Abstract
Using a luciferase reporter assay, we previously demonstrated that a Z-DNA-forming sequence of alternating thymine–guanine repeats in the human heme oxygenase-1 gene (HO-1) promoter is involved in nuclear factor erythroid-derived 2 (NF-E2)–related factor 2 (Nrf2)-mediated HO-1 promoter activation. However, the actual Z-DNA formation in this native genomic locus has not been experimentally demonstrated. To detect Z-DNA formation in vivo, we generated a construct containing the Z-DNA-binding domain of human adenosine deaminase acting on double-stranded RNA 1 fused with enhanced green fluorescence protein, designated as the Z-probe. A chromatin immunoprecipitation assay using an anti-GFP antibody showed that the Z-probe detects the well-characterized Z-DNA formation in the CSF1 promoter. Using this detection system, we demonstrated that the glutathione-depleting agent, diethyl maleate, induced Nrf2-dependent Z-DNA formation in the HO-1 promoter, but not in the thioredoxin reductase 1 gene promoter. Moreover, a time course analysis revealed that Z-DNA formation precedes HO-1 transcriptional activation. Concurrent with Z-DNA formation, nucleosome occupancy was reduced, and the recruitment of RNA polymerase II was enhanced in the HO-1 promoter region, suggesting that Z-DNA formation enhances HO-1 gene transcription. Furthermore, Nrf2-induced BRG1 recruitment to the HO-1 promoter temporarily occurred simultaneously with Z-DNA formation. Thus, these results implicate Nrf2-dependent Z-DNA formation in HO-1 transcriptional activation and suggest the involvement of BRG1 in Z-DNA formation.
INTRODUCTION
Watson and Crick proposed a structural model of DNA as a right-handed double helix (1), called B-DNA. However, the first single-crystal X-ray structure of DNA, i.e. hexamer dCGCGCG, showed a left-handed double helix (2). The phosphate–sugar backbone of left-handed DNA exhibits a zigzag arrangement, and is thereby called Z-DNA (2). DNA regions containing alternating purine–pyrimidine repeat sequences typically adopt a Z-DNA structure and are called Z-DNA-forming sequences. Because Z-DNA-forming sequences are often identified in gene promoter regions (3), it has been proposed that Z-DNA plays a role in nuclear processes, such as transcription regulation and nucleosome positioning (4,5). Z-DNA formation has been detected under high salt conditions in vitro (6), but in vivo, Z-DNA formation and its physiological roles in nuclear processes are largely unknown.
Heme oxygenase catalyzes the conversion of heme into biliverdin, iron and carbon monoxide (7). Two isoforms of heme oxygenase have been characterized: an inducible enzyme (heme oxygenase-1, HO-1) and a constitutive isoform (heme oxygenase-2, HO-2) (7). HO-1 gene expression is sensitive to the substrate heme (8) and various environmental stressors, such as cadmium (9), lipopolysaccharide (10), nitric oxide (11) and oxidative stress (12). The 5′-flanking region of the human HO-1 gene contains several binding sites for transcription factors, such as the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) responsive element, cadmium responsive element and antioxidant responsive element (ARE). In the human HO-1 gene locus, the regions at −4 and −9 kb upstream of the transcription start site, called enhancer 1 (E1) and enhancer 2 (E2), respectively (13,14), contain multiple ARE sites (15). The AREs in these enhancers overlap with TPA responsive elements and respond to multiple environmental stressors. Therefore, AREs in the HO-1 gene enhancers are also referred to as stress responsive elements (16). On oxidative stress, the nuclear factor erythroid-derived 2(NF-E2)–related factor 2 (Nrf2)–small Mafs heterodimer binds to E1 and E2 stress responsive elements and up-regulates HO-1 gene expression (17).
Nrf2 belongs to the CNC (Cap‘n’collar) transcription factor family, which is characterized by a highly conserved basic region leucine-zipper structure (18). On exposure to oxidative stress or electrophiles, Nrf2 translocates to the nucleus, where it binds to AREs via heterodimerization with small Maf proteins and activates the expression of >100 target genes, including HO-1 and the thioredoxin reductase 1 gene (TXNRD1) (19).
Previously, we showed that BRG1 (Brahma-related gene 1), a core ATPase subunit of chromatin remodeling complexes, interacts with Nrf2 and selectively induces Nrf2-dependent HO-1 gene expression (20,21). In addition, we demonstrated that the Nrf2-BRG1 complex enhances human HO-1 gene promoter activity, which partially depends on the presence of thymine–guanine (TG) repeats in the promoter sequence (20). However, it remains unknown whether the native human HO-1 gene promoter forms Z-DNA.
In this study, we developed a strategy for the detection of Z-DNA formation in cultured cells. Using this method, we demonstrated that the human HO-1 gene promoter adopts a Z-DNA structure in response to the Nrf2 activator diethyl maleate (DEM). Furthermore, the dynamics of Z-DNA formation during HO-1 gene transcriptional activation were examined.
MATERIALS AND METHODS
Cell culture
Human adrenal carcinoma SW13, 293 T-REx-3xFLAG-human Nrf2/Z-probe* cells were maintained in Dulbecco’s modified Eagle medium (Sigma-Aldrich) containing 10% fetal bovine serum and 100 U/ml penicillin–streptomycin (Gibco). Human cervix carcinoma HeLa cells were cultured in RPMI 1640 medium (Sigma-Aldrich) containing 10% fetal bovine serum and 100 U/ml penicillin–streptomycin. The cells were cultured at 37°C with 5% CO2 and saturated humidity.
Construction of plasmids
To construct a mammalian Z-probe expression vector, a cDNA fragment encoding human adenosine deaminase acting on double-stranded RNA 1 (ADAR1) Zα was PCR amplified using SW480 cDNA as a template with the following primers: 5′-GGG GTA CCG CCA CCA TGG TGC TGA GTA TCT ACC AAG ATC-3′ and 5′-CCG CTC GAG TCC GCT GTG CTG GTT CCA AGC-3′. A double-stranded DNA fragment encoding the SV40 nuclear localization signal (NLS) was generated after annealing the following sense and antisense strands, respectively: 5′-CCG CTC GAG CCT CCT AAG AAG AAG CGC AAA GTC GAG GGA TCC CG-3′ and 5′-CGG GAT CCC TCG ACT TTG CGC TTC TTC TTA GGA GGC TCG AGC GG-3′. The enhanced green fluorescence protein (EGFP) cDNA fragment was PCR amplified using the pEGFP-N1 vector (Clontech) as a template with the following primers: 5′-GAA GAT CTA TGG TGA GCA AGG GCG AGG A-3′ and 5′-GGG GGC CCT TAC TTG TAC AGC TCG TCC A-3′. These three DNA fragments were digested with following restriction enzymes: Zα: KpnI–XhoI; SV40 NLS: XhoI–BamHI; and the EGFP fragment: BglII–ApaI. The digestion products were ligated and cloned into pcDNA3 to generate pcDNA-Zα-NLS-EGFP. Subsequently, to insert (Gly4–Ser)3 linker amino acid residues between Zα and NLS in the pcDNA-Z-probe, the following annealed double-stranded oligonucleotides were ligated using the In-Fusion Cloning Kit (Clontech): 5′-GGT GGA GGA GGT TCT GGA GGC GGT GGA AGT GGT GGC GGA GGT AGC-3′. To introduce N173A and Y177A mutations (22) in the Z-probe (Zmut-probe), PCR-based site-directed mutagenesis was performed using the following primers: 5′-TGG GAC TCC GAA GAA AGA AAT CGC TCG AGT TTT AGC CTC CCT GGC AAA GAA GG-3′ and 5′-CCT TCT TTG CCA GGG AGG CTA AAA CTC GAG CGA TTT CTT TCT TCG GAG TCC CA-3′. To construct the recombinant Z-probe or Zmut-probe expression vectors, the Zα or Zmut-probe DNA fragments were PCR amplified using pcDNA-Z-probe or pcDNA-Zmut-probe templates, respectively, with the following primers: 5′-CGG GAT CCA AAT GGT GCT GAG TAT CT-3′ and 5′-GGC GTC GAC TTA CTT GTA CAG CTC GTC CA-3′. These fragments were digested with BamHI and SalI and cloned into the BamHI–SalI site of pGEX-4T-3 to generate pGEX-Z-probe or pGEX- Zmut-probe, respectively. The sequences of all constructs were confirmed through DNA sequencing. The detailed diagram of Z-probes, including the residue numbers and boundaries between the Zα domain of human ADAR and other domains, is shown in Supplementary Figure S1.
Gel shift assay
A 5′-Cy5-labeled annealed oligonucleotide of 18 TG repeats (TG18) was used as a probe. Non-labeled TG18 repeat, GC18 repeat and TA18 repeat oligonucleotides were used as competitors. Recombinant GST-Z-probe, GST-Zmut-probe and GST-NLS-EGFP proteins were purified using glutathione sepharose 4B (GE Healthcare). The purified proteins were analyzed using SDS–PAGE and stored at −30°C until further use. The oligonucleotide probe was incubated in the binding buffer containing 20 mM Tris–HCl (pH 8.5), 10 mM MgCl2, 1 mM dithiothreitol (DTT), 225 mM KCl and 1 mM cobalt hexamine chloride at room temperature for 30 min, and subsequently, the recombinant proteins were mixed with probe solutions at room temperature for 30 min. The samples were subjected to 4.5% acrylamide/0.5× Tris-Borate EDTA (TBE) gel and electrophoresed at 65 volts for 150 min at 4°C. The signals were detected using an FLA9000 Fluorescence imager (GE Healthcare).
Fluorescence microscopic observation
The mammalian Z-probe, Zmut-probe or NLS-EGFP expression vectors were transiently transfected into HeLa cells using FuGENE HD reagent (Roche). After transfection, the cells were fixed with 3.7% formaldehyde at 4°C for 15 min, and the nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). The Z-probes and nuclei were visualized using the AF6500 Fluorescence imaging system (Leica).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as previously described, with few modifications (23). Briefly, the cells were fixed with 1% formaldehyde for 10 min at room temperature. Subsequently, glycine was added to a final concentration of 0.125 M, and the cells were incubated for 5 min at room temperature. The fixed cells were sonicated and centrifuged, and the supernatant fraction was used as the cell lysate in the ChIP analysis. Anti-Nrf2 (Santa Cruz, H300), anti-BRG1 (Abcam, ab4081), anti-FLAG M2 (SIGMA) and anti-Pol II (Millipore, clone CTD4H8) antibodies were used. Normal rabbit IgG (Millipore) was used as a negative control. These antibodies were immobilized to Dynabeads Protein G (Life Technologies) and used for the immunoprecipitation reactions. The precipitated DNA fragments were detected using real-time PCR with the following primers: HO-1 E2 enhancer region (HO-1-E2): 5′-CCC TGC TGA GTA ATC CTT TCC-3′ and 5′-GGC GGT GAC TTA GCG AAA AT-3′; HO-1 gene promoter region (HO-1-Pro): 5′-GCC AGA AAG TGG GCA TCA G-3′ and 5′-CTG AGG ACG CTC GAG GGA G-3′; HO-1 Exon 3 region (HO-1-Ex3): 5′-CAC CAA GTT CAA GCA GCT CTA C-3′ and 5′-CTT CTA TCA CCC TCT GCC TGA C-3′; TXNRD1 ARE region (TXNRD1-ARE): 5′-TGC ACG AGG AGT GGA TTT CTG CTT-3′ and 5′-GCT GCA AAT GCC GGA GTG AAG AAA-3′; and the γ-globin gene (HGB2) promoter region (HGB2-Pro): 5′-TGG CTA GGG ATG AAG AAT AAA AGG-3′ and 5′-ATT GAT AAC CTC AGA CGT TCC AGA AG-3′.
Z-DNA detection
The expression plasmids were transiently transfected into SW13 and HeLa cells using FuGENE HD (Roche). The siRNA against human Nrf2 was synthesized at QIAGEN using the following sequence: 5′-AAG AGU AUG AGC UGG AAA AACdTdT-3′ (24). At 24 h after plasmid transfection, siRNA (final 20 nM) was transfected using Lipofectamine RNAi Max reagent according to the manufacturer’s protocol (Life Technologies). At 24 h after siRNA transfection, the cells were exposed to 100 µM of DEM. The 293T-REx-3xFLAG-human Nrf2/Z-probe* cells were treated with 1 µg/ml tetracycline (Tet). The anti-GFP antibody (Clontech) was immobilized to Dynabeads Protein G and used in the immunoprecipitation experiments. Precipitated DNA fragments were detected using real-time PCR with the following primers: CSF1 TG repeat region (CSF1-TG): 5′-CGC AGA AGA CAG AGG GTG AC-3′ and 5′-GGC ATG TGG TTT ATG GGA AA-3′; −3600 upstream from the CSF1 transcription start site (CSF1-3600): 5′-ACA AGG GCA TTC AGT CCA AA-3′ and 5′-AGA CAG CGT GAA GGG TGG TA-3′; CSF1 Exon 4 region (CSF1-Ex4): 5′-TCA GAG ATA ACA CCC CCA ATG-3′ and 5′-CTT CAT AAT CCT TGG TGA AGC A-3′; and HO-1 TG repeat region (HO-1-TG): 5′-CTC TGG AAG GAG CAA AAT CAC A-3′ and 5′-GGC CAT AGG ACT TTT AGA GAA AAC A-3.
Nucleosome occupancy detection
The protocol for nucleosome occupancy detection was modified from a previously described method (25). Briefly, formaldehyde-fixed cells were permeabilized with 5 mM PIPES–KOH (pH 8.0), 85 mM KCl and 1 mM DTT containing 0.5% NP40, and the chromatin was digested with 2000 U of micrococcal nuclease (New England BioLabs) for 10 min at 37°C. The digestion was terminated after the addition of 10 mM EDTA, and the cells were lysed and centrifuged. The supernatant fraction was used in the ChIP assay. Antibodies against histone H3 (Millipore, clone A3S) or normal mouse IgG (Millipore) were immobilized to Dynabeads Protein G and used for immunoprecipitation experiments. The HO-1 TG repeat region and TXNRD1 ARE region in the precipitated DNA fragments were detected using real-time PCR with the primers described above.
Gene expression analysis
The cDNAs were synthesized using PrimeScript® RTase (TaKaRa Bio) and total RNA as a template. The real-time PCR analyses were performed using SYBR® Premix Ex Taq™ II (Perfect Real Time; Takara Bio) and a CFX Real-Time PCR Detection System (BioRad). The cyclophilin A gene was used as an internal control with the following primers: 5′-ATG CTG GAC CCA ACA CAA AT-3′ and 5′-TCT TTC ACT TTG CCA AAC ACC-3′. The gene expression was measured using real-time PCR with the following primers: CSF1: 5′-GTT TGT AGA CCA GGA ACA GTT GAA-3′ and 5′-TGT ACC AGG AGA AAT GCC TTC-3′; HO-1: 5′-CCA GCA ACA AAG TGC AAG ATT C-3′ and 5′-TCA CAT GGC ATA AAG CCC TAC AG-3′; and TXNRD1: 5′-ACA CAA AGC TTC AGC ATG TCA-3′ and 5′-CAA TTC CGA GAG CGT TCC-3′.
Statistical analysis
The results were expressed as the means ± standard error of the mean (SEM), and the statistical significance was determined using Dunnett’s test for multiple-parameter comparisons or two-tailed unpaired Student’s t-test for two-parameter comparisons. P < 0.05 was considered statistically significant.
RESULTS
The recombinant Z-probe binds to a Z-DNA-forming sequence in vitro
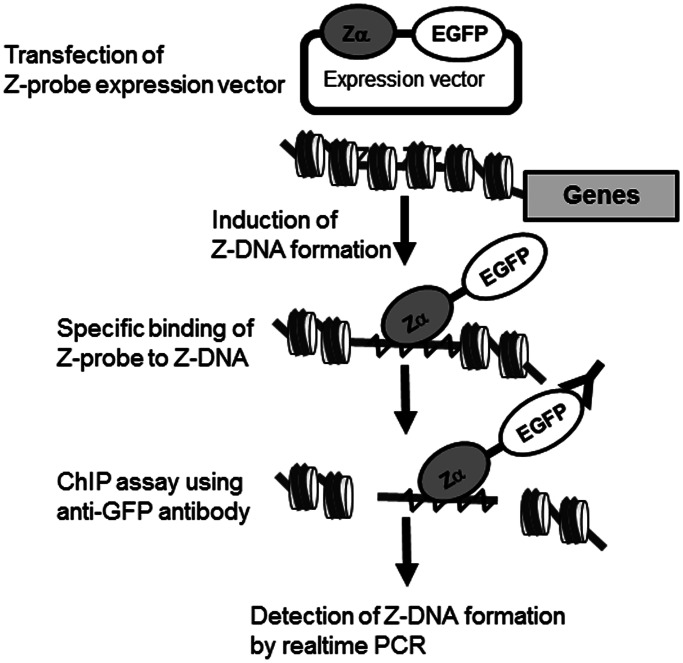
The Z-DNA-forming sequences, such as d(CG)n and d(TG)n, easily form Z-DNA structures under high salt conditions in vitro, but the in vivo formation of Z-DNA structures in the human HO-1 TG repeat region has not been demonstrated. Thus, we developed a method for the detection of Z-DNA structures on genomic DNA in cultured cells. Figure 1 illustrates our strategy for the detection of Z-DNA formation in genomic DNA. We generated a fusion protein consisting of the Zα domain of human ADAR1, a well-characterized Z-DNA-binding domain (26), (Gly4–Ser)3 flexible linker residues (Ln), an NLS and EGFP, which was referred to as the Z-probe (Figure 1 and Supplementary Figure S1). A similar probe was previously used to detect the distribution of Z-DNA in the human genome (27), but in the present study, the Z-probe was fused with EGFP to visualize the localization of the probe in the living cultured cells and serve as an antigen for the ChIP assay using an anti-GFP antibody. After ChIP, the Z-DNA formation was quantitatively measured using real-time PCR. As a negative control, we generated a Zmut-probe carrying N173A and Y177A mutations (numbers corresponds to human ADAR1 residues) in the Zα domain of the Z-probe, which disrupt Z-DNA binding (22) (Supplementary Figure S1 and Figure 2A and B).
Figure 1.
Strategy for the detection of genomic Z-DNA formation. To detect Z-DNA formation, a mammalian expression vector was constructed containing the ADAR1 Z-DNA-binding domain Zα, a (Gly4–Ser)3 flexible linker, an NLS and EGFP, called the Z-probe. The cultured cells were transfected with the Z-probe expression vector and subjected to an appropriate stress. Subsequently, Z-DNA formation was quantitatively measured using a ChIP assay with an anti-GFP–specific antibody, followed by real-time PCR using specific primer sets.
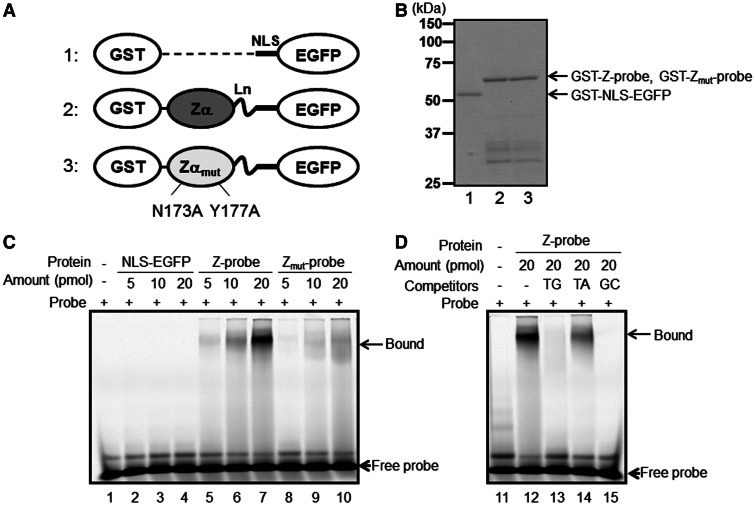
Figure 2.
The recombinant Z-probe binds to a Z-DNA-forming sequence in vitro. (A) A schematic figure of the recombinant proteins. 1: NLS-EGFP; 2: Z-probe; 3: Zmut-probe. The proteins were expressed as GST fusion proteins and purified using glutathione-conjugated beads. Zα: human ADAR1 Zα domain; Zαmut: the Zα domain possessing N173A and Y177A mutations corresponding to human ADAR1 residue numbers; Ln: (Gly4–Ser)3 linker residues. Detailed information is provided as Supplemental Figure S1. (B) SDS–PAGE analysis of the purified recombinant proteins. 1: GST-NLS-EGFP; 2: GST-Z-probe; 3: GST-Zmut-probe. A gel shift assay was performed using Cy5-labeled d(TG/AC)18 double-stranded oligonucleotides as probes. (C) The purified GST-NLS-EGFP vector (lanes 2–4), GST-Z-probe (lanes 5–7) and GST-Zmut-probe (lanes 8–10) were incubated with a Cy5-labeled oligonucleotide probe. Protein–DNA complexes were separated using native PAGE, and the fluorescence signals were detected using Typhoon FLA9000. (D) The GST-Z-probe (lanes 12–15) was incubated with excess non-labeled competitors (lanes 13–15) and mixed with a Cy5-labeled probe. Subsequently, the protein–DNA complexes were separated using native PAGE, and the fluorescence signals were detected. Competitors: TG: d(TG/AC)18 (lane 13); TA: d(TA/AT)18 (lane 14); GC: d(GC/CG)18 (lane 15).
To validate whether the Z-probe specifically binds to Z-DNA, we generated a GST fusion protein of the Z-probe and performed a gel shift assay using double-stranded TG18 repeats as a probe (Figure 2). Consistent with the previous result (22), the recombinant Z-probe, but not the Zmut-probe, strongly and specifically bound to the TG18 repeats (Figure 2C). The binding of the Z-probe to TG18 repeats was inhibited after the addition of excess TG18 or GC18 repeat oligonucleotides, which form Z-DNA. In contrast, the addition of TA18 repeats did not inhibit Z-probe binding to Z-DNA as efficiently as TG18 or GC18 repeats (Figure 2D). These results indicate that the recombinant Z-probe specifically binds to Z-DNA in vitro.
The Z-probe specifically binds to the TG repeats of the CSF1 promoter in cultured cells
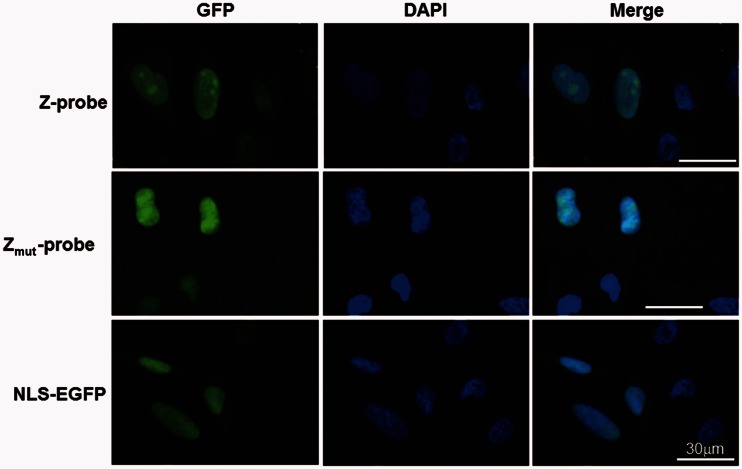
To detect Z-DNA formation in cultured cells, we constructed a mammalian expression plasmid for the Z-probe and investigated the localization of the Z-probe in HeLa cells. The fluorescent microscopic observation revealed that the Z-probe and Zmut-probe showed similar localization patterns in the nuclei (Figure 3).
Figure 3.
Z-probes are localized in the nucleus. HeLa cells were transfected with the expression vectors for the Z-probe, Zmut-probe or NLS-EGFP. The cells were observed using a fluorescence microscope at 24 h after transfection. The Z-probe, Zmut-probe and NLS-EGFP proteins are shown in green. The nuclei were stained with DAPI (blue).
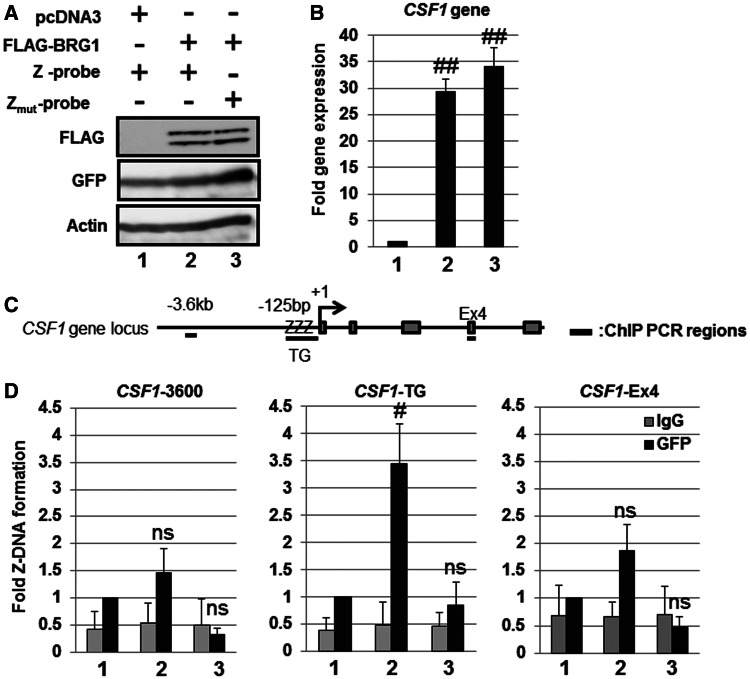
To confirm whether the Z-probe binds to Z-DNA in cultured cells, we performed a ChIP assay using an anti-GFP antibody. It has been reported that the human colony stimulating factor 1 gene (CSF1) promoter contains a TG repeat sequence and adopts a Z-DNA structure in a BRG1-dependent manner in SW13 cells (28,29). Thus, we examined the CSF1 promoter region as a positive control for Z-DNA formation. We confirmed the overexpression of FLAG-BRG1 through immunoblot analysis using the anti-FLAG antibody (Figure 4A) and detected the BRG1-dependent induction of CSF1 gene expression in SW13 cells (Figure 4B). BRG1 significantly induced the binding of the Z-probe, but not the Zmut-probe, to the CSF1-TG repeat sequence (Figure 4C and D). In addition, BRG1 did not enhance the binding of the Z-probe to either the CSF1-3600 or CSF1-Ex4 regions (Figure 4D). These results indicate that the Z-probe specifically detects Z-DNA formation in the CSF1 promoter in cultured cells.
Figure 4.
Detection of BRG1-dependent Z-DNA formation in the human CSF1 promoter using the Z-probe. (A) SW13 cells were transfected with the Z-probe or Zmut-probe together with the BRG1 expression vector as indicated in the figure. Whole-cell lysates were separated using SDS–PAGE, and the protein expression was analyzed through immunoblotting using specific antibodies as indicated in the figure. (B) SW13 cells were transfected with BRG1 together with the Z-probe (lanes 1 and 2) or Zmut-probe (lane 3) expression vectors, and the CSF1 gene expression was analyzed using real-time PCR. The value of lane 1 was arbitrarily set as 1, and the relative expression levels were expressed as the means ± SEM of four independent assays. (C) A schematic figure of the human CSF1 gene locus and regions detected using ChIP analysis. +1: transcription start site; ZZZ: CSF1-TG repeat region; Ex4: CSF1 Exon 4 region. (D) The SW13 cells were transfected with the Z-probe or Zmut-probe together with BRG1 expression vectors, and subsequently, a ChIP assay was performed using an anti-GFP antibody (black bars). Normal rabbit IgG was used as a negative control (gray bars). Fold Z-DNA formation was measured using real-time PCR with specific primer sets for the upstream region of the CSF1 promoter (CSF1-3600), the CSF1-TG or CSF1-Ex4 regions. The anti-GFP ChIP value in lane 1 is set as 1, and the relative binding values are expressed as the means ± SEM of four independent assays. #P < 0.05 and ##P < 0.01 compared with the value of lane 1 (Dunnett’s test for multiple parameter comparisons); ns: no significant difference. The lane numbers correspond to the cells transfected in (A).
Human HO-1 gene promoter forms Z-DNA in response to Nrf2-activating agent DEM
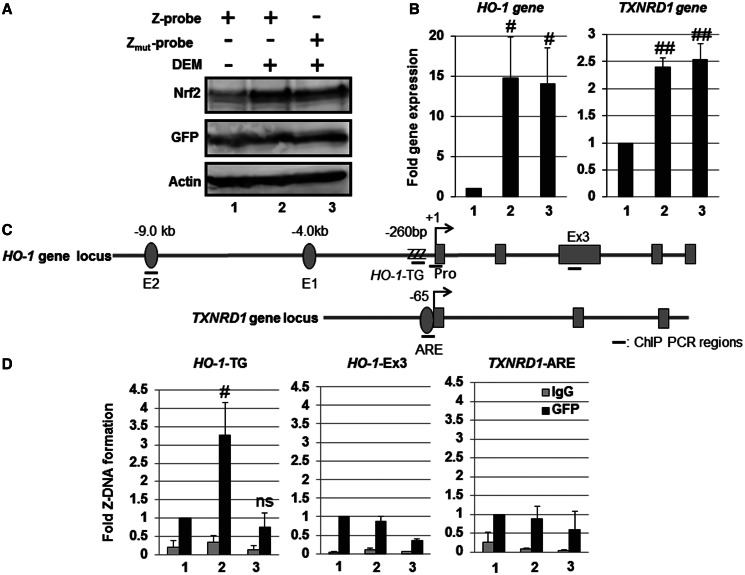
To investigate Z-DNA formation in the human HO-1-TG repeat region, we transiently transfected Z-probe expression plasmids into HeLa cells, and the cells were exposed to DEM. As expected, DEM induced Nrf2 protein expression and HO-1 and thioredoxin reductase 1 gene (TXNRD1) expression in HeLa cells (Figure 5A and B). The ChIP analysis demonstrated that DEM treatment significantly induced Z-probe binding to the human HO-1-TG repeats, but not to either of the HO-1-Ex3 or TXNRD1 ARE regions (Figure 5C and D). In contrast, Zmut-probe binding to the HO-1-TG region was not induced (Figure 5D). These results indicate that Z-DNA formation in the human HO-1 gene promoter is associated with Nrf2 activation in HeLa cells.
Figure 5.
Human HO-1 gene promoter adopts a Z-DNA form in response to Nrf2-activating agent DEM. HeLa cells were transfected with the Z-probe (lanes 1 and 2) or Zmut-probe (lane 3) expression vectors. At 24 h after transfection, the cells were untreated (lane 1) or treated with 100 µM DEM for 3 h (lanes 2 and 3). (A) Whole-cell lysates were separated using SDS–PAGE, and the protein expression was analyzed through immunoblotting using specific antibodies, as indicated in the figure. (B) The gene expression was analyzed through real-time PCR using primers specific for the HO-1 and TXNRD1 genes. The value of lane 1 is set as 1, and the relative expression values are expressed as the means ± SEM of five independent assays. (C) A schematic figure of the human HO-1 and TXNRD1 gene loci and regions as detected through ChIP analysis. E2: E2 enhancer; E1: E1 enhancer; +1: transcription start site; ZZZ: HO-1-TG repeat region; Pro: HO-1 gene promoter region; Ex3: HO-1 Exon 3 region; TXNRD1-ARE: TXNRD1 ARE region. (D) HeLa cells were transfected with the Z-probe or Zmut-probe, and subsequently, the cells were exposed to 100 µM DEM for 3 h. After DEM treatment, a ChIP assay was performed using an anti-GFP antibody (black bars). Normal rabbit IgG was used as a negative control (gray bars). The amount of precipitated DNA was measured through real-time PCR using specific primer sets for the HO-1-TG, HO-1-Ex3 and TXNRD1 ARE regions. The anti-GFP ChIP value of lane 1 is set as 1, and the relative bindings are expressed as the means ± SEM of five independent assays. #P < 0.05 and ##P < 0.01 compared with the value of lane 1 (Dunnett’s test for multiple parameter comparisons); ns: no significant difference. The lane numbers correspond with the treatments in (A).
Z-DNA formation in the HO-1-TG region precedes HO-1 gene induction
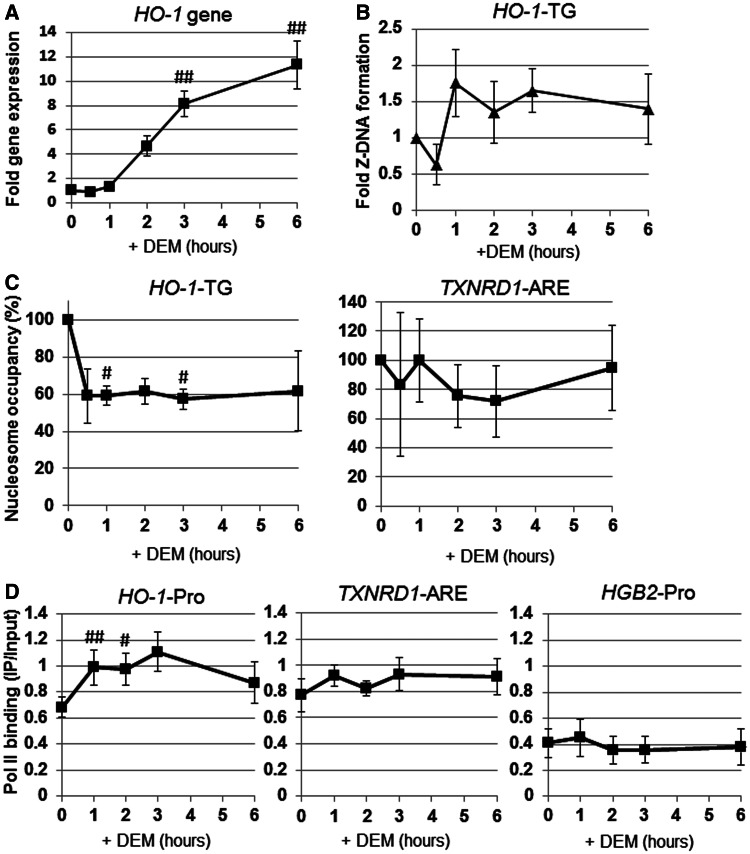
Using a luciferase reporter analysis of the HO-1 gene promoter construct, we previously showed that the Z-DNA-forming TG repeat sequence enhances HO-1 transcription (20). Therefore, we examined the effects of Z-DNA formation on HO-1 gene expression. The apparent induction of HO-1 gene expression occurred at 2 h after DEM treatment and continued to increase over 6 h (Figure 6A). Z-DNA formation was induced as early as 1 h after DEM treatment and remained relatively constant during the time course (Figure 6B). These results indicate that Z-DNA formation in the HO-1-TG region precedes HO-1 gene induction.
Figure 6.
Dynamics of Z-DNA formation, Pol II recruitment and nucleosome occupancy on the HO-1 regulatory region in response to DEM. HeLa cells were transfected with the Z-probe expression vector, and at 24 h after transfection, the cells were exposed to 100 µM DEM for various amounts of time as indicated. (A) HO-1 gene expression was analyzed through real-time PCR using an HO-1-specific primer set. (B–D) The ChIP assays were performed using anti-GFP, anti-Pol II and anti-H3 antibodies. Z-DNA formation was measured through real-time PCR using a specific primer set for the HO-1 TG repeat region (HO-1-TG; B). Nucleosome occupancy was measured through real-time PCR using specific primer sets for the HO-1 TG repeat (HO-1-TG) and the TXNRD1 ARE (TXNRD1-ARE) regions (C). The value (%) at 0 h is set as 100, and the relative changes are expressed as the means ± SEM of three independent assays. Pol II recruitment was measured through real-time PCR using specific primer sets for the HO-1 gene promoter (HO-1-Pro), TXNRD1 gene in the ARE region (TXNRD1-ARE) and γ-globin promoter region (HGB2-Pro) (D). The values are shown in IP/input, and the relative changes are expressed as the means ± SEM of three independent assays. #P < 0.05 and ##P < 0.01 compared with the value at 0 h (Dunnett’s test for multiple parameter comparisons).
We hypothesized that if the Z-DNA formation participates in transcription activation, nucleosome occupancy on the HO-1-TG region would be reduced. Thus, we performed ChIP assay using anti-histone H3 for the detection of nucleosome occupancy. For this purpose, we used micrococcal nuclease digestion instead of sonication before immunoprecipitation, as previously described (25). As shown in Figure 6C, we demonstrated that nucleosome occupancy in the HO-1-TG region, but not in the TXNRD1-ARE, was rapidly reduced in response to DEM treatment. Moreover, Pol II recruitment was significantly induced after DEM treatment in the HO-1 gene promoter region, but not in the TXNRD1 ARE region (Figure 6D). These results suggest that Z-DNA formation is actively involved in the transcription regulation of the HO-1 gene.
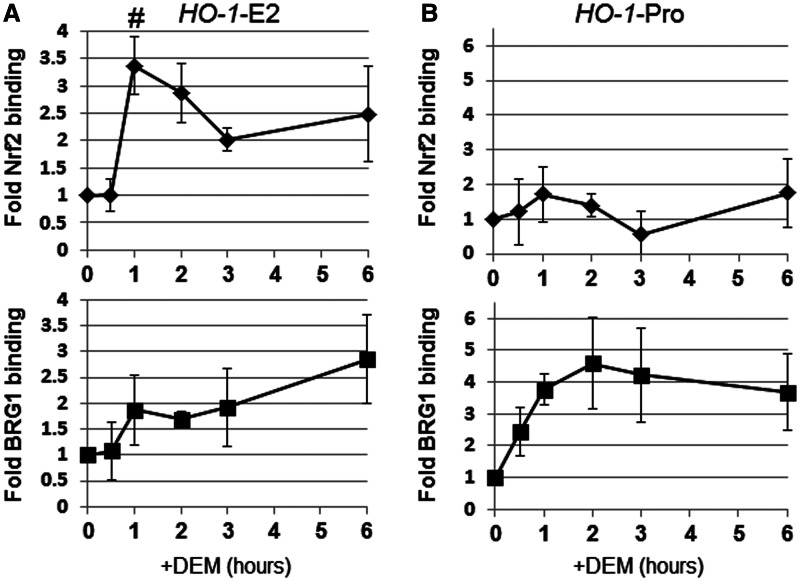
We previously demonstrated that BRG1 interacts with Nrf2, selectively inducing HO-1 gene expression (20,21). Thus, we analyzed the dynamics of Nrf2 and BRG1 recruitment to the HO-1 regulatory region. Nrf2 binding to the HO-1-E2 enhancer region was significantly induced after DEM treatment, with peak Nrf2 binding observed 1 h after DEM administration (Figure 7A). Along with Nrf2 enhancer binding, the BRG1 binding to the HO-1-E2 enhancer was observed at 1 h after DEM treatment and gradually increased until 6 h (Figure 7A). Although we did not detect a significant change in Nrf2 binding to the HO-1 gene promoter, DEM induced BRG1 binding to this region, with peak binding observed after 2 h (Figure 7B). These results are consistent with our previous report that BRG1 is positively involved in Z-DNA formation (20).
Figure 7.
Dynamics of Nrf2 and BRG1 on the HO-1 regulatory region in response to DEM. HeLa cells were transfected with the Z-probe expression vector, and at 24 h after transfection, the cells were treated with 100 µM DEM for various amounts of time, as indicated. ChIP assays were performed using anti-Nrf2 and anti-BRG1 antibodies. Nrf2 and BRG1 binding were measured through real-time PCR using specific primer sets for the HO-1 E2 enhancer (HO-1-E2; A) or HO-1 gene promoter (HO-1-Pro; B) regions. The value at 0 h is set as 1, and the relative changes are expressed as the means ± SEM of four independent assays. #P < 0.05 compared with the value at 0 h (Dunnett’s test for multiple parameter comparisons).
Nrf2 activation is associated with Z-DNA formation in the human HO-1 gene promoter
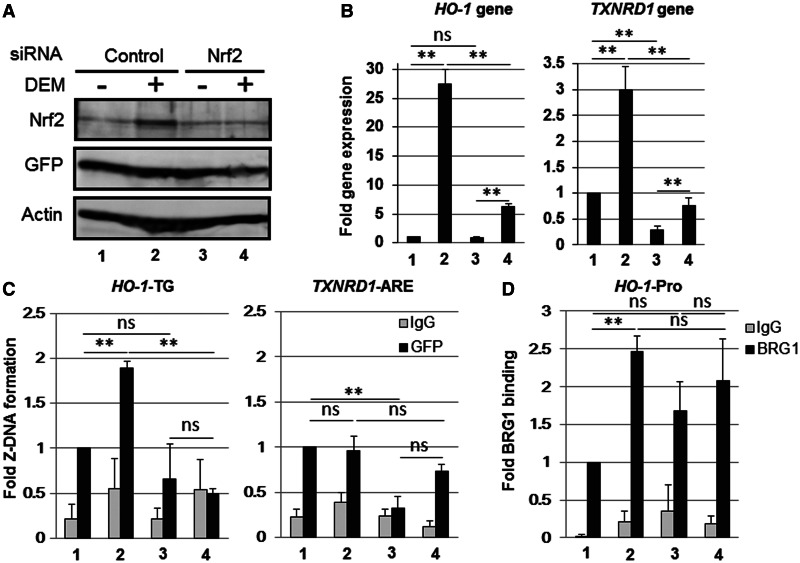
To determine whether Nrf2 is required for DEM-inducible Z-DNA formation in the human HO-1-TG repeat region, we knocked down endogenous Nrf2 using Nrf2-specific siRNA. The effect of Nrf2 siRNA was confirmed through immunoblot analysis using an Nrf2-specific antibody (Figure 8A). The Nrf2 knockdown significantly and effectively reduced the gene expression of both HO-1 and TXNRD1 in response to DEM treatment (Figure 8B). DEM-induced Z-DNA formation in the HO-1-TG region was significantly reduced through Nrf2 knockdown compared with control siRNA transfection (Figure 8C). In the TXNRD1 ARE region, we did not detect any DEM-inducible Z-DNA formation in control siRNA–transfected cells (Figure 8C). BRG1 binding to the HO-1 gene promoter was induced after DEM treatment in control siRNA cells, but not in Nrf2 knockdown cells (Figure 8D).
Figure 8.
Nrf2 knockdown reduces the DEM-induced Z-DNA formation in the human HO-1-TG region. HeLa cells were transfected with the Z-probe expression vector, and at 24 h after transfection, the cells were transfected with control siRNA (Control) or Nrf2-specific siRNA (Nrf2) and treated with 100 µM DEM for 3 h. (A) Whole-cell lysates were separated using SDS–PAGE, and the protein expression was analyzed through immunoblotting using specific antibodies, as indicated in the figure. (B) HO-1 and TXNRD1 gene expression were analyzed through real-time PCR using specific primer sets. The value of lane 1 is arbitrarily set as 1, and the relative expressions are expressed as the means ± SEM of three independent assays. (C and D) A ChIP assay was performed using anti-GFP (C) or anti-BRG1 antibodies (black bars; D). Normal rabbit IgG was used as a negative control (gray bars). The Z-DNA formation was measured through real-time PCR using specific primer sets for the HO-1-TG and TXNRD1 ARE regions (C). BRG1 binding was measured through real-time PCR using a specific primer set for the HO-1 gene promoter region (HO-1-Pro; D). The anti-GFP ChIP (C) or anti-BRG1 ChIP (D) values of lane 1 are set as 1, and the relative bindings are expressed as the means ± SEM of three independent assays. **P < 0.01(two-tailed unpaired Student’s t-test). The lane numbers correspond with the treatments in (A).
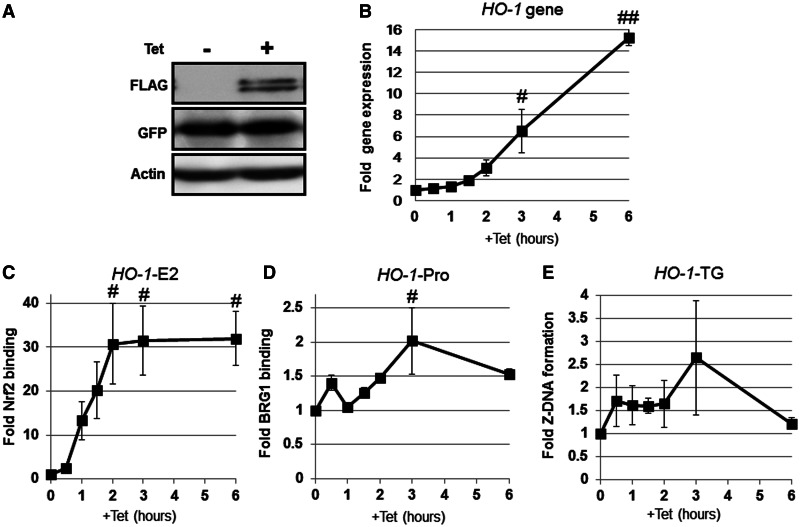
To investigate whether Nrf2 activation is sufficient for the induction of Z-DNA formation in the human HO-1-TG repeat region, we examined Z-DNA formation in the HO-1 gene promoter in 293T-REx-FLAG-human Nrf2/Z-probe* cells, possessing a stably integrated Z-probe* and Tet-inducible Nrf2 cassettes. Note that the Z-probe* protein has no flexible linker between Zα and NLS (Supplementary Figure S1), but similar to the Z-probe, Z-probe* specifically detects Z-DNA formation both in vitro and in vivo (data not shown). Tet administration to the cells induced FLAG-Nrf2 protein expression (Figure 9A) and HO-1 gene expression (Figure 9B). Tet treatment also induced Nrf2 binding to the HO-1-E2 enhancer region (Figure 9C). Moreover, Tet significantly enhanced BRG1 binding to the HO-1 gene promoter at 3 h (Figure 9D). Consistent with BRG1 recruitment, we detected a substantial induction of Z-DNA formation in the HO-1-TG repeat region from 0.5 to 3 h after Tet administration, although we did not detect statistical significances (Figure 9E). Taken together, these results indicate that Z-DNA formation in the TG repeat region is associated with Nrf2 activation.
Figure 9.
Nrf2 activation is sufficient to induce Z-DNA formation in the human HO-1-TG region. 293T-REx-FLAG-human Nrf2 cells stably expressing the Z-probe* were treated with Tet for the indicated periods of time. (A) Whole-cell lysates were separated using SDS–PAGE, and the protein expression was analyzed through immunoblotting using specific antibodies, as indicated in the figure. (B) The HO-1 gene expression after Tet administration was analyzed through real-time PCR using specific primer sets. The value at time 0 h was arbitrarily set as 1, and the relative expression values are expressed as the means ± SEM of three independent assays. (C) Nrf2 binding to the human HO-1 E2 enhancer (HO-1-E2) was analyzed through ChIP using an anti-FLAG antibody. (D) BRG1 recruitment to the human HO-1 gene promoter (HO-1-Pro) was analyzed through ChIP using an anti-BRG1 antibody. (E) Z-DNA formation was measured through real-time PCR using a specific primer set for the HO-1 TG repeat region (HO-1-TG). The value at 0 h is arbitrarily set as 1, and the relative bindings are expressed as the means ± SEM of three independent assays. #P < 0.05 and ##P < 0.01, compared with the values at 0 h (Dunnett’s test for multiple parameter comparisons).
DISCUSSION
In this study, we developed a Z-DNA detection system using a Z-probe and successfully detected the induction of Z-DNA formation in the human HO-1-TG repeat region in cultured cells (Figures 4–6 and 9). These results showed that the human HO-1-TG repeat in the genome adopts a Z-DNA structure in response to DEM treatment.
Although these results suggest that Nrf2 induces Z-DNA formation in the HO-1 gene promoter, the possibility that Z-DNA is constitutively formed in the HO-1-TG region and that Z-DNA is detected after Nrf2 activation cannot be excluded, as the surrounding chromatin structure is relaxed after Nrf2 activation, allowing the Z-probe to access the Z-DNA structure. To avoid the effect of the 3D chromatin structure in Z-DNA detection, we fixed the cells with formaldehyde in the absence of Z-probe and subsequently sonicated the chromatin before precipitating the Z-DNA through GST-affinity purification using the recombinant GST-Z-probe or immunoprecipitation assay using the anti-Z-DNA antibody. Unfortunately, we did not detect the precipitation of both HO-1-TG region and CSF1-TG region in BRG1-overexpressed SW13 cells using either method (data not shown). However, a significant amount of Pol II was bound to the HO-1 gene promoter before DEM administration compared with the γ-globin promoter (HGB2-Pro), which is exclusively expressed in the erythroid cell linage (Figure 6D). This result suggests that the HO-1 gene promoter region is in a relatively open structure, as previously reported in mouse cells (30), and these regions are accessible to the Z-probe, even before DEM administration.
We detected slight, but significant, binding of Z-probe to non–Z-DNA-forming sequences of the CSF1 gene (i.e. CSF1-3600 and CSF1-Ex4) in an N173- and Y177-dependent manner (Figure 4D). Ha et al. (31) previously reported that the Zα domain of ADAR1 recognizes non–CG-repeat Z-DNA-forming sequences, such as d(CACGTG)2 and d(CGGCCG)2. Because the binding of the Z-probe to non–Z-DNA-forming sequences is not significantly enhanced by BRG1, we propose that CSF1-3600 and CSF1-Ex4 regions form weak Z-DNA structures that are not augmented through BRG1. These notions should be clarified in the future.
We demonstrated that Nrf2 is necessary and sufficient for the Z-DNA formation at the HO-1 gene promoter (Figures 8 and 9). Because we previously indicated that BRG1 was an important co-factor of Nrf2 in the induction of Z-DNA formation (20), we also investigated the role of BRG1 using a BRG1 knockdown. However, we could not clarify the precise role of BRG1 in Z-DNA formation, as Nrf2 protein expression was substantially attenuated after BRG1 knockdown in HeLa cells (Supplementary Figure S2). Thus, the role of BRG1 in Z-DNA formation in the HO-1-TG region requires further clarification.
We showed that the human HO-1 gene promoter region transiently adopts a Z-DNA structure in response to Nrf2 activation treatment (Figures 6 and 9). Interestingly, the Z-DNA formation of the human HO-1 gene promoter preceded HO-1 mRNA induction (Figures 6 and 9). Thus, although some reports have suggested that Z-DNA-forming sequences are involved in the down-regulation of gene expression (32,33), we propose that Z-DNA formation in the HO-1 gene promoter preferentially functions as an enhancer of HO-1 gene expression. We previously reported that Nramp1/Slc11A1 is one of the genes regulated through Nrf2 expression in mouse macrophages (34). Importantly, HIF1α induces Z-DNA formation in the Nramp1 promoter (35). In addition, TPA induces Z-DNA formation in the Nramp1 gene promoter, and ATF3-dependent BRG1 recruitment is essential for the activation of Nramp1 gene expression in HL60 cells (36). Wittig et al. (37) previously reported that Z-DNA is formed in the c-myc regulatory region of the human genome using permeated nuclei of U937 cells and an anti-Z-DNA antibody. These authors also showed that the amount of Z-DNA in the c-myc region is diminished within a relatively short period after treatment with the differentiation agents TPA and vitamin D3, which are associated with the down-regulation of c-myc gene expression (37). Collectively, the results from the present and other studies indicate that Z-DNA formation is associated with the increased expression of the HO-1, Nramp1 and c-myc genes, although the detailed molecular mechanism of this induction remains elusive.
Because the HO-1 gene is rapidly and strongly induced compared with other Nrf2 target genes, including the TXNRD1 gene, we propose that the HO-1 gene is regulated through a unique molecular mechanism. In the present study, we showed that Z-DNA formation in the human HO-1 gene promoter is associated with Nrf2 activation (Figures 6–9). In addition, Wang et al. reported that Z-DNA blocks nucleosome formation, suggesting that Z-DNA formation in the gene promoter is sufficient to demarcate the boundaries of neighboring nucleosomes, producing transcriptionally favorable locations for the TATA box near the nucleosomal DNA entry site of Pol II (5). Indeed, we showed that nucleosome occupancy at the HO-1-TG region was rapidly reduced in response to DEM treatment (Figure 6C). Therefore, we propose that Z-DNA formation in the human HO-1-TG region is one of the mechanisms underlying strong HO-1 gene induction.
Although the correlation between susceptibility to disease and the TG-repeat length polymorphism in promoters has been reported in several genes, including HO-1 for emphysema and Nramp1 for type I diabetes, respectively (4,38), the precise biological function of the Z-DNA-forming sequence remains unclear. In this study, we developed a method for the detection of Z-DNA and demonstrated that Z-DNA formation in the human HO-1 gene promoter is associated with Nrf2 activation. Therefore, the human HO-1 gene promoter is a good model system to understand Z-DNA formation and how Z-DNA-forming sequences regulate gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
MEXT/JSPS KAKENHI [23790319 to A.M.], [20117010 to K.I.]; Priority Research Designated by the President of Hirosaki University. Funding for open access charge: Karoji Memorial Fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Ms Fumiko Tsukidate for technical assistance.
REFERENCES
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 3.Schroth GP, Chou PJ, Ho PS. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J. Biol. Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 4.Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front. Biosci. 2007;12:4424–4438. doi: 10.2741/2399. [DOI] [PubMed] [Google Scholar]
- 5.Wong B, Chen S, Kwon JA, Rich A. Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2007;104:2229–2234. doi: 10.1073/pnas.0611447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thamann TJ, Lord RC, Wang AH, Rich A. The high salt form of poly(dG-dC).poly(dG-dC) is left-handed Z-DNA: raman spectra of crystals and solutions. Nucleic Acids Res. 1981;9:5443–5457. doi: 10.1093/nar/9.20.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maines MD. The heme oxygenase system: a regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- 8.Hill-Kapturczak N, Sikorski E, Voakes C, Garcia J, Nick HS, Agarwal A. An internal enhancer regulates heme- and cadmium-mediated induction of human heme oxygenase-1. Am. J. Physiol. Renal Physiol. 2003;285:F515–F523. doi: 10.1152/ajprenal.00137.2003. [DOI] [PubMed] [Google Scholar]
- 9.Maeshima H, Sato M, Ishikawa K, Katagata Y, Yoshida T. Participation of altered upstream stimulatory factor in the induction of rat heme oxygenase-1 by cadmium. Nucleic Acids Res. 1996;24:2959–2965. doi: 10.1093/nar/24.15.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camhi SL, Alam J, Otterbein L, Sylvester SL, Choi AM. Induction of heme oxygenase-1 gene expression by lipopolysaccharide is mediated by AP-1 activation. Am. J. Respir. Cell Mol. Biol. 1995;13:387–398. doi: 10.1165/ajrcmb.13.4.7546768. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Hara E, Suzuki H, Sasano H, Shibahara S. Expression of heme oxygenase isozyme mRNAs in the human brain and induction of heme oxygenase-1 by nitric oxide donors. J. Neurochem. 1996;67:482–489. doi: 10.1046/j.1471-4159.1996.67020482.x. [DOI] [PubMed] [Google Scholar]
- 12.Vile GF, Basu-Modak S, Waltner C, Tyrrell RM. Heme oxygenase 1 mediates an adaptive response to oxidative stress in human skin fibroblasts. Proc. Natl Acad. Sci. USA. 1994;91:2607–2610. doi: 10.1073/pnas.91.7.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alam J. Multiple elements within the 5' distal enhancer of the mouse heme oxygenase-1 gene mediate induction by heavy metals. J. Biol. Chem. 1994;269:25049–25056. [PubMed] [Google Scholar]
- 14.Alam J, Camhi S, Choi AM. Identification of a second region upstream of the mouse heme oxygenase-1 gene that functions as a basal level and inducer-dependent transcription enhancer. J. Biol. Chem. 1995;270:11977–11984. doi: 10.1074/jbc.270.20.11977. [DOI] [PubMed] [Google Scholar]
- 15.Reichard JF, Motz GT, Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi AM, Alam J. Heme oxygenase-1: function, regulation, and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am. J. Respir. Cell Mol. Biol. 1996;15:9–19. doi: 10.1165/ajrcmb.15.1.8679227. [DOI] [PubMed] [Google Scholar]
- 17.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 18.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 19.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, Yamamoto M. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol. Cell. Biol. 2006;26:7942–7952. doi: 10.1128/MCB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Hosoya T, Maruyama A, Nishikawa K, Maher JM, Ohta T, Motohashi H, Fukamizu A, Shibahara S, Itoh K, et al. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem. J. 2007;404:459–466. doi: 10.1042/BJ20061611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 23.Maruyama A, Nishikawa K, Kawatani Y, Mimura J, Hosoya T, Harada N, Yamamoto M, Itoh K. The novel Nrf2-interacting factor KAP1 regulates susceptibility to oxidative stress by promoting the Nrf2-mediated cytoprotective response. Biochem. J. 2011;436:387–397. doi: 10.1042/BJ20101748. [DOI] [PubMed] [Google Scholar]
- 24.Hosoya T, Maruyama A, Kang MI, Kawatani Y, Shibata T, Uchida K, Warabi E, Noguchi N, Itoh K, Yamamoto M. Differential responses of the Nrf2-Keap1 system to laminar and oscillatory shear stresses in endothelial cells. J. Biol. Chem. 2005;280:27244–27250. doi: 10.1074/jbc.M502551200. [DOI] [PubMed] [Google Scholar]
- 25.Laptenko O, Beckerman R, Freulich E, Prives C. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc. Natl Acad. Sci. USA. 2011;108:10385–10390. doi: 10.1073/pnas.1105680108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herbert A, Schade M, Lowenhaupt K, Alfken J, Schwartz T, Shlyakhtenko LS, Lyubchenko YL, Rich A. The Zalpha domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic Acids Res. 1998;26:3486–3493. doi: 10.1093/nar/26.15.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Xiao J, Li J, Lu L, Feng S, Dröge P. Human genomic Z-DNA segments probed by the Z alpha domain of ADAR1. Nucleic Acids Res. 2009;37:2737–2746. doi: 10.1093/nar/gkp124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Mulholland N, Fu H, Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell. Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, Igarashi K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl Acad. Sci. USA. 2004;101:1461–1466. doi: 10.1073/pnas.0308083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha SC, Choi J, Hwang HY, Rich A, Kim YG, Kim KK. The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ alpha(ADAR1) Nucleic Acids Res. 2009;37:629–637. doi: 10.1093/nar/gkn976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothenburg S, Koch-Nolte F, Rich A, Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl Acad. Sci. USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards SF, Sirito M, Krahe R, Sinden RR. A Z-DNA sequence reduces slipped-strand structure formation in the myotonic dystrophy type 2 (CCTG) x (CAGG) repeat. Proc. Natl Acad. Sci. USA. 2009;106:3270–3275. doi: 10.1073/pnas.0807699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harada N, Kanayama M, Maruyama A, Yoshida A, Tazumi K, Hosoya T, Mimura J, Toki T, Maher JM, Yamamoto M, et al. Nrf2 regulates ferroportin 1-mediated iron efflux and counteracts lipopolysaccharide-induced ferroportin 1 mRNA suppression in macrophages. Arch. Biochem. Biophys. 2011;508:101–109. doi: 10.1016/j.abb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Bayele HK, Peyssonnaux C, Giatromanolaki A, Arrais-Silva WW, Mohamed HS, Collins H, Giorgio S, Koukourakis M, Johnson RS, Blackwell JM, et al. HIF-1 regulates heritable variation and allele expression phenotypes of the macrophage immune response gene SLC11A1 from a Z-DNA forming microsatellite. Blood. 2007;110:3039–3048. doi: 10.1182/blood-2006-12-063289. [DOI] [PubMed] [Google Scholar]
- 36.Xu YZ, Thuraisingam T, Marino R, Radzioch D. Recruitment of SWI/SNF complex is required for transcriptional activation of the SLC11A1 gene during macrophage differentiation of HL-60 cells. J. Biol. Chem. 2011;286:12839–12849. doi: 10.1074/jbc.M110.185637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittig B, Wolfl S, Dorbic T, Vahrson W, Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada N, Yamaya M, Okinaga S, Nakayama K, Sekizawa K, Shibahara S, Sasaki H. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to emphysema. Am. J. Hum. Genet. 2000;66:187–195. doi: 10.1086/302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.