Abstract
The choice between double-strand break (DSB) repair by either homology-directed repair (HDR) or non-homologous end-joining (NHEJ) is tightly regulated. Defects in this regulation can induce genome instability and cancer. 53BP1 is critical for the control of DSB repair, promoting NHEJ and inhibiting the 5’ end resection needed for HDR. Using dysfunctional telomeres and genome-wide DSBs, we identify Rif1 as the main factor used by 53BP1 to impair 5’ end resection. Rif1 inhibits resection involving CtIP, BLM, and Exo1, limits accumulation of BRCA1/BARD1 complexes at sites of DNA damage, and defines one of the mechanisms by which 53BP1 causes chromosomal abnormalities in Brca1-deficient cells. These data establish Rif1 as an important contributor to the control of DSB repair by 53BP1.
53BP1 can influence the type of DNA repair at DSBs (1) as seen in immunoglobulin gene rearrangements (2-4) and in the fusion of telomeres rendered dysfunctional through the removal of the shelterin protein TRF2 (5), where 53BP1 enhances the mobility of damaged telomeres, thus potentially promoting the chance of telomeretelomere encounters. In Brca1-deficient cells, 53BP1 enhances aberrant NHEJ events that create lethal radial chromosomes in response to poly(ADP-ribose) polymerase PARP1 inhibitors (PARPi) (6). In this setting, 53BP1 may favor NHEJ-mediated mis-rejoining by blocking the DSB resection needed for HDR (6, 7). 53BP1 was shown to impede 5’ end resection at dysfunctional telomeres lacking all shelterin proteins and similarly, telomeres lacking only TRF2 show evidence of 53BP1-dependent protection from resection (5, 8). Based on the finding that an allele of 53BP1 (53BP128A) lacking all potential ATM/ATR kinase S/TQ target sites did not support immunoglobulin Class Switch Recombination (CSR) and failed to generate radial chromosomes in Brca1-deficient cells (7), it appears that these functions of 53BP1 involve interacting partner(s) modulated by the S/TQ sites. One candidate 53BP1-interacting factor is Rif1, which localizes to DSBs and dysfunctional telomeres, in a manner that is dependent on ATM signaling (9-11). Rif1 was originally identified as part of the telomeric complex in budding yeast (12) and was recently shown to inhibit resection at yeast telomeres (13, 14). In contrast, mammalian Rif1 has no known function at functional telomeres but contributes to the intra-S phase checkpoint, facilitates recovery from replication stress, and affects replication timing (10, 15-17).
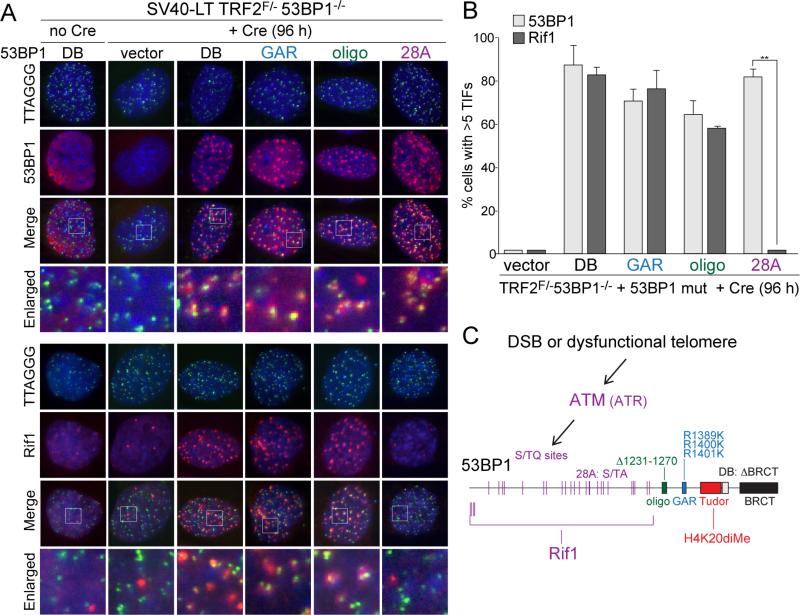
We introduced 53BP128A and other 53BP1 mutant alleles (7) into immortalized TRF2F/-53BP1-/- mouse embryo fibroblasts (MEFs) and induced telomere dysfunction by deletion of TRF2 (Fig. 1A,B). The results showed that the S/TQ sites were required for the accumulation of Rif1 at deprotected telomeres, whereas the GAR, BRCT, and oligomerization domains of 53BP1 were not (Fig. 1A-C; fig. S1). The functional significance of the Rif1-53BP1 interaction was addressed using a telomere-based assay system that previously uncovered the role of 53BP1 in stimulating telomeric NHEJ and protecting telomere ends from 5’ resection (5, 8). Using TRF2/Rif1 conditional double knockout MEFs, we documented a significant reduction in the incidence and rate of telomere fusions in cells lacking Rif1 (Fig. 2A-C; fig. S2A). This reduced NHEJ rate was not due to changes in cell cycle progression or diminished activation of the ATM kinase pathway by the deprotected telomeres (fig. S2B-G).
Figure 1. Rif1 recruitment requires the S/TQ ATM/ATR target sites of 53BP1.
(A) Detection of 53BP1 and Rif1 at dysfunctional telomeres in Cre-treated SV40-LT immortalized TRF2F/-53BP1-/- MEFs expressing 53BP1 mutant alleles (shown in (C)). IF for 53BP1 and Rif1 (red) was combined with telomeric TTAGGG FISH (green). Blue: DAPI DNA stain. (B) Quantification of 53BP1 and Rif1 Telomere Dysfunction Induced Foci (TIFs; (21)) detected as in (A). Data represent means of 3 experiments ±SDs (≥70 cells/experiment). ** indicates p value <0.05 (two-tailed paired Student's t-test). (C) Schematic of the 53BP1 mutant alleles and the role of the N-terminal S/TQ sites in the recruitment of Rif1.
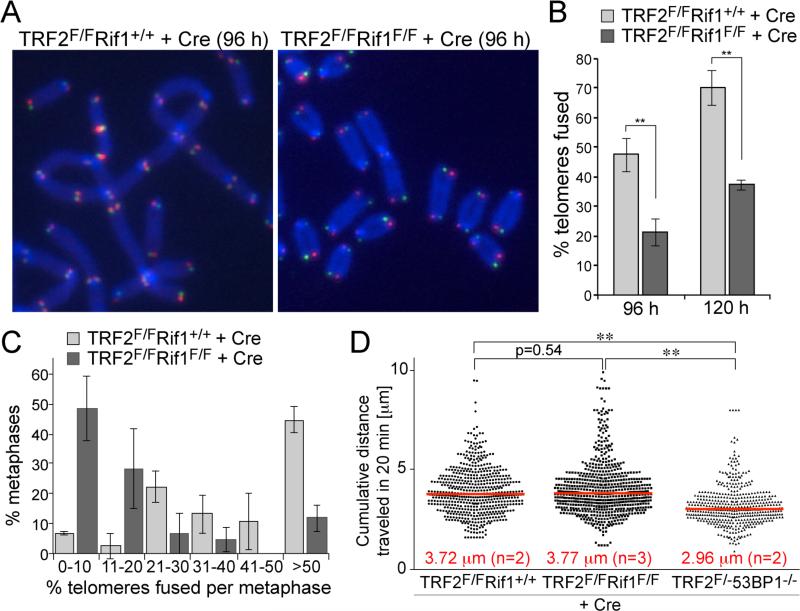
Figure 2. Rif1 promotes telomeric NHEJ without affecting telomere mobility.
(A) Metaphase chromosomes of Cre-treated SV40-LT immortalized TRF2F/FRif1+/+ and TRF2F/FRif1F/F MEFs showing NHEJ-mediated telomere fusions detected by CO-FISH. Telomeres synthesized by leading-end DNA synthesis are in red; lagging-end telomeres in green. (B) Quantification of telomere fusions as determined in (A) at 96 and 120 h post Cre. Data represent means of three independent experiments ± SDs (> 3000 telomeres/experiment). ** indicates p value <0.01 based on two-tailed paired Student's t-test. (C) Distributions of telomere fusions per metaphase at 96 h after Cre for experiments shown in (B). (D) Distribution of cumulative distances traveled by mCherry-53BP11220-1711 foci in the indicated cell lines. Red lines represent medians. ** indicates p values < 0.0001 (two-tailed Mann-Whitney test).
As 53BP1 increases the mobility of dysfunctional telomeres, we determined whether Rif1 contributes to this aspect of 53BP1 by live-cell imaging of mCherry fused to the 53BP1 Tudor domain, which targets this marker to dysfunctional telomeres (fig. S2H). As expected, traces of the mCherry marker demonstrated that 53BP1-deficiency reduced the mobility of dysfunctional telomeres (Fig. 2D). In contrast, absence of Rif1 did not affect the mobility of the deprotected telomeres. Thus, Rif1 is not required for the 53BP1-dependent increase in the mobility of dysfunctional telomeres.
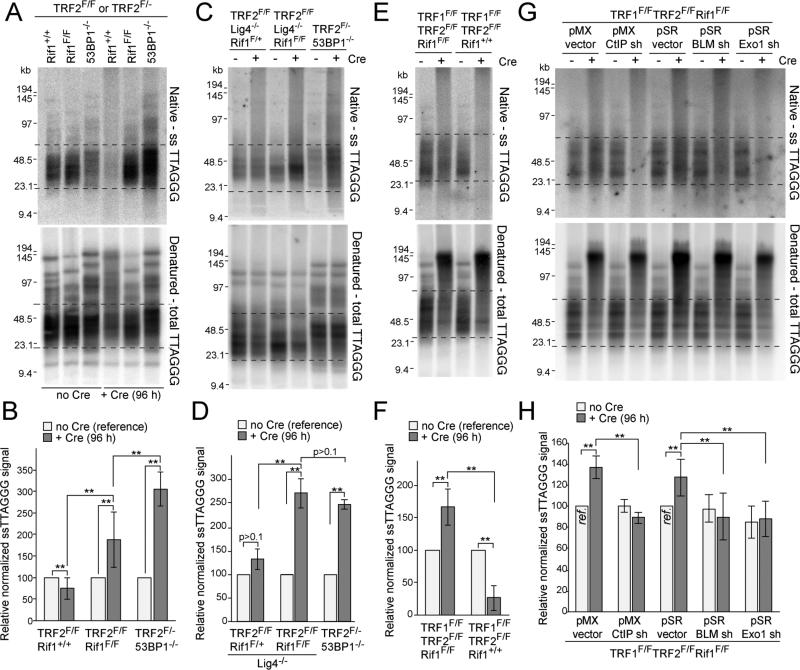
We next determined whether Rif1 contributes to the inhibition of 5’ end resection by 53BP1. When TRF2 is deleted from cells lacking 53BP1, there is a 2-3 fold increase in the telomeric 3’ overhang signal (5) which can be detected based on annealing a telomeric oligonucleotide to native telomeric DNA (Fig. 3). As expected, deletion of TRF2 resulted in the removal of the overhangs concomitant telomere fusion, whereas the overhang signal increased 3-fold when TRF2 was deleted from 53BP1-deficient cells in which telomeric NHEJ is rare and 5’ end resection is uninhibited (Fig. 3A,B). Deletion of TRF2 from Rif1-deficient cells also resulted in an increase in the overhang signal (Fig. 3B). However, the increase was less compared to that observed in the 53BP1-deficient cells. As the difference might be due the lower rate of telomere fusions in the 53BP1-deficient setting, we generated immortalized TRF2F/FLig4-/-Rif1F/F cells, which, owing to the absence of DNA ligase IV, have the same low telomere fusions rates as TRF2F/- 53BP1-/- cells (5). When NHEJ was blocked, the telomeric overhang increase in the Rif1-deficient cells was the same as that which occurred in the 53BP1-deficient cells (Fig. 3C,D). The increase in overhang signal was demonstrably due to 3’ terminal sequences since the signal was removed by digestion with the E. coli 3’ exonuclease ExoI (fig. S3A). These data suggest that Rif1 is the main factor acting downstream of 53BP1 to inhibit the resection at telomeres that lack TRF2 protection.
Figure 3. Rif1 blocks 5’ end resection at dysfunctional telomeres.
(A) Telomeric overhang assays on TRF2F/FRif1+/+, TRF2F/FRif1F/F and TRF2F/-53BP1-/- MEFs. Native in-gel hybridization of MboI/AluI digested DNA with end-labeled [AACCCT]4 (top) and re-hybridization with the same probe after denaturation in situ (bottom). Dashed lines represent the bulk of free (unfused) telomeres used for quantification. (B) Quantification of overhang assays as in (A). Overhang signals in no Cre samples was set at 100%. (C, D) Overhang assays on TRF2F/FRif1F/+Lig4-/-, TRF2F/FRif1F/FLig4-/- and TRF2F/-53BP1-/- MEFs and quantification as in (B). (E, F) Overhang assays on TRF1F/FTRF2F/FRif1+/+ and TRF1F/FTRF2F/FRif1F/F MEFs and quantification. (G, H) Overhang assays to measure dependency on CtIP, BLM, and Exo1 and quantification. Cells infected with either pMX or pSR with or without the indicated shRNAs and treated with Cre for 96 h. Samples with empty vectors and no Cre (ref.) were used as references. Data in (B,D,F,H) represent means of ≥3 experiments ±SDs. ** indicates p values <0.05 (two-tailed paired Student's t-test). MEFs are SV40-LT immortalized.
At telomeres that are deprived of both TRF1 and TRF2 and therefore lack all shelterin proteins, 53BP1 blocks extensive 5’ end resection that involves CtIP, BLM, and Exo1 (8). To test the ability of Rif1 to inhibit resection at such shelterin-free telomeres, we generated immortalized TRF1F/FTRF2F/FRif1F/F MEFs. As expected, deletion of TRF1 and TRF2 resulted in frequent telomere fusions and nearly complete loss of the telomeric overhang signal when Rif1 was present (Fig. 3E,F). When Rif1 was co-deleted with TRF1 and TRF2, telomere fusions were also frequent, resulting in most telomeric restriction fragments shifting to a higher MW (Fig. 3E). However, the telomeres that had not fused at the time-point analyzed showed a notable increase in overhang signal (Fig. 3E,F). This increase in the signal was diminished when cells were treated with shRNAs against CtIP, BLM, or Exo1 (Fig. 3G,H; fig. S3B,C). Thus, like 53BP1, Rif1 inhibits 5’ end resection that involves CtIP, BLM, and Exo1.
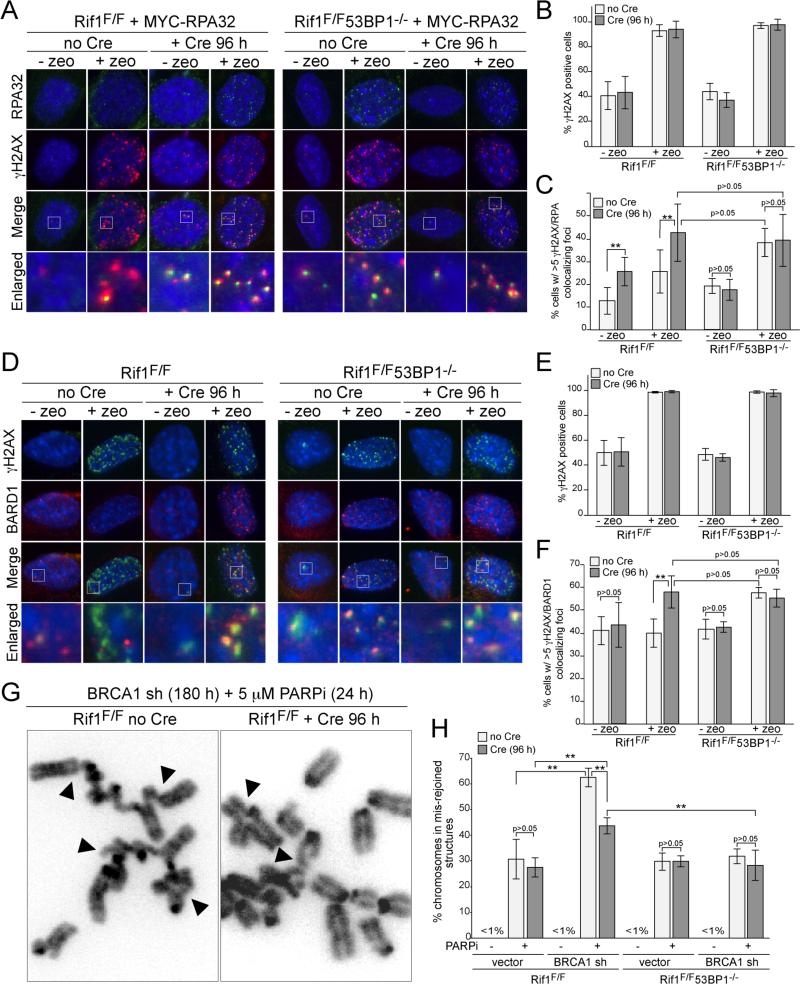
We next asked whether Rif1 affects resection at zeocin-induced DSBs by monitoring the formation of RPA foci (Fig. 4A; fig. S4A). The absence of either Rif1, 53BP1, or both did not affect zeocin-induced γ-H2AX foci or the basal level of cells containing γ-H2AX foci, which likely represent replicating cells (Fig. 4A,B). However, in zeocin-treated cells, the absence of either Rif1 or 53BP1 resulted in a significant increase in γ-H2AX foci that co-localized with RPA (Fig. 4A,C). When either 53BP1 or Rif1 were absent, there also was a significant increase in γ-H2AX foci containing RPA in cells not treated with zeocin, presumably reflecting a higher level of resection at stalled replication forks (Fig. 4A,C). Examination of the RPA/γ-H2AX foci in zeocin-treated Rif1/53BP1 double knockout cells indicated that Rif1 and 53BP1 are epistatic in this regard since the induction of RPA/γ-H2AX foci in absence of 53BP1 was the same as in Rif1-deficient cells and the absence of both Rif1 and 53BP1 did not further increase the response (Fig. 4C). The simplest interpretation of this data is that Rif1 is the main factor acting downstream of 53BP1 to block 5’ end resection at the zeocin-induced DSBs.
Figure 4. Rif1 inhibits resection at DSBs and promotes radial formation.
(A) IF for γ-H2AX (red) and MYC-RPA32 (green) in Cre-treated SV40-LT immortalized Rif1F/F and Rif1F/F53BP1-/- cells expressing MYC-RPA32 treated with zeocin (100 ug/ml, for 1 h; 2 h prior to analysis). (B) Percentage of γ-H2AX positive cells in experiments as in (A). (C) Percentage of cells (as in A) scored positive when containing at least five γ-H2AX foci co-localizing with RPA. (D) IF for γH2AX (green) and BARD1 (red) in Rif1F/F and Rif1F/F53BP1-/- MEFs. Cells and treatment as in (A). (E) Percentage of γ-H2AX positive cells in experiments in (D). (F) Percentage of cells in (D) containing >5 BARD1/γ-H2AX colocalizing foci. (G) Examples of mis-rejoined and radial chromosomes (arrowheads) in BRCA1sh/PARPi-treated Rif1F/F cells with or without Cre. (H) Percentages of chromosomes that are mis-rejoined in the indicated genotypes and treatments. Data in (B,C), (E,F) and (H) are means of 3-5 experiments ±SDs. ** indicates p values <0.05 (two-tailed paired Student's t-test).
Since the 53BP1/Rif1 control affects CtIP, which is thought to be delivered by a complex containing BRCA1 (18-20), we also determined whether 53BP1 and Rif1 had an effect on the presence of the BRCA1 at zeocin-induced DSBs. Using an antibody to the constitutive BRCA1 partner BARD1, we found that absence of Rif1 or 53BP1 resulted in a significant increase in the accumulation of BRCA1 complexes at zeocin-induced DSBs (Fig. 4E-F). Consistent with the data above, Rif1 and 53BP1 were again epistatic in this regard. The absence of 53BP1 resulted in the same phenotype as absence of Rif1 and the double knockout did not show an additional increase in the incidence of BARD1 foci (Fig. 4E,F). The absence of Rif1 also resulted in an increase in the presence of BARD1 at dysfunctional telomeres (fig. S4B-D).
As 53BP1 mediates the formation of mis-rejoined and radial chromosomes in PARPi-treated Brca1-deficient cells, we asked to what extent Rif1 is responsible for this effect. Cells lacking Rif1, 53BP1, or both were treated with a BRCA1 shRNA and the PARP inhibitor and mis-rejoined chromosomes were quantified (Fig. 4G, H). The data show the previously documented decrease in the frequency of chromosome mis-rejoining when 53BP1 is absent. Interestingly, absence of Rif1 also lowers the frequency of chromosome mis-rejoining but the effect is significantly less than for 53BP1. Thus, the formation of mis-rejoined chromosomes in PARPi-treated Brca1-deficient cells is due to two distinct attributes of 53BP1, one of which requires Rif1 function.
These data identify Rif1 as the major factor acting downstream of 53BP1 in the control of 5’ end resection. In contrast, Rif1 does not appear to be required for the ability of 53BP1 to promote an increase in the mobility of dysfunctional telomeres. The intermediate effect of Rif1 on the fusion of dysfunctional telomeres can explained based on these two observations. The increased resection of dysfunctional telomeres in absence of Rif1 is likely to be responsible for the mild inhibition of NHEJ. However, in the absence of 53BP1, the effect of increased resection is combined with a defect in the induction of the mobility of the dysfunctional telomeres, resulting in a more severe blockade to NHEJ. Similarly, we propose that Rif1 deletion leads to partial rescue of chromosome mis-rejoining in PARPi/BRCA1sh-treated cells because the control of 5’ end resection is only one of multiple mechanisms by which 53BP1 acts. One possibility is that the other mechanism used by 53BP1 in this context, similar to what happens at dysfunctional telomeres, involves the induction of DSB mobility that increases the chance that DSB mis-rejoining occurs.
Supplementary Material
Acknowledgments
We thank Devon White for expert mouse husbandry and Michel Nussenzweig for providing mutant 53BP1 constructs. MZ was supported by a Brno PhD talent fellowship and by a grant from the Czech Science Foundation to Dr. Ctirad Hofr (P205/12/0550) (see SOM for detailed acknowledgement). This work was supported by a grant from the Breast Cancer Research Foundation to TdL. TdL is an ACS Research Professor.
REFERENCES
- 1.Noon AT, Goodarzi AA. DNA Repair (Amst) 2011;10:1071. doi: 10.1016/j.dnarep.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Ward IM, Minn K, van Deursen J, Chen J. Mol Cell Biol. 2003;23:2556. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morales JC, et al. J Biol Chem. 2003;278:14971. doi: 10.1074/jbc.M212484200. [DOI] [PubMed] [Google Scholar]
- 4.Difilippantonio S, et al. Nature. 2008;456:529. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimitrova N, Chen YC, Spector DL, de Lange T. Nature. 2008;456:524. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunting SF, et al. Cell. 2010;141:243. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bothmer A, et al. Mol Cell. 2011;42:319. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sfeir A, de Lange T. Science. 2012;336:593. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, Blackburn EH. J Cell Biol. 2004;167:819. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Genes Dev. 2004;18:2108. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huen MS, et al. Mol Cell. 2010;37:854. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy CF, Sussel L, Shore D. Genes Dev. 1992;6:801. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 13.Anbalagan S, Bonetti D, Lucchini G, Longhese MP. PLoS Genet. 2011;7:e1002024. doi: 10.1371/journal.pgen.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonetti D, et al. PLoS Genet. 2010;6:e1000966. doi: 10.1371/journal.pgen.1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buonomo S, Wu Y, Ferguson D, de Lange T. J Cell Biol. 2009;187:385. doi: 10.1083/jcb.200902039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornacchia D, et al. EMBO J. 2012;31:3678. doi: 10.1038/emboj.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamazaki S, et al. EMBO J. 2012;31:3667. doi: 10.1038/emboj.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartori AA, et al. Nature. 2007;450:509. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. J Biol Chem. 1998;273:25388. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 20.Wong AK, et al. Oncogene. 1998;17:2279. doi: 10.1038/sj.onc.1202150. [DOI] [PubMed] [Google Scholar]
- 21.Takai H, Smogorzewska A, de Lange T. Curr Biol. 2003;13:1549. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.