Abstract
The lack of a consensus bacterial species concept greatly hampers our ability to understand and organize bacterial diversity. Operational taxonomic units (OTUs), which are clustered on the basis of DNA sequence identity alone, are the most commonly used microbial diversity unit. Although it is understood that OTUs can be phylogenetically incoherent, the degree and the extent of the phylogenetic inconsistency have not been explicitly studied. Here, we tested the phylogenetic signal of OTUs in a broad range of bacterial genera from various phyla. Strikingly, we found that very few OTUs were monophyletic, and many showed evidence of multiple independent origins. Using previously established bacterial habitats as benchmarks, we showed that OTUs frequently spanned multiple ecological habitats. We demonstrated that ecological heterogeneity within OTUs is caused by their phylogenetic inconsistency, and not merely due to ‘lumping’ of taxa resulting from using relaxed identity cut-offs. We argue that ecotypes, as described by the Stable Ecotype Model, are phylogenetically and ecologically more consistent than OTUs and therefore could serve as an alternative unit for bacterial diversity studies. In addition, we introduce QuickES, a new wrapper program for the Ecotype Simulation algorithm, which is capable of demarcating ecotypes in data sets with tens of thousands of sequences.
INTRODUCTION
The question of how and whether life organizes itself into discrete species units is key to our understanding of how diversity originates and is maintained. This question is a particularly challenging one for microbiologists because unlike plant and animal biologists, we can seldom directly observe phenotypic traits that may predict a microorganism’s ecological niche. Indeed, of the many millions of estimated bacterial taxa, the vast majority is uncultivable and is known only by DNA sequences (1–4). This greatly hampers our ability to meaningfully classify microbes based on their phenotypes. A key step in overcoming this challenge is developing methods to organize bacterial DNA sequences into biologically and ecologically meaningful taxonomic units. Even whether bacterial species exist at all is still a matter of some debate among microbiologists (5–8). In the absence of a consensus species concept, the most frequently used practice for organizing fine scale bacterial diversity is to cluster sequences solely on the basis of DNA sequence similarity at a conserved locus. Sequence clusters delineated in this manner are termed operational taxonomic units (OTUs).
OTUs, clustered based on the 16S rRNA gene, have been widely used to approximate bacterial species. Although OTUs are expedient for quickly clustering large numbers of bacterial sequences, they have several significant limitations as a unit of diversity. First, the similarity cut-off used to define OTUs is arbitrary. Defining species by 97% 16S identity is a commonly used rule of thumb (9,10), but species so defined are known to encompass large diversity in genome content, physiology and ecology (4,11,12) and are expected to underestimate the total diversity present when compared with the accepted 70% DNA–DNA hybridization threshold (13). Tighter thresholds such as 99 or 100% identity have been proposed (14) to address this issue, but this only serves to highlight the fundamental problem inherent in selecting a universal cut-off. It has been shown that regardless of the cut-off used, OTUs will not correspond directly to existing taxonomic units (15). This is because different lineages evolve at different rates; therefore, no universal cut-off will capture equivalent units of diversity across all bacterial lineages.
Another problem with OTUs based strictly on identity is that they do not take phylogenetic information into account, as pointed out previously (16–21). Because different lineages evolve at different rates, sequence similarity alone is inadequate to infer evolutionary relationships. This problem has been well illustrated in the context of gene function prediction based solely on sequence similarity (22,23). It is therefore expected that similarity-based OTUs will contain sequences with mixed phylogenetic signal such that an OTU is not guaranteed to be monophyletic. However, the extent and degree to which this phylogenetic inconsistency exists among OTUs remains largely unknown and has not been explicitly investigated. Recent studies indicate that bacterial ecological traits in general are phylogenetically conserved (17,18,24–31). Assuming phylogenetic niche conservatism, we hypothesize that the lack of phylogenetic consistency among OTUs will be associated with ecological heterogeneity within OTUs.
These problems with OTUs have led to the development of several alternative methods for species demarcation, all incorporating phylogeny and having a common ground on evolutionary theory. For example, the general mixed Yule-coalescent (GMYC) model delineates the species boundary by identifying the transition point from speciation to coalescent events using a likelihood framework (19–21,32). Ecotype Simulation (ES) (33) and AdaptML (34) aim to demarcate DNA sequences into ecologically cohesive clades (or ecotypes). ES identifies ecotypes by comparing the observed pattern of sequence diversity in a bacterial community to those of simulated communities ‘evolved’ based on the Stable Ecotype Model (8,35). AdaptML, by contrast, demarcates ecotypes by inferring the evolutionary history of habitat transitions. It identifies an ecotype as the largest clade whose members share an inferred habitat.
AdaptML and ES have both been successful in the past at predicting bacterial ecotypes from environmental DNA sequences. Ecotypes predicted by ES have been confirmed as ecologically distinct in isolates from natural communities of Bacillus sampled from desert canyons in Israel (33) and Death Valley (7), communities of Synechococcus from hot springs in Yellowstone (11,36,37) and in clinical and environmental isolates of Legionella pneumophila (38). AdaptML meanwhile has been used to demarcate ecologically distinct clusters in marine communities of Vibrio (34) and Desulfobulbus (39), as well as desert soil Bacillus (7).
By incorporating evolutionary models, GMYC, ES and AdaptML carry at least two advantages over OTU clustering. First, they do not require the selection of an arbitrary sequence identity cut-off. Second, these methods will always demarcate species as clades with a single evolutionary origin.
ES carries the additional advantage of generating predictions about the rates of ecotype formation and periodic selection within a clade. Periodic selection occurs when an individual within an ecotype gains a selective advantage within its ecological niche and carries all or nearly all of its genotype to fixation within that ecotype (selective sweeps) (40,41). This process results in genetic cohesion within ecotypes, analogous to the cohesion provided by interbreeding among members of an animal or plant species. Ecotype formation occurs when an individual changes its ecological niche, thereby releasing itself from the cohesive force of periodic selection within its parental ecotype. Ecotype formation is therefore analogous to sexual isolation and speciation among macroorganisms (8,42). Estimating the rates of periodic selection and ecotype formation therefore can shine light on the evolutionary and ecological processes that drive microbial diversity.
Our primary aim for this study was to investigate the degree and extent to which OTUs are phylogenetically and ecologically inconsistent. We compared and contrasted OTUs with ecotypes using both 16S rRNA and protein-coding genes. We found surprisingly extensive phylogenetic inconsistency among OTUs, to the extent that only a small minority of the OTUs comprised monophyletic clades. We also found a large amount of ecological inconsistency among OTUs. Specifically, when tested against habitats defined by their ecological parameters, OTUs were much more likely to span multiple habitats and less efficient in explaining the ecological variation than were ecotypes.
In addition, we introduce QuickES, a modified version of ES that runs much faster than the original version. Although an approximation of the complete algorithm, this version is capable of generating rough ecotype estimates for many thousands of sequences, making ecotypes a practical alternative unit for microbial diversity studies involving large sequence datasets.
MATERIALS AND METHODS
Data set, sequence alignment and classification
The 16S rRNA data set consisted of 116 391 near full-length Sanger-sequenced bacterial 16S rRNA sequences sampled from 21 different skin sites of 10 human subjects (43). The 16S rRNA sequences were aligned using the PyNAST algorithm in QIIME (44) and classified to the genus level using RDP Classifier, version 2 (45) at the default settings. We focused on the 10 most abundant genera within the skin data set for subsequent analyses. These genera made up ∼85% of the data set and spanned a broad taxonomic range, including the phyla Actinobacteria (Propionibacterium and Corynebacterium), Firmicutes (Staphylococcus, Streptococcus and Anaerococcus), Bacteroidetes (Cloacibacterium) and Proteobacteria (Diaphorobacter, Aquabacterium, Acidovorax and Acinetobacter).
In addition to the 16S rRNA gene, we also analysed protein-coding genes including 1025 hsp60 Sanger sequences of the genus Vibrio sampled from a coastal marine environment (34) and 132 psaA Sanger sequences of the genus Synechococcus sampled from the effluent channel of Mushroom Spring in Yellowstone National Park (36). The hsp60 encodes a heat shock protein, whereas psaA encodes a photosynthetic reaction centre protein. Protein-coding sequences were aligned by their amino acid sequences using MUSCLE (46) and then converted back to a DNA alignment using in-house scripts. All sequences were retrieved from Genbank, along with the sampling data for each sequence.
Phylogenetic analysis
Maximum-likelihood (ML) trees used as input for the ES and AdaptML analyses were generated using FastTree (47), with the gtr and gamma model engaged. The gtr and gamma model was chosen as the best model by JModelTest (48). For the assessment of OTU monophyly, we generated additional maximum likelihood trees and maximum-parsimony trees using RAxML (49), and neighbour-joining trees using QuickTree (50). To control for uncertainty in the tree topology, we only considered clades with >80% bootstrap support in our monophyly tests. An OTU was classified as monophyletic if all its members shared a single common ancestor to the exclusion of other OTUs. Otherwise, it was classified as paraphyletic, so long as the last common ancestor of all sequences of the OTU had at least 80% bootstrap support, and not all the descendants of that ancestral node were from the same OTU. We noted, however, that paraphyly, so defined, encompassed a broad array of phylogenetic patterns. These patterns ranged from OTUs that were very close to being monophyletic, (i.e. classed as paraphyletic due only to one or two divergent sequences within an otherwise monophyletic clade), to OTUs that were spread across the entire phylogeny. To distinguish between these extremes, we computed a ‘Paraphyly Index’ (PI) (Supplementary Figure S1) for each OTU defined by the formula:
Where NOTU is the number of sequences belonging to the OTU, and Nclade is the total number of sequences that are descendants of the last common ancestor of the OTU. A PI of zero indicates a monophyletic group, whereas a PI >0 indicates some degree of paraphyly.
Although we did not specifically classify any OTUs as polyphyletic based on these criteria, we were able to determine based on direct observation of phylogenies that many OTUs were in fact polyphyletic. That is, they were not explainable by a single evolutionary origin.
OTUs
Unless otherwise specified, OTUs were generated with the Uclust de novo clustering algorithm using QIIME (44). For the skin 16S rRNA data set, OTUs were generated using identity cut-offs of 99.5, 99 and 97%. For the protein-coding Vibrio hsp60 and Synechococcus psaA data sets, OTUs were generated using cut-offs of 100, 99, 97, 95, 90 and 85%. It has been shown that different OTU clustering algorithms can return very different outputs (51). Therefore, to ensure the robustness of our analysis of OTU monophyly, additional software [MOTHUR (52) and Clusterer (53)] and additional clustering algorithms (nearest neighbour and average neighbour clustering) were also used to cluster the sequences. In each case, the default clustering parameters were used for all analyses, except to modify the cut-off value, or the clustering algorithm as indicated.
Demarcation of ecotypes using ES
We used ES (33) version 0.6 to demarcate ecotypes on two abundant genera within the human skin data (Aquabacterium and Diaphorobacter), as well as on the Vibrio hsp60 and Synechococcus psaA sequences data. ES is an algorithm for predicting ecologically homogeneous populations (ecotypes) from sequence data alone, without the need for inputting any ecological data, or for selecting any similarity cut-off value [see Koeppel et al. (33) for a full description of the ES algorithm]. Briefly, ES operates by simulating the evolution of a set of sequences based on parameterized values for the number of ecotypes, as well as the rates of ecotype formation, periodic selection and genetic drift. It uses a maximal likelihood framework to estimate values for each of these four parameters by fitting the simulated sequences to the observed diversity curve (see Supplementary Figure S2 for examples). Using the best parameter solutions, ES then demarcates ecotypes onto a phylogeny generated from the same set of sequences, by selecting the most inclusive clades consistent with being a single ecotype.
The full version of ES is only capable of analysing ∼200 sequences at once within a reasonable time frame. As all of these genera contained many more sequences, we used a divide-and-conquer approach. Using a guiding tree, we subdivided the sequences into clades containing fewer than 200 sequences and ran ES separately on each clade. We then demarcated ecotypes on the entire tree by finding the most inclusive clades consistent with being a single ecotype (33).
QuickES
To speed up the ES and make it practical to analyse thousands of sequences, we modified the original ES algorithm and created a version called QuickES. QuickES approximates the ecotype estimation process but still carries the key advantages of the standard ES ecotypes over OTUs, in that its ecotypes are always monophyletic, and there is no need to select an identity cut-off. The improved speed was achieved in two primary ways.
First, we used a divide-and-conquer approach similar to the one described earlier in the text. In this case, we subdivided the sequences into clades such that each clade was the most inclusive possible clade in which at least 90% of the descendants belonged to the same OTU (99% cut-off). Subdividing the data set into clades dramatically improved the speed of analysis because ES scales poorly as more sequences are added.
The second improvement in computation speed came from using a rougher estimate of the periodic selection and ecotype formation rate parameters. From the set of clades obtained in the divide-and-conquer step, we selected those clades that contained between 25 and 200 sequences. Each of these moderately sized clades was then analysed using a truncated version of the brute force search algorithm from the original ES, to glean a rough estimate of the parameter values necessary for ecotype demarcation. The very time consuming hill-climbing algorithm that ES uses to refine the estimates further was eliminated. Eliminating this step markedly increases the speed but decreases the reliability of the rate estimates; therefore, QuickES should not be used for the final rate estimation but only for ecotype demarcations when data sets are too large to analyse with the full ES algorithm. Having obtained rough estimates of the rate parameters for several clades, we then generated global rate estimates by computing the mean of the rates from the clades analysed. Then, following the basic demarcation protocol from the original ES (33), we demarcated ecotypes for every clade, by finding the most inclusive subclades consistent with being a single ecotype, given the global rate estimates. QuickES is freely available software and can be downloaded from http://wolbachia.biology.virginia.edu/WuLab/Software.html. Detailed instructions of running QuickES are described in the Supplementary Methods.
Demarcation of ecotypes using AdaptML
We used AdaptML (34) to demarcate ecotypes for the marine Vibrio hsp60 data set. Our AdaptML analysis of Vibrio returned habitats virtually identical to those of Hunt et al. (34), with the exception that we had seven habitats instead of six. This is likely due to slight variations in tree topology resulting from using different tree-building algorithms. We refer to the seventh habitat as HG (Hunt et al. habitats are HA- HF). The habitat-learning and clustering steps of AdaptML were performed using the default settings.
Benchmarking the performance of OTUs and ecotypes in explaining the ecological variance in the vibrio data set
We used the methods described in (19) to compare OTUs and ecotypes in their ability to account for the ecological variation in the Vibrio data set. Vibrio sequences were associated with two ecological measurements: the size of the particle from which the Vibrio sequences were isolated (<1 μm, 1–5 μm, 5–63 μm, >63 μm) and the season when the samples were collected (spring and fall) (34). We first transformed the particle size and season categories into quantitative variables using multiple correspondence analysis implemented in the ‘ade4’ package (54). Then we carried out redundancy analysis implemented in the ‘vegan’ package in R to estimate the amount of variation in the ecological parameters that could be explained when sequences were grouped by either OTUs or ecotypes. Statistical significance was assessed using permutation tests. Akaike information criterion was used to evaluate the performance of the different species delineation models.
Estimation of periodic selection and ecotype formation rates
The complete ES algorithm generates estimates of the rates of periodic selection and ecotype formation. We performed ES analyses individually on all major subclades of the genera Aquabacterium, Diaphorobacter and Vibrio and tested whether the mean rates of ecotype formation and periodic selection were different between genera using Student’s t-tests. Rates within the same genus were grouped into high, medium and low categories using the Tukey–Kramer test as described in Supplementary Methods. ES estimated rates in units of events per nucleotide substitution. Rates were log-transformed before statistical analysis so as to more closely approximate a normal distribution.
RESULTS
Extensive microdiversity within clades
Consistent with our expectations, and with findings observed in other microbial habitats, we observed extensive microdiversity in the data sets we analysed (Supplementary Figure S2). The number of 16S rRNA OTUs in each genus showed a dramatic flare-up between the 98 and 99% cut-off levels (Supplementary Figure S2A–E). The presence of such a ‘hockey-stick’ pattern has previously been observed in natural bacterial populations (55,56) and is considered typical of human associated microbial populations (57,58). This pattern is consistent with the Stable Ecotype Model (8,33), which predicts that ephemeral microdiversity should be present within bacterial communities, as bacteria undergo neutral divergence between periodic selective sweeps.
The protein-coding sequences of the Vibrio and Synechococcus data sets displayed similar flare-ups, though at slightly lower sequence identity thresholds (∼97% in both cases) (Supplementary Figure S2 Fand G). This reflects the more rapid evolution of the protein-coding hsp60 and psaA genes compared with the 16S rRNA gene.
Extensive and pronounced paraphyly and polyphyly among OTUs
To deal with the potential uncertainty in the tree topology, we only considered clades that were well supported (bootstrap values ≥80) in our monophyly analyses. Our analysis of the predominant skin bacterial genera revealed that strikingly few OTUs are monophyletic (Table 1). At the 97% identity level, no >75% of the OTUs were monophyletic groups in any of the genera analysed. At the 99% identity level, fewer than 60% of OTUs in each genus were monophyletic. The percentages were far smaller in most genera. The results were even more remarkable among larger OTUs (those containing at least 50 sequences): <67% of the large OTUs in each genus were monophyletic at the 97% cut-off. At the 99% cut-off, fewer than 25% of the large OTUs in each genus were monophyletic groups. In fact, in five of the 10 genera analysed (Acidovorax, Acinetobacter, Aquabacterium, Cloacibacterium and Diaphorobacter), none of the large 99% OTUs were monophyletic.
Table 1.
Phylogenetic heterogeneity among 16S rRNA OTUs of skin data set
| Genus | OTU identity threshold | OTUs >1 Sequence |
OTUs >50 Sequences |
||
|---|---|---|---|---|---|
| Number of OTUs | % Monophyletic | Number of OTUs | % Monophyletic | ||
| Acidovorax | 97% | 2 | 0.00% | 1 | 0.00% |
| 99% | 3 | 0.00% | 1 | 0.00% | |
| 99.5% | 4 | 0.00% | 2 | 0.00% | |
| Acinetobacter | 97% | 8 | 12.50% | 3 | 0.00% |
| 99% | 31 | 25.81% | 8 | 0.00% | |
| 99.5% | 45 | 31.11% | 1 | 0.00% | |
| Anaerococcus | 97% | 31 | 51.61% | 4 | 0.00% |
| 99% | 64 | 37.50% | 4 | 25.00% | |
| 99.5% | 71 | 11.27% | 3 | 0.00% | |
| Aquabacterium | 97% | 5 | 60.00% | 1 | 0.00% |
| 99% | 5 | 60.00% | 1 | 0.00% | |
| 99.5% | 12 | 41.67% | 1 | 0.00% | |
| Cloacibacterium | 97% | 2 | 50.00% | 1 | 0.00% |
| 99% | 7 | 57.14% | 2 | 0.00% | |
| 99.5% | 44 | 11.36% | 3 | 0.00% | |
| Corynebacterium | 97% | 53 | 33.96% | 22 | 13.64% |
| 99% | 160 | 25.00% | 44 | 2.27% | |
| 99.5% | 323 | 18.89% | 45 | 0.00% | |
| Diaphorobacter | 97% | 1 | 0.00% | 1 | 0.00% |
| 99% | 1 | 0.00% | 1 | 0.00% | |
| 99.5% | 11 | 18.18% | 1 | 0.00% | |
| Propionibacterium | 97% | 3 | 66.67% | 3 | 66.67% |
| 99% | 7 | 42.86% | 4 | 25.00% | |
| 99.5% | 88 | 23.86% | 3 | 0.00% | |
| Staphylococcus | 97% | 6 | 16.67% | 3 | 0.00% |
| 99% | 59 | 18.64% | 16 | 12.50% | |
| 99.5% | 230 | 24.78% | 12 | 0.00% | |
| Streptococcus | 97% | 12 | 75.00% | 5 | 60.00% |
| 99% | 50 | 22.00% | 7 | 14.29% | |
| 99.5% | 127 | 14.96% | 4 | 0.00% | |
This table displays the number of monophyletic OTUs in each genus at three different identity thresholds (97, 99 and 99.5%). Only OTUs containing at least two sequences and meeting the support criteria were considered, as a single sequence is monophyletic by definition. The effect was more pronounced among larger OTUs (OTUs containing at least 50 sequences, right-hand columns).
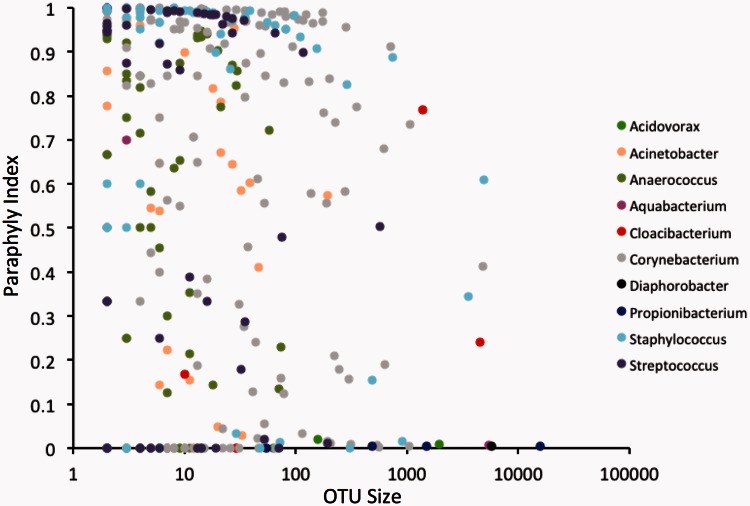
To measure the degree of paraphyly among these OTUs, we computed the PI (see ‘Materials and Methods’ section for details) for all of the 99% OTUs in each genus. A PI of 0 indicates a monophyletic group, whereas a PI close to 1 indicates substantial paraphyly. Surprisingly, large numbers of OTUs of all sizes across all genera had mixed phylogenetic signal, some extensively (i.e. their PI was close to 1.0, Figure 1). We observed similar patterns among OTUs clustered at 97 and 99.5% thresholds (Supplementary Figure S3). This result suggests that phylogenetic incoherence among OTUs is far more pronounced and pervasive than is generally recognized.
Figure 1.
OTU paraphyly is pervasive and pronounced. This graph plots OTU size against PI for all 99% 16S rRNA OTUs among 10 genera. PI values of 0.0 indicate monophyletic groups, whereas a PI close to 1 indicates substantial paraphyly. Genus classifications of OTUs are colour coded as indicated in the key.
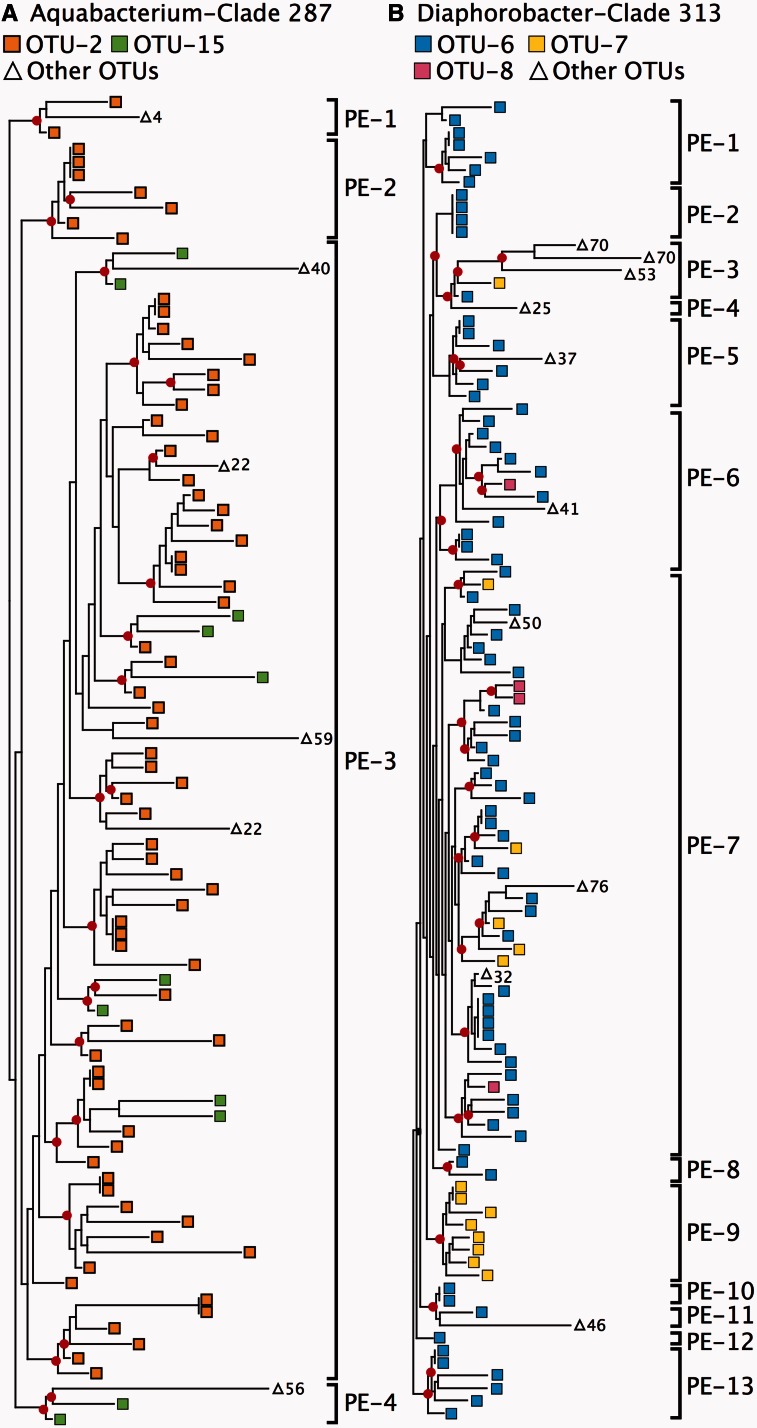
Given the extreme deviation from monophyly that we observed with the PI scores, we mapped several OTUs onto phylogenies to observe their phylogenetic patterns directly. We observed that many of the OTUs appear to be polyphyletic because they required multiple independent evolutionary origins to explain their distribution across the phylogeny (Figure 2). Extensive paraphyly was observed, regardless of the phylogenetic or OTU clustering methods used (Table 2). Putative ecotypes, by contrast, always mapped to monophyletic clades (Figure 2) because phylogenetic information was taken into account during the demarcation process.
Figure 2.
Extensive paraphyly and polyphyly among OTUs. Maximum likelihood trees of representative subclades of the genera (A) Aquabacterium and (B) Diaphorobacter. OTU generated using the 99% identity cut-off are shown with the putative ecotypes (PE) demarcated by ES. Internal nodes with >80% bootstrap support are highlighted with red circles.
Table 2.
Phylogenetic heterogeneity of OTUs is robust to methodology
| Clustering method | OTU cut-off | n | Phylogenetic method |
|||
|---|---|---|---|---|---|---|
| FastTree (ML) | RAxML (ML) | RAxML (MP) | QuickTree (NJ) | |||
| QIIME (Uclust) | 97% | 5 | 60.00% | 40.00% | 40.00% | 50.00% |
| 99% | 5 | 60.00% | 60.00% | 40.00% | 60.00% | |
| 99.5% | 12 | 41.67% | 60.00% | 45.45% | 80.00% | |
| MOTHUR (farthest neighbour) | 97% | 9 | 33.33% | 33.33% | 25.00% | 37.50% |
| 99% | 41 | 14.63% | 15.79% | 15.79% | 17.65% | |
| 99.5% | 306 | 10.53% | 10.44% | 10.34% | 14.34% | |
| MOTHUR (nearest neighbour) | 97% | 1 | 0.00% | 0.00% | 0.00% | 0.00% |
| 99% | 3 | 66.67% | 66.67% | 66.67% | 66.67% | |
| 99.5% | 3 | 33.33% | 50.00% | 33.33% | 33.33% | |
| MOTHUR (average neighbour) | 97% | 2 | 50.00% | 50.00% | 50.00% | 50.00% |
| 99% | 5 | 60.00% | 40.00% | 60.00% | 60.00% | |
| 99.5% | 31 | 48.15% | 58.33% | 60.87% | 77.78% | |
| Clusterer (farthest neighbour) | 97% | 11 | 18.18% | 18.18% | 18.18% | 20.00% |
| 99% | 43 | 20.93% | 17.95% | 17.50% | 28.13% | |
| 99.5% | 227 | 13.24% | 14.36% | 14.78% | 15.57% | |
| Clusterer (nearest neighbour) | 97% | 1 | 0.00% | 0.00% | 0.00% | 0.00% |
| 99% | 5 | 40.00% | 40.00% | 40.00% | 40.00% | |
| 99.5% | 7 | 14.29% | 16.67% | 14.29% | 14.29% | |
| Clusterer (UPGMA) | 97% | 4 | 25.00% | 25.00% | 25.00% | 25.00% |
| 99% | 9 | 33.33% | 22.22% | 22.22% | 33.33% | |
| 99.5% | 21 | 45.00% | 52.63% | 47.37% | 47.37% | |
This table displays the percentage of monophyletic 16S rRNA OTUs in the genus Aquabacterium at three different identity thresholds (97, 99 and 99.5%). As in Table 1, only OTUs containing at least two sequences and meeting support criteria were considered in the percentage computations, though the total number of OTUs is displayed in the n column. Different phylogenetic methods (columns) and different OTU clustering algorithms (rows) were tested. Cells display the percentage of OTUs that were monophyletic clades.
ML, Maximum likelihood; MP, Maximum parsimony; NJ, Neighbour joining.
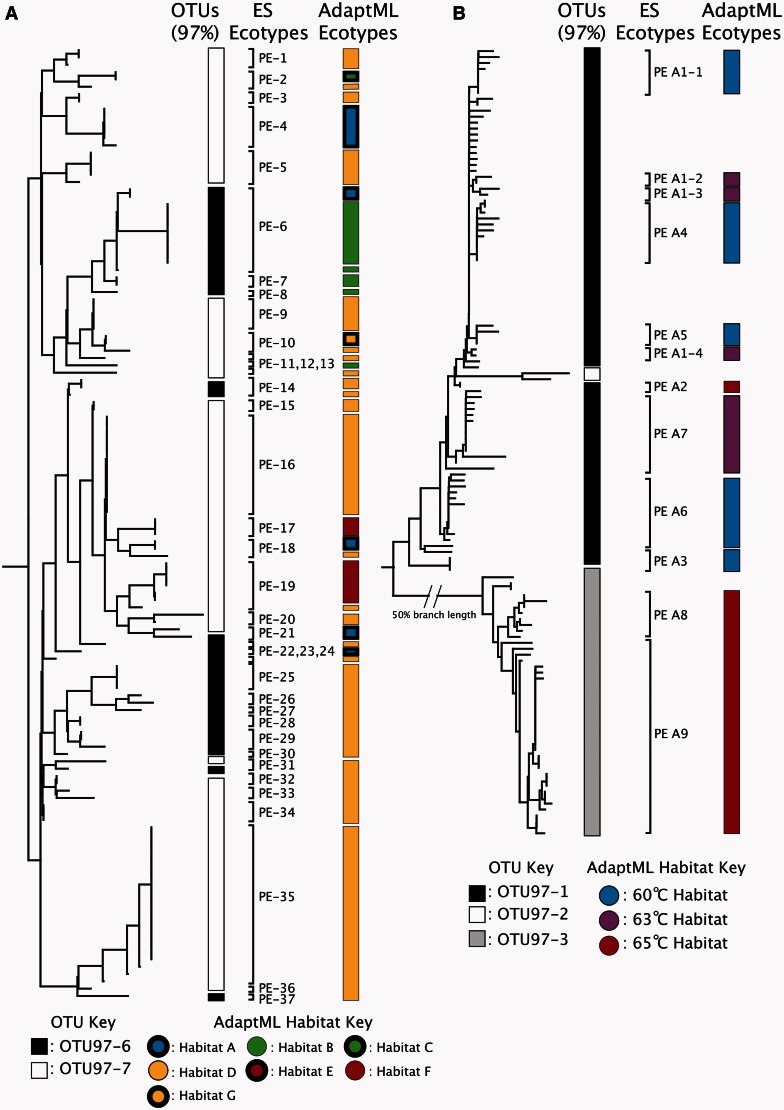
We next checked whether the phylogenetic heterogeneity we had observed among OTUs based on 16S rRNA also existed in OTUs of protein-coding genes. OTUs in the marine Vibrio data set, clustered based on similarity at the hsp60 locus, showed a similar, if less extreme phylogenetic pattern. There was considerably more monophyly among OTUs in this data set than in the skin data set. Over 80% of the OTUs at all cut-offs except 99% were monophyletic (Supplementary Table S1). At the 99% cut-off, however, only 74.56% of the OTUs were monophyletic groups. Although the majority of the non-monophyletic OTUs were paraphyletic (i.e. explainable by a single evolutionary origin), several OTUs did appear to be polyphyletic (Figure 3A) (see 97% OTU-6 and OTU-7). The Yellowstone Synechococcus psaA sequences clustered into only three OTUs (97% cut-off). Two of the three were monophyletic, but the third and largest was paraphyletic (Figure 3B).
Figure 3.
OTUs and Ecotypes show distinct habitat associations. An ML tree of a subset of the Vibrio hsp60 sequences (A) and a neighbour-joining tree of the full set of Synechococcus psaA sequences (B). OTUs, ES ecotypes and AdaptML ecotypes are shown. Note that the formatting in the OTU column and the AdaptML column is different. In the OTU column, all leaves marked by the same color belong to the same OTU. In the AdaptML column, different colours denoted different habitats. Each distinct colour bar is its own ecotype, whereas bars of the same colour are ecotypes co-occurring in the same habitat.
Extensive ecological heterogeneity among OTUs
It has been established that OTUs contain a great deal of ecological diversity (11). However, this is usually assumed to be a result of defining OTUs too broadly. Under this line of reasoning, it might be assumed that simply narrowing the breadth of OTUs, either by using a more stringent identity cut-off or by using a marker less conserved than 16S rRNA, can address this problem in microbial ecology studies. We have undercut this reasoning by demonstrating that OTUs are phylogenetically inconsistent. Our results suggest that the problem runs deeper than simply using a cut-off that is too relaxed. We hypothesized that the lack of phylogenetic consistency among OTUs will be associated with ecological heterogeneity within OTUs, even when a more rapidly evolving protein-coding gene was used to cluster them.
We used the previously established marine Vibrio (34) and hot spring Synechococcus (36) AdaptML habitat types as benchmarks for determining ecological homogeneity. We reasoned that populations belonging to a single habitat could be considered more ecologically homogeneous than populations spanning multiple habitats. We observed that in the Vibrio data, OTUs (97% identity cut-off at the hsp60 locus) frequently spanned several of the habitats predicted by AdaptML (Figure 3A).
For the entire Vibrio data set, we found that only 55.1% of the OTUs belonged to just one AdaptML habitat. In comparison, 77.2% of ecotypes generated by ES were either identical to or nested within the AdaptML ecotypes, as expected (35) (and see ‘Discussion’ section). A Student’s t-test confirmed that the mean number of habitats spanned by Vibrio OTUs was significantly greater than the mean number spanned by Vibrio ES ecotypes (P < 0.0002). This indicated that ES ecotypes showed greater ecological homogeneity than the 97% OTUs in the Vibrio data set.
The Synechococcus analysis showed a similar pattern. Our ES analysis predicted 12 non-singleton ES ecotypes in the Synechococcus data set (Figure 3B), which appear to be ecologically distinct based on the temperature of the sample site (36). Representatives of several ecotypes were also found to be predominant at different depths of the microbial mat, suggesting potential ecological distinctness based on light and O2 concentrations (36). Each of the ES ecotypes corresponded to a single AdaptML habitat, though, as expected, some AdaptML ecotypes contained multiple ES ecotypes (Figure 3B). In comparison, the largest OTU spanned all three AdaptML habitats.
The size of an OTU is expected to have a large impact on its level of ecologically heterogeneity. For example, for the same data set, 97% OTUs will be larger than 99% OTUs (i.e. containing more sequences) and thus ecologically will be more heterogeneous. To control for the potential size difference between OTUs and ecotypes, we followed the approach of Powell et al. (19) and benchmarked the performance of OTUs of different cut-offs and ecotypes in explaining the ecological variance in the Vibrio data set. Our results show that ecotypes (ES or AdaptML) significantly outperformed OTUs in accounting for variation in the ecological parameters (sampling particle size and season) (Table 3). This was the case, regardless of the size of the OTUs (at 97, 99 or 100% identity cut-offs). Although 100% OTUs explained the most variance, as expected, it came with a high cost associated with the large number of classes. Ecotypes most efficiently explained the variance according to the Akaike information criterion (δAIC = 181 for AdaptML and δAIC = 49 for ES against 100% OTUs, respectively). It is not surprising that AdaptML performed better than ES in this test because AdaptML uses both the sequences and the ecological information to demarcate ecotypes, whereas ES only uses sequences. Our results are in agreement with the Powell et al. study (19) showing that evolutionary theory-based approaches outperform operational approaches in producing ecologically meaningful diversity units.
Table 3.
Performance of OTUs and ecotypes in explaining ecological variation in the Vibrio data set
| Model | Number of classes | Variance explained | AIC |
|---|---|---|---|
| OTU 97% | 68 | 42%* | 764 |
| OTU 99% | 187 | 60%* | 649 |
| OTU 100% | 382 | 73%* | 639 |
| ES | 190 | 62%* | 590 |
| QuickES | 156 | 58%* | 609 |
| AdaptML | 214 | 69%* | 458 |
The variance explained was calculated by dividing the constrained inertia by the total inertia. The two methods that returned the lowest AIC scores are highlighted in bold. Asterisk denotes the significance of P ≤ 0.005 by permutation tests.
AIC, Akaike information criterion.
Universal identity cut-offs fail to capture all putative ecotypes
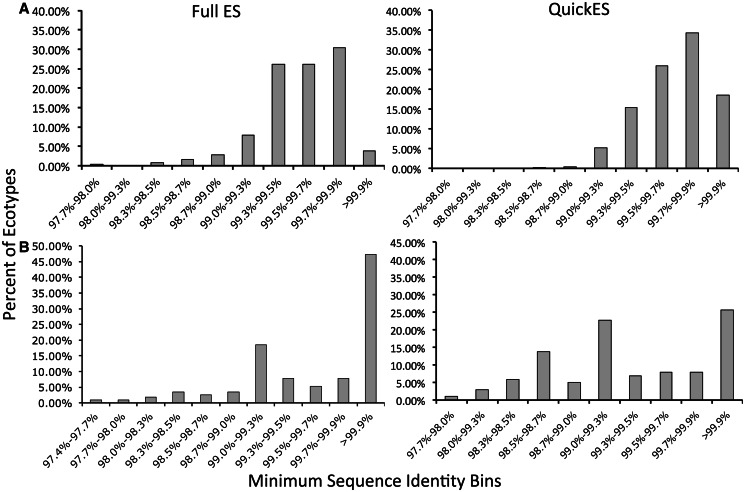
Having shown that 97% cut-off OTUs were ecologically heterogeneous, we next sought to determine whether any single identity cut-off could have generated OTUs similar to the putative ecotypes designated by the ES algorithm. We binned the ecotypes by their minimum pairwise sequence identity and plotted the number of ecotypes in each sequence identity bin for the Aquabacterium and Vibrio genera (Figure 4). Figure 4 shows that ecotypes display a wide range of sequence identities, and no universal OTU cut-off can be applied to capture all these ecotypes. This held true, regardless of whether the complete ES method or QuickES was used. For example, in the Aquabacterium genus, a large majority (94.2%) of the ecotypes had a minimum pairwise sequence identity ≥99% (Figure 4A left panel). This means that if these sequences were clustered into OTUs using a 99% identity cut-off, many of those OTUs might contain sequences from multiple ecotypes. At the other end of the spectrum are the 14 ecotypes (5.8%) whose minimum sequence identity is <99%. A 99% identity OTU cut-off would subdivide these ecotypes into multiple OTUs.
Figure 4.
Putative ecotypes are not captured by any single sequence identity cut-off. Graphs display the number of ecotypes whose minimum pairwise sequence identity fall into each of the displayed bins in the skin Aquabacterium (A) and the marine Vibrio (B) data sets.
Rates of periodic selection vary within and between genera
One advantage of ES over other methods is that it can be used to gain insight on the processes of microbial diversification. We used the complete ES to compare the rates of periodic selection and ecotype formation within and between genera. We generated estimates of the rates of periodic selection and ecotype formation for subclades within each genus. Consistent with the Stable Ecotype Model, we found that, in each case, the rate of periodic selection was estimated to be greater than the rate of ecotype formation. In Aquabacterium and Diaphorobacter, the median rate of periodic selection among subclades was around twice the median rate of ecotype formation (2.04 times and 1.50 times greater, respectively). Interestingly, in Vibrio, the median rate of periodic selection was 26.5 times higher than the median rate of ecotype formation (Table 4). Our analysis revealed no statistically significant differences between the Vibrio, Aquabacterium and Diaphorobacter genera in their rates of ecotype formation (Student’s t-test: Vibrio-Diaphorobacter: P ≤ 0.298; Vibrio-Aquabacterium: P ≤ 0.097; Aquabacterium-Diaphorobacter: P ≤ 0.997). However, we did detect significant differences in the rate of periodic selection between genera. Specifically, both Vibrio and Aquabacterium showed a significantly higher rate of periodic selection than Diaphorobacter (Student’s t-test: P < 0.0059 and P < 0.0139, respectively).
Table 4.
Ecotype formation and periodic selection rates of three genera
| Genus | n | Ecotype formation rate |
Periodic selection rate |
||||
|---|---|---|---|---|---|---|---|
| Mean | Standard deviation | Median | Mean | Standard deviation | Median | ||
| Aquabacterium | 20 | 0.179 | 0.092 | 0.458 | 6.969 | 18.950 | 0.934 |
| Diaphorobacter | 19 | 0.166 | 0.130 | 0.340 | 0.707 | 0.472 | 0.510 |
| Vibrio | 10 | 0.312 | 0.370 | 0.105 | 88.467 | 239.166 | 2.780 |
ES (original version) estimates of the mean and median periodic selection and ecotype formation rates for skin Aquabacterium and Diaphorobacter, and for marine Vibrio. The n values are the number of subclades for which the rates were independently calculated.
There were surprisingly large variations in the rates of periodic selection and ecotype formation (Supplementary Figures S4 and S5) within each genus. For example, both Aquabacterium and Vibrio contained clades with greatly elevated periodic selection rates. In Aquabacterium, the rate estimated for one clade was nearly 8-fold higher than that of the other clades (Supplementary Figure S4B). In Vibrio, two clades showed rates 16-fold and 160-fold higher above the average, respectively (Supplementary Figure S4C). The rate of ecotype formation within Vibrio was also highly variable, with one group of clades showing ecotype formation rates as much as 10-fold higher than the rest of the genus (Supplementary Figure S5C).
Validation of QuickES
We tested QuickES on a three-gene concatenation of sequences from soil Bacillus simplex and Bacillus subtilis/licheniformis. These sequences were isolated from the Negev Desert in Israel and had previously been analysed using the complete ES algorithm (33). For each data set, we ran three QuickES trials, selecting the results from the trial whose parameter solutions gave the best likelihood value (best of three). We then repeated this procedure twice, resulting in a total of three replicate runs. Supplementary Figures S6 and S7 show that ecotypes demarcated in three QuickES runs were very similar to each other and also to those predicted by the full analysis.
We also benchmarked the performance of QuickES ecotypes in its ability to explain the ecological variance in the Vibrio dataset (Table 3). Grouping sequences by QuickES ecotypes accounted for 58% of ecological variation (P ≤ 0.005). QuickES ecotypes were much better at explaining the ecological variance than all the OTUs we tested (97%, 99% and 100%, δAIC ≥ 30), although they performed slightly worse than the ecotypes demarcated with the complete ES (δAIC = 19).
DISCUSSION
Although great strides have been made in our understanding of bacterial diversity in recent years, the field has been challenged by the lack of an agreed on unit of diversity analogous to the biological species of animals and plants. Although identity-based OTUs provide a ‘quick and dirty’ approach to quantifying bacterial diversity and are very useful, they are no substitute for coherent and meaningful units of bacterial ecology and evolution (8,59,60).
Our results illustrate several problems with using OTUs to approximate bacterial species. Although these problems are generally known, we have shown here that they are far more serious than has previously been appreciated. First, paraphyly and polyphyly among OTUs are far more extensive and pronounced than expected. We observed this pattern in a wide variety of genera spanning multiple bacterial phyla, indicating that this problem is widespread and not taxon specific. Our results showing significant numbers of paraphyletic and polyphyletic OTUs at various sequence identity thresholds further reinforce the point that simply narrowing the identity cut-off will not correct the phylogenetic inconsistency among OTUs. Although the acceptability of paraphyletic taxa in systematics is debatable (8,61,62), polyphyletic groups are generally considered to be unacceptable as true taxa. This is because polyphyletic groups by definition lack a single evolutionary origin. Most modern species concepts require individuals within the same species to share a single evolutionary lineage (63,64) and therefore a single evolutionary origin.
Second, we show that extensive phylogenetic inconsistency is associated with extensive ecological heterogeneity in OTUs. This is distinct from ecological heterogeneity caused by using OTUs that are too broadly defined. Just as phylogenetic inconsistency will persist, regardless of identity cut-off, simply narrowing the identity cut-off might not produce ecologically coherent OTUs. We demonstrated that ecotypes outperform OTUs in explaining the ecological variance in the sequence data. This finding is particularly relevant to human microbiome research, in which 16S rRNA OTUs are commonly used to compare the composition of microbial communities between healthy and diseased individuals (65). The 16S rRNA OTUs at 97 (66,67), 98 (58) and 99% (43,68) identity cut-offs have been used as units of microbial diversity. Our results suggest that such OTUs may not be ecologically homogeneous and therefore can be problematic for association studies. Specifically, the use of ecologically heterogeneous units could add noise when investigating the associations between OTUs and health states, resulting in false negatives.
An additional problem with OTUs is that the identity cut-off is arbitrarily defined and subjectively applied. The threshold selected can greatly affect estimates of both the numbers and composition of species in a community. Ecotypes, as predicted by either ES or AdaptML, require no arbitrary cut-off, and therefore can be compared consistently across taxa. We demonstrated that ecotypes can be variable in the amount of sequence diversity they contain such that no single universal identity threshold will accurately capture the ecotypes in a community. We calculated alpha diversity (diversity within habitats) and beta diversity (diversity among habitats) indices for the skin data set using both OTUs and ecotypes (data not shown). The overall alpha diversity values measured using ecotypes are higher than those measured using 16S rRNA OTUs (97 or 99% cut-off), suggesting that the routinely used 16S rRNA units might underestimate the diversity. However, we noticed no apparent or significant differences between beta diversity measures derived from ecotypes and OTUs.
One benefit of the complete ES algorithm over other methods is that it provides estimates of the rates of ecotype formation and periodic selection. These rate estimates can be used to gain insight into the mode and tempo of bacterial diversification. Periodic selection events purge the genetic diversity within a population and lead to adaptation within a species lineage (anagenesis) (40,41). Ecotype formation (cladogenesis) occurs when an individual becomes sufficiently ecologically distinct that it is no longer vulnerable to periodic selection events occurring in its parent population. Determining the relative rates of periodic selection and ecotype formation can help us distinguish between different models of bacterial speciation and evolution (8,69). For example, the Stable Ecotype Model proposes recurring periodic selective sweeps between rare ecotype formation events, and therefore predicts that anagenesis is the most dominant mode of adaptive evolution (8). In contrast, the Species-Less Model features a high rate of species turnover, with frequent cladogenesis and almost no anagenesis, because each species is likely to go extinct before its first periodic selection event (69). Our ES analyses indicate that periodic selection happens much more frequently than ecotype formation, suggesting that at least in the bacterial lineages analysed in our study, anagenesis is the dominant mode of adaptive evolution.
For a lineage that is recently formed, either as a result of invading a new habitat or via horizontal transfer of a niche-expanding gene (35), we might expect an elevated rate of anagenesis as the lineage adapts to the conditions of its new environment. Highly elevated periodic selection rates like those we observed in some subclades of Vibrio and Aquabacterium therefore might be an indication of their recent expansion into a novel ecological niche. This is consistent with a punctuated equilibrium mode of evolution, in which evolutionary change occurs in rare but rapid bursts following major changes in a lineage’s environment (70). This hypothesis could be tested using comparative genomic and ecological analyses between sister clades with high and low periodic selection rates, for example, to look for genes that are undergoing positive selection.
Capable of processing only hundreds of sequence at a time, the original version of ES would be unable to practically analyse massive next generation sequencing data sets. We have demonstrated here that QuickES, by using a divide and conquer approach coupled with parameter approximation, can demarcate ecotypes in datasets with tens of thousands of sequences. QuickES generates only very rough estimates of the periodic selection and ecotype formation rates, and we do not recommend QuickES be used to draw conclusions of this type. In addition to the QuickES package introduced here, a new version of the ES algorithm is under development that should allow the full ES algorithm to directly analyse many thousands of sequences at once (Frederick M. Cohan, Danny Krizanc, personal communication). AdaptML can already analyse thousands of sequences simultaneously but requires ecological data to estimate ecotypes. ES needs only sequence data to predict ecotypes and therefore is advantageous when little or no ecological data is available. When possible, the best practice is to use both AdaptML and ES together to take advantages of their complementary benefits, with AdaptML demarcating putative ecotypes based on known ecological parameters, and ES then subdividing them based on evolutionary models, as we and others have demonstrated (7,36,37). Because ecotypes incorporate evolutionary and ecological models, they are evolutionarily and ecologically more consistent than OTUs. Given the numerous advantages of ecotypes over OTUs, we advocate for using ecotype as an alternative unit of microbial diversity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–7 and Supplementary Methods.
FUNDING
Gordon and Betty Moore Foundation Grant #1660 and by the University of Virginia. Funding for open access charge: University of Virginia start-up funds for the Wu lab.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Frederick M. Cohan for discussions and feedback regarding QuickES.
REFERENCES
- 1.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc. Natl Acad. Sci. USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the sargasso sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 3.Giovannoni SJ, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437:343–348. doi: 10.1038/nature04158. [DOI] [PubMed] [Google Scholar]
- 4.Staley JT. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- 6.Staley JT. Universal species concept: pipe dream or a step toward unifying biology? J. Ind. Microbiol. Biotechnol. 2009;36:1331–1336. doi: 10.1007/s10295-009-0642-8. [DOI] [PubMed] [Google Scholar]
- 7.Connor N, Sikorski J, Rooney AP, Kopac S, Koeppel AF, Burger A, Cole SG, Perry EB, Krizanc D, Field NC, et al. Ecology of speciation in the genus bacillus. Appl. Environ. Microbiol. 2010;76:1349–1358. doi: 10.1128/AEM.01988-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohan FM, Perry EB. A systematics for discovering the fundamental units of bacterial diversity. Curr. Biol. 2007;17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994;44:846–849. [Google Scholar]
- 10.Schloss PD, Handelsman J. Toward a census of bacteria in soil. PLoS Comput. Biol. 2006;2:e92. doi: 10.1371/journal.pcbi.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward DM, Bateson MM, Ferris MJ, Kuhl M, Wieland A, Koeppel A, Cohan FM. Cyanobacterial ecotypes in the microbial mat community of mushroom spring (yellowstone national park, wyoming) as species-like units linking microbial community composition, structure and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1997–2008. doi: 10.1098/rstb.2006.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goris J, Konstantinidis K, Klappenbach J, Coenye T, Vandamme P, Tiedje J. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 13.Pedros-Alio C. Marine microbial diversity: can it be determined? Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today. 2006;33:152–155. [Google Scholar]
- 15.Schloss PD, Wescott S. Assessing and improving methods used in operational taxonomic unit-based for 16s rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc. Natl Acad. Sci. USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell JR, Monaghan MT, Öpik M, Rillig MC. Evolutionary criteria outperform operational approaches in producing ecologically relevant fungal species inventories. Mol. Ecol. 2011;20:655–666. doi: 10.1111/j.1365-294X.2010.04964.x. [DOI] [PubMed] [Google Scholar]
- 20.Barraclough TG, Hughes M, Ashford-Hodges N, Fujisawa T. Inferring evolutionarily significant units of bacterial diversity from broad environmental surveys of single-locus data. Biol. Lett. 2009;5:425–428. doi: 10.1098/rsbl.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell JR. Accounting for uncertainty in species delineation during the analysis of environmental DNA sequence data. Methods Ecol. Evol. 2012;3:1–11. [Google Scholar]
- 22.Eisen JA. Phylogenomics: Improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 23.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 24.Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 2004;70:5057–5065. doi: 10.1128/AEM.70.9.5057-5065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- 26.Danon M, Franke-Whittle IH, Insam H, Chen Y, Hadar Y. Molecular analysis of bacterial community succession during prolonged compost curing. FEMS Microbiol. Ecol. 2008;65:133–144. doi: 10.1111/j.1574-6941.2008.00506.x. [DOI] [PubMed] [Google Scholar]
- 27.Fulthorpe RR, Roesch LF, Riva A, Triplett EW. Distantly sampled soils carry few species in common. ISME J. 2008;2:901–910. doi: 10.1038/ismej.2008.55. [DOI] [PubMed] [Google Scholar]
- 28.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009;3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philippot L, Bru D, Saby NPA, Čuhel J, Arrouays D, Šimek M, Hallin S. Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ. Microbiol. 2009;11:3096–3104. doi: 10.1111/j.1462-2920.2009.02014.x. [DOI] [PubMed] [Google Scholar]
- 30.Pointing SB, Chan Y, Lacap DC, Lau MC, Jurgens JA, Farrell RL. Highly specialized microbial diversity in hyper-arid polar desert. Proc. Natl Acad. Sci. USA. 2009;106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koeppel AF, Wu M. Lineage-dependent ecological coherence in bacteria. FEMS Microbiol. Ecol. 2012;81:574–582. doi: 10.1111/j.1574-6941.2012.01387.x. [DOI] [PubMed] [Google Scholar]
- 32.Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. [DOI] [PubMed] [Google Scholar]
- 33.Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, Ward DM, Rooney AP, Brambilla E, Connor N, Ratcliff RM, et al. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc. Natl Acad. Sci. USA. 2008;105:2504–2509. doi: 10.1073/pnas.0712205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 35.Cohan FM, Koeppel AF. The origins of ecological diversity in prokaryotes. Curr. Biol. 2008;18:R1024–R1034. doi: 10.1016/j.cub.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Becraft ED, Cohan FM, Kuhl M, Jensen SI, Ward DM. Fine-scale distribution patterns of Synechococcus ecological diversity in the microbial mat of mushroom spring, yellowstone national park. Appl. Environ. Microbiol. 2011;77:7689–7697. doi: 10.1128/AEM.05927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melendrez MC, Lange RK, Cohan FM, Ward DM. Influence of molecular resolution on sequence-based discovery of ecological diversity among synechococcus populations in an alkaline siliceous hot spring microbial mat. Appl. Environ. Microbiol. 2010;77:1359–1367. doi: 10.1128/AEM.02032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohan FM, Koeppel A, Krizanc D. Sequence-based discovery of ecological diversity within Legionella. In: Cianciotto N, Edelstein BS, Fields DF, Geary TG, editors. Legionella: State of the Art 30 Years After Recognition. Washington, DC, USA: ASM Press; 2006. pp. 367–376. [Google Scholar]
- 39.Oakley BB, Carbonero F, van der Gast CJ, Hawkins RJ, Purdy KJ. Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME J. 2010;4:497. doi: 10.1038/ismej.2009.146. [DOI] [PubMed] [Google Scholar]
- 40.Koch AL. The pertinence of the periodic selection phenomenon to prokaryote evolution. Genetics. 1974;77:127. doi: 10.1093/genetics/77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin B. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics. 1981;99:1. doi: 10.1093/genetics/99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohan FM. What are bacterial species? Annu. Rev. Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 43.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price MN. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 49.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood–based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 50.Howe K, Bateman A, Durbin R. QuickTree: building huge neighbor-joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- 51.Sun Y, Cai Y, Huse SM, Knight R, Farmerie WG, Wang X, Mai V. A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Brief. Bioinform. 2011;13:107–121. doi: 10.1093/bib/bbr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klepac-Ceraj V, Ceraj I, Polz MF. CLUSTERER: extendable java application for sequence grouping and cluster analyses. Online J. Bioinf. 2006;7:15–21. [Google Scholar]
- 54.Dray S, Dufour A. The ade4 package: implementing the duality diagram for ecologists. Mol. Biol. Evol. 2007;22:1–22. [Google Scholar]
- 55.Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- 56.Brown M, Fuhrman J. Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat. Microb. Ecol. 2005;41:15–23. [Google Scholar]
- 57.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 59.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, et al. Opinion: Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 60.Ward DM, Cohan FM, Bhaya D, Heidelberg JF, Kuhl M, Grossman A. Genomics, environmental genomics and the issue of microbial species. Heredity. 2008;100:207–219. doi: 10.1038/sj.hdy.6801011. [DOI] [PubMed] [Google Scholar]
- 61.Cavalier-Smith T. Deep phylogeny, ancestral groups and the four ages of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:111–132. doi: 10.1098/rstb.2009.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horandl E, Stuessy TF. Paraphyletic groups as natural units of biological classification. Taxon. 2010;59:1641–1653. [Google Scholar]
- 63.de Queiroz K. Different species problems and their resolution. Bioessays. 2005;27:1263–1269. doi: 10.1002/bies.20325. [DOI] [PubMed] [Google Scholar]
- 64.de Queiroz K. Species concepts and species delimitation. Syst. Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 65.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl Acad. Sci. USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 69.Cohan FM. Are species cohesive?—a view from bacteriology. In: Walk ST, Feng PCH, editors. Population Genetics of Bacteria: A Tribute to Thomas S. Whitman. Washington, DC, USA: ASM Press; 2011. pp. 43–65. [Google Scholar]
- 70.Gould SJ, Eldredge N. Puctuated equilibrium: the tempo and mode of evolution reconsidered. Paleobiology. 1977;3:115–151. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.