Abstract
MicroRNAs (miRNAs) are key mediators of post-transcriptional gene regulation. The miRNA precursors are processed by the endonucleases Drosha and Dicer into a duplex, bound to an Argonaute protein and unwound into two single-stranded miRNAs. Although alternative ways to generate miRNAs have been discovered, e.g. pre-miRNA cleavage by Ago2 or cleavage products of snoRNAs or tRNAs, all known pathways converge on a double-stranded RNA duplex. Exogenous single-stranded siRNAs (ss-siRNAs) can elicit an effective RNA interference reaction; recent studies have identified chemical modifications increasing their stability and activity. Here, we provide first evidence that endogenous, unmodified, single-stranded RNA sequences are generated from single-stranded loop regions of human pre-miRNA hairpins, the so called loop-miRs. Luciferase assays and immunoprecipitation validate loop-miR activity and incorporation into RNA-induced silencing complexes. This study identifies endogenous miRNAs that are generated from single-stranded regions; hence, it provides evidence that precursor-miRNAs can give rise to three distinct endogenous miRNAs: the guide strand, the passenger strand and the loop-miR.
INTRODUCTION
Small RNAs are important post-transcriptional regulators of gene expression [reviewed in (1)]. Endogenously, microRNAs (miRNAs) are transcribed mostly by RNA polymerase as long primary microRNA transcripts, the pri-miRNA. These long pri-miRNA precursors are processed by the RNase III enzyme Drosha and its cofactor DiGeorge syndrome critical region gene 8 (DGCR8) into hairpin precursors, the pre-miRNA (2,3). Dicer and its cofactor TRBP (the human immunodeficiency virus transactivating response RNA-binding protein) cleave the pre-miRNA hairpin separating the loop from the double-stranded stem forming an miRNA duplex (4). Dicer and the miRNA duplex form a ternary complex with Argonaute (Ago) proteins (5). The duplex is unwound, giving rise to the active single-stranded miRNA in the RNA-induced silencing complex (RISC) that targets mRNAs for degradation or translational inhibition (6,7). In general, the mature miRNA with the less stable base pair at the 3′-end is selected as guide strand (8,9). However, evidence is mounting that for some miRNAs, both strands of the miRNA duplex can act as active miRNAs and contribute to physiological and pathological processes (10,11).
Recently, alternative miRNA processing pathways have also been described: mirtrons (12–14), snoRNA- and tRNA-derived miRNAs (15,16) are produced independently of the nuclear microprocessor complex of Drosha/DGCR8. Mirtrons are derived from small introns that form a pre-miRNA hairpin after splicing and debranching of the lariat. Double-stranded regions of other classes of RNA, such as snoRNAs or tRNAs, could be processed directly by Dicer or by currently unknown RNases. In turn, the biogenesis of miR-451 depends on Drosha-mediated cleavage but does not require Dicer processing (17–19). Instead, pre-miR-451 is loaded directly into Ago proteins and is further processed by the slicer function of Ago2 into an Ago2-cleaved pre-miRNA or ac-pre-miRNA (20) and then further trimmed by a currently unknown exonuclease to its final length (17–19).
Exogenously, short interfering RNA (siRNA) or short hairpin RNA (shRNA) can be delivered into the cell as an miRNA duplex or hairpin, respectively, to silence specific target genes. Despite the different biogenesis pathways of endogenous and ectopic small RNAs, they all share one common feature: in the end, they form a double-stranded short RNA duplex that gives rise mostly to one active guide strand representing the mature small RNA. This step is believed to stabilize the miRNA by protecting it from ubiquitous RNases degrading single-stranded RNAs. Nonetheless, the non-selective loading of abundant RNA oligonucleotides of the correct length has been shown in multiple systems, including recombinant Argonaute proteins (21–24). Exogenous single-stranded siRNAs have been shown to be able to elicit an RNA interference (RNAi) response ‘bypassing’ the microRNA biogenesis pathway (21,25). However, the significance of the bypass Argonaute loading pathway for endogenous small RNAs remains to be elucidated. Only recently, chemical modifications to stabilize and activate these ss-siRNAs in vivo have been described (26,27). Given this observation, we asked whether endogenous unmodified miRNAs could also be generated from single-stranded regions. Indeed, we discovered loop-miRs, which are active miRNAs of moderate abundance derived from the single-stranded loop region of selected pre-miRNA hairpins. While these have been previously emerging in deep sequencing libraries of small RNAs of vertebrates and flies (28–30), they have so far been overlooked as mere byproducts of miRNA biosynthesis. Here, we show that selected human pre-miRNAs can give rise to stable, Ago2-binding and active mature miRNAs derived from the hairpin loop, which we call loop-miRs.
MATERIALS AND METHODS
Cell lines, transfection, knockdown and treatments
A549 and HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS) and glutamine (Sigma, Taufkirchen, Germany) at 37°C in 5% CO2. HepG2 cells were cultured in Roswell Park Memorial Institute medium (RPMI) with 10% FCS and glutamine (Sigma) at 37°C in 5% CO2. For plasmid transfection, cells were seeded to reach 90% confluence at the day of transfection, and 3 µg of plasmid DNA was transfected with 10 µl of polyethylenimine (PEI) in per well in six-well plates. Transcription was inhibited by tris-buffered saline (TBS buffer) adding media containing actinomycin D (10 µg/ml in dimethylsulfoxid (DMSO); Applichem Darmstadt) or the appropriate control, and A549 cells were lysed in 1 ml of TRIzol (Invitrogen) at the indicated time points.
RNA extraction, quantitative reverse transcriptase–polymerase chain reaction, northern blotting and fractionation
Cells were lysed in 1 ml of TRIzol (Invitrogen) at the indicated time points. RNA was isolated according to the manufacturer’s recommendation with minor modifications. miRNA expression was detected by stem–loop TaqMan assays as previously described (31). miRNA northern blotting was performed as previously described (20). Nuclear and cytoplasmic fractionation was performed according to previously published protocols (32). Primer and probe sequences are listed in Supplementary Table S3. For the comparative quantification of the miR-219-loop and the miR-219-5p guide strand, 1 and 5 fmol synthetic RNA oligonucleotides were subjected to quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR), and the expression values in different cell lines were corrected for the different amplification efficiency of the respective primer pairs (Figure 4B).
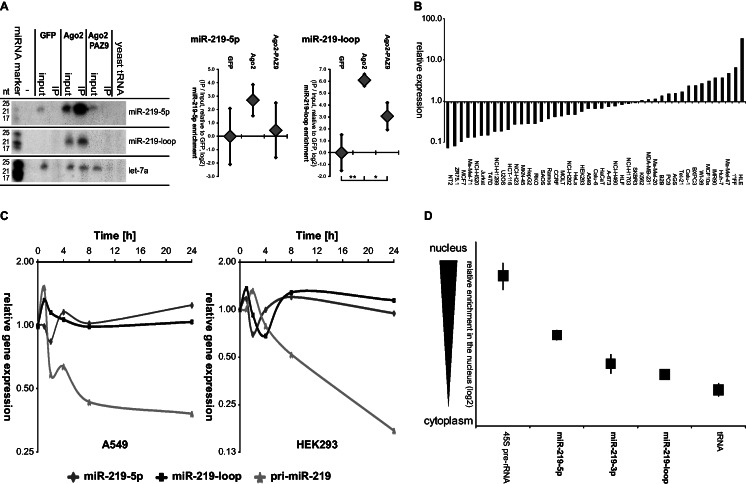
Figure 4.
miR-219-2-loop binds to Ago proteins and is stably expressed in the cytoplasm of a cell. (A) HEK293 cells were transiently co-transfected with miR-219-2 and FLAG-tagged GFP, Ago2 or the binding-defective mutant Ago2-PAZ9. Ago2-binding of the miRNAs was verified by co-immunoprecipitation (IP) using anti-FLAG agarose beads. miRNA expression was subsequently detected by northern blot analysis. Exposure times of the blots: miR-219-loop = 1 day; miR-219-5 p and let-7a = 2 days. The miRNA let-7a served as endogenous control. The input depicts 10% of the IP lysate volume. To quantify the enrichment in the IP, input and IP were subjected to qRT–PCR. Shown is the average enrichment of IP/input normalized to the negative control GFP in five experiments (±SEM). (B) Endogenous mature 5p and loop-miR expression was determined in 45 different cell lines by qRT–PCR analysis. Displayed is the ratio of miR-219-loop to miR-219-5p guide strand expression normalized for the qPCR primer efficiency. (C) miRNA expression in HEK293 and A549 cells treated with the transcription inhibitor actinomycin D was determined by qRT–PCR analysis at the indicated time points. Depicted is the mean expression of three independent experiments as compared with the appropriate DMSO-treated cells. (D) HEK293 cells were fractionated and nuclear and cytoplasmic RNA was isolated. Endogenous pri-miRNA and mature 5p, 3p and loop-miR expression of miR-219-2 was determined by qRT–PCR analysis. The 45S pre-rRNA expression served as nuclear marker, whereas tRNA was used as a cytoplasmic marker. Depicted is the mean of three independent experiments (±SEM) of the ratio between expressions in the nuclear/cytoplasmic fraction in log2 scale (Supplementary Table S5).
Expression plasmid cloning and mutagenesis
miRNA expression plasmids were cloned into the pcDNA3.1D-TOPO vector (Invitrogen) by TOPO cloning using the primers listed in Supplementary Table S3. Luciferase constructs were generated using the psiCheck2 vector (Promega, Mannheim, Germany). Cloning or sources of other expression plasmids were described previously (20). Primer sequences are listed in Supplementary Table S3.
Luciferase assays
HEK293 cells were seeded to reach 80% confluence at the day of transfection and transfected with 1.2 μg of plasmid DNA and 4.0 μl of PEI (1 mg/ml) in a 24-well plate. Each transfection contained 200 ng of psiCheck2-Firefly expression construct, including the 3′-untranslated region (3′-UTR) of interest, 500 ng of expression plasmid for the miRNA of interest and 500 ng of Ago2 expression plasmid. After 72 h, cells were lysed in 150 μl of passive lysis buffer (Promega, Mannheim). Twenty microliters of lysate was subsequently analyzed using 50 μl of LAR II substrate or Stop&Glow (1:3 dilutions) to determine Firefly and Renilla luciferase activity, respectively (Dual Luciferase Reporter Assay System; Promega). Each sample was analyzed in quadruplicates, and each transfection was carried out at least three times. Firefly activity was normalized to Renilla activity, and mean values plus standard error of mean are depicted. Reporter constructs encoding luciferase fused to a 3′-UTR with mismatches in the seed region served as negative controls. Sequences are listed in Supplementary Table S3.
Co-immunoprecipitation
For co-immunoprecipitation (Co-IP) experiments, one 15-cm plate of 80% confluent HEK293 cells per sample was transfected with appropriate plasmids for 2 days. Cells were washed twice with phosphate-buffered saline (PBS) and lysed in 350 μl of lysis buffer [150 mM KCl, 25 mM Tris–HCl (pH 7.5), 2 mM ethylenediaminetetraacetic acid (EDTA), 0.5% NP-40, 1 mM NaF, 0.5 mM dithiothreitol (DTT) and 2 mM 4-(2-aminoethyl)-benzensulfonylfluorid (AEBSF)] for 5 min on ice. Lysates were cleared by centrifugation at 17 000g for 10 min at 4°C and incubated with 20 μl of anti-FLAG M2 agarose beads (Sigma) rotating at 4°C for 2 h. All IP samples were washed three times with 1.5 ml of IP wash buffer (300 mM NaCl, 50 mM Tris (pH 7.5), 0.05% NP-40 and 5 mM MgCl2) and once with PBS. For the detection of proteins, beads were boiled in protein sample buffer. For the detection of associated RNAs, beads were lysed in 1 ml of TRIzol (33).
RESULTS
miRNA precursors can generate three distinct single-stranded mature miRNAs
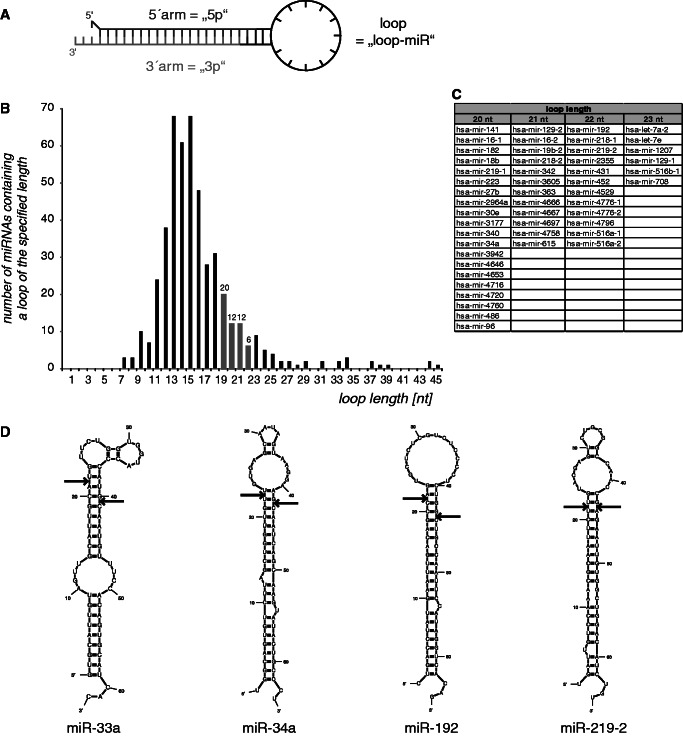
In general, one strand of an miRNA duplex is assumed to be incorporated into the RISC and is, hence, termed the guide strand. In contrast to this generally accepted model, the passenger strand of some miRNA duplexes can also be incorporated into the RISC (34–39). In some cases, both arms of the pre-miRNA hairpin are found in significant amounts (40). Furthermore, single-stranded, 5′-phosphorylated and chemically modified RNA oligonucleotides could effectively mediate RNAi (26,27). Therefore, we wanted to elucidate whether endogenous pre-miRNAs could give rise to even more active molecules than the guide and the passenger strand, as well as to determine whether endogenous miRNAs could also be generated from single-stranded RNA sequences. We hypothesized that the loop region of pre-miRNAs after Dicer cleavage—if of the right size—could also be active miRNAs. We termed these hypothetical molecules ‘loop-miRs’, reflecting their origin from the pre-miRNA loop and distinguishing them from the 5′-arm (5p) and 3′-arm (3p) of the hairpin (Figure 1A). To identify the length of human pre-miRNA loops, precursor sequences were analyzed using the miRNA sequence repository miRBase (version 18) (41). Of the 480 annotated human precursor-miRNAs with defined 5p and 3p sequences, 50 displayed a loop size 20–23 nt corresponding to the length of most endogenous mature miRNAs. The minimum loop length consisted of 8 nt, even though 4 nt would be sufficient to form a loop (Figure 1B and C). Notably, this corroborates data from artificial precursors verifying that Dicer cleavage is supported by a larger loop region (42,43). We analyzed the predicted structure of selected examples with silencing activity (Figure 3) using the publicly available software Mfold (44) (Figure 1D), confirming that the loop regions are single-stranded.
Figure 1.
Loop-miR structures. (A) Hypothetically, miRNA precursors could be processed into three single-stranded short RNA molecules: the 5′-arm (5p), the 3′-arm (3p) and the loop (loop-miR). (B) The length of the loop of 480 pre-miRNAs was determined using miRBase (release 18). (C) miRNAs with loop lengths of 20–23 nt are displayed. (D) Selected examples of loop-miR-generating pre-miRNA structures were predicted using Mfold, verifying single-stranded loop regions.
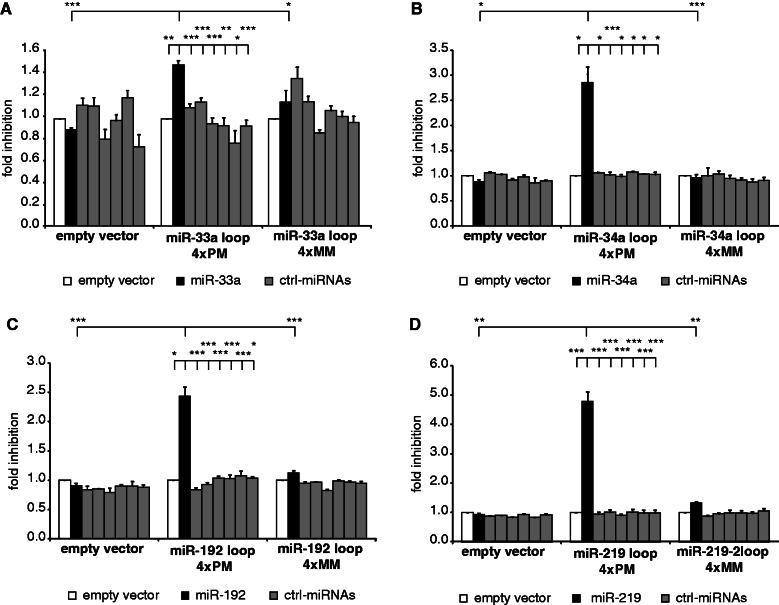
Figure 3.
Loop-miRs can actively repress luciferase activity. HEK293 cells were co-transfected with the miRNA of interest and a reporter construct encoding luciferase fused to four perfectly matched binding sites (4× PM) for the loop region of (A) miR-33a, (B) miR-34a, (C) miR-192 and (D) miR-219-2. To verify the specificity of the assay, control miRNAs with divergent seed regions were transfected, as well (gray bars). Furthermore, constructs with mismatches in the seed region of the binding sites (4× MM) served as negative control. Depicted is the mean fold inhibition (+SEM) of the respective targets compared with control/empty vector transfection. Statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001.
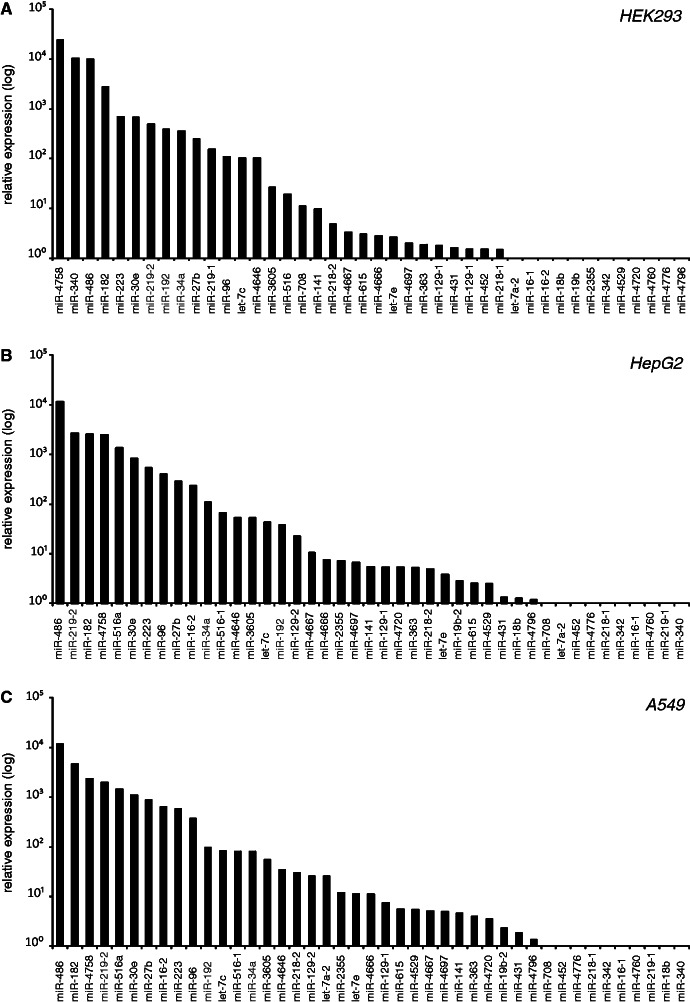
To test the expression of these potential loop-miRs (Figure 1C), we performed qRT–PCR analysis in three different human cell lines: immortalized embryonic kidney cells HEK293 (Figure 2A), liver cancer line HepG2 (Figure 2B) and lung cancer line A549 (Figure 2C). Indeed, we found detectable expression of many of the predicted loop-miRs representing a novel class of pre-miRNA processing products.
Figure 2.
Expression analysis of potential loop-miR candidates. Endogenous mature loop-miR expression was determined by qRT–PCR analysis in (A) HEK293, (B) HepG2 and (C) A549 cells. Depicted is the mean relative expression (log-scale) of two independent experiments per cell line. miR-1207, miR-3177, miR-3942 and miR-4716 were excluded from this study because of unspecific detection by the qRT–PCR. The loop-miRs miR-34a, miR-192 and miR-219-2 were selected for further investigation.
Loop-miRs are active molecules
After validating loop-miR expression, we asked whether loop-miRs could act as active miRNAs—serving in this sense as a ‘guide strand’ and targeting complementary mRNA transcripts. We selected three candidate loops, which were well-expressed, as well as effectively overexpressed on transfection (miR-34a-loop, miR-192-loop and miR-219-2-loop). Furthermore, a genome-wide deep sequencing analysis validated the existence of several loop-miRs (Supplementary Table S1) and additionally identified novel loop-miRs that had not been included in the in silico analysis (Figure 1B) because of shorter loop regions or non-determined terminal nucleotides of the passenger strand. In total, loops of 19–24 nt in length were sequenced for 20 miRNAs in HEK293, HepG2 or A549 cells (Supplementary Table S1). From this analysis, we included the miR-33 a-loop into the activity analysis because of its high expression. Notably, the read count for miR-33a-loop was higher than for the miR-33a-5p and miR-33a-3p guide and passenger strands. Of the 20 loops (Supplementary Table S1), four were more abundant than the respective guide and passenger strand, whereas additional nine were more abundant than the passenger strand. If the loop-miRs were just by-products of miRNA biogenesis, one would expect them to be of similar or lower abundance than passenger strands, as they are hypothetically released and, hence, exposed to nucleases before the passenger strand, which is forming a duplex with the guide strand. Thus, these data suggest that these loops are likely not only processing ‘noise’. Although these loops are of limited abundance, they might still be of biological relevance, especially considering the fact that most miRNAs derived from double-stranded regions with known functions are found in vanishingly small read counts: 94% of the 5p and 3p miRNAs from miRBase-annotated pre-miRNA are found with <1000 reads in these libraries.
To analyze loop-miR activity, we created reporter constructs encoding luciferase fused to a 3′-UTR with four binding sites fully complementary to the respective loop-miR of interest. Reporter constructs encoding luciferase fused to a 3′-UTR containing binding sites with two mismatches in the seed region served as negative controls (Figure 3). Loop-miRs were overexpressed, and their activity was determined using dual luciferase assays compared with a set of non-related control miRNAs. Loop-miR expression specifically and significantly inhibited expression of targets with perfectly matched binding sites for the loop of miR-33a, miR-34a, miR-192 and miR-219 (black bars), whereas mismatched targets were not significantly affected and the overexpression of control miRNAs with divergent seed regions (gray bars) did not impact the luciferase activity (Figure 3A–D) (t-test; all P-values can be found in Supplementary Table S4). The loop-miRs of miR-182 and miR-486 did not possess a detectable activity in luciferase assays (data not shown). As miR-219-loop showed the strongest activity, further studies were focused on this specific loop-miR.
MiR-219-2-loop binds to Argonaute-2 and is stably expressed in the cytoplasm
Active miRNA guide strands bind to Ago proteins and form a RISC to perform target silencing. To test whether loop-miRs act in the same manner, we analyzed whether miR-219-loop bound to the major human Ago protein, Ago2. After immunoprecipitation of epitope-tagged human Ago2, miR-219 was determined by northern blotting (Figure 4A, Supplementary Figure S1). The green fluorescent protein (GFP) and the binding-defective Ago2-PAZ9 mutant served as negative controls. This result was validated using qRT–PCR (Figure 4A). Both, miR-219-5p and miR-219-loop, displayed a significant enrichment, verifying Ago2 binding and thus incorporation into the RISC of both, the miR-219 guide and loop strand miRNAs.
To determine the generality of miR-219-loop expression, endogenous guide strand and loop-miR expression was determined by qRT–PCR in a broad panel of 45 human cell lines. Notably, the loop-miR expression was comparable with the expression of the miR-219-5p guide strand (Figure 4B).
To compare the expression of the loop-miRs to other miRNA species of known physiological or pathological importance, the expression of the three most active loop-miRs, as well as several mirtrons and miRNA guide and passenger strands, was determined (Supplementary Table S2). The expression of the three loop-miRs tested was even higher than the expression of the mirtrons (Supplementary Figure S2A) and other guide and passenger strands with already known and published functions (Supplementary Figure S2B), indicating that their expression level is in a physiologically meaningful range.
miRNAs have been described to be highly stable molecules with long half-lives (2,45,46). To determine the stability of miR-219-loop and to exclude that loops of pre-miRNAs are degraded rapidly, cells were treated with the RNA polymerase inhibitor actinomycin D, and miRNA expression was analyzed at different time points after transcriptional inhibition. The pri-miR-219 served as an unstable marker to verify proper transcriptional inhibition. The miR-219-2 guide strand was stable for >24 h (Figure 4C), validating previously published results (46). Remarkably, the loop was not degraded either within the first 24 h of transcriptional inhibition, indicating a long half-life and stability.
miRNA precursor hairpins are generated by Drosha cleavage and exported into the cytoplasm. Subsequent Dicer cleavage and RISC formation are thought to take place in the cytoplasm where the most mature miRNAs also act, whereas only few of them are re-imported into the nucleus [for review: (1)]. Based on this analogy to active guide and passenger strands, we hypothesized that mature loop-miRs should be generated and localized in the cytoplasm. To validate this hypothesis, HEK293 cells were biochemically fractionated into the nuclear and cytoplasmic compartments, total RNA was isolated and the expression of a primary miRNA and the 5p, 3p and loop-miR was detected by qRT–PCR analysis and compared with the nuclear marker 45S pre-rRNA and the cytoplasmic marker tRNA. As expected, endogenous miR-219-5p and -3p were enriched in the cytoplasm. Interestingly, the miR-219-loop (Figure 4D and Supplementary Table S5), as well as the other loop-miRs, was enriched in the cytoplasmic fraction (Supplementary Figure S3).
Taken together, this study provides evidence that, on Dicer cleavage, pre-miRNAs can give rise to three distinct endogenous single-stranded active mature miRNAs: not only are the guide strands and passenger strands derived from the miRNA duplex, but a novel class of miRNAs also derive from the pre-miRNA single-stranded loop region, referred to as ‘loop-miRs’. Loop-miRs are moderately abundant molecules that are localized in the cytoplasm, incorporated into Ago2-containing RISCs and function as active miRNAs. Importantly, this is the first class of active endogenous unmodified miRNAs that are generated from a single-stranded region.
DISCUSSION
This study identified a new pathway generating mature miRNAs from single-stranded RNA regions. These loop-miRs are endogenous small RNAs—in contrast to the previously described exogenous and chemically modified ss-siRNAs (21,25–27). We discovered that—with different frequencies—multiple mature and active miRNAs with distinct seed regions and target specificities can be generated from one precursor hairpin: guide, passenger and loop strands. Future studies will unravel the physiological or pathological role, as well as the endogenous target genes, of this newly discovered class of endogenous loop-miRs.
Loop-derived RNAs have been found in deep sequencing studies before, but they have only been used to validate the origin of guide and passenger strand miRNAs from Dicer-mediated pre-miRNA hairpin processing and were overlooked during the analysis (e.g. in miRDeep and miRDeep2). Our results suggest for the first time that these could be active, stable, cytoplasmic miRNAs that in rare cases are even of similar abundance as the guide strand. Notably, loop-derived, but uncharacterized, RNAs have also been found in other species, such as Drosophila (28–30); raising the interesting hypothesis that loop-miR activity could also be conserved throughout evolution. Nonetheless, each of these RNA fragments will need to be individually studied for its functional activity.
These data also point toward a RISC loading mechanism in human cells that seems to be uncoupled from miRNA duplex unwinding. One important question in this field is how the endonuclease-deficient Ago proteins could get loaded with microRNA duplices with perfectly matched stems, although they cannot cleave the miRNA passenger strand during the unwinding process. Previous studies on ss-siRNAs (21,25–27), the fact that miRNAs are in huge excess over Ago proteins (47), as well as our data regarding loop-miRs, could indicate that Ago loading or exchange is possible at the single-stranded stage independent of small RNA duplex binding and unwinding.
These findings also have immediate implications for the design of shRNAs for RNA interference: the loop region of these shRNAs should be designed to avoid potentially giving rise to a mature and active miRNA that might cause off-target effects. Hence, the loop regions of shRNAs should be <18 nt or >25 nt, the range of currently annotated miRNAs. Notably, the miR-30e hairpin indeed gives rise to a loop-miR of 20 nt in length that is detectable by qRT–PCR, whereas the miR-30 family has been used as an shRNA-backbone with a 19-nt loop region (48,49).
Finally, based on our data one could speculate that other nucleases than Dicer should be able to generate small active RNAs from single-stranded regions. Indeed, these can be found in small RNA deep sequencing studies but are usually discarded when filtering the reads for originating from a hairpin structure (40,50). Based on our study, these short RNAs should be scrutinized for their potential functional significance based on their abundance, stability and ability to bind to Ago2. Hence, the discovery of loop-miRs generated from single-stranded RNA regions and nevertheless loaded into RISCs might open an entirely new source of small gene regulatory RNAs.
NOTE ADDED IN PROOF
While our manuscript was in the review process, another manuscript came to our attention describing the discovery of loop-miRs in mammals and flies from the laboratory of Eric Lai (51).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–3, Supplementary Methods and Supplementary References [52–68].
FUNDING
Helmholtz Society [VH-NG-504]; DKFZ-MOST-Cooperation Program [Ca-135]; Marie Curie Program of the European Union [239308]; German Research Foundation [TRR77 TP B03]. Funding for open access charge: Research Group ‘Molecular RNA Biology & Cancer’, Institute of Pathology, University Hospital Heidelberg—Institutional Funding.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Agnes Hotz-Wagenblatt and the DKFZ Genomics & Proteomics Core Facility for deep sequencing and data analysis and Nina Schuermann and all members of the Diederichs laboratory for helpful discussions. The authors are grateful to Mimi Chong for editing the manuscript.
REFERENCES
- 1.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell. Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 2.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 3.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 5.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 10.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 21.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 22.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 23.Chorn G, Klein-McDowell M, Zhao L, Saunders MA, Flanagan WM, Willingham AT, Lim LP. Single-stranded microRNA mimics. RNA. 2012;18:1796–1804. doi: 10.1261/rna.031278.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 26.Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, Crooke ST, Prakash TP, Corey DR. Single-stranded RNAs Use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, Seth PP, et al. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grund SE, Polycarpou-Schwarz M, Luo C, Eichmuller SB, Diederichs S. Rare drosha splice variants are deficient in MicroRNA processing but do not affect general MicroRNA expression in cancer cells. Neoplasia. 2012;14:238–248. doi: 10.1593/neo.111586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol. Immunol. 2011;48:1084–1090. doi: 10.1016/j.molimm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J. Mol. Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pass HI, Goparaju C, Ivanov S, Donington J, Carbone M, Hoshen M, Cohen D, Chajut A, Rosenwald S, Dan H, et al. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer Res. 2010;70:1916–1924. doi: 10.1158/0008-5472.CAN-09-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–959. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 40.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 42.Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat. Struct. Mol. Biol. 2011;18:1153–1158. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- 43.Feng Y, Zhang X, Graves P, Zeng Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA. 2012;18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 47.Janas MM, Wang B, Harris AS, Aguiar M, Shaffer JM, Subrahmanyam YV, Behlke MA, Wucherpfennig KW, Gygi SP, Gagnon E, et al. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA. 2012;18:2041–2055. doi: 10.1261/rna.035675.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paddison PJ, Cleary M, Silva JM, Chang K, Sheth N, Sachidanandam R, Hannon GJ. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 49.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 50.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 51.Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev. 2013;27:778–792. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11, Unit 11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YJ, Bae SW, Yu SS, Bae YC, Jung JS. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J. Bone. Miner Res. 2009;24:816–825. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 56.Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 57.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 58.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong P, Kaneuchi M, Watari H, Hamada J, Sudo S, Ju J, Sakuragi N. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol. Cancer. 2011;10:99. doi: 10.1186/1476-4598-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L, Li Y, Cheng M, Huang D, Zheng J, Liu B, Ling X, Li Q, Zhang X, Ji W, et al. A functional polymorphism at microRNA-629-binding site in the 3′-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis. 2012;33:338–347. doi: 10.1093/carcin/bgr272. [DOI] [PubMed] [Google Scholar]
- 61.Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, Wood CG, Wu X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- 62.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl Acad. Sci. USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J. Hepatol. 2009;50:453–460. doi: 10.1016/j.jhep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 65.Salvi A, Sabelli C, Moncini S, Venturin M, Arici B, Riva P, Portolani N, Giulini SM, De Petro G, Barlati S. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 66.Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patterson EE, Holloway AK, Weng J, Fojo T, Kebebew E. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer. 2011;117:1630–1639. doi: 10.1002/cncr.25724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuchenbauer F, Mah SM, Heuser M, McPherson A, Ruschmann J, Rouhi A, Berg T, Bullinger L, Argiropoulos B, Morin RD, et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood. 2011;118:3350–3358. doi: 10.1182/blood-2010-10-312454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.