In these models, the enhancer can be in proximity to the IGF2 promoters, despite it being in a different looping domain. The enhancer being in a different looping domain to the IGF2 promoters may be a mechanism whereby IGF2 can be activated, even though it is located in domain with a broad H3K27me3 enrichment and low nucleosome accessibility. Interestingly, CTCF-DS is enriched for the enhancer mark, H3K4me1. If CTCF-DS is an enhancer, then it may be acting via its associations with CTCF-AD or the ICR in addition to mobilizing the downstream H19 enhancers.
The locus-wide epigenetic changes that we observed after a reduction in DNA methylation at the ICR are still consistent with previous studies showing that the chromatin structure of the IGF2-H19 domain is determined by the ICR (49). DNA methylation at the ICR may be maintaining the whole locus in a closed chromatin conformation that is not constrained by the looping topography. Indeed, we found that the nucleosome accessibility is low in both the ubiquitous and the dynamic looping domains when the ICR is methylated. On DNA demethylation and CTCF recruitment to the ICR, the nucleosome accessibility increases at the whole locus. Unless 5-AzaCdR specifically targets nucleosome accessibility, DNA demethylation at the ICR is sufficient to reorganize nucleosomes at the wider locus. It is conceivable that the increased recruitment of CTCF may have a role in increasing the nucleosome accessibility through its role in nucleosome phasing (50,51).
The effects of 5-AzaCdR treatment at a given locus may not necessarily be directly owing to demethylation. For example, 5-AzaCdR treatment in lung cancer cell lines has been shown to upregulate expression of the CTCF paralogue, BORIS/CTCFL (49), which can modulate chromatin (52). BORIS/CTCFL has also been shown to bind preferentially to the methylated ICR at the IGF2-H19 locus in cancer cell lines (52–54). More recently, it has been demonstrated that unlike CTCF which is commonly found associated with phased nucleosomes, BORIS/CTCFL is associated with active promoters that are ‘nucleosome-free’ or enriched for the H3.3 histone variant (55). If BORIS/CTCFL binds to the ICR in JEG3 cells, it is possible that after 5-AzaCdR treatment BORIS/CTCFL is replaced by CTCF.
The objective of epigenetic therapy is to reverse the pathological gene expression patterns in malignant cells. As more epigenetic mechanisms of gene silencing become known, the number of targets for intervention increases. However, the large amount of interplay and cross-talk between epigenetic components requires that the effects of such therapies are monitored at well-characterized epigenetically regulated loci. The effect of chromatin topology on the regulation of long range gene expression is beginning to be appreciated. Future experiments looking at shorter time points of 5-AzaC treatment in cell lines will start to address the role of CTCF (and possibly BORIS/CTCFL) in nucleosome positioning in relation to chromatin loop formation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1–3.
FUNDING
Cancer Research UK (CRUK); University of Cambridge and Hutchison Whampoa Limited. Funding for open access charge: CRUK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Lovorka Stojic for her technical advice. They also thank Dr Kelly Holms and Ms Malwina Niemczyk for their comments on this manuscript.
REFERENCES
- 1.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uribe-Lewis S, Woodfine K, Stojic L, Murrell A. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert. Rev. Mol. Med. 2011;13:e2. doi: 10.1017/S1462399410001717. [DOI] [PubMed] [Google Scholar]
- 3.Kelly T, De Carvalho D, Jones P. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010;28:1069–2147. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan P, Frankhouser D, Murphy M, Tam HH, Rodriguez B, Curfman J, Trimarchi M, Geyer S, Wu YZ, Whitman SP, et al. Genome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemia. Blood. 2012;120:2466–2474. doi: 10.1182/blood-2012-05-429175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murrell A. Setting up and maintaining differential insulators and boundaries for genomic imprinting. Biochem. Cell Biol. 2011;89:469–478. doi: 10.1139/o11-043. [DOI] [PubMed] [Google Scholar]
- 6.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol. Syst. Biol. 2010;6:426. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 14.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nativio R, Sparago A, Ito Y, Weksberg R, Riccio A, Murrell A. Disruption of genomic neighbourhood at the imprinted IGF2-H19 locus in Beckwith-Wiedemann syndrome and Silver-Russell syndrome. Hum. Mol. Genet. 2011;20:1363–1374. doi: 10.1093/hmg/ddr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin Is Required for Higher-Order Chromatin Conformation at the Imprinted IGF2-H19 Locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 26.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 27.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 28.Szabo P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 29.Delaval K, Wagschal A, Feil R. Epigenetic deregulation of imprinting in congenital diseases of aberrant growth. Bioessays. 2006;28:453–459. doi: 10.1002/bies.20407. [DOI] [PubMed] [Google Scholar]
- 30.Murrell A, Ito Y, Verde G, Huddleston J, Woodfine K, Silengo MC, Spreafico F, Perotti D, De Crescenzo A, Sparago A, et al. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PLoS One. 2008;3:e1849. doi: 10.1371/journal.pone.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeschnigk M, Albrecht B, Buiting K, Kanber D, Eggermann T, Binder G, Gromoll J, Prott EC, Seland S, Horsthemke B. IGF2/H19 hypomethylation in Silver-Russell syndrome and isolated hemihypoplasia. Eur. J. Hum. Genet. 2008;16:328–334. doi: 10.1038/sj.ejhg.5201974. [DOI] [PubMed] [Google Scholar]
- 32.Vu TH, Nguyen AH, Hoffman AR. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum. Mol. Genet. 19:901–919. doi: 10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer. 2005;5:210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 34.Barletta JM, Rainier S, Feinberg AP. Reversal of loss of imprinting in tumor cells by 5-aza-2'-deoxycytidine. Cancer Res. 1997;57:48–50. [PubMed] [Google Scholar]
- 35.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.di Pietro M, Lao-Sirieix P, Boyle S, Cassidy A, Castillo D, Saadi A, Eskeland R, Fitzgerald RC. Evidence for a functional role of epigenetically regulated midcluster HOXB genes in the development of Barrett esophagus. Proc. Natl Acad. Sci. USA. 2012;109:9077–9082. doi: 10.1073/pnas.1116933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charalambous M, Menheniott TR, Bennett WR, Kelly SM, Dell G, Dandolo L, Ward A. An enhancer element at the Igf2/H19 locus drives gene expression in both imprinted and non-imprinted tissues. Dev. Biol. 2004;271:488–497. doi: 10.1016/j.ydbio.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 38.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu TH, Hoffman AR. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Niu B, Hu JF, Ge S, Wang H, Li T, Ling J, Steelman BN, Qian G, Hoffman AR. Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J. Cell Biol. 2011;193:475–487. doi: 10.1083/jcb.201101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishihara K, Hatano N, Furuumi H, Kato R, Iwaki T, Miura K, Jinno Y, Sasaki H. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in Igf2/H19 imprinting. Genome Res. 2000;10:664–671. doi: 10.1101/gr.10.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 42.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McEwen KR, Ferguson-Smith AC. Distinguishing epigenetic marks of developmental and imprinting regulation. Epigenetics Chromatin. 2010;3:2. doi: 10.1186/1756-8935-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Litt MD, Hansen RS, Hornstra IK, Gartler SM, Yang TP. 5-Azadeoxycytidine-induced chromatin remodeling of the inactive X-linked HPRT gene promoter occurs prior to transcription factor binding and gene reactivation. J. Biol. Chem. 1997;272:14921–14926. doi: 10.1074/jbc.272.23.14921. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 47.Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Noushmehr H, Han H, Andreu-Vieyra C, Liang G, Jones PA. Gene Reactivation by 5-Aza-2′-Deoxycytidine-Induced Demethylation Requires SRCAP-Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions. PLoS Genet. 2012;8:e1002604. doi: 10.1371/journal.pgen.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, Fischette MR, Adnani MT, Loukinov DI, Vatolin S, Risinger JI, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–7774. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 50.Kanduri M, Kanduri C, Mariano P, Vostrov AA, Quitschke W, Lobanenkov V, Ohlsson R. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol. Cell. Biol. 2002;22:3339–3344. doi: 10.1128/MCB.22.10.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen P, Bar-Sela G, Sun L, Bisht KS, Cui H, Kohn E, Feinberg AP, Gius D. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol. Cell. Biol. 2008;28:6720–6729. doi: 10.1128/MCB.00568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen P, Cui H, Bisht KS, Sun L, Patel K, Lee RS, Kugoh H, Oshimura M, Feinberg AP, Gius D. CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region. Cancer Res. 2008;68:5546–5551. doi: 10.1158/0008-5472.CAN-08-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sleutels F, Soochit W, Bartkuhn M, Heath H, Dienstbach S, Bergmaier P, Franke V, Rosa-Garrido M, van de Nobelen S, Caesar L, et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics Chromatin. 2012;5:8. doi: 10.1186/1756-8935-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.