Abstract
An important requirement for achieving many goals of synthetic biology is the availability of a large repertoire of reprogrammable genetic switches and appropriate transmitter molecules. In addition to engineering genetic switches, the interconnection of individual switches becomes increasingly important for the construction of more complex genetic networks. In particular, RNA-based switches of gene expression have become a powerful tool to post-transcriptionally program genetic circuits. RNAs used for regulatory purposes have the advantage to transmit, sense, process and execute information. We have recently used the hammerhead ribozyme to control translation initiation in a small molecule-dependent fashion. In addition, riboregulators have been constructed in which a small RNA acts as transmitter molecule to control translation of a target mRNA. In this study, we combine both concepts and redesign the hammerhead ribozyme to sense small trans-acting RNAs (taRNAs) as input molecules resulting in repression of translation initiation in Escherichia coli. Importantly, our ribozyme-based expression platform is compatible with previously reported artificial taRNAs, which were reported to act as inducers of gene expression. In addition, we provide several insights into key requirements of riboregulatory systems, including the influences of varying transcriptional induction of the taRNA and mRNA transcripts, 5′-processing of taRNAs, as well as altering the secondary structure of the taRNA. In conclusion, we introduce an RNA-responsive ribozyme-based expression system to the field of artificial riboregulators that can serve as reprogrammable platform for engineering higher-order genetic circuits.
INTRODUCTION
Cellular systems have acquired versatile RNA-dependent regulatory platforms based on several different principles such as the RNA interference pathway mediating gene silencing in most eukaryotes (1), the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) machinery providing a form of acquired immunity in prokaryotes (2) and diverse classes of riboswitches that generally feature ligand-dependent, RNA-based regulation of gene expression (3). Riboswitches are mainly found within the 5′-untranslated region (UTR) of bacterial mRNAs, hence constituting intramolecular (or in cis) regulation. Ligand-binding to the aptamer domain of a riboswitch typically controls the architecture of an expression platform affecting transcription, translation or splicing of the message (3). An exceptional activity is catalyzed by the glmS riboswitch, which acts an allosterically controlled ribozyme promoting self-cleavage of the glmS mRNA in presence of glucosamine-6-phosphate (4).
The often modular as well as hierarchical architectures of functional RNAs (5) have inspired synthetic biologists to construct artificial genetic switches via the re-assembly of certain RNA domains for a wide range of tasks. For example, RNA-based devices for controlling transcription (6,7), translation (8–10), splicing (11) and primary microRNA (pri-miRNA) processing (12) have been realized. The use of aptamers has been proven powerful for the construction of ligand-dependent RNA switches (13–15). Importantly, aptamers can be evolved to sense virtually any kind of ligand including ions, small molecules and proteins (16). An improved concept for the construction of artificial riboswitches is the use of the full-length hammerhead ribozyme (HHR) as expression platform. In bacteria, a ligand-dependent self-cleaving ribozyme is found as a riboswitch in the glmS mRNA (4). On the other hand, HHRs are found in many species (17,18). Although their role in the life cycle of plant viroids seems obvious, the function of cleavage-competent versions in higher organism is unknown (19). Apart from their occurrence in nature, HHRs have been used as expression platforms to engineer artificial riboswitches (20). In particular, the Schistosoma mansoni HHR, which is extensively characterized in vitro, has been used to control gene expression in cellular systems including Escherichia coli (8), Saccharomyces cerevisiae (21) and mammalian cells (22). Ligand-dependent ribozymes, also called aptazymes, display an increased modularity, as they can be transferred between different RNA classes. In addition, their switching property is often maintained even when transferred from one organism to another one (10,20). Hence, aptazymes are powerful gene control elements whose toolbox is constantly increasing. The reprogramming of ligand selectivity yielded switches that are triggered by theophylline, thiamine pyrophosphate, p-aminophenylalanine, tetracycline and guanine (8,21,23–25). Besides controlling translation initiation through the insertion of the aptazyme within the UTR of an mRNA (8,22,26), allosteric HHRs can also be attached to other functional RNA classes to control tRNA (27), 16S ribosomal subunit (28) and pri-miRNA function (12). Our group recently demonstrated that the combination of aptazyme-based genetic switches for mRNAs and tRNAs enables the construction of Boolean logic operators inside cells (29).
As described, ligand-dependent ribozymes provide a powerful tool for the construction of post-transcriptionally controlled genetic networks in prokaryotic and eukaryotic organisms (24,29,30). However, for facile and broad interconnectivity, it would be beneficial to have RNA switches that sense universally producible signals. RNA fulfills this requirement best among all biomolecules: i is synthesized by all host cells via transcription and its specific binding to another RNA is governed by basic base-pairing rules. These characteristics confer RNA the ubiquitous advantage to act as reprogrammable transmitter molecule (31). Natural systems have evolved intricate networks and wide-spread mechanisms such as RNAi and CRISPR, which rest upon non-coding RNAs as trans-regulatory agents (32,33). In addition, many bacterial small RNAs (sRNA) operate in trans to modulate gene expression. Often, hybridization of the sRNA to a cis-encoded response element within a target mRNA results in the formation of an RNA–RNA complex, either blocking translation or influencing the stability of the message. In many cases, the base-pairing reaction is facilitated by structurally defined RNA motifs and catalyzed by protein cofactors.
As synthetic biology approaches the stage of programming increasingly complex biological functions (34–37), a huge repertoire of easily reprogrammable genetic switches and transmitter molecules will become indispensable. Although trans-acting RNAs (taRNAs) are the ideal choice as trigger of artificial genetic switches at the post-transcriptional level and as connector of individual switches, so far only few synthetic systems using these advantages have been reported. The sRNAs have been successfully applied as transmitter molecules within artificial genetic circuits (36,38), allowing for the construction of higher-order genetic networks including a cellular counter (36) and a genetic switchboard (35). However, structure–function relationships of sRNAs are only poorly understood, which makes the rational design of artificial sRNAs difficult (39). Collins and coworkers reported artificial trans-acting sRNAs able to de-repress translation initiation of a cis-repressed mRNA in E. coli (40). In this design the ribosomal binding site (RBS) of a reporter gene is sequestered by an antisense helix. Binding of an artificial sRNA melts the inhibitory helix within the cis-repressed mRNA and leads to initiation of translation.
Although RNA-responsive HHRs have been studied in vitro (41), they have never been successfully applied within living organisms. Breaker and Penchovsky (42) rationally engineered allosteric ribozymes, which are controlled by hybridization to added oligonucleotides. Multiple input switches performing Boolean logic computation were constructed, as well as cascaded switches in which the cleavage product of the first ribozyme served as molecular transmitter and triggered a cascaded ribozyme switch. The concept beautifully depicts the power of rationally designed RNA that serve as transmitter and receptor molecules for performing user-defined tasks. In the present study, we engineered HHRs to sense small taRNAs in E. coli. Our riboregulatory system is characterized by a HHR-based artificial expression platform in which the catalytic activity of the ribozyme controls translation initiation of a reporter gene.
MATERIALS AND METHODS
Plasmid construction
All plasmids are based on the pGDR11 plasmid (43). Standard molecular cloning procedures were performed as described in literature (44). Phusion Hot Start 2 Polymerase (NEB) was used for polymerase chain reaction (PCR) amplification of DNA, endonucleases were purchased from NEB, and all ligations were performed with the Quick ligation kit (NEB). Pac1 and Not1 restriction sites were introduced into the pGDR11 backbone by PCR using the primers 5′-CTCTTCTTAATTAAGGGTGCGCATGATCTAGAGC-3′ and 5′-CTCTTCGCGGCCGCCCAACGCTGCCCGAAATTCC-3′. A cassette containing the gene encoding araC and the PBad promoter was amplified from the pBAD18a vector (45) using the primers 5′-CTCTTCTTAATTAAATGAGCGGATACATATTTGAATGTATTTAG-3′ and 5′-CTCTTCGCGGCCGCCGGTGATGTCGGCGATATAGGC-3′ and introduced via the Pac 1 and Not1 restriction sites into the pGDR11 backbone. The enhanced green fluorescent protein (eGFP) gene controlled by a taRNA-responsive (TR)-HHR was constructed under control of the PBad promoter using Spe1 and Sac2 restriction sites, and the taRNA was inserted under control of the Isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter via Bgl2 and Avr2 restriction sites. Successful molecular cloning was verified by DNA sequencing (GATC). All sequences are given in the supplementary material.
E. coli strain and growth conditions
All experiments were conducted with the E. coli Top10 (Invitrogen) strain (F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ−). All plasmids were introduced by electroporation. Bacterial cultures were grown aerobically in Luria-Bertani (LB) medium supplemented with 100 µg/ml carbenicillin.
Gene expression and quantification
For eGFP expression measurement, single colonies were first outgrown to stationary phase in LB medium. The next day a 1% bacterial suspension was regrown for 2–3 h at 200 rpm and 37°C in an Infors HT Ecotron shaker using Erlenmeyer flasks. Cultures were diluted to an OD600 of 0.1 and induced with transcriptional inducers. In all experiments (except Figure 3E–G and Supplementary Figures S1 and S2), fixed concentrations of the inducers (1 mM IPTG and 20 µM arabinose) were used. In the other experiments, IPTG and arabinose concentrations, as indicated in the figure or written in the figure legend, were added to the culture medium. Cultures were incubated with shaking at 200 rpm and 37°C in 50 ml centrifuge tubes for 18 h. In all, 100 µl of each culture were transferred into a 96-well microtiter plate and OD600, and the fluorescence of the expressed eGFP was measured with a Tecan Infinite® M200 plate reader (excitation wavelength: 488 nm, emission wavelength 535 nm). Fluorescence values were corrected by dividing with the OD600 values. An equally treated culture, which did not express any eGFP, was used for subtraction of background fluorescence. All experiments were performed in triplicates, and error bars represent standard deviations. In Figure 3E–G, eGFP expression values were normalized to an equally treated culture expressing the TR-HHR-eGFP fusion transcript and taRNA V1-M3.
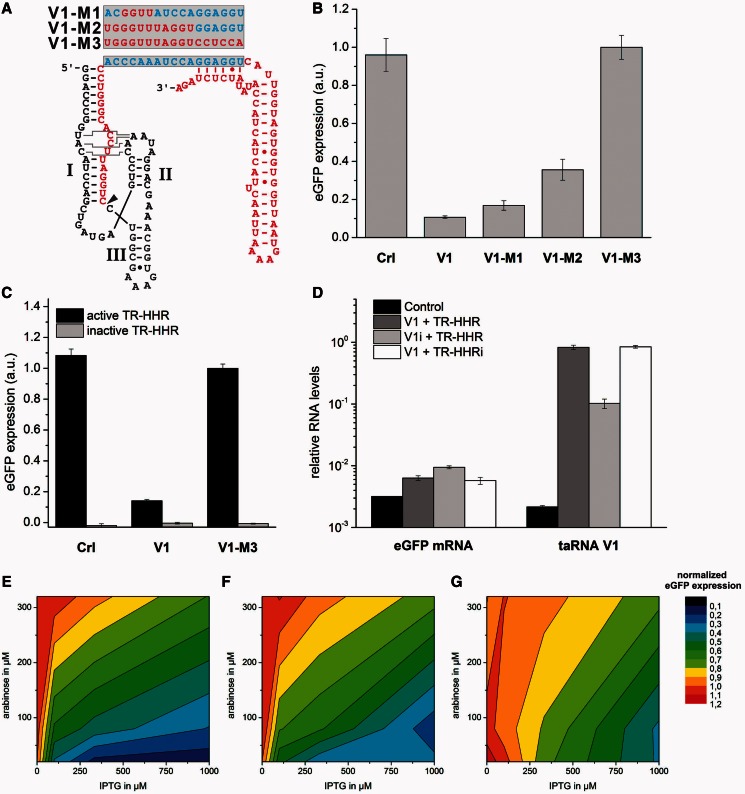
Figure 3.
The sRNA-mediated repression of translation depends on the formation of the RNA–RNA hybrid structure. (A) Schematic illustration of artificial taRNA variants with reduced number of complementary nucleobases to the taRNA responsive element was constructed. The constructs V1-M1 and V1-M2 display partial hybridization capability (blue nucleobases), whereas the construct V1-M3 is non-complementary at all. The site of 5′-processing is marked by an arrow head. All taRNA constructs were engineered under control of an IPTG-inducible promoter. (B) The influence of the taRNAs shown in (A) on the TR-HHR activity was examined. Bacterial cultures of the E. coli Top10 strain were induced with 20 µM arabinose and 1 mM IPTG. Transformants were cultivated in LB medium at 37°C, and eGFP expression of outgrown bacterial cultures was measured. Error bars represent the standard deviation of experiments performed in triplicates. (C) The inactivation of the TR-HHR results in non-detectable eGFP expression levels and was not influenced by the co-expressed taRNAs Crl, V1 and V1-M3. The assay was performed as described earlier in the text. (D) Analysis of eGFP mRNA and taRNA levels by semi-quantitative RT-PCR. Bacterial cultures were induced with 20 µM arabinose and 1 mM IPTG. Results indicate a high excess of taRNA over eGFP mRNA transcripts and an accumulation of taRNA V1 through the 5′-processing reaction of the HHR. (E–G) Heat maps of observed repression of translation by the constructs (E) V1, (F) V1-M1 and (G) V1-M2 as function of transcriptional induction of the reporter construct and the taRNAs. Data were normalized to the construct V1-M3. Transcription of the reporter gene was induced by the addition of 20, 80 and 320 µM arabinose, and transcription of the taRNA variants was induced by the addition of 0, 100, 333 and 1000 µM IPTG. The translational repressed state is indicated in blue, whereas the unrepressed state is shown red. Bacterial cultures of the E. coli Top10 strain were induced with arabinose and IPTG as indicated. Transformants were cultivated in LB medium at 37°C, and eGFP expression of outgrown bacterial cultures was determined.
Quantification of RNA levels
Bacterial cultures were induced with 20 µM arabinose and 1 mM IPTG as described earlier in the text and grown for 2 h. Bacterial cultures carried plasmids as described in Figure 3D including a control with the empty pGDR11 vector. Total RNA was extracted using RNAzol RT® (Sigma) and an additional purification with the Direct-zol™ RNA MiniPrep Kit (Zymo Research). The reverse transcription reaction was performed with 250 ng total RNA and random hexamer priming using the Verso cDNA kit (Thermo Scientific) in a total volume of 10 µl for 60 min at 55°C. Real-time PCR analysis was performed on a Light-Cycler II 480 instrument (Roche). Each reaction mixture was prepared using Phusion Hot Start Polymerase II (NEB) for amplification and SYBR green (Sigma) for detection in a total volume of 10 µl. The following primers were used for the amplification reaction of the taRNA (F: 5′-CTGGATTCCACGGGTACC-3′, R: 5′-CCAGGCGTTTAAGCCTAGGAAG-3′), the eGFP mRNA (F: 5′-GAAGGAGATATACCATGGGCCATCA-3′, R: 5′-GCTGAACTTGTGGCCGTTTAC-3′) and ssrA (46) (reference gene; F: 5′-ACGGGGATCAAGAGAGGTCAAAC-3′, R: 5′-CGGACGGACACGCCACTAAC-3′). The threshold cycles were determined using the ligthcycler 480 software SP4. RNA levels were calculated assuming a static PCR efficiency of two for each primer pair and determined relative to the expression of the genomically encoded ssrA gene.
RESULTS AND DISCUSSION
Rational design of TR-HHRs
Within this study, we provide a framework in which HHRs sense small taRNAs as inputs. Previous studies successfully demonstrated that ribozyme activity can be controlled with short oligonucleotides in vitro (41,42,47). However, these studies were based on a minimal variant of the HHR, which displays drastically reduced catalytic activity in vivo. Our riboregulatory system is based on the full-length HHR of S. mansoni (48,49), which exhibits enhanced activity at physiological magnesium concentrations (50,51). The full-length HHR of S. mansoni can be used to control bacterial translation initiation by the sequestration of the ribosomal binding site within an extended stem I of the HHR (8,25). Only cleavage of the HHR liberates the ribosomal binding site resulting in binding of the small ribosomal subunit and robust expression of the downstream coding reporter gene. In contrast, the inactivation of the HHR by an A to G mutation within the catalytic core results in non-detectable reporter gene expression owing to a sequestered ribosomal binding site. The attachment of an aptamer sensor domain to stem 3 of the HHR enables small molecule-dependent control of gene expression. The schematic outline of the riboregulatory system reported in this study is shown in Figure 1. An HHR serving as the expression platform controls translation initiation of an eGFP reporter mRNA as previously described. An additional domain containing a TR element is attached to stem 3. In the absence of the taRNA input, the HHR folds into a catalytically active conformation, which results in liberation of the RBS and translation initiation. However, in case of the expression of a small taRNA, an RNA–RNA interaction between the target HHR and the taRNA should form. In the initial recognition step, the single-stranded seed region of the taRNA pairs with the loop region of stem 3 of the HHR. Subsequently, stem 3 is melted, which inhibits HHR catalysis, hence repressing translation initiation of the reporter gene.
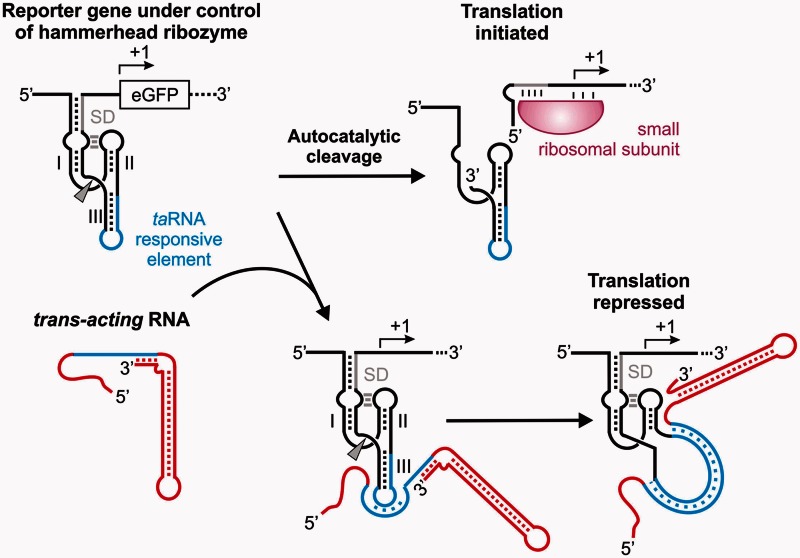
Figure 1.
Mechanism of trans-acting sRNA controlling a hammerhead ribozyme. Expression of an eGFP reporter gene is post-transcriptionally controlled by a TR-HHR. An extended stem 1 of the TR-HHR sequesters the ribosomal binding site. In addition, stem 3 harbors a TR element (blue), which in the absence of a taRNA folds into a catalytically active ribozyme conformation. Autocatalytic cleavage (marked by an arrowhead) frees the RBS resulting in binding of the small ribosomal subunit and initiation of translation. Upon expression of a trans-acting RNA (red), an RNA–RNA hybrid between the complementary sequences (blue) within the taRNA and the TR-HHR is formed. A single-stranded seed region of the taRNA hybridizes to the complementary nucleotides located in the loop region of stem 3. Full hybridization unzips stem 3 of the TR-HHR rendering the HHR catalytically inactive and repressing translation.
In our previous studies, small molecule triggers were used to control the aptazyme switches. Theophylline and thiamine were delivered by addition to the growth medium (25). In the present study, the taRNAs serving as triggers controlling the RNA switch are transcribed by the host genetic machinery. Studies on naturally occurring sRNA reported that a sufficiently high excess of the sRNA compared with target RNA concentrations is needed for efficient interactions with a complementary sequence (52,53). Therefore, we designed a reporter system, which allowed for the transcriptional titration of both the taRNA and the target mRNA levels (see Figure 2A and B). Both transcripts are transcribed from a single plasmid, with the reporter gene being under control of an arabinose-inducible promoter and the taRNA being under control of a synthetic IPTG-inducible promoter. As mentioned, our ribozyme switches were inspired by the riboregulatory system previously reported by Collins and coworkers (40). We made use of sequence domains of the RR12 variant, which most efficiently induced gene expression. We first engineered stem 3 of the HHR in a way that it contained a TR element while sustaining HHR activity in the absence of any input. The secondary structure of the TR-HHR is shown in Figure 2A. Importantly, the sequence attached to stem 3 is identical to the inhibitory helix of crR12, which includes the stem loop structure for the initial recognition of the taRNA seed region. The first kissing interaction with the taRNA is believed to be promoted by a YUNR motif, which is located within the loop of the TR element (40). YUNR motifs adopt a defined structure and are critical elements within natural antisense RNA systems (54).
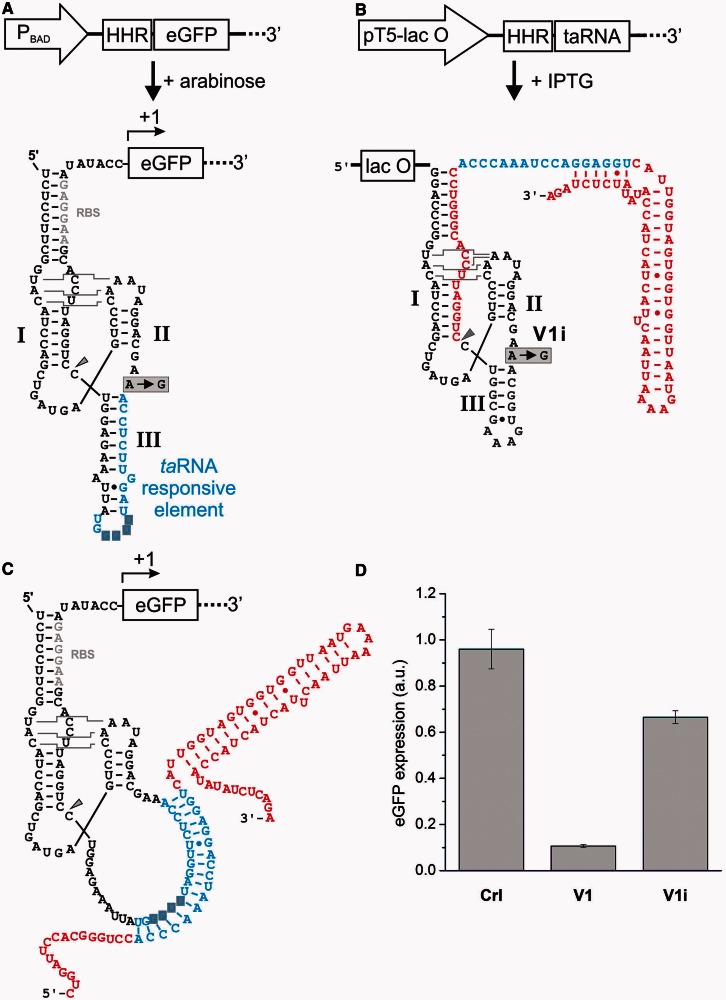
Figure 2.
Engineering of TR-HHR-eGFP fusions and artificial taRNAs. (A) The reporter gene, in which cleavage of the TR-HHR controls translation initiation of eGFP, was transcriptionally controlled by an arabinose-inducible PBAD promoter. The secondary structure of the highly modular HHR-based genetic switches is shown. The TR-HHR serves as the expression platform to which multiple functional RNA domains are attached. Control of translation initiation is obtained through sequestration of the RBS (gray nucleotides) by an extended stem 1. A domain harboring a taRNA responsive element (blue) can be attached to stem 3. The YUNR motif located in the loop of stem 3 is shaded in gray. (B) The generation of the artificial taRNA V1 is transcriptionally controlled by an IPTG-inducible promoter. Previous studies reported that extended 5′ single-stranded regions impede taRNA function. Therefore, a 5P-HHR was attached to the taRNA for 5′-processing. The autocatalytic cleavage reaction (marked by an arrow head) results in a single-stranded region made-up of 17 non-complementary nucleotides. Within the construct V1i, the 5′-processing reaction is eliminated by the A to G point-mutation within the catalytic core of the 5P-HHR. (C) The hybrid complex between the taRNA responsive element of the TR-HHR and the processed taRNA V1 is shown. (D) The influence of the taRNAs V1 and V1i in comparison with a control RNA (Crl) with no complementarity to the TR-HHR was examined. Bacterial cultures of the E. coli Top10 strain were induced with 20 µM arabinose and 1 mM IPTG. Transformants were cultivated in LB medium at 37°C, and reporter gene expression of outgrown bacterial cultures was measured. Errors bars represent the standard deviation of experiments performed in triplicates.
First, we performed control experiments in which we demonstrate that neither translation nor HHR activity was impaired upon the production of a non-complementary sRNA from an IPTG-inducible promoter (see Supplementary Figure S1). However, we observed that reporter gene expression was generally affected upon addition of IPTG to the culture medium (see Supplementary Figure S2). We reasoned that this observation should not impede our study because most experiments were performed at fixed concentrations of transcriptional inducers and normalization of the reporter gene expression to a control proofed as a convenient method to overcome this obstacle. In the initial study by Collins and coworkers, it was reported that an extended 5′ single-stranded region of the taRNA can impair its function (40). As a lac-operator site is located downstream of the transcription start site, it is part of the nascent transcript (see Figure 2B). To circumvent any inhibitory impact of the operator sequence on taRNA function, we inserted a constitutively active HHR into the taRNA, which 5′-processed the nascent transcript. To distinguish the two HHRs in the two RNAs, we termed the HHR within the taRNA 5P-HHR (for 5′-processing-HHR), and the HHR based in the target mRNA was termed TR-HHR (for TR-HHR).
To determine whether gene expression can be controlled by RNA–RNA interactions mediated by a taRNA binding to a TR-HHR, we next expressed a taRNA targeting the TR-HHR. Based on the previously reported sRNA taR12 (40), we designed a taRNA, termed V1, which contains a 5′-processing 5P-HHR (Figure 2B). The taRNA V1 contains a sequence complementary to the TR element of the TR-HHR; interaction of both RNAs should form a 17 bp helix (see Figure 2C). Bacterial cultures expressing V1 exhibited a 10-fold drop of reporter gene expression (Figure 2D). Expression of an unrelated control sRNA did not perturb gene expression, demonstrating that the taRNA effect is sequence specific. In addition to the control expression of an unrelated taRNA, we next examined whether the 5′-processing of the taRNA is necessary. For this purpose, we introduced an A to G mutation (V1i; see Figure 2B and D) within the catalytic core of the 5P-HHR. Expression of the taRNA containing such an inactivated 5P-HHR resulted in <2-fold reduced gene expression, demonstrating that 5′-processing resulted in an improved repression. We assume that mainly two factors contribute to this phenomenon. First, cleavage results in optimized conformational and dynamical properties of the seed region (55). A shorter 5′ terminus allows the seed region to move more freely facilitating exploration of the conformational space by rotating about its hinge, potentially resulting in more efficient recognition of the TR element. Second, HHR cleavage of transcripts was reported to result in prolonged lifetimes of the resulting 3′-cleavage fragment in E. coli (23). We performed semi-quantitative reverse transcription PCR for determining taRNA and eGFP mRNA levels. The results indicate a high excess of taRNA transcript levels over eGFP mRNA transcript levels and an accumulation of taRNAs, which are 5′-processed by the active 5P-HHR in comparison with an inactive mutant of the 5P-HHR (Figure 3D). Primary transcripts possess a triphosphorylated 5′-terminus from which a pyrophosphate is removed in a rate-limiting enzymatic reaction (56). Monophosphorylated transcripts are less stable because they are endonucleolytically degraded by ribonucleases. In contrast, ribozyme cleavage results in a 3′-cleavage product with a 5′-terminal hydroxyl group that is not recognized by cellular RNAses in E. coli, thus increasing the half-life of the transcript.
Mutational and structural characterization of the TR-HHR interaction
To validate that translational repression was mediated by the interaction between the predicted nucleotides, we experimentally tested the influence of mutations affecting the formation of the RNA–RNA duplex structure. We designed three variants of taRNA V1 with a consecutively reduced number of complementary nucleotides to the TR element within the TR-HHR (see Figure 3A). For the construct taRNA V1-M1, the four nucleotides targeting the YUNR motif were altered; for taRNA V1-M2, the 11 nucleotide-long single-stranded seed region was mutated, and in the case of taRNA V1-M3, all 17 complementary nucleotides were mutated. The results (see Figure 3B) demonstrate that translational repression is dependent on the number of complementary nucleotides. For taRNA V1-M1, a 6-fold repression of gene expression, and for taRNA V1-M2, a 3-fold repression was observed, whereas for the non-complementary construct taRNA V1-M3, eGFP expression levels comparable with expression of the control RNA were measured. This experiment proves that the predicted nucleotides are directly involved in the formation of the RNA duplex, thereby inhibiting TR-HHR activity. The higher the degree of complementarity, the more efficiently was the taRNA directed to the TR element. Interestingly, only six complementary nucleotides to the taRNA responsive element, as found in taRNA V1-M2, were sufficient for a 3-fold decrease in reporter gene expression relative to the non-complementary taV1-M3 construct. This observation supports the idea of a thermodynamically driven process for duplex formation, as suggested by an earlier study (40). A strong hybridization free energy was found necessary for efficient repression, but that additional structural features determine RNA–RNA interaction because an extended 5′ tail of the taRNA fully impaired taRNA function (40). A model in which additional structural features rule the efficiency of trans-interaction between two complementary RNA strands was also suggested by other studies (7,31). We included the new data in Figure 3C and Supplementary Figure S1 together with the following explanation in the manuscript: Next, we performed an important control experiment in which we inactivated the cleavage activity of the TR-HHR by the A to G point mutant in the catalytic core (TR-HHRi). The eGFP expression levels were not distinguishable from background fluorescence values (see Figure 3C), demonstrating that the ribosomal binding site is entirely sequestered by its antisense strand in the non-cleaved state. In contrast, the constitutively active TR-HHR results in reporter gene expression reaching values close to a wt eGFP expression construct, which is not controlled by a HHR (see Supplementary Figure S1 for comparison). Noteworthy, none of the investigated taRNAs (see Figure 3C) had any influence on eGFP expression when the TR-HHR regulatory module was inactivated. Hence, the trans-acting system displays a maximum regulatory range reaching from zero to full gene expression.
We next investigated the dependency of the strength of repression of gene expression from the concentrations of the taRNAs V1, V1-M1 and V1-M2 relative to the target mRNA concentration (see Figure 3E–G). For this purpose, different concentrations of arabinose and IPTG, the first inducing the target mRNA and the latter the transcription of the taRNAs, were added. Reporter gene expression was measured and normalized to a culture expressing the taRNA V1-M3 construct. Greater 3-fold translational repression was only observed for the constructs V1 and V1-M1 when arabinose concentrations were <80 µM. Interestingly, 100 µM IPTG already allowed for a 3-fold repression by taRNA V1, whereas much higher IPTG concentrations were required for efficient repression by V1-M1. In the case of V1-M2, even strong transcriptional induction of the taRNA did not allow for efficient repression. These results demonstrate that the higher the number of base-pair interactions of the taRNA to its target, the more reduced is the need for a high excess of taRNA over its target mRNA. According to theoretical and biochemical studies, key features influencing natural sRNA-mediated repression are the ratio of the transcription rates of the sRNA to its target, the stability of the two transcripts and the rate of complex formation (52,57). Most efficient repression was observed at strong transcriptional induction of the taRNA V1 and weak production of the target mRNA. Thus, a sufficiently high excess of taRNA over target mRNA is necessary for efficient inhibition of the TR-HHR activity. This is in accordance with the assumed mechanism, in which the control of TR-HHR catalytic activity with a taRNA requires a stable complex between both RNAs.
Alterations within the secondary structure of the taRNA impede riboregulatory function
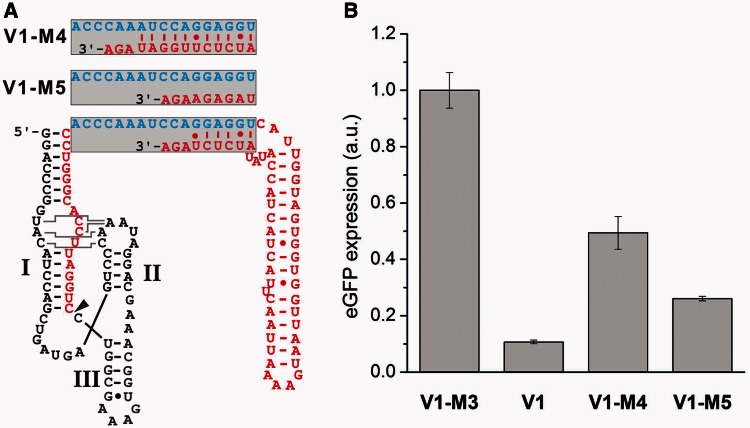
Besides the observation that the nature of the 5′ tail is important (40), there is only little knowledge available how riboregulation is dependent on or hampered by secondary structures of the taRNA. We next examined the structure–function relationship of taRNA V1 by altering the accessibility of the single-stranded seed region. We constructed two mutants based on taRNA V1 (see Figure 4A): The construct taRNA V1 is characterized by a single-stranded seed region of 11 nucleotides and six 3′ terminal nucleotides, which are sequestered by an antisense helix. In construct taRNA V1-M4, five consecutive nucleobases of the originally single-stranded seed region were engaged in a duplex, which was formed by an extended antisense strand. In contrast, taRNA V1-M5 displayed a fully single-stranded seed region made up of 17 complementary nucleotides to the taRNA responsive element. Both mutants V1-M4 and V1-M5 were not able to repress eGFP expression as efficiently as taRNA V1 (see Figure 4B). V1-M4 displayed only 2-fold and V1-M5 4-fold repression. One could speculate that the introduced structural alterations reduce the accessibility of the seed region and thereby disturb the hybridization reaction of the two RNAs. Our results suggest that taRNA V1 adopts a highly defined structure, which is required for efficient duplex formation. Already slight structural distortions lead to a significant loss of function. Interestingly, the taRNAs V1-M4 and V1-M5 both displayed a weaker repression than the taRNA V1-M1, although in the latter, the complementary nucleotides to the YUNR motif were mutated. This result suggests that imperfect base pairing of a taRNA to its target mRNA is better tolerated than changes of key secondary structures. Apparently, the perfect positioning of some nucleobases in space is sufficient for initiating complex formation. Indeed, many natural sRNAs do not require full complementarity for repressing target mRNA translation. For example, the sRNA SgrS forms imperfect duplexes with its target mRNAs ptsG and sopD (58,59). Interestingly, the substitution of a single nucleotide within sgrS is sufficient to abrogate regulatory function.
Figure 4.
Structure–function relationship of taRNAs with modified seed regions. (A) Structural modifications were introduced into the single-stranded seed region by either reducing (V1-M5) or extending (V1-M4) the amount of complementary bases to the hybridization domain (blue) at the 3′ terminus of the taRNA. The variants V1-M4 and V1-M5 are both fully complementary to the taRNA responsive element. The site of 5′-processing is marked by an arrow head. All taRNA constructs were engineered under control of an IPTG-inducible promoter. (B) The influence of the taRNAs shown in A on TR-HHR activity was examined. Bacterial cultures of the E. coli Top10 strain were induced with 20 µM arabinose and 1 mM IPTG. Transformants were cultivated in LB medium at 37°C, and reporter gene expression of outgrown bacterial cultures was measured. Error bars represent the standard deviation of experiments performed in triplicates.
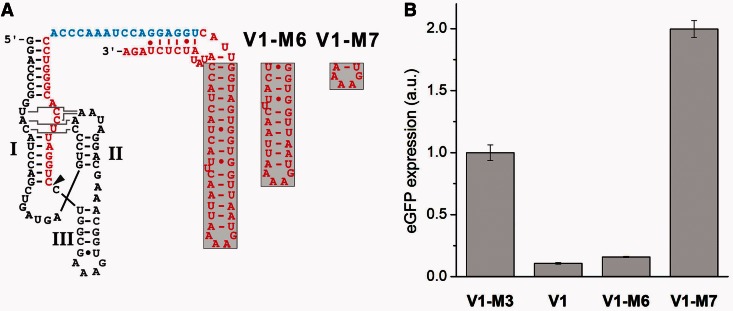
We performed an additional experiment in which we investigated the necessity for the 3′-terminal helix of the taRNA. The helix is not directly involved in the duplex formation with the TR element of the target mRNA. We designed two taRNAs with shortened helices, termed taRNA V1-M6 and V1-M7 (see Figure 5A). Reporter gene expression revealed that a partial reduction of the helix length is tolerated and does not lead to a loss of function of the taRNA (see Figure 5B). However, when the helix was further shortened, the taRNA lost its regulatory activity completely. We observed a 2-fold increase in eGFP expression compared with V1-M3. A comparable phenomenon was observed by Yokobayashi and coworkers, when they constructed artificial sRNAs against natural mRNA target sequences (60). The wild-type MicF sRNA and some of their artificial sRNAs, which repressed translation of their target mRNA OmpF::GFPuv, surprisingly activated the expression of an OmpC::GFPuv fusion mRNA >2-fold. In contrast, cross-reactivity between the micF sRNA and the OmpC mRNA was not observed in a different study (61). Yokobayashi and coworkers assumed that non-specific factors or indirect effects contributed to this observation (60).
Figure 5.
Influence of truncated taRNAs. (A) The helical region outside of the hybridization domain was shortened. The variants V1-M6 and V1-M7 are both fully complementary to the taRNA responsive element. The site of 5′-processing is marked by an arrow head. (B) The influence of the taRNAs shown in A on the TR-HHR activity was examined. Bacterial cultures of the E. coli Top10 strain were induced with 20 µM arabinose and 1 mM IPTG. Transformants were cultivated in LB medium at 37°C, and eGFP expression of outgrown bacterial cultures was measured. Error bars represent the standard deviation of experiments performed in triplicates.
CONCLUSION
In this study, we rationally designed an sRNA-responsive riboswitch that relies on a catalytically active HHR as expression platform. The riboregulatory system displayed a robust switching behavior, which was dependent on the degree of complementarity, the stability of the taRNA, the ratio of taRNA production rate over target mRNA production rate and secondary structural features. To our knowledge, this is the first time that ribozyme activity was regulated by a taRNA to control gene expression in vivo. A ubiquitous advantage of RNA-based genetic switches rests upon the ease to construct orthogonally acting switches (31,62). Independently acting switches are of great importance for future synthetic biology applications because their combination within one host cell allows for parallel information processing. Importantly, orthogonal variants of riboregulator RR12, which was the foundation for our HHR-based riboregulatory system, were recently reported by Collins and coworkers (35). These variants may also be transferrable to the HHR-based riboregulatory system and might enable the construction of orthogonal switches. In general, making use of RNA as regulatory molecule has multiple advantages over protein factors. For example, RNA production is time- and cost-efficient, and genetic encryption requires limited genomic space compared with protein-based regulatory networks (39). In addition, the modular architecture of functional nucleic acid macromolecules makes RNA a perfect substrate for the rational programming of cellular function (63). Recent progress enabled the predictable modeling of regulatory RNAs based on computer-aided design, which extensively increased the synthetic biology toolbox (23,31).
The system reported in this study might be of particular interest for future synthetic biology applications owing to several reasons. First, the sRNAs used in this study were originally designed to act as activators of gene expression in E. coli (40). We rationally designed an artificial HHR-based expression platform in a way that the same sRNAs act as inhibitors of gene expression. Hence, artificial riboregulatory circuits can now be engineered in which a single sRNA can target multiple mRNAs and either repress or induce gene expression. Second, besides controlling translation initiation in principle, the taRNA-dependency could be incorporated in HHR switches developed by our group and others to control tRNA activation (27), 16S ribosomal subunit integrity (28) and pri-miRNA processing (12). In addition, RNA-responsive HHRs could be used to act as sensors of endogenous mRNA or ncRNA levels. As the mediation of RNA cleavage is a drastic response to RNA binding, the general mechanism of riboregulation should be transferable to other organisms. For example, HHR-based RNA switches have been successfully applied to control mRNA stability and pri-miRNA processing in mammalian cells (10,12,22). Here, the stability of the mRNAs under control of a HHR switch is regulated. This feature should prove highly advantageous if more than one RNA-based switch will be connected to each other: In response to a sensed input, a given RNA switch would change its own abundance. This change in the concentration of the specific RNA could then be sensed by a second switch. In turn, the abundance of the switch 2 RNA would vary in response to the concentration of switch 1 RNA, demonstrating immediate interconnection of the abundance of two individual RNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–2 and Supplementary DNA Sequences.
FUNDING
Funding for open access charge: J.S.H. gratefully acknowledges the VolkswagenStiftung for funding a Lichtenberg Professorship and the Fonds der Chemischen Industrie for financial support.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Astrid Joachimi for excellent technical assistance and members of the Hartig group for helpful discussions.
REFERENCES
- 1.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 2.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 3.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 5.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Wang S, Shepard JR, Shi H. An RNA-based transcription activator derived from an inhibitory aptamer. Nucleic Acids Res. 2010;38:2378–2386. doi: 10.1093/nar/gkp1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl Acad. Sci. USA. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angewandte Chemie. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 9.Saito H, Kobayashi T, Hara T, Fujita Y, Hayashi K, Furushima R, Inoue T. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 2010;6:71–78. doi: 10.1038/nchembio.273. [DOI] [PubMed] [Google Scholar]
- 10.Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D′Amato RJ, Mulligan RC. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 11.Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007;35:4179–4185. doi: 10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar D, An CI, Yokobayashi Y. Conditional RNA interference mediated by allosteric ribozyme. J. Am. Chem. Soc. 2009;131:13906–13907. doi: 10.1021/ja905596t. [DOI] [PubMed] [Google Scholar]
- 13.Vinkenborg JL, Karnowski N, Famulok M. Aptamers for allosteric regulation. Nat. Chem. Biol. 2011;7:519–527. doi: 10.1038/nchembio.609. [DOI] [PubMed] [Google Scholar]
- 14.Wieland M, Hartig JS. Artificial riboswitches: synthetic mRNA-based regulators of gene expression. ChemBioChem. 2008;9:1873–1878. doi: 10.1002/cbic.200800154. [DOI] [PubMed] [Google Scholar]
- 15.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 16.Famulok M, Mayer G, Blind M. Nucleic acid aptamers-from selection in vitro to applications in vivo. Acc. Chem. Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 17.de la Pena M, Garcia-Robles I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA. 2010;16:1943–1950. doi: 10.1261/rna.2130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seehafer C, Kalweit A, Steger G, Graf S, Hammann C. From alpaca to zebrafish: hammerhead ribozymes wherever you look. RNA. 2011;17:21–26. doi: 10.1261/rna.2429911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann IT, Hartig JS. Artificial ribozyme-based regulators of gene expression. Synlett. 2011:1486–1494. [Google Scholar]
- 21.Wittmann A, Suess B. Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol. Biosyst. 2011;7:2419–2427. doi: 10.1039/c1mb05070b. [DOI] [PubMed] [Google Scholar]
- 22.Auslander S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 2010;6:807–814. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 23.Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 24.Nomura Y, Kumar D, Yokobayashi Y. Synthetic mammalian riboswitches based on guanine aptazyme. Chem. Commun. 2012;48:7215–7217. doi: 10.1039/c2cc33140c. [DOI] [PubMed] [Google Scholar]
- 25.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed. Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa A, Maeda M. An artificial aptazyme-based riboswitch and its cascading system in E. coli. ChemBioChem. 2008;9:206–209. doi: 10.1002/cbic.200700478. [DOI] [PubMed] [Google Scholar]
- 27.Berschneider B, Wieland M, Rubini M, Hartig JS. Small-molecule-dependent regulation of transfer RNA in bacteria. Angew. Chem. Int. Ed. Engl. 2009;48:7564–7567. doi: 10.1002/anie.200900851. [DOI] [PubMed] [Google Scholar]
- 28.Wieland M, Berschneider B, Erlacher MD, Hartig JS. Aptazyme-mediated regulation of 16S ribosomal RNA. Chem. Biol. 2010;17:236–242. doi: 10.1016/j.chembiol.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Klauser B, Saragliadis A, Auslander S, Wieland M, Berthold MR, Hartig JS. Post-transcriptional Boolean computation by combining aptazymes controlling mRNA translation initiation and tRNA activation. Mol. Biosyst. 2012;8:2242–2248. doi: 10.1039/c2mb25091h. [DOI] [PubMed] [Google Scholar]
- 30.Ketzer P, Haas SF, Engelhardt S, Hartig JS, Nettelbeck DM. Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res. 2012;40:e167. doi: 10.1093/nar/gks734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mutalik VK, Qi L, Guimaraes JC, Lucks JB, Arkin AP. Rationally designed families of orthogonal RNA regulators of translation. Nat. Chem. Biol. 2012;8:447–454. doi: 10.1038/nchembio.919. [DOI] [PubMed] [Google Scholar]
- 32.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 34.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 35.Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proc. Natl Acad. Sci. USA. 2012;109:5850–5855. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl Acad. Sci. USA. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 41.Komatsu Y, Yamashita S, Kazama N, Nobuoka K, Ohtsuka E. Construction of new ribozymes requiring short regulator oligonucleotides as a cofactor. J. Mol. Biol. 2000;299:1231–1243. doi: 10.1006/jmbi.2000.3825. [DOI] [PubMed] [Google Scholar]
- 42.Penchovsky R, Breaker RR. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat. Biotechnol. 2005;23:1424–1433. doi: 10.1038/nbt1155. [DOI] [PubMed] [Google Scholar]
- 43.Peist R, Koch A, Bolek P, Sewitz S, Kolbus T, Boos W. Characterization of the aes gene of Escherichia coli encoding an enzyme with esterase activity. J. Bacteriol. 1997;179:7679–7686. doi: 10.1128/jb.179.24.7679-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 45.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou K, Zhou L, Lim Q, Zou R, Stephanopoulos G, Too HP. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011;12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burke DH, Ozerova ND, Nilsen-Hamilton M. Allosteric hammerhead ribozyme TRAPs. Biochemistry. 2002;41:6588–6594. doi: 10.1021/bi0201522. [DOI] [PubMed] [Google Scholar]
- 48.Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell. Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 51.De la Pena M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine E, Hwa T. Small RNAs establish gene expression thresholds. Curr. Opin. Microbiol. 2008;11:574–579. doi: 10.1016/j.mib.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franch T, Petersen M, Wagner EG, Jacobsen JP, Gerdes K. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 1999;294:1115–1125. doi: 10.1006/jmbi.1999.3306. [DOI] [PubMed] [Google Scholar]
- 55.Eichhorn CD, Feng J, Suddala KC, Walter NG, Brooks CL, 3rd, Al-Hashimi HM. Unraveling the structural complexity in a single-stranded RNA tail: implications for efficient ligand binding in the prequeuosine riboswitch. Nucleic Acids Res. 2012;40:1345–1355. doi: 10.1093/nar/gkr833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 57.Mitarai N, Benjamin JA, Krishna S, Semsey S, Csiszovszki Z, Masse E, Sneppen K. Dynamic features of gene expression control by small regulatory RNAs. Proc. Natl Acad. Sci. USA. 2009;106:10655–10659. doi: 10.1073/pnas.0901466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papenfort K, Podkaminski D, Hinton JC, Vogel J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl Acad. Sci. USA. 2012;109:E757–E764. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 60.Sharma V, Yamamura A, Yokobayashi Y. Engineering artificial small RNAs for conditional gene silencing in Escherichia coli. ACS Synth. Biol. 2012;1:6–13. doi: 10.1021/sb200001q. [DOI] [PubMed] [Google Scholar]
- 61.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dixon N, Duncan JN, Geerlings T, Dunstan MS, McCarthy JE, Leys D, Micklefield J. Reengineering orthogonally selective riboswitches. Proc. Natl Acad. Sci. USA. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.