Abstract
Uncontrolled cell proliferation and cytoskeletal remodeling are responsible for tumor development and ultimately metastasis. A number of studies have implicated microRNAs in the regulation of cancer cell invasion and migration. Here, we show that miR-23b regulates focal adhesion, cell spreading, cell-cell junctions and the formation of lamellipodia in breast cancer (BC), implicating a central role for it in cytoskeletal dynamics. Inhibition of miR-23b, using a specific sponge construct, leads to an increase of cell migration and metastatic spread in vivo, indicating it as a metastatic suppressor microRNA. Clinically, low miR-23b expression correlates with the development of metastases in BC patients. Mechanistically, miR-23b is able to directly inhibit a number of genes implicated in cytoskeletal remodeling in BC cells. Through intracellular signal transduction, growth factors activate the transcription factor AP-1, and we show that this in turn reduces miR-23b levels by direct binding to its promoter, releasing the pro-invasive genes from translational inhibition. In aggregate, miR-23b expression invokes a sophisticated interaction network that co-ordinates a wide range of cellular responses required to alter the cytoskeleton during cancer cell motility.
INTRODUCTION
Deregulation of cell cycle progression genes is crucial to tumor growth, and additional genetic modifications are required to promote motile and invasive behaviors in cancer cells, leading to tumor dissemination and metastasis.
The cytoskeleton controls cell motility, invasion and adhesion. Three members of the Rho family of small GTPases, RhoA, Rac1 and cell division cycle 42, are crucial in regulating the molecular pathways implicated in cell migration (1,2), controlling cytoskeletal dynamics, cell adhesion, morphology and motility (1). This is achieved via effector proteins that regulate actin-based structures, and the p21-activated kinases (PAKs) are among the best characterized effectors of cell division cycle 42 and Rac1 (3). In addition, several growth factor receptor tyrosine kinases activate PAKs through Rho GTPases leading to cytoskeletal remodeling (4–6). Furthermore, in breast cancer (BC), the HER-2 pathway is able to regulate the actin cytoskeleton and cell motility via PAK1 and PAK2 activation and distinct downstream signaling mechanisms (7).
MicroRNAs (miRNAs) are small, single-stranded RNAs that regulate gene expression (8), and their dysregulation can therefore contribute to cell motility and metastasis by affecting relevant transcripts (9–13).
Using a bioinformatic approach, we identified miR-23b as a miRNA implicated in cytoskeletal remodeling and motility, and we further demonstrated this finding through a number of experimental techniques. In addition, using RNA-sequencing (RNA-seq) and luciferase reporter assays, we validated a subset of cytoskeletal genes as direct targets of miR-23b in BC, implicating a direct role of this miRNA in cytoskeletal remodeling and motility. Notably, when AP-1 is activated by growth factors, it binds directly to the miR-23b promoter, thereby reducing its expression, which indicates that this event contributes to motility and metastasis in BC.
MATERIALS AND METHODS
Cell culture
BC cell lines (MCF-7, MDA-MB-231 and MDA-MB-468) and colon cancer HCT116 cell lines were maintained in Dulbecco’s modified Eagle’s medium and McCoy’s medium, respectively, supplemented with 10% FCS, 1% penicillin/streptomycin and 2% glutamine. MDA-MB-231 cells isolated from distant sites were maintained as previously described (14). Stable transfected MDA-MB-231 cell clones were expanded in G-418 (Roche Applied Science, Burgess Hill, UK).
Plasmid constructions
The miR-23b sponges were constructed by annealing, purifying and cloning oligonucleotides containing six tandem bulged miRNA binding motifs, into the HindIII and BamHI sites of the pEGFP-C1 plasmid (Contech, Saint-Germain-en-Laye, France), as 3′UTR of the EGFP mRNA. For 3′UTRs reporter construction, the 3′UTR of Pak2 or Pak1 were PCR-amplified from human genomic DNA and cloned into the pMIR-REPORT Firefly Luciferase vector. The miR-23b sensors were constructed as for miR-23b sponges, but into the SpeI and HindIII sites of the pMIR-REPORT Firefly Luciferase vector. The ANXA2, ARHGEF6, CFL2, LIMK2, PIK3R3, PLAU and TLN2 3′UTRs cloned into pLightSwitch_3′UTR GoClone vectors were used (Switch Gear Genomics, Menlo Park CA, USA). The indicated mutagenized miR-23b seed-containing luciferase reporter vector was created with a QuickChange II or II XL Site-Directed Mutagenesis Kit (Agilent Technology, Edinburgh, UK) according to the manufacturer’s instructions. All plasmid sequences were verified to be free of mutations by direct sequencing. The sequences of all primers used for plasmid construction and site-directed mutagenesis are reported in Supplementary Table S1.
Transfections, reporter assays and cell treatments
Cells were plated in 6-well plates at 50% confluence and transfected with either the miRNA mimics (5 nM), miRNA inhibitors (100 nM) (Applied Biosystems, Warrington, UK) or siRNA oligonucleotides (40 nM) for 48 h using HiPerFect Transfection Reagent (Qiagen, Crawley, UK). For experiments of miRNA over-expression for 9 days, cells were plated in 6-well plates at 30% confluence and transfected with the miRNA mimics (5 nM); after 72 h of transfection, cells were split and re-transfected with additional miRNAs mimics; this protocol was repeated every 3 days for up to 9 days. Transfection of GFP-sponge-expressing plasmids was performed using Lipofectamine 2000 (Life Technologies Ltd, Paisley, UK). For 3′UTR reporter assays, cells were co-transfected with Renilla Luciferase vector, the indicated miRNA precursors or plasmids and 3′UTR-p-MIR REPORT Firefly Luciferase vectors in 24-well plates and analyzed as previously described (15). Cells co-transfected with pLightSwitch_3UTR GoClone vectors, and the indicated miRNA precursors or plasmids in 24 well plates were lysed using a passive lysis buffer (Promega, Southampton, UK) and processed with the LightSwitch Assay System (Switchgear Genomics Menlo Park CA, USA) according to manufacturer’s instructions. Luciferase activity detection was performed using a GLOMAX 96 Microplate luminometer (Promega). For EGF treatment, MDA-MB-468 cells were plated in triplicate in normal growth medium. After 24 h, EGF (25 ng/ml) was added for the indicated time points in three independent biological replicates.
Immunofluorescence and cell–cell junction quantification
Cells were fixed with 4% paraformaldehyde and permeabilized in 0.2% Triton-X-100 for 30 min. F-actin was detected with phalloidin-Alexa Fluor® 488 (Invitrogen), and nuclei were visualized with TO-PRO-3 (Invitrogen). For E-cadherin and focal adhesion (FA) visualization, cells were stained either with anti-E-cadherin antibody (HECD-1) (M106, Takara Bio Inc.) or anti-Vinculin antibody (V9264, Sigma-Aldrich) and imaged using a confocal microscope. To analyze cell–cell junction linearity, the junction lenghts and distances between vertices were manually traced and measured by using LSM 5 Image Browser (Carl Zeiss MicroImaging, Inc.). To highlight the area of FAs, the images taken were inverted in black-and-white color and analyzed using NHI ImageJ software for FA area quantification. To study -lamellipodial formation, cells were imaged using ImageXpress Micro® microscope (Molecular Devices), and lamellipodia number was manually counted in a blind fashion (100 cells per each condition in the three independent experiments).
In vitro migration and invasion assays
For cell-tracking assays, cells were seeded in 24-well tissue culture plates at a final density of 1 × 104. After 48 h of transfection, time-lapse sequences were digitally recorded at intervals of 20 min for 24 h using the ImageXpress Micro® microscope (Molecular Devices). Cell trajectories were determined by following the centroid of the nuclei using the ImageJ plugin ‘MTrackJ’ (http://www.imagescience.org/meijering/software/mtrackj/), and speed of cell body movement was calculated for mean cell migratory speed. A 50–100 cells per each condition were analyzed, and three independent experiments were performed. For transwell migration assays, 5 × 104 cells were seeded atop uncoated membranes with 8.0 μm pores (BD Biosciences). Cell were plated in serum free-medium and allowed to migrate toward a complete growth medium for 9 h. For 3D Collagen-I invasion assays, 7 × 104 cells were plated in black-walled 96-well plates and allowed to attach. Collagen-I gels (n = 5 gels per condition, 100 µl) were prepared as described (16) at a final concentration of 2.3 mg/ml and added atop the cells. After polymerization and equilibration for 1 h at 37°C and 5% CO2, collagen gels were coated with 100 µl of cell culture medium supplemented with 10% FCS as chemoattractant, and cell invasion was allowed for 16–24 h. Collagen gels containing invaded cells were then fixed and stained, and z sections were taken by confocal microscopy. Invasion indexes were calculated as the number of cells at 40 μm divided by those at 0 μm. Three independent experiments were performed.
Cell growth and Annexin V-apoptotic assays
MCF7 and MDA-MB-231 cells growth was analyzed by performing an SRB assay. Cells were seeded in 96-well plates and transfected with the indicated molecules. The SRB cell viability assay was performed 48 h after transfections as described previously (17). To assay the apoptotic properties of miR-23b, MDA-MB-231 were transfected with the indicated microRNA precursor mimics and the cell death control siRNAs (20 nM) (Qiagen) used as positive control. After 48 h, detached cells were combined with adherent cells after lifting with trypsin-EDTA, stained with Annexin V/PE apoptosis detection kit (BD Biosciences) according to manufacturer’s protocol and analyzed using a FACSCanto II flow citometer (BD Biosciences). Apoptotic cells were represented by high PE-Annexin V fluorescence signals.
Spreading assays
Spreading assays was performed as previously described (18).
Patients and samples
To investigate the potential role of miR-23b in BC metastasis, we prospectively measured their expression in primary BCs and corresponding lymph node metastasis from patients with lymph node positive BC. Between January 2008 and February 2010, we obtained primary breast tumors and corresponding lymph node metastases from 66 consecutive individuals eligible for chemotherapy at Imperial College Healthcare NHS Trust, London, UK. All 132 samples were formalin-fixed and paraffin-embedded. The clinico-pathologic characteristics are listed in Supplementary Table S2. Prospective written consent was obtained in accordance with ethical guidelines.
Statistical analysis
Data are presented as mean ± standard error of the mean calculated using Graph Prism software. Student’s t-test and Chi-Square test were used for comparison, with P < 0.05 considered significant.
Sequence data were submitted to Gene Expression Omnibus database, accession number GSE37918.
RESULTS
Bioinformatic analyses reveal that miR-23b is involved in cytoskeletal remodeling and adhesion
Using a bioinformatic approach and an extensive literature review, we identified candidate miRNAs involved in cell motility and invasion. We selected miR-23b, as it reduces hepatocellular and cervical cancer cell motility through the regulation of PLAU and c-Met (19) and reduces experimental metastasis formation in colorectal cancer (20). However, its role in BC and the subsequent pathways regulated remain largely unknown. Nine hundred and thirty-eight randomly chosen genes and 938 potential miR-23b targets [identified using TargetScan (21)] were analyzed using DAVID (http://david.abcc.ncifcrf.gov/home.jsp) for gene ontology (GO) annotation and pathway enrichment analysis (22,23). This method suggested that miR-23b may regulate pathways involved in cancer, cytoskeletal remodeling, cell-cell junctions and cell adhesion (Supplementary Figure S1).
The miR-23b expression in cancer cell lines enhances cell–cell adhesion
We transfected epithelial-like (MCF-7) and mesenchymal-like (MDA-MB-231) BC cells with synthetic miR-23b precursor or a negative control (miR-n.c.). Ectopic expression of miR-23b increased epithelial characteristics in MCF-7 cells, as demonstrated by tighter colony morphology with marked cell–cell junctions (Figure 1A), independent of E-cadherin (CDH1) expression (Figure 1B). However, as miR-23b over-expression did not induce these characteristics in mesenchymal-like MDA-MB-231 cells (Figure 1C), and in addition, CDH1 expression did not change upon miR-23b over-expression, we hypothesized that such effects were unlikely to be due to re-expression of adhesion molecules during the mesenchymal-to-epithelial transition, but more likely due to miR-23b affecting the tension of existing cell–cell adhesions.
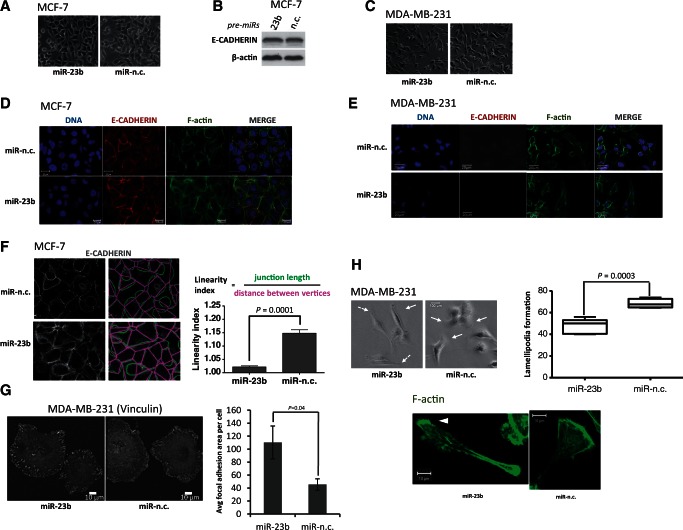
Figure 1.
miR-23b expression affects cytoskeletal remodeling as predicted by bioinformatic analyses. (A) Phase-contrast images of MCF-7 cells upon transfection with miR-23b and miR-n.c. precursors (5 nM) for 48 h. (B) Western blot showing E-cadherin and β-actin expression in MCF-7 cells transfected with miR-23b and miR-n.c. precursors (5 nM) for 48 h. β-actin was used as a loading control. (C) Phase-contrast images of MDA-MB-231 cells upon transfection with miR-23b and miR-n.c. precursors (5 nM) for 48 h. (D) From left to right: TO-PRO-3 staining of DNA, immunofluorescence staining of E-cadherin (1:1000), Alexa Fluor 488 phalloidin (1:500) staining of F-actin and overlay of the three images of MCF-7 cells transfected with miR-23b and miR-n.c. precursors (5 nM) for 9 days. Scale bar = 20 µm (E) From right to left: TO-PRO-3 staining of nuclei, immunofluorescence staining of E-cadherin (1:1000), Alexa Fluor 488 phalloidin (1:500) staining of F-actin and overlay of the three images of MDA-MB-231 cells transfected with miR-23b and miR-n.c. precursors (5 nM) for 9 days. Scale bar = 20 µm. (F) Quantification of junction linearity. Junction length (green) and distance between vertices (pink) were measured and a linearity index was calculated by dividing the two measures (right). This index decreases in miR-23b over-expressing MCF-7 cells. Scale bar = 20 µm. 150–250 junctions per condition in two independent experiments were counted. Data are mean ± s.e.m. (P = 0.0001, Student's t-test). (G) Immunofluorescence staining of MDA-MB-231 using Vinculin antibody (1:400); images were false colored in a black-and-white fashion to highlight FAs. After transfection with miR-23b and miR-n.c. precursors (5 nM) for 48 h the cells were left to adhere on coverslips coated with 50 uM of collagen I for 1 h before staining. Scale bar = 10 µm. The bottom graph shows the FA size per treated cell. Quantification of FA size is represented as the average of FA area per cell. Data are shown as mean values ± s.e.m. from three independent experiments (P = 0.04, Student's t-test). (H) Phase-contrast images of MDA-MB-231 cells transfected with the indicated miRNA precursors (5 nM) for 48 h. Lamellipodial structures are indicated by continuous arrows; lack of lamellipodia is indicated by discontinuous arrows. The box plot on the right shows the number of lamellipodia formed after treating the cells with the indicated precursors. Scale bar = 100 µm. 50–150 cells per condition in three independent experiments were counted. Data are mean ± s.e.m. (P = 0.0003, Student's t-test). The image at the bottom shows typical lamellipodia in MDA-MB-231 cells transfected with the indicated precursors (5 nM) for 48 h and stained with Alexa Fluor 488 phalloidin.
Next, we aimed to induce mesenchymal-to-epithelial transition via prolonged (9 days) miR-23b over-expression, using immunofluorescence to examine CDH1 and cell junction formation (24) (Figure 1D and E). The miR-23b over-expressing-MCF-7, but not MDA-MB-231 cells, overall showed more orderly and stable tight and adherens junctions compared with controls, confirming that miR-23b leads to an enhanced tension between existing cell–cell junctions (Figure 1D and E). The quantification of this effect is expressed as a linearity index, defined by the ratio of junction length to the distance between vertices (25), and supported that junctions in miR-23b-expressing MCF-7 cells were straighter compared with controls (Figure 1F).
The miR-23b expression increases FA size and cell spreading and modulates the formation of lamellipodia
We stained MDA-MB-231 cells for the FA marker vinculin (26) to assess the effect of miR-23b over-expression on their ability to form FAs. FAs were significantly larger in cells after over-expressing miR-23b compared with miR-n.c.-transfected controls (Figure 1G). Remarkably, we also noticed that the number of cells forming lamellipodia was significantly less after miR-23b over-expression and at most these formed only small narrow protrusions (Figure 1H).
To assess the effects of miR-23b on cell-spreading capacity, we plated MDA-MB-231 cells for 30, 60 and 120 min (Figure 2) on Type I collagen. The miR-23b-transfected cells spread more than controls and after 30 min contained lamellar protrusions, stress fiber-like actin bundles and punctate peripheral FAs, whereas the controls remained rounded with no FAs or F-actin bundles (Figure 2).
Figure 2.
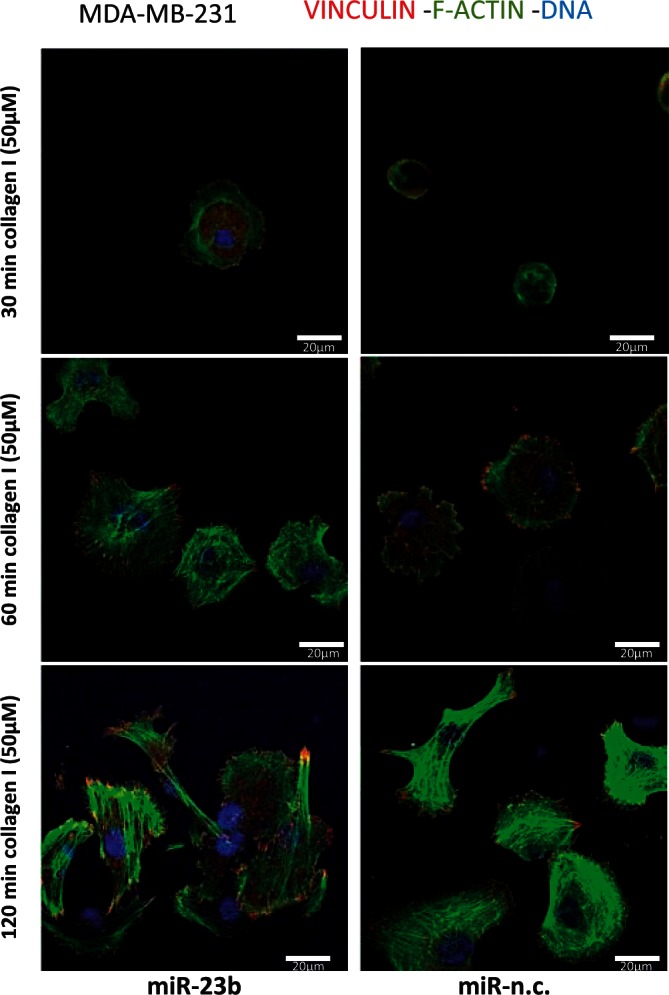
miR-23b expression modulate spreading capabilities on ECM in MDA-MB-231 cells. For spreading assays, MDA-MB-231 cells were transfected with miR-23b and miR-n.c. precursors (5 nM) for 48 h and seeded on collagen I (50 µM) matrices for 30, 60 and 120 min. Cells were than fixed and stained for anti-Vinculin and phalloidin to visualize FAs and F-actin, respectively. Nuclei are visualized with TO-PRO-3 stain (blue); scale bar = 20 µm.
The miR-23b regulates breast cancer cell motility and invasion
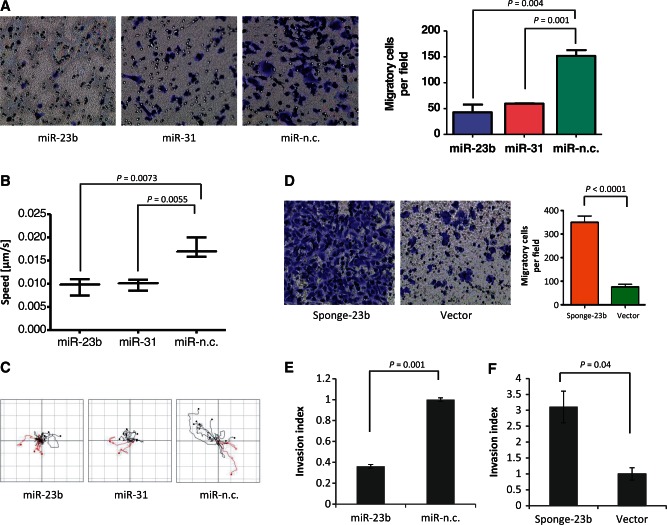
We found that miR-23b over-expression did not affect the proliferation rate or promote apoptosis in cultured MDA-MB-231 or MCF-7 cells (Supplementary Figure S2A and B). However, miR-23b transfection markedly reduced in vitro migration of highly motile MDA-MB-231 cells compared with miR-n.c., effects which were similar to those seen in miR-31 over-expressing positive controls (Figure 3A). The miR-31 reduces MDA-MB-231 cell motility and experimental metastasis in BC (13) and accordingly miR-23b and miR-31 both reduced cell migratory speed (Figure 3B and Supplementary Movies S1–S3) and shortened migration trajectory lines (Figure 3C). Next, we developed MDA-MB-231 cell lines that stably expressed an EGFP-miR-23b sponge construct (Supplementary Figure S3A), enabling them to inhibit endogenous miR-23b. In particular, clone 10 reduced miR-23b levels by up to 70% (Supplementary Figure S3B). Co-expression of the sponge vector and a sensor vector [that contains six perfectly interacting sites for miR-23b (Supplementary Figure S3D)] increases luciferase activity compared with empty vector control (Supplementary Figure S3E), demonstrating the efficiency of the sponge vector in the inhibition of miR-23b activity. Notably, the inhibition of miR-23b significantly increased MDA-MB-231 cell migration (Figure 3D) and increased lamellipodia size (Supplementary Figure S3C), which was consistent with the effects of miR-23b over-expression on lamellipodia as described earlier in the text (Figure).
Figure 3.
mir-23b inhibits MDA-MB-231 cell motility and invasion. (A) Transwell migration assays were performed for 9 h after transfecting MDA-MB-231 cells with the indicated precursors (5 nM) for 48 h. The graph on the right indicates cell migration expressed as percentage of the average of migratory cells per field (n = 5 fields per transfection). Data are mean of three experiments ± s.e.m. performed in triplicates (P = 0.004, P = 0.001, Student's t-test). (B) Motility of MDA-MB-231 cells expressing the indicated precursors was examined for 24 h by phase-contrast, time-lapse microscopy. Cell speed of 50–100 cells per condition was quantified and speed averages are presented as mean of three experiments ± s.e.m. (P = 0.0001, Student's t-test). (C) Plots show overlays of representative trajectories described by pre-miR-expressing MDA-MB-231 cells during time-lapse motility assays. (D) Transwell migration assays of MDA-MB-231 cells stably expressing the indicated constructs (Vector refers to pEGFP-C1 empty vector) were performed for 9 h. The graph on the right indicates cell migration expressed as a percentage of the average of migratory cells per field (n = 5 fields per transfection). Data are mean of three experiments ± s.e.m. performed in triplicates (P < 0.0001 Student's t-test). (E) Invasion of MDA-MB-231 cells transfected with miR-23b and miR-n.c. precursors (5 nM) for 48 h was assessed in collagen-I-3D-matrices (2.3 mg/ml) for 16 h. Confocal z sections were collected from each well at 0 μm (bottom of the well) and 40 μm and invasion indexes were calculated as the number of cells at 40 μm divided by those at 0 μm. Invasion indexes are mean of three experiments ± s.e.m. performed in quintuplicates (P = 0.001 Student's t-test). (F) Invasion assays using MDA-MB-231 cells stably expressing the miR-23b-sponge construct or the parental control pEGFP-C1 plasmid vector were performed as in (E). Invasion indexes are mean of three experiments ± s.e.m. performed in quintuplicates (P = 0.04, Student's t-test).
To evaluate invasive capacity, we tested MDA-MB-231 migration in 3D matrices. The miR-23b over-expression and silencing significantly reduced and increased cell invasion, respectively (Figure 3E and F).
The miR-23b directly targets PAK2 and increases myosin light chain II phosphorylation
We implicated miR-23b in cytoskeletal regulation and selected PAK2 as its candidate target based on our TargetScan analysis (Figure 4A and Supplementary Figure S4A) and its previously described role in cytoskeletal remodeling (7,27). Subsequently, we found that silencing PAK2 reduced motility and increased myosin light chain (MLC) II phosphorylation in MDA-MB-231 cells (Supplementary Figure S4C and D). Interestingly, PAK3 was not detected in three epithelial cancer cell lines (data not shown), consistent with its restricted tissue expression pattern (28). The miR-23b specifically reduced PAK2 levels in all tested cell lines, but not PAK1 (which is not a predicted miR-23b target; Figure 4B and Supplementary Figure S4B). Accordingly, MLC II phosphorylation was upregulated on miR-23b over-expression, which is important for regulating cytoskeletal reorganization and cell migration (Figure 4C). Furthermore, PAK2 levels increased in MCF-7 cells transiently transfected with miR-23b sponge construct and in MDA-MB-231 stably transfected with miR-23b sponge construct (i.e. after reduction of endogenous miR-23b levels; Figure 4D and Supplementary Figure S4E). Luciferase reporter assays next demonstrated that miR-23b regulates PAK2, but not PAK1, by direct interaction with its 3′UTR (Supplementary Figure S4F–I).
Figure 4.
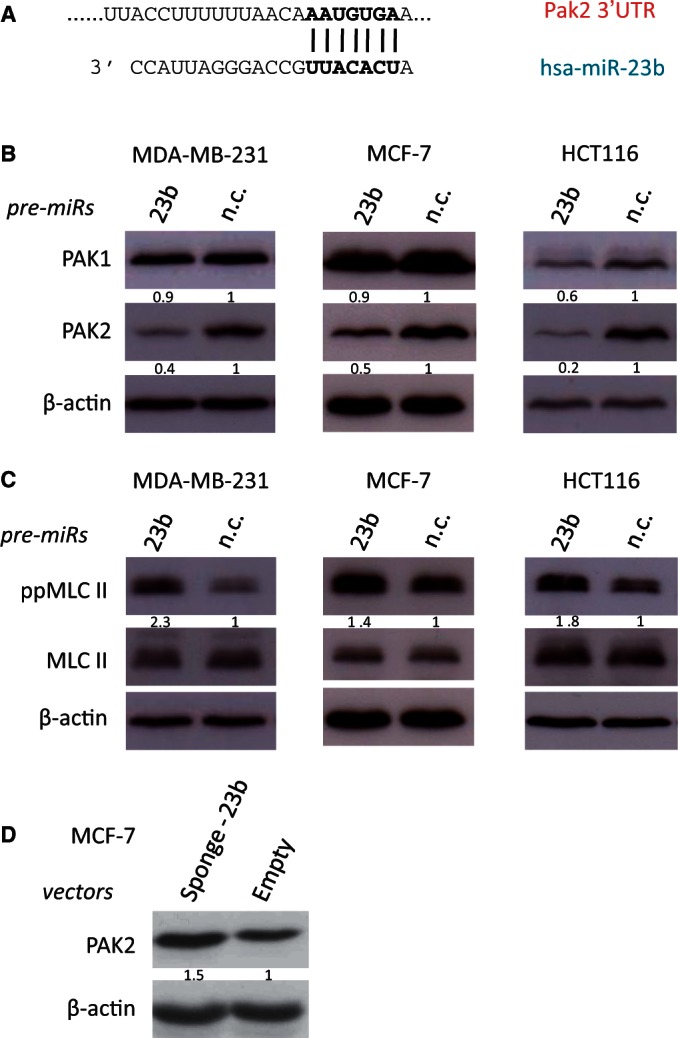
miR-23b targets PAK2 kinase, leading to induction of MLC II phosphorylation. (A) Predicted duplex formation between human Pak2 3′UTR (top) and has-miR-23b (bottom). (B) Western blots showing PAK1 and PAK2 levels after transfection for 48 h of the indicated miRNA precursors (5 nM) in breast cancer MDA-MB-231 and MCF-7 cells and colon cancer HCT116 cells. (C) Western blots showing MLC II levels and phosphorylation of MLC II at Thr18/Ser19 position, after transfection for 48 h of miR-23b and miR-n.c. precursors (5 nM) in the same cancer cell lines described in (B). (D) Western blots showing PAK2 levels after transient transfection of MCF-7 cells with mi-23b-sponge construct and pEGFP-C1 parental control (2 µg). β-actin was used as a loading control. Fold changes in protein expression levels were normalized for β-actin using ImageJ software are shown underneath each relative protein plot.
AP-1 directly interacts with the miR-23b promoter and transcriptionally suppresses its expression
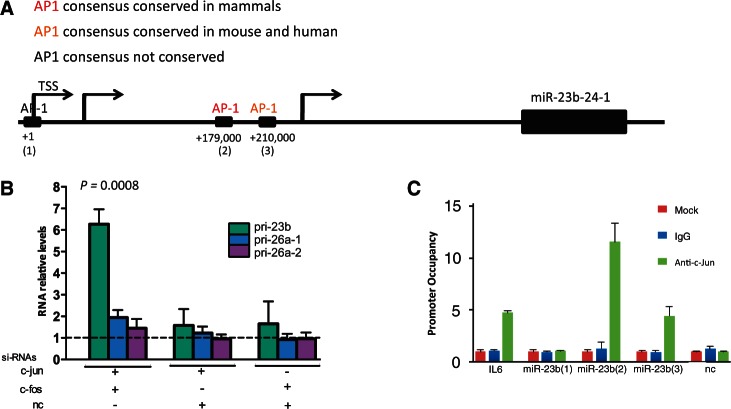
The transcription factor AP-1 is activated through EGFR and HER-2 signaling pathways. It subsequently recognizes TPA-responsive elements (TREs) in promoter and enhancer regions of target genes involved in cell motility and growth (29) and is composed of c-JUN and c-FOS heterodimers (Supplementary Figure S5A) (30). Analysis of the transcription unit (TU) for transcription factors that may regulate this miRNA gene, revealed conserved and less conserved TREs along miR-23b TU (Figure 5A). The miR-23b is encoded in the complex TU, Chromosome 9 open reading frame 3 (C9orf3; Figure 5A), and c-MYC regulates its expression by directly interacting with its upstream transcriptional start site (31,32).
Figure 5.
AP-1 transcriptionally suppresses miR-23b expression levels directly binding with its promoter. (A) Schematic representation of miR-23b TUs showing conserved, less conserved and non-conserved AP-1-specific TREs. TSS: transcription start site. (B) Relative levels of pri-mir-23b, pri-mir-26a-1 and pri-mir-26a-2 were measured by RT-qPCR after co-silencing of c-jun and c-fos, silencing of c-jun or c-fos by using siRNAs (20 nM) in MDA-MB-231 cells for 48 h. Data are presented relative to the siRNA-n.c. (40 nM) single transfection (dotted line). Data are mean of three experiments ± s.e.m. (P = 0.0008, Student's t-test). (C) MDA-MB-231 cells were processed for ChIP assays and RT-qPCR was performed. The c-JUN-interaction site genomic regions are presented, expressed as promoter occupancy. MiR-23b (1), (2), (3) represent each AP-1-specific TREs present in the TU of miR-23b promoter, respectively. IL6: interleuchin-6 promoter region; n.c.: a genomic region that does not contain AP-1-specific TREs. Data are mean of two independent experiments ± s.e.m.
To determine whether AP-1 regulates miR-23b transcription, we next silenced c-FOS and c-JUN in MDA-MB-231 cells. Although individual gene silencing had little effect, co-silencing of both genes significantly increased primary (pri-) and mature miR-23b levels, thus implicating AP-1 in the regulation and reduction of miR-23b expression (Figure 5B and Supplementary Figure S5B). In addition, c-JUN-c-FOS co-silencing reduced PAK2 levels and greatly increased MLC II phosphorylation (Supplementary Figure S5C). Next, chromatin immunoprecipitation revealed that AP-1 is directly involved in the transcriptional inhibition of miR-23b, as c-JUN directly bound to at least two evolutionarily conserved TREs in the TU of C9orf3 (Figure 5A and C). c-JUN also bound to the IL-6 gene promoter, used as positive control (33), but not a genomic DNA region that lacks the TREs (Figure 5C).
As c-JUN is transcriptionally activated by EGF (34), we exposed EGFR positive MDA-MB-468 BC cells to EGF. We found that c-Jun mRNA increased within 20 min and for up to 2 h (Supplementary Figure S5D), as expected (34), whereas miR-23b levels concomitantly decreased (Supplementary Figure S5E), thus indicating that EGF reduces miR-23b expression through c-JUN.
The action of miR-23b on the cytoskeleton is mediated by regulation of a subset of cytoskeletal genes
By treating BC cells with a siRNA against PAK2, we obtained ∼94% silencing of the gene (Supplementary Figure S4D). On the other hand, we showed that miR-23b treatment reduced PAK2 levels to ∼60% (Figure 4B). As miR-23b induced a stronger reduction of MDA-MB-231 motility compared with the silencing of PAK2 alone (Figure 3B and Supplementary Figure S4C), this miRNA must also mediate this effect through the regulation of other cytoskeletal genes. With this hypothesis in mind, we performed RNA-seq of miR-23b-over-expressing MCF-7 cells and MDA-MB-231 cells stably transfected with miR-23b sponge vector. The miR-23b expression in MCF-7 cells affected levels of 7.4% of transcripts, whereas in MDA-MB-231 cells stably transfected with the sponge vector, gene expression changed for 22.9% (Supplementary Table S3). As miRNAs repress gene expression, we selected downregulated genes from miR-23b-over-expressing MCF-7 and upregulated genes from miR-23b-sponge MDA-MB-231 (Supplementary Table S4) and performed GO term enrichment analysis (Supplementary Figure S6). Consistent with our findings, we observed enrichment in cytoskeletal organization genes in both cases (Supplementary Figure S6A and B). To further evaluate miR-23b targets, we intersected genes downregulated in MCF-7 cells over-expressing miR-23b with those genes upregulated in MDA-MB-231 cells containing sponge vectors that stably reduce miR-23b and combined them with those genes derived from a TargetScan analysis (Supplementary Table S4). We reasoned that the intersection would contain a highly enriched list of direct targets and considered a cutoff expression change of 1.2-fold sufficient, as miRNAs regulate transcripts by promoting destabilization through deadenylation (35,36), although mediating a stronger effect on protein translation (37). Higher fold changes may result in omitting many relevant targets. Furthermore, the impact of miRNAs on gene targets is variable and usually mild (38,39). Seventy percent of previously described miR-23b gene targets were among the intersection representing those genes downregulated in MCF-7 and/or upregulated in MDA-MB-231 and those in the TargetScan list (Supplementary Tables S4). Accordingly, we found that PAK2 is within this intersection (Supplementary Table S4). From this gene list, we selected a group of pro-metastatic genes for further validation.
Cytoskeletal genes are direct targets of miR-23b in BC
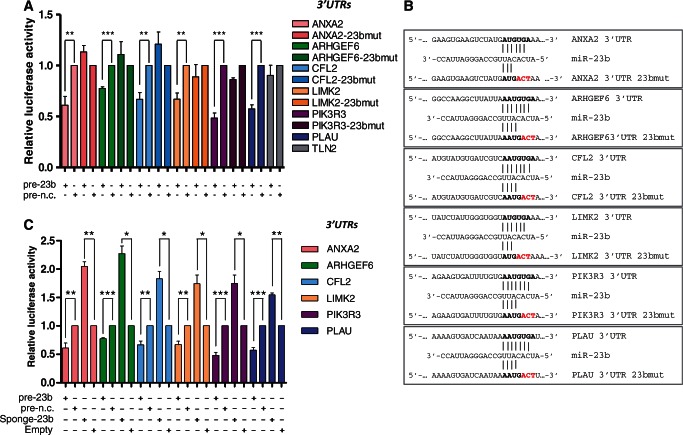
This finding was further validated performing RT-qPCR on 15 pro-metastatic genes involved in cytoskeletal remodeling from our RNA-seq analysis, that contain ‘seeds’ for miR-23b interaction (TargetScan analysis). Accordingly, these were all found to be downregulated in MCF-7 over-expressing miR-23b and upregulated in MDA-MB-231 stably transfected with miR-23b sponge vector (Supplementary Figure S7). Next, we selected seven genes (LIMK2, ARHGEF6, CFL2, PIK3R3, PLAU, ANXA2, TLN2) that are well described in the literature as promoters of cell motility through regulation of cytoskeletal remodeling pathways (19,40–43) and performed 3′UTR luciferase reporter assays (Figure 6). We could demonstrate the direct regulation by miR-23b of six of seven of these using both over-expression and inhibition of miR-23b (Figure 6A and C). In addition, site-directed mutagenesis precisely defined the sites of miR-23b interaction within their 3′UTRs (Figure 6A and B).
Figure 6.
miR-23b targets ANXA2, ARHGEF6, CFL2, LIMK2, PIK3R3 and PLAU mRNAs by directly interacting with their relative 3′UTRs. (A) and (C), Relative luciferase activity levels were measured after 24 h from co-transfection of MCF-7 cells with the indicated 3′UTR-luciferase reporter constructs either with miR-23b or miR-n.c. precurors (5 nM) or with mi-23b-sponge construct or pEGFP-C1 parental control (150 ng). Data are mean of three independent experiments (each of them performed in triplicate) ± s.e.m (*P < 0.05; **P ≤ 0.006; ***P ≤ 0.0005). (B) Representation of miR-23b and the indicated target mRNA 3′UTR heteroduplexes: complementary nucleotides of the miR-23b seed sequence (in bold) and the mutant nucleotides of the miR-23b binding site (in red).
The miR-23b expression is inversely correlated with breast cancer metastases
We hypothesized that loss of miR-23b may be involved in BC spread and therefore examined miR-23b levels in MDA-MB-231 cells that had been inoculated into the mammary fat pads of nude mice and allowed to form metastatic deposits (44,45). We found higher miR-23b levels in the primary tumor cells at the inoculated sites, compared with metastatic cells at distant sites, such as the adrenal glands, bones and lungs (Figure 7A). Subsequently, we measured miR-23b levels in a small number of paired primary and metastatic BC patient samples. Accordingly, miR-23b levels were higher in primary tumors, compared with their corresponding lymph-node metastases (Figure 7B). We next validated this in a larger patient cohort (n = 132; 66 primary tumors and matching metastases) and again confirmed significantly higher mean expression of miR-23b in primary tumors than in the metastases (Figure 7C).
Figure 7.
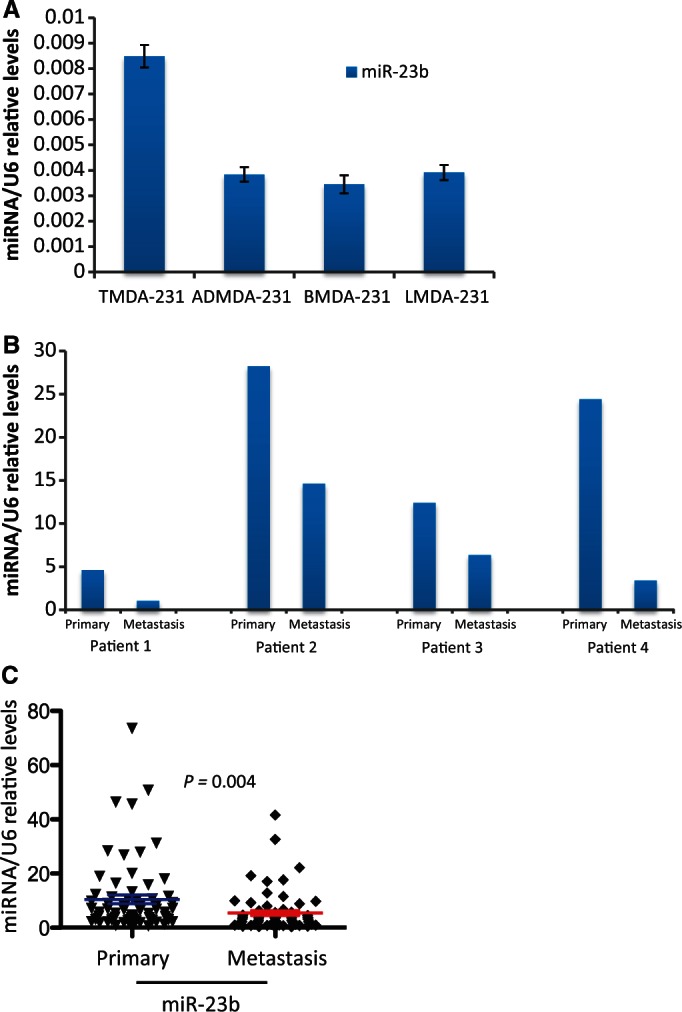
mir-23b expression is inversely correlated with metastasis. (A) RT-qPCR shows relative expression levels of miR-23b in MDA-MD-231 breast cancer cell lines that formed the primary tumor at the inoculation site (TMD-231) and cells that metastasized to the adrenal gland (AMD-231), bone (BMD-231) and lung (LMD-231) from the primary tumor in the fat pad of nude mice. Data are mean of triplicate samples in one experiment ± s.e.m. (B) miR-23b relative levels in four pairs of human primary breast cancer samples and their associated lymph-node metastasis. Data are relative levels of miR-23b in the primary tumor and matched lymph-node metastasis. (C) Scatter plot shows the mean miR-23b expression levels in 66 primary breast tumors and their corresponding metastatic specimens (P = 0.004, Student's t-test).
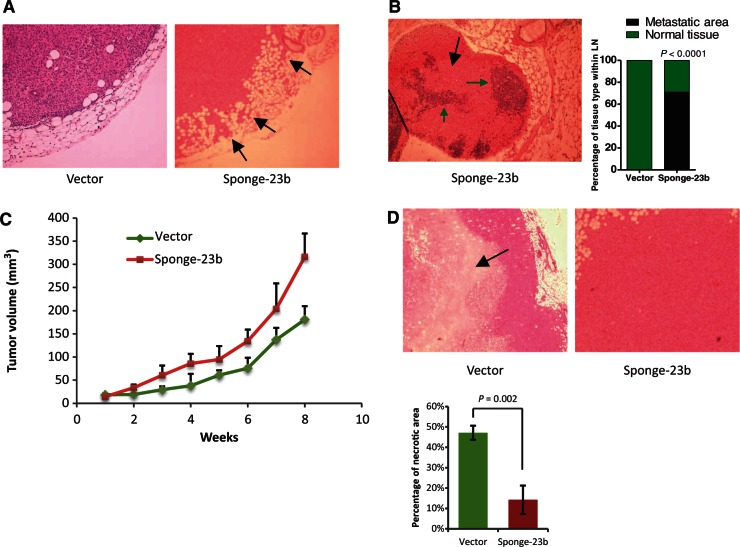
The miR-23b inhibition increases experimental metastasis and tumor growth in vivo
We further investigated the role of miR-23b in an experimental metastasis model. We injected MDA-MB-231 cells, stably transfected with miR-23b sponge or empty vector control constructs, into the mammary fat pad of BALB/c Nude mice and allowed tumor growth up to 300 mm2 (n = 7 per treatment). We observed that local tumor invasion into the surrounding fat was dramatically increased by long-term miR-23b inhibition (Figure 8A). Extensive metastatic deposits were only detected in draining lymph nodes of mice injected with stably transfected miR-23b sponge cells, indicating that reduced miR-23b activity enhanced the ability of these cells to spontaneously metastasize (Figure 8B). Our in vitro findings showed that neither expression (Supplementary Figure S2A) nor inhibition of miR-23b (Supplementary Figure S8A) significantly altered cell growth, but miR-23b inhibition surprisingly increased tumor growth in vivo (Figure 7C). Normally, xenografts derived from MDA-MB-231 injection into the mammary fat pad of immunocompromised mice have extensive central necrotic areas (46); however, we saw that miR-23b downregulation dramatically reduced tumor necrosis without significantly affecting the number of tumor microvessels (Figure 8D and Supplementary Figure S8B). This could explain the discrepancies seen in cell growth rate in vivo and in vitro following miR-23b inhibition.
Figure 8.
miR-23b inhibition increases tumor growth by reducing cell necrosis and increasing local invasion in vivo. (A) H&E stain of GFP-miR-23b-Sponge expressing- or GFP expressing-MDA-MB-231 cell primary tumors 2 months after orthotopic injection. Black arrows indicate regions of invasion into the surrounding fat. (B) Representation of a lymph node from a mouse injected with GFP-miR-23b-Sponge expressing-MDA-MB-231 cells H&E stained. The black arrow indicates metastatic areas whereas green arrows indicate residual normal tissue characterized by the presence of smaller nuclei (n = 7 per treatment). A quantification of the tissue type within the lymph nodes is shown in the graph on the right (P = 0.0001, Chi-Square test). (C) Line chart showing changes in tumor volume (mm3) over time (weeks) after injection of 1 × 106 GFP-miR-23b-Sponge expressing- or GFP expressing-MDA-MB-231 cells into the mammary fat pad of BALB/c nude mice (n = 7 per treatment). (D) H&E stain of MDA-MB-231 cell primary tumors 2 months after orthotopic injection. Arrow indicates region of necrosis (n = 7 per treatment). The graph below shows a quantification of necrosis expressed as percentage of necrotic areas formed within the primary tumors (P = 0.002, Student's t-test).
DISCUSSION
The current study focused on miR-23b because, although its involvement in some cancers is well-known, its global functions and role in BC are poorly defined. Using a pathway enriched analysis, we have suggested a role for miR-23b in cytoskeletal remodeling. We subsequently demonstrated this experimentally and validated a group of cytoskeletal genes as its direct targets.
The miR-23b over-expression increased epithelial characteristics in MCF-7 cells, but not in mesenchymal-like cells where CDH1 is not expressed. This is probably because miR-23b was unable to promote the formation of new junctions or increase CDH1 levels (Figure 1B), but rather enhanced the tension of existing ones. CDH1 is the principal mediator of adherens junction formation between cells and although intracellular it is anchored to the actin cytoskeleton through α- and β-catenin (47). Recently, Zhang et al. (20) reported that miR-23b can inhibit migration and metastasis in colorectal cancer cells, indicating a similar role may exist for it in other tumor types. They showed that miR-23b increases epithelial characteristics through the upregulation of CDH1 (20). However, we could not demonstrate CDH1 upregulation on over-expression of miR-23b, neither at the RNA nor the protein level. This indicates that miR-23b probably confers epithelial characteristics in both tissues types by different pathways. Importantly, our RNA-seq analysis showed that miR-23b over-expression increased the expression of the cell–cell adhesion molecule Nectin1, which probably contributes to the formation of adherens junctions, as its downregulation in MCF-7 cells mediates EMT (48). In adition, LMO7 is involved in the association between CDH1 and Nectin1 at the cell–cell junction level (49) and was upregulated by miR-23b expression (Supplementary Table S3).
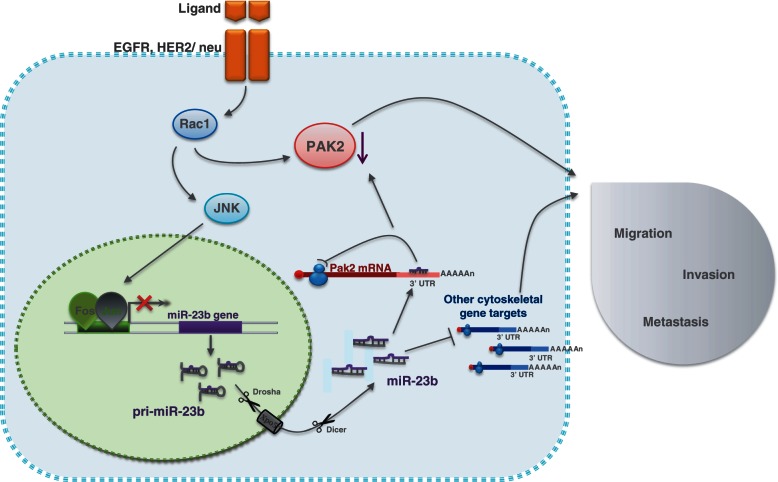
The HER family of tyrosine kinase receptors is frequently over-expressed in breast and other types of cancers. Ligands such as EGF and heregulin can bind to them and increase the metastatic potential of cancer cells (50). AP-1 is a transcription factor formed by the interaction between c-FOS and c-JUN that is activated by HER-2 signaling and increases metastasis by regulating pro-metastatic genes (51). Here, we show that miR-23b is a miRNA transcriptionally suppressed by AP-1 (Figures 5 and 9). On the other hand, EGFR stimulation by EGF decreased the levels of mature miR-23b (Supplementary Figure S5 and Figure 9). The over-expression of miR-23b consequently regulates cytoskeletal dynamics and decreases cell motility and invasion in vitro. These results are consistent with the strong down-regulation of cell motility exerted by miR-23b over-expression in MCF-7 cells, recently shown by Zhang et al.(20). This indicates that its downregulation by AP-1 is an important event that then leads to the acquisition of a more invasive phenotype for the cancer cells. Importantly, miR-23b is part of a cluster composed of miR-23b, miR-27b and miR-24-1 that are transcribed as a unique primary transcript (52). This may indicate that AP-1 mediates its effect by regulating all of the miRNA components of this cluster, but this observation requires further investigation.
Figure 9.
Schematic representation of the mechanism of action of miR-23b in BC cells.
Furthermore, the inhibition of miR-23b, by stably transfecting BC cells with a miR-23b sponge construct (i.e. loss-of-function), increases spontaneous metastasis in the lymph nodes of orthotopically injected nude mice. This demonstrated that the downregulation of miR-23b is an important event that mediates metastatic formation in vivo. Notably, miR-23b levels are reduced in MDA-MB-231 cells that have metastasized to distant organs (e.g. adrenal glands, bones and lungs; Figure 7A) after creation of orthotopic BC xenografts in nude mice. More importantly, we found miR-23b expression is significantly downregulated in lymph node metastases in a large patient cohort of paired BC samples (Figure 7B and C).
GO terms analysis of our RNA-seq data, obtained after miR-23b perturbation in BC cells, demonstrated that this miRNA regulates pathways involved in cytoskeletal remodeling, which is an essential mechanism that modifies tumor cells giving them more motile characteristics during the metastatic process. Accordingly, we showed here that miR-23b inhibits a large number of cytoskeletal genes, of which PAK2 (7,27), LIMK2 (40), ARHGEF6 (41), CFL2, PIK3R3 (42), PLAU (19) and ANXA2 (43) are regulated by direct interaction of miR-23b with their 3′UTRs (Figure 6 and Supplementary Figure S4). Moreover, in contrast to the other genes, luciferase levels of PAK2 3′UTR did not increase after the miR-23b sponge transfection. This suggests that the regulation exerted by miR-23b on PAK2, binding to the analyzed site, makes a minor contribution to the regulation of PAK2, indicating that other mechanisms, indirectly mediated by miR-23b, maybe involved. Interestingly, miR-23b over-expression dramatically reduced, and conversely its inhibition increased, lamellipodia size, which may explain changes in BC cell motility and invasiveness. Furthermore, miR-23b expression also increased FA size and the phosphorylation of MLC II, which regulates cell contractility. Importantly, these data indicate the mechanism of action of miR-23b in BC cells and demonstrate that its downregulation induces tumor growth, regulates cell–cell junctions and increases cell motility and invasion. Replacing miR-23b in BC patients may thus represent a novel therapeutic strategy to prevent tumor progression and metastatic spread.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. Supplementary Tables 1–6, Supplementary Figures 1–8, Supplementary Methods, Supplementary Movie 1–3 and Supplementary References [49–53].
FUNDING
Association for International Cancer Research, the Rosetrees Trust, The Joseph Ettedgui Charitable Foundation, the Breast Cancer Campaign and the National Insitute for Health Research (in part). The authors are indebted to the support of Michael and Lotty Hunter. Funding for open access charge: Breast Cancer Campaign.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to the advice, expertise and time of Erik Sahai and Vikash Reebye. They thank Harikrishna Nakshatri for the MDA-MB-231 cells derived from metastatic loci in nude mice. We would like to acknowledge support of the Imperial BRC and ECMC.
REFERENCES
- 1.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 2.Michiels F, Habets GG, Stam JC, van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 3.Molli PR, Li DQ, Murray BW, Rayala SK, Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–2555. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menard RE, Mattingly RR. Cell surface receptors activate p21-activated kinase 1 via multiple Ras and PI3-kinase-dependent pathways. Cell. Signal. 2003;15:1099–1109. doi: 10.1016/s0898-6568(03)00087-1. [DOI] [PubMed] [Google Scholar]
- 5.Tsakiridis T, Taha C, Grinstein S, Klip A. Insulin activates a p21-activated kinase in muscle cells via phosphatidylinositol 3-kinase. J. Biol. Chem. 1996;271:19664–19667. doi: 10.1074/jbc.271.33.19664. [DOI] [PubMed] [Google Scholar]
- 6.Bagheri-Yarmand R, Vadlamudi RK, Wang RA, Mendelsohn J, Kumar R. Vascular endothelial growth factor up-regulation via p21-activated kinase-1 signaling regulates heregulin-beta1-mediated angiogenesis. J. Biol. Chem. 2000;275:39451–39457. doi: 10.1074/jbc.M006150200. [DOI] [PubMed] [Google Scholar]
- 7.ConiglioS J, Zavarella S, Symons MH. Pak1 and Pak2 mediate tumor cell invasion through distinct signaling mechanisms. Mol. Cell. Biol. 2008;28:4162–4172. doi: 10.1128/MCB.01532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs—the micro steering wheel of tumour metastases. Nat. Rev. Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 11.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. The microRNAs miR–373 and miR-520c promote tumour invasion and metastasis. Nat. Cell. Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 12.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson L, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Patel JB, Appaiah HN, Burnett RM, Bhat-Nakshatri P, Wang G, Mehta R, Badve S, Thomson MJ, Hammond S, Steeg P, et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 2011;30:1290–1301. doi: 10.1038/onc.2010.510. [DOI] [PubMed] [Google Scholar]
- 15.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl Acad. Sci. USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper S, Marshall JF, Sahai E. Tumor cell migration in three dimensions. Methods Enzymol. 2006;406:625–643. doi: 10.1016/S0076-6879(06)06049-6. [DOI] [PubMed] [Google Scholar]
- 17.Zhao DH, Hong JJ, Guo SY, Yang RL, Yuan J, Wen CY, Zhou KY, Li CJ. Aberrant expression and function of TCF4 in the proliferation of hepatocellular carcinoma cell line BEL-7402. Cell Res. 2004;14:74–80. doi: 10.1038/sj.cr.7290205. [DOI] [PubMed] [Google Scholar]
- 18.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell. Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salvi A, Sabelli C, Moncini S, Venturin M, Arici B, Riva P, Portolani N, Giulini SM, De Petro G, Barlati S. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, Sun C, Ma M, Huang Y, Xi JJ. Genome—wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat. Commun. 2011;2:554. doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otani T, Ichii T, Aono S, Takeichi M. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 2006;175:135–146. doi: 10.1083/jcb.200605012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 27.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat. Rev. Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 28.Kreis P, Barnier JV. PAK signalling in neuronal physiology. Cell. Signal. 2009;21:384–393. doi: 10.1016/j.cellsig.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 30.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 31.Lundgren P, Johansson L, Englund C, Sellstrom A, Mattsson MO. Expression pattern of glutamate decarboxylase (GAD) in the developing cortex of the embryonic chick brain. Int. J. Dev. Neurosci. 1997;15:127–137. doi: 10.1016/s0736-5748(96)00068-8. [DOI] [PubMed] [Google Scholar]
- 32.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ndlovu N, Van Lint C, Van Wesemael K, Callebert P, Chalbos D, Haegeman G, Vanden Berghe W. Hyperactivated NF-{kappa}B and AP-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells. Mol. Cell. Biol. 2009;29:5488–5504. doi: 10.1128/MCB.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaerli P, Jaggi R. EGF-induced programmed cell death of human mammary carcinoma MDA-MB-468 cells is preceded by activation AP-1. Cell. Mol. Life Sci. 1998;54:129–138. doi: 10.1007/s000180050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 37.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 38.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 39.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson EO, Chang KH, Ghosh S, Venkatesh C, Giger K, Low PS, Shah K. LIMK2 is a crucial regulator and effector of Aurora-A-kinase-mediated malignancy. J. Cell Sci. 2012;125:1204–1216. doi: 10.1242/jcs.092304. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberger G, Gal A, Kutsche K. AlphaPIX associates with calpain 4, the small subunit of calpain, and has a dual role in integrin-mediated cell spreading. J. Biol. Chem. 2005;280:6879–6889. doi: 10.1074/jbc.M412119200. [DOI] [PubMed] [Google Scholar]
- 42.Xu L, Wen Z, Zhou Y, Liu Z, Li Q, Fei G, Luo J, Ren T. MicroRNA-7-regulated TLR9 signaling-enhanced growth and metastatic potential of human lung cancer cells by altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt pathway. Mol. Biol. Cell. 2013;24:42–55. doi: 10.1091/mbc.E12-07-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu B, Zhang F, Yu M, Zhao P, Ji W, Zhang H, Han J, Niu R. Up-regulation of Anxa2 gene promotes proliferation and invasion of breast cancer MCF-7 cells. Cell Prolif. 2012;45:189–198. doi: 10.1111/j.1365-2184.2012.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helbig G, Christopherson KW, 2nd, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE, Nakshatri H. NF-kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J. Biol. Chem. 2003;278:21631–21638. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 45.Patel JB, Appaiah HN, Burnett RM, Bhat-Nakshatri P, Wang G, Mehta R, Badve S, Thomson MJ, Hammond S, Steeg P, et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 30:1290–1301. doi: 10.1038/onc.2010.510. [DOI] [PubMed] [Google Scholar]
- 46.Amstalden van Hove ER, Blackwell TR, Klinkert I, Eijkel GB, Heeren RM, Glunde K. Multimodal mass spectrometric imaging of small molecules reveals distinct spatio-molecular signatures in differentially metastatic breast tumor models. Cancer Res. 70:9012–9021. doi: 10.1158/0008-5472.CAN-10-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukata M, Kaibuchi K. Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat. Rev. Mol. Cell Biol. 2001;2:887–897. doi: 10.1038/35103068. [DOI] [PubMed] [Google Scholar]
- 48.Vetter G, Saumet A, Moes M, Vallar L, Le Bechec A, Laurini C, Sabbah M, Arar K, Theillet C, Lecellier CH, et al. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting Nectin-1 and StarD10 messengers. Oncogene. 2010;29:4436–4448. doi: 10.1038/onc.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Ooshio T, Irie K, Morimoto K, Fukuhara A, Imai T, Takai Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells. J. Biol. Chem. 2004;279:31365–31373. doi: 10.1074/jbc.M401957200. [DOI] [PubMed] [Google Scholar]
- 50.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 51.Ozanne BW, McGarry L, Spence HJ, Johnston I, Winnie J, Meagher L, Stapleton G. Transcriptional regulation of cell invasion: AP-1 regulation of a multigenic invasion programme. Eur. J. Cancer. 2000;36:1640–1648. doi: 10.1016/s0959-8049(00)00175-1. [DOI] [PubMed] [Google Scholar]
- 52.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a∼27a∼24-2 cluster and its implication in human diseases. Mol. Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fritah A, Saucier C, Mester J, Redeuilh G, Sabbah M. p21WAF1/CIP1 selectively controls the transcriptional activity of estrogen receptor alpha. Mol. Cell. Biol. 2005;25:2419–2430. doi: 10.1128/MCB.25.6.2419-2430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.