Abstract
Homologous recombination affects myriad aspects of genome evolution, from standing levels of nucleotide diversity to the efficacy of natural selection. Rates of crossing over show marked variability at all scales surveyed, including species-, population-, and individual-level differences. Even within genomes, crossovers are nonrandomly distributed in a wide diversity of taxa. Although intra- and intergenomic heterogeneities in crossover distribution have been documented in Drosophila, the scale and degree of crossover rate heterogeneity remain unclear. In addition, the genetic features mediating this heterogeneity are unknown. Here we quantify fine-scale heterogeneity in crossover distribution in a 2.1-Mb region of the Drosophila melanogaster X chromosome by localizing crossover breakpoints in 2500 individuals, each containing a single crossover in this specific X chromosome region. We show 90-fold variation in rates of crossing over at a 5-kb scale, place this variation in the context of several aspects of genome evolution, and identify several genetic features associated with crossover rates. Our results shed new light on the scale and magnitude of crossover rate heterogeneity in D. melanogaster and highlight potential features mediating this heterogeneity.
Keywords: crossover, recombination, Drosophila melanogaster
HOMOLOGOUS recombination is critical for proper chromosome segregation during meiosis. In spite of this vital function, rates of meiotic recombination are highly variable at all scales. Previous research in a diversity of taxa indicates substantial heterogeneity in recombination rate within species. In some cases, this heterogeneity can be partitioned simply, such as differences manifested between males and females. Drosophila represents an extreme example of such variation, as Drosophila males do not normally undergo meiotic recombination (Morgan 1912). Less extreme but equally notable differences in meiotic recombination rate between the sexes have also been documented in a wide variety of taxa, including humans (Broman et al. 1998), mice (Dietrich et al. 1996), dogs (Neff et al. 1999), zebrafish (Singer et al. 2002), sheep (Crawford et al. 1995), and wallabies (Zenger et al. 2002).
Intraspecific variation in recombination rate independent of sex has been found as well, in a similarly broad range of taxa. Early work in Drosophila revealed differences in rates of crossing over among strains (e.g., Brooks and Marks 1986), and this population-level variability in recombination rate has since been recapitulated in many other systems, including Arabidopsis (e.g., Sanchez-Moran et al. 2002) and mice (e.g., Dumont et al. 2009). Extensive work in humans similarly indicates substantial variability in recombination rate among populations and individuals (Broman et al. 1998; Lynn et al. 2002; Fearnhead and Smith 2005; Neumann and Jeffreys 2006; Graffelman et al. 2007; Coop et al. 2008).
Finally, many species show evidence of coarse-scale intragenomic heterogeneity in recombination rate. For example, depressions in levels of crossing over in euchromatin adjacent to centromeric regions are found in Drosophila (Beadle 1932) and other metazoans (Rahn and Solari 1986; Kipling et al. 1994; Mahtani and Willard 1998), plants (Sherman and Stack 1995; Round et al. 1997; Harushima et al. 1998; Haupt et al. 2001; Anderson et al. 2003), and yeast (Nakaseko et al. 1986; Lambie and Roeder 1988). Species with heteromorphic sex chromosomes show chromosome-level heterogeneity in recombination rate, with the Y chromosome lacking recombination, excepting pseudoautosomal regions that retain homology with the X chromosome.
In addition to broad-scale variation in rates of crossing over, fine-scale distributions of crossover events are heterogeneous within genomes in a similarly impressive array of taxa. In humans, for instance, it is estimated that up to 80% of all crossover events occur in 10–20% of the genomic sequence (Myers et al. 2005), illustrating extreme heterogeneity in crossover distribution in this species. The recombinational landscape of Western chimpanzees is also dominated by recombination hotspots (Auton et al. 2012), as are those of mice (Smagulova et al. 2011) and yeast (Mancera et al. 2008). Intragenomic crossover rate heterogeneity is seen in dog genomes as well; although 80% of crossovers occur in 46% of the sequence, crossover rates fluctuate five orders of magnitude across the genome (Axelsson et al. 2012).
In Drosophila, rates of crossing over are clearly heterogeneous at a fine scale (Cirulli et al. 2007; Kulathinal et al. 2008; Singh et al. 2009; Stevison and Noor 2010; Comeron et al. 2012). However, the reported magnitude of the fluctuations in crossover rates varies substantially, with 3- to 20-fold variation reported in Drosophila melanogaster (Singh et al. 2009; Comeron et al. 2012) and 20- to 40-fold variation in D. pseudoobscura (Cirulli et al. 2007; Kulathinal et al. 2008). These differences may reflect population-level variation or interspecific divergence in the distribution of crossovers or may instead reflect differences in experimental design among studies. Indeed, these studies had major differences in the experimental approaches used, the number of meioses surveyed, the proportion of the genome surveyed, and the marker density/window size. Thus, the magnitude of fluctuations in crossover rates within and between Drosophila species remains an exciting and open question.
Moreover, the genetic basis of crossover distribution in Drosophila remains unknown. Although recombination appears to be under polygenic control, with the total amount of crossing over and the distribution of crossovers potentially being mediated by different factors (e.g., Brooks and Marks 1986), these factors have yet to be identified. This contrasts with our understanding of the genetic basis of crossover distribution in humans and mice, as the first known determinant of recombination hotspots in metazoans was recently discovered (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). This remarkable discovery implicates Prdm9 in determining the locations of meiotic recombination hotspots in both humans and mice. Importantly, Prdm9 appears to lack an ortholog in Drosophila (Oliver et al. 2009), indicating that crossover distribution in Drosophila is mediated by other, yet unknown genetic factors.
To begin to address these outstanding questions, we generated a fine-scale map of crossover distribution of a 2.1-Mb region of the D. melanogaster X chromosome. The goal of this study was to survey a large number of crossover events in a small physical span such that we can gain maximal resolution in the magnitude of fine-scale fluctuation in crossover rates as well as potentially gain insight into genetic features mediating crossover distribution. Using a combination of classical genetics and multiplex next-generation sequencing, we inferred breakpoints for 2500 crossover events localized to the region between the X chromosome genes garnet and scalloped. Our results indicate fluctuations in crossover rate spanning nearly two orders of magnitude at a 5-kb scale, with these fluctuations dampening as we increase window size to 10, 20, 50, and 100 kb. We also explore the effects of this fine-scale heterogeneity on genome evolution. Our results underscore the importance of scale in studies of crossover rate heterogeneity and highlight potential genetic features mediating this heterogeneity.
Methods
Fly strains
The double-mutant strain of D. melanogaster used in this experiment contained two X-linked recessive mutations with visible phenotypes, corresponding to mutations in the garnet (g) and scalloped (sd) genes. This strain was constructed by crossing a g50e mutant line (stock no. 101116 from the Drosophila Genetic Resource Center) with an sd1 mutant line (stock no. 1027 from the Bloomington Drosophila Stock Center) and screening for recombinant phenotypes in the F2 generation. We then created an extracted X chromosome line by crossing our g-sd line to the FM7a balancer stock. Thus, our g-sd stock is isogenic for the X chromosome. The wild-type line used in this study (UgX54A) corresponds to an X-extracted line from a Ugandan population of D. melanogaster (collected originally in 2005 as described in Pool and Aquadro 2006). We screened for inversions by crossing these two parental lines and performing standard polytene chromosome squashes of the F1 progeny; these lines appear to be inversion-free relative to each another on the X and the autosomes.
Experimental crosses
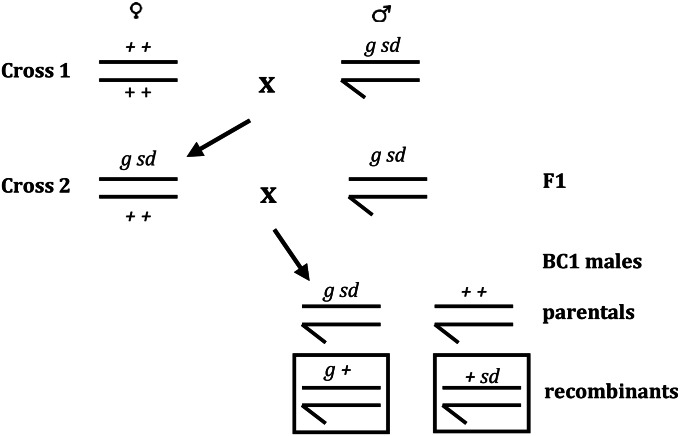
We used a two-step crossing scheme to generate the recombinant males used in this study (Figure 1). All crosses were set up in bottles on standard yeast–glucose media in glass bottles and involved 20 virgin females and 20 males. A total of 448 bottles of crosses were established in the first round; these crosses were conducted at 21°. There were a total of 569 bottles of crosses established in the second round, which all involved virgin females 24–36 hr old. These crosses were incubated at 25° for the first 5 days and were held at 21° thereafter. The initial incubation temperature (25°) was chosen because crossover frequency appears to increase above 22° (Ashburner 1989). Although we expect that this temperature may lead to an overall increase in rates of crossing over, we do not expect that it will generate spatial heterogeneity in crossover distribution. Rather, we expect that the increased temperature may simply amplify the underlying heterogeneity in crossover distribution. The later shift to 21° was necessitated by incubator space limitations. Parents were cleared on day 4, and the progeny were scored on day 18, where day 0 is the day the crosses were set up. Males were counted and scored for the recombinant phenotype. In total, 92,105 males were scored for the recombinant phenotype, and 6716 recombinant males were collected (2483 +sd males and 4233 g+ males).
Figure 1.
Schematic representation of our crossing scheme. Boxed BC1 progeny correspond to the two recombinant phenotypes identified by our cross; these individuals have a single crossover between garnet (g) and scalloped (sd).
DNA preparation
Recombinant males were grouped by phenotype into pools of 100, yielding 66 pools (24 pools of +sd males and 42 pools of g+ males). Pools were flash-frozen in liquid nitrogen and subsequently pulverized in 96-well plates on a high-speed microplate shaker, using a galvanized steel BB. Genomic DNA was then extracted from each pool, using the QIAGEN (Valencia, CA) DNeasy Blood and Tissue kit. Genomic DNA was sheared using sonication and genomic DNA libraries suitable for Illumina single-end sequencing were prepared following a standard protocol (available upon request). Each library was barcoded with one of twelve 3-bp barcodes (ACT, ATA, AAG, GGA, TTG, GCG, TAA, TGT, GAT, TCC, CAC, and AGC). DNA from each genomic library was quantified using PicoGreen, and DNAs from 11 different libraries (each with a different barcode) were combined in equal amounts. This procedure thus yielded six superpools of DNA, each of which consisted of DNA from 1100 different recombinant males.
We also extracted DNA from pools of single males representing different relative proportions of the two parental lines used in our two-step crossing scheme. We constructed eight such pools, consisting of 95:5, 85:15, 70:30, 55:45, 40:60, 25:75, 15:85, and 5:95 g-sd:wild-type flies. DNA was extracted from each of these eight pools, using the methodology applied to the recombinant males (described above), and barcoded genomic DNA libraries suitable for single-end Illumina sequencing were prepared as above. DNA from each genomic library was quantified using PicoGreen, and DNAs from these eight different libraries were combined in equal amounts in a single superpool.
These seven DNA samples (six recombinant male superpools and one parental mixture superpool) were enriched for the X chromosome region between the genes garnet and scalloped (coordinates in release 5.2 are 13,621,236–15,719,755), using a custom NimbleGen Comparative Genomic Hybridization Array (OID26736). This array contains 385,000 oligos, each of which map to the target region of interest. The seven DNA samples were individually hybridized to an array at the Cornell Microarray core, following standard NimbleGen hybridization protocol. The resulting DNA, enriched for the 2.1-Mb X chromosome region between garnet and scalloped, was sequenced using Illumina single-end sequencing (86-bp reads) at the Cornell Life Sciences Core Laboratories Center. The parental mixture was sequenced in a single lane twice on separate flow cells. The six recombinant superpools were sequenced in a single flow cell, with one lane per superpool. For five of these superpools, reduced read yields for this first run led us to resequence these same samples on a separate flow cell.
SNP identification
Individual reads were sorted by barcode such that each read was assigned to one of the 66 recombinant or 8 parental mixture pools; only those reads matching the barcode exactly were retained for analysis. Overall we observed approximately threefold variation in representation among barcodes within a single superpool (data not shown), suggesting that there is no marked bias toward or away from any of the barcodes used in our analysis. The retained reads were then mapped to the D. melanogaster genome, using the Burrows–Wheeler Aligner (BWA) (Li and Durbin 2009) with default parameters. Only reads mapping uniquely to the genome were retained for analysis. SAMtools (Li et al. 2009) was used to generate alignment files in pileup format. These pileup files were used for subsequent bioinformatic analysis.
Our strategy was to identify a small set of well-behaved, parentally informative single nucleotide polymorphisms (SNPs) based exclusively on the sequence data from the parental mixture pools. We restricted ourselves to this subset of SNPs because differential binding of our parental genotypes to the hybridization array has the potential to adversely affect our inference of allele frequency in each recombinant pool and thus compromise our estimation of the fine-scale distribution of crossover events in this interval. We thus sought to identify the subset of SNPs that did not appear to suffer from this hybridization bias. This was accomplished using a series of heuristic filters. First, we combined the reads across parental mixtures and included only those sites at which the two most frequent nucleotides made up at least 95% of the total reads. From these, we excluded any site at which any of the eight parental mixture pools had no coverage. The remaining sites were filtered using two indices of goodness-of-fit. The first index was the P-value obtained via logistic regression in which the observed counts were fitted to the mixing proportions; we employed a stringent threshold of 10−8. The second index was a chi-square-like statistic formed by comparing the SNP frequencies observed in each sequencing run (8 pools × 2 runs per pool for a total of 16) to their respective mixing proportions; here we required a value less than one. The two indexes are similarly motivated yet complementary, as only the former uses read count information. Both enabled us to polarize the SNPs from the mixing proportions and thus infer parental genotypes. A total of 451 SNPs survived our filters (Supporting Information, Figure S1)—these were then used to estimate allele frequencies in the 66 recombinant pools.
Allele frequency estimation
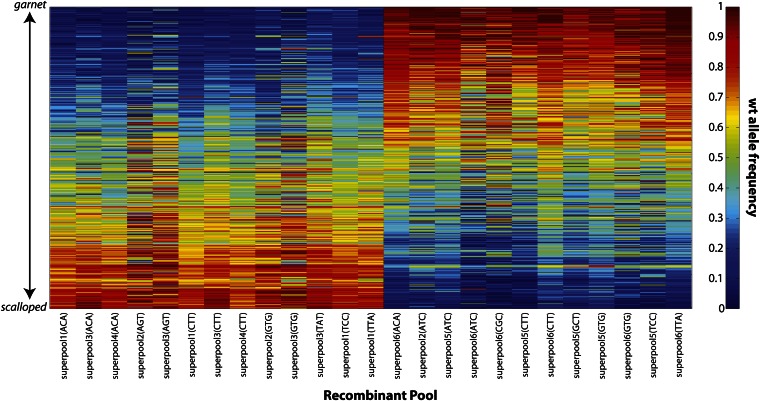
For each of the 66 recombinant pools, read counts were used to estimate allele frequencies at the 451 sites identified from the parental mixtures (Figure S2). These allele frequency estimates were then used as a diagnostic to disqualify a subset of recombinant pools. Specifically, because each pool was constructed to be homogenous with respect to recombinant type (either +sd or g+), there was an expectation that allele frequency would vary monotonically with position. In 41 of the 66 pools, we deemed this expectation to be strongly violated, either due to insufficient coverage (inflating the variance of allele frequency estimates) or otherwise. Sequence coverage appears to contribute substantially to whether or not a given pool was included; average depth of coverage of included pools was 148 (median 120) vs. 97 (median 96) for the excluded pools. Of the remaining 25 pools, 13 and 12 were composed of +sd and g+ recombinants, respectively. Allele frequency estimates at each of the 451 SNPs for only these surviving 25 pools are presented in Figure 2. We proceeded to infer individual recombination landscapes for each.
Figure 2.
Heat map depicting the frequency of the wild-type (wt) parental allele at each of 451 SNPs for each of the 25 pools of 100 recombinant males included in the final analysis. Each column represents a pool, with the top of the plot representing the 5′ end of the g-sd region and the bottom representing the 3′ end. Note that the expectation is that allele frequencies should start at 0 (blue) and increase to 1 (red) from top to bottom or vice versa, depending on the recombinant phenotype class comprising the pool. The pools are ordered such that all of the g+ pools are presented first followed by the +sd pools. Within each phenotype class, the ordering of pools is arbitrary. Within each phenotype class each pool is labeled by its superpool identification number (1–6) and barcode.
Recombination landscapes
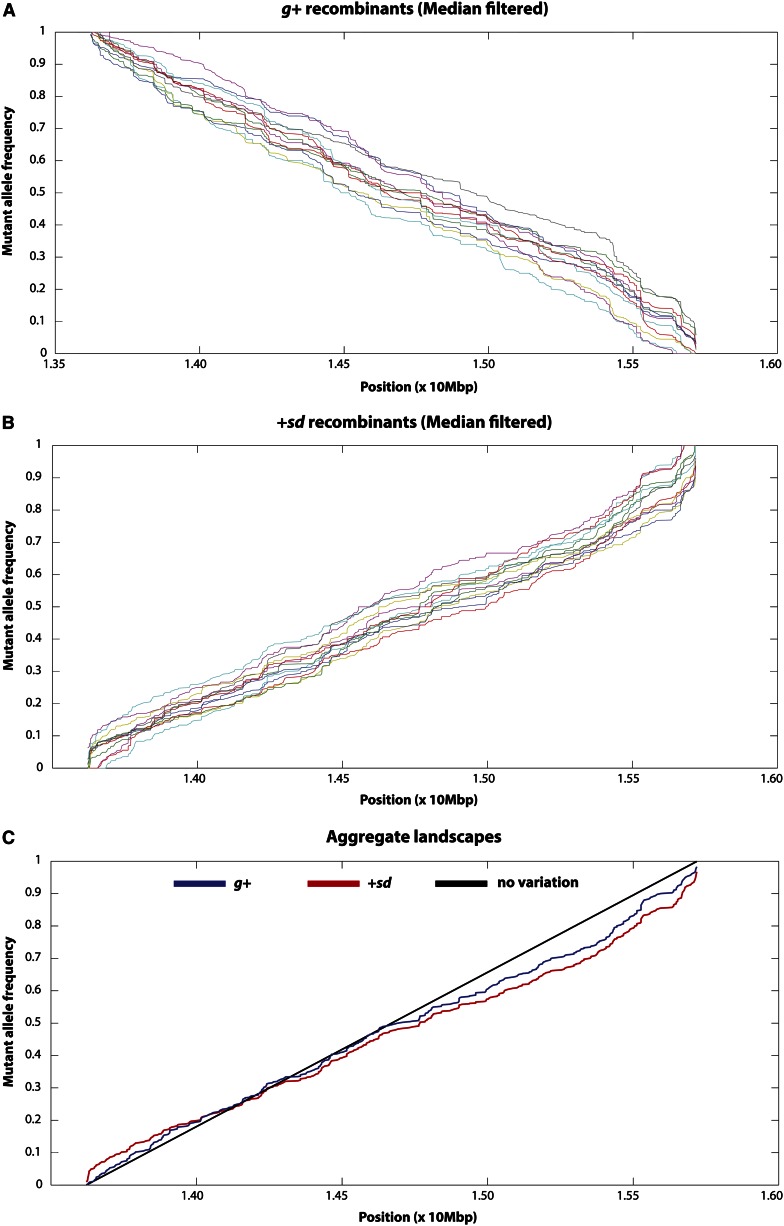
Even after choosing a select set of SNPs and excluding a majority of the recombinant pools, there remained substantial variability in allele frequency estimates. For this reason, we chose a robust approach for inferring recombination landscapes. Specifically, we used a median filter on the allele frequency estimates from each pool. The allele frequency at each SNP was estimated as the median frequency within a 2k + 1 SNP window (including itself, k 5′ SNPs, and k 3′ SNPs). For SNPs too close to either the garnet or the scalloped locus, the window was completed by assuming no recombination outside the g-sd interval. For each recombinant pool, k was chosen to be the minimum value for which the median-smoothed allele frequency estimates varied monotonically across the g-sd interval (File S1). In this way, recombination landscapes were inferred for each pool, using the change in (median-smoothed) estimated allele frequency as a function of physical distance (Figure 3). Figure S3 illustrates the relationship between the raw data and the median-smoothed data. By averaging the median-smoothed allele frequency estimates at each site across the 13 +sd pools, a single composite landscape was obtained. A similar composite was obtained for the 12 g+ recombinant pools. The allele frequencies of these composites were averaged to obtain a single recombination landscape for the g-sd interval. We then linearly interpolated allele frequency between each SNP and inferred fine-scale crossover distribution based on changes in allele frequency in 5-, 10-, 20-, 50-, and 100-kb windows (File S1). At each granularity, a chi-square test was used to compare the inferred crossover distribution to what would be expected in the absence of recombination rate heterogeneity.
Figure 3.
Median-filtered allele frequency landscape of the frequency of the g-sd parental allele as a function of physical position for each of the pools surviving our stringent filtering criteria for (A) the 12 g+ pools and (B) the 13 +sd pools. The aggregate landscapes (averaged across pools within recombinant phenotype classes) are depicted in C. In this aggregate landscape plot, the black line depicts uniform distribution of crossover events across this interval. The aggregate landscape for the +sd recombinant individuals is plotted in red, and the aggregate landscape for the g+ recombinant individuals is plotted in blue. Note that this latter landscape has been reflected for ease of comparing the allele frequency landscapes between the two recombinant phenotype classes.
Genomic correlates
To correlate local recombination rate with various aspects of genomic context, we examined local codon bias, GC content, gene density, repeat content, nucleotide diversity, nucleotide divergence, and sequence motifs. To estimate codon bias, we retrieved the sequences of all genes located in the g-sd region based on Release 5.4 of the D. melanogaster genome. We concatenated all exonic sequence within individual genes and estimated codon bias for each gene, using a standalone implementation of codonW (downloaded from http://codonw.sourceforge.net). We used the codons defined as preferred in D. melanogaster to estimate the frequency of optimal codons (FOP) for each gene in this region. We assigned genes to intervals based on physical position; if the midpoint of the gene was included in a given interval, it was assigned to that interval.
Overall GC content for a given window was calculated by simply counting the incidence of G’s and C’s and dividing it by the total number of nucleotides in that window. We used percentage of cover by coding sequence (CDS) as an indicator of gene density. Annotations for this 2.1-Mb region on the X chromosome were kindly provided by R. Kulathinal (personal communication). These annotations are based on annotations of Release 5.4 of the D. melanogaster genome as well as information from the RedFly database and a curated database of footprinting literature. For each window, we calculated the number of bases annotated as protein coding and used this to estimate the percentage of CDS cover for each window. To estimate repeat density, we first used RepeatMasker (Smit et al. 1996–2010) to identify repetitive regions within the g-sd region. We used the RepeatMasker output to estimate the fraction of each window that contained repetitive sequence. We estimated simple repeat cover, low complexity sequence cover, transposable element cover, and total repeat cover (the sum of those three classes).
Nucleotide diversity was estimated in each interval, using population resequencing data from 20 lines of D. melanogaster from Uganda (Singh et al. 2013). We restricted our analysis of polymorphism to third codon positions and included only windows containing at least 500 such sites. We estimated both average pairwise differences among alleles π (Nei 1987) and Watterson’s estimator ϑw (Watterson 1975). To estimate divergence, we generated pairwise alignments between D. melanogaster and D. sechellia for the garnet-scalloped region by parsing the net.axt files from the University of California, Santa Cruz genome browser. This net.axt file was based on Release 3 of the D. melanogaster genome and release 1 of the D. sechellia genome. We used these alignments to calculate Dxy (Nei 1987).
Finally, we searched for motifs enriched in high recombination areas, using Multiple Em for Motif Elicitation (MEME) (Bailey and Elkan 1994). We used a “discriminative” motif discovery approach, which attempts to identify motifs specific to regions of interest and not specific to other, similar sequences. We searched for motifs enriched in the three 10-kb windows with the highest estimated crossover rate relative to the three 10-kb windows with the lowest crossover rate. We also explored the correlation between estimated recombination rate and previously reported sequence motifs as well as several simple repeats, using a custom Perl script.
Results and Discussion
We quantified the fine-scale distribution of crossover events in the 2.1-Mb region separating the genes garnet (g) and scalloped (sd) on the X chromosome of D. melanogaster. We focused on this region in part because broad-scale estimates of crossing-over rates are high (2–7 cM/Mb) and in part because the visible markers bounding this region are extremely easy to visualize, thus facilitating a large-scale screen for recombinant individuals. We crossed a wild-type X-extracted line of D. melanogaster from Uganda to g-sd double-mutant males (Figure 1) (Both the wild-type X and the g-sd-bearing X were made isogenic by extraction with the FM7a balancer). The resulting F1 females were doubly heterozygous for our visible markers, and because crossing over in Drosophila takes place only in females, it is crossover events in these F1 females that we ultimately scored. The F1 females were backcrossed to g-sd males and the male progeny from this cross (BC1 males) were scored for the recombinant phenotype. Because males were hemizygous for the X chromosome, recessive mutations are exposed and recombinant males should carry one visible mutation but not the other (Figure 1). Other chromosomes were not controlled, but they will assort randomly and so our study integrates over all possible background genotypes on these uncontrolled chromosomes. This approach facilitates identifying individual males with an odd number of crossovers between garnet and scalloped. Given crossover interference in Drosophila (Sturtevant 1913; Muller 1916), we do not believe that double and triple crossovers play a significant role at this physical scale (Cirulli et al. 2007; Stevison and Noor 2010).
In total, we screened 92,105 BC1 males for the recombinant phenotype. This screen yielded 6176 recombinant males, 2483 of the +sd phenotype class of 4233 of the g+ phenotype class. This significantly deviates from the expected 1:1 ratio (P < 0.0001, G-test), although the reason underlying this deviation remains unclear. One unlikely possibility is phenotyping error; the sd mutation was noted to have incomplete penetrance in our BC1 progeny, which could result in +sd recombinant males being overlooked in this screen. In addition, natural variation in eye color might result in yielding false positive g+ recombinant individuals. However, given our approach for inferring recombination maps (see the section Inference of fine-scale crossover distribution), we do not believe that these phenotyping challenges will introduce artificial heterogeneity in fine-scale rates of crossing over. That is, including false positive g+ individuals will introduce noise; although this will make our statistical inference more challenging, it should not falsely introduce a signal of crossover rate heterogeneity. Similarly, as long as the putatively overlooked +sd recombinant males are not biased with respect to crossover location, failing to include these individuals in our screen simply reduces our sample size. Other possible explanations for the deviation from the expected 1:1 ratio include segregation distortion or viability defects associated with the sd chromosome. Consistent with a viability defect associated with the sd chromosome, we also recovered an excess of wild-type vs. g-sd (nonrecombinant) male progeny (67,137 wild type vs. 24,698 g-sd). However, again assuming that the viability effects of the sd phenotype are independent of crossover location, this should not compromise our estimation of fine-scale crossover rates.
In spite of the inequality of the two recombinant phenotype classes, the overall genetic distance between garnet and scalloped revealed by our screen was 7.3 cM, in close agreement with the published map distance of 7.1 cM (Lindsley and Grell 1967). This is arguably inconsistent with segregation distortion, as this would yield a systematic reduction in the estimated genetic distance between our two visible markers. However, this is potentially consistent with phenotyping error; if our rates of false positive g+ identification are similar to our rates of overlooking +sd recombinants, this would yield a genetic distance in our screen similar to the true map distance. This is also consistent with viability defects associated with the sd chromosome, as this would yield similar reductions in both the +sd (recombinant) and g-sd (nonrecombinant) phenotype classes. Note that the deviations from 1:1 expectation are similar in both the recombinant (1.7:1 g+:+sd) and the nonrecombinant (2.7:1 ++:g-sd) phenotype classes. We thus believe that the most likely explanation for the skew in recombinant recovery toward the g+ class is a viability defect associated with the sd chromosome, but we cannot exclude a contribution of some degree of phenotyping error.
Sequencing
We sequenced our parental lines in eight pools composed of different proportions of the double-mutant and wild-type parents (Materials and Methods). In all, 41,914,472 eighty-six-base-pair single-end Illumina reads were obtained across all pools (2,768,127–7,302,199 reads per pool). On average, >80% of these reads map uniquely to the D. melanogaster genome (77–89% in each pool). Of these uniquely mapping reads, 84% map to the g-sd region (76–87% per pool), suggesting that the Nimblegen targeted enrichment was highly efficacious. We used these 28,327,993 uniquely mapping reads localized to the g-sd region for SNP identification (Materials and Methods). Our stringent approach yielded 451 SNPs differentiating our parental lines. The median distance between adjacent SNPs is 2.6 kb (average = 4.6 kb), indicative of a high density of informative markers for inference of crossover distribution.
The 66 pools of recombinant males were also sequenced using single-end 86-bp Illumina reads. In total, 255,415,020 reads were obtained (66,846–13,437,128 per pool). Over 72% (155,910,649 reads) of uniquely mapping reads localize to our target region (40–94% per pool). This reflects 6088× coverage of the g-sd region across all 66 pools (3–524× per pool).
Inference of fine-scale crossover distribution
We inferred the crossover distribution based on allele frequency changes within each pool of recombinants across the g-sd interval. The intuition behind this inference is as follows. For a pool of 100 g+ recombinant males, the frequency of the g-sd allele should start at 1 at the 5′ end of this region and should decrease monotonically across the 2.1-Mb region with a final g-sd allele frequency of 0 at the 3′ end of the g-sd region. The reciprocal is of course true for the +sd recombinant males. Thus, we can estimate the fraction of crossover events localizing to individual windows by tracking the change in allele frequency between adjacent windows. Within each phenotype class, allele frequency estimates of the g-sd allele as a function of physical distance across pools were highly similar (Figure 3). We thus averaged across pools to create an aggregate +sd landscape and an aggregate g+ landscape. Importantly, the smoothed composite landscapes based on the 13 +sd pools and 12 g+ pools were highly correlated (r = 0.61, P < 0.001; Figure 3). This strongly supports the conjecture that potential phenotyping error, segregation distortion, and/or viability defects associated with the +sd recombinant phenotype do not compromise our ability to infer fine-scale crossover distribution. Given the strong concordance between these two composite landscapes, we averaged across these landscapes to generate a single allele frequency landscape. Linear interpolation of allele frequencies between SNPs facilitated estimating rates of crossing over in windows of arbitrary size; for our analyses we estimated crossing-over frequencies in windows of 5, 10, 20, 50, and 100 kb.
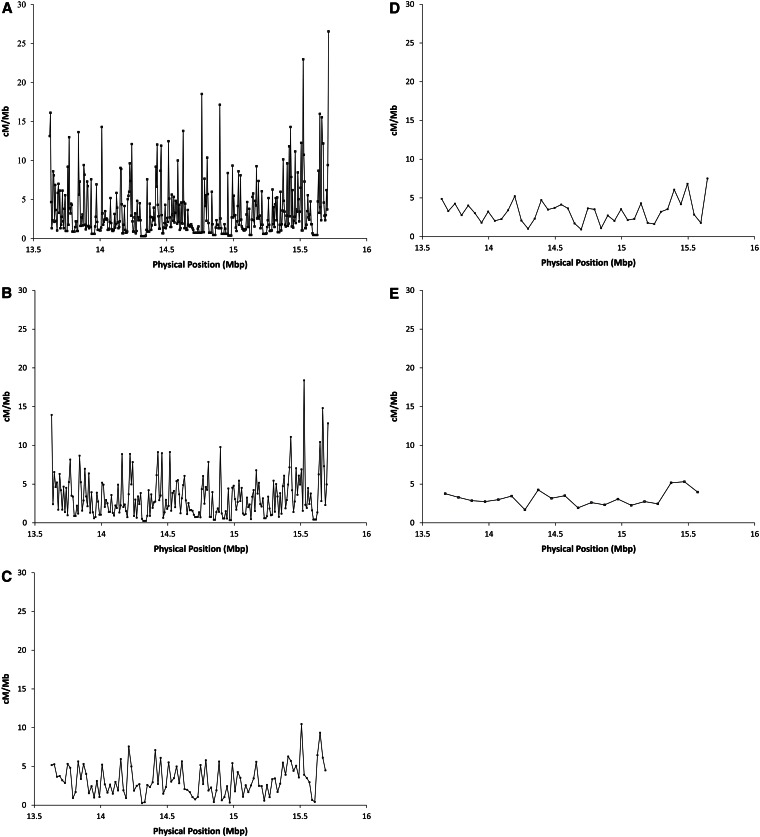
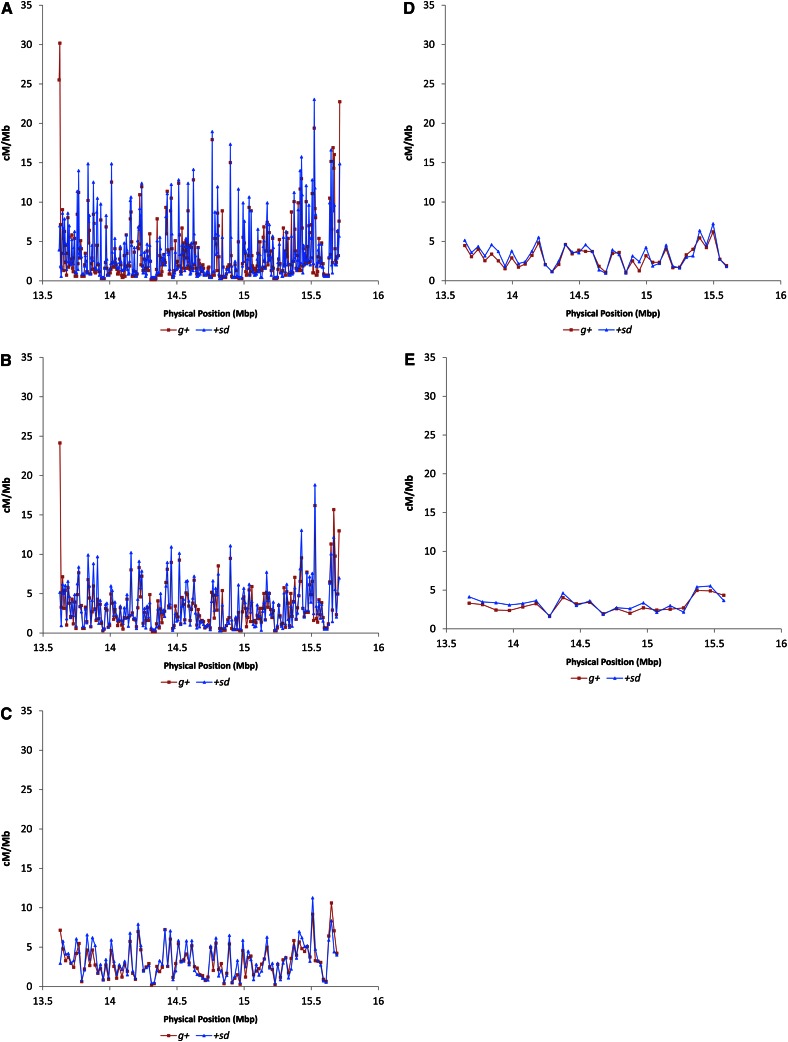
There is significant heterogeneity in crossover distribution at all physical scales [P < 0.001, χ2-test, all window sizes (Materials and Methods)]. The magnitude of the fluctuations in crossover rates varies as a function of window size (Figure 4). At the 5-kb scale, rates of crossing vary >90-fold (estimated as the maximum observed crossover rate divided by the minimum observed crossover rate). Increasing window size to 10 kb reduces the magnitude of crossover rate heterogeneity to 72-fold, and window sizes of 20, 50, and 100 kb show further dampening of crossover rate variation (41-, 8-, and 3-fold, respectively). This is to be expected, given that increasing window size leads to smoothing of the local crossover distribution. Importantly, we do not believe that the increased magnitude of crossover rate fluctuation with smaller window size is a function of sampling error. Repeating this scale analysis with each recombinant phenotype class separately yields highly similar recombinant landscapes between the g+ vs. +sd samples (Figure 5). The estimated crossover rates for the two classes of flies are highly and significantly correlated (r = 0.84, 0.82, 0.90, 0.91, and 0.90 for the 5-, 10-, 20-, 50-, and 100-kb scales, respectively; P < 0.001, all comparisons).
Figure 4.
Crossover rate (cM/Mbp) as a function of physical position estimated in nonoverlapping windows of (A) 5 kb, (B) 10 kb, (C) 20 kb, (D) 50 kb, and (E) 100 kb.
Figure 5.
Crossover rate (cM/Mbp) as a function of physical position estimated in nonoverlapping windows of (A) 5 kb, (B) 10 kb, (C) 20 kb, (D) 50 kb, and (E) 100 kb. The two recombinant phenotype classes are presented separately, with the recombinational landscape derived from g+ flies presented in red and the landscape derived from the +sd flies in blue.
It should be noted that we are not arguing that any particular window size represents the best fit to our empirical data. Rather, we argue that the degree to which crossover rate is heterogeneous depends heavily on window size and moreover, that it remains to be determined which window size is most informative in the context of organismal fitness and function.
The observed magnitude of fluctuations in rates of crossing over is largely consistent with previous work. Analysis of crossover distribution of a 1.2-Mb region between the genes white and echinus on the D. melanogaster X chromosome revealed 3.5-fold variation with markers spaced 40–300 kb apart (Singh et al. 2009). Similarly, analysis of crossover distribution in a 2-Mb region of D. pseudoobscura revealed 40-fold variation in rates of crossing over, using markers 2–40 kb apart (Cirulli et al. 2007); this degree of heterogeneity agrees well with our results at a 50-kb scale. This comparison is particularly interesting given that rates of crossing over are elevated in D. pseudoobscura relative to D. melanogaster (Hamblin and Aquadro 1999; Ortiz-Barrientos et al. 2006). That we see similar fluctuations in rates of crossing over in D. melanogaster might suggest that the degree to which crossover rates are heterogeneous throughout the genome is conserved in Drosophila even if genome-wide levels of crossing over have diverged. However, whole-genome analysis of crossover rates in D. melanogaster reveals 15- to 20-fold variation at the 100-kb scale even in noncentromeric regions (Comeron et al. 2012), which is far greater than what we observe here at a comparable scale. This could result from differences in rates of crossing over among strains (Brooks and Marks 1986; Comeron et al. 2012), or the reduction in fine-scale heterogeneity in crossing-over rate could be specific to the g-sd region.
Sequence motifs
Although it is clear from this work and others that the distribution of crossovers in Drosophila is significantly nonuniform (e.g., Cirulli et al. 2007; Kulathinal et al. 2008; Singh et al. 2009; Stevison and Noor 2010), the genetic basis of crossover distribution remains unknown. We used MEME (Bailey and Elkan 1994) to identify possible sequence motifs associated with crossover locations in this region. Of the motifs identified as enriched in the three 10-kb windows with the highest rates of crossing over relative to the three 10-kb windows with the lowest rates of crossing over, none showed a significant correlation between incidence within a window and crossover rate across the entire g-sd region at any window size. However, we do note that simple repeats CAA, TAT, and CA as well as poly(A) stretches are significantly positively correlated with recombination rate at the 5-, 10-, and 20-kb scales (r > 0.1, P < 0.03 in all cases). The CA repeat and poly(A) stretches have also independently been found to be associated with crossover locations in D. melanogaster (Comeron et al. 2012). These authors also noted that poly(A) stretches are associated with crossover events in yeast (Mancera et al. 2008), and the dinucleotide CA repeat stimulates homologous recombination in human cells in culture (Wahls et al. 1990). In addition, we find significant correlations between motif incidence and recombination rate for the motif AAAC[CG]AAA, a core of a previously identified motif in D. melanogaster (Comeron et al. 2012), as well as the motif ATGGAAA, which has also been previously implicated with crossover distribution in D. melanogaster (Miller et al. 2012). The former is correlated with recombination rates at the 5-, 10-, and 20-kb scales (r > 0.095, P < 0.05, all scales) while the latter is significantly correlated with recombination rates at all scales except the 50-kb scale (r > 0.13, P < 0.006, all scales). We found no other previously reported motifs or short repeats associated with rates of crossing over in D. melanogaster or D. pseudoobscura (Cirulli et al. 2007; Kulathinal et al. 2008; Stevison and Noor 2010; Comeron et al. 2012; Miller et al. 2012), humans (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010), or dogs (Axelsson et al. 2012) to be correlated with recombination rates at multiple window sizes.
The complete lack of overlap between sequence motifs associated with rates of crossing over between D. melanogaster and D. pseudoobscura may suggest that sequence motifs mediating crossover distribution have diverged significantly between species. The limited overlap among all motifs currently or previously correlated with crossover rates within D. melanogaster (Comeron et al. 2012; Miller et al. 2012) is more puzzling and may indeed suggest that in contrast to humans and mice (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010), crossover distribution is not largely determined by a single sequence motif in D. melanogaster. Instead, crossover distribution could be determined by multiple sequence motifs or some other aspect of genomic context such as chromatin structure. However, that motifs AAAC[CG]AAA and ATGGAAA have now been identified in two independent studies using different strains indicates that these motifs may play some role in crossover distribution in D. melanogaster and certainly merit further investigation.
Genomic context
Although our recombination landscapes show clear heterogeneity in fine-scale crossover distribution, the functional and evolutionary consequences of this heterogeneity remain unknown. To investigate the implications of this heterogeneity for genome evolution, we examined the correlation between local crossover rate and several aspects of genomic context including GC content, codon bias, coding sequence content, and repeat content. GC content was not significantly correlated with crossover rate for any window size (Table 1). This is unexpected given that previous work reported a significant relationship between these two features on the X chromosome of D. melanogaster, a relationship that does not appear to be exclusively driven by GC content in regions of severely depressed rates of crossing over (Singh et al. 2005). However, previous analysis of the fine-scale correlation between GC content and crossover rate (Singh et al. 2009) also failed to recover the expected negative correlation between these two features. This may reflect the reduced sample size of the fine-scale studies or may suggest that the correlation between GC content and recombination rate is apparent only in the context of broad-scale recombination rate estimation.
Table 1. Pearson’s correlation coefficient of local crossover rate and genomic context.
| 5 kb | 10 kb | 20 kb | 50 kb | 100 kb | |
|---|---|---|---|---|---|
| GC content | −0.092 | −0.021 | −0.075 | −0.13 | −0.049 |
| FOP | −0.12 | −0.14 | −0.098 | −0.14 | −0.082 |
| CDS cover | 0.087 | 0.079 | 0.017 | 0.19 | 0.15 |
| Total repeat cover | −0.041 | 0.013 | 0.031 | 0.11 | 0.26 |
| ϑw | −0.057 | 0.035 | −0.011 | 0.063 | −0.31 |
| π | −0.013 | 0.010 | 0.0011 | 0.13 | −0.16 |
| Dxy | 0.15 | 0.0099 | 0.068 | 0.17 | 0.17 |
Boldface type indicates significance at the 5% level.
In contrast, codon bias (as measured by FOP; see Materials and Methods) is significantly negatively correlated with recombination rate as has been previously reported for the X chromosome (Singh et al. 2005, 2009) at the 5- and 10-kb scales (r < −0.12, P < 0.04, both scales; Table 1). Although these two parameters are also negatively correlated to a similar degree at the 20-, 50-, and 100-kb scales, these correlations are not statistically significant, perhaps owing to the limited number of windows with these increased window sizes. This thus suggests that the X chromosome-wide negative correlation between recombination rate and codon bias is recapitulated at a fine scale. Why the X chromosome differs from the autosomes in the direction of this correlation remains a puzzle, particularly since the X chromosome correlation differs from the expectations based on the supposed increased efficiency of selection in regions of high recombination.
We recover no significant association between crossover rate and gene density (measured as the fraction of each window annotated as coding sequence; Table 1), which is consistent with previous results from D. pseudoobscura (Kulathinal et al. 2008). In addition, these data are consistent with the recent finding in D. melanogaster of no enrichment of crossover events in genic vs. intergenic sequences (Comeron et al. 2012). Repeat cover was not significantly correlated with crossover rate at any scale (Table 1), echoing findings in D. pseudoobscura and D. persimilis (Kulathinal et al. 2008; Stevison and Noor 2010), indicating that this aspect of genomic context may not be relevant for crossover distribution at a fine scale in Drosophila.
The relationship between nucleotide polymorphism and crossover rate enjoys a rich empirical and theoretical history in Drosophila. The positive correlation between these two parameters was first discovered by Begun and Aquadro (1992) and has since been confirmed at both coarse and fine scales in D. melanogaster and other Drosophilids (e.g., Begun et al. 2007; Shapiro et al. 2007; Kulathinal et al. 2008; Sackton et al. 2009; Stevison and Noor 2010). This reduction in polymorphism with decreased recombination has been argued to result from genetic hitchhiking (Maynard Smith and Haigh 1974; Gillespie 2000), background selection (Charlesworth et al. 1993), or some combination of both (Kim and Stephan 2000). In spite of the strong expectation, we fail to recover a significant association between third codon position diversity and local recombination rate at any scale (Table 1). Although consistent with previous fine-scale dissection of crossover rate variation and its correlates on the D. melanogaster X chromosome (Singh et al. 2009), this lack of association between polymorphism and recombination in our data is somewhat challenging to explain. One possibility is that we do not have sufficient data at the relevant physical scale to recover this association. If the well-described correlation between recombination rate and nucleotide diversity is driven by genetic hitchhiking, for instance, and the molecular footprint of a selective sweep is on the order of 50–100 kb, then perhaps no correlation between recombination rate and diversity would be observed at 5- to 20-kb scales. Previous work reporting a significant correlation between diversity and recombination rate was based on window sizes of 50 kb–1.5 Mb (Begun et al. 2007; Kulathinal et al. 2008; Sackton et al. 2009; Stevison and Noor 2010); although we have data at the 50- and 100-kb scales, these data are limited and thus the lack of observed correlation at these scales may reflect sample size limitations.
Another possible confounding factor is that of timescale. Although crossover rate appears to be conserved among closely related species in Drosophila (at the ≥50-kb scale) (Begun et al. 2007; Stevison and Noor 2010), little is known regarding the stability of the recombination landscape at smaller physical scales. If this landscape is more labile evolutionarily, this too could contribute to our lack of observed correlation between nucleotide diversity and recombination rate, as such a correlation results from repeated hitchhiking or background selection events over evolutionary time.
Recent evidence tentatively supports a weak relationship between interspecific divergence and local crossover rate at the scale of entire chromosomes or genome-wide (Begun et al. 2007; Kulathinal et al. 2008), although ancestral polymorphism may be driving this observation for at least one of these reports (Noor 2008). We find no significant association between crossover rate and divergence with D. sechellia at any scale (Table 1). Given our sample size and the purported weakness of the correlation, it is not clear whether this reflects a lack of association or a lack of statistical power.
Future directions
Moving forward, it is clear that much work is needed to determine the genetic basis of crossover distribution in this species in particular and Drosophila in general. In addition, as we continue to refine our understanding of the scale at which crossover rates are heterogeneous, it will remain important to determine the relevant scale to consider in light of genome evolution and the effects of genetic hitchhiking and background selection. Furthermore, the scale of crossover rate heterogeneity that is most important for organismal fitness and function is an important unknown and merits careful future consideration.
Supplementary Material
Acknowledgments
The authors thank A. Kelkar, D. Cloutier, D. Fister G. Chi, J. Duggal, E. Kwan, H. Flores, and V. Bauer Dumont for their assistance with phenotype scoring. The authors also thank two anonymous reviewers whose thoughtful comments markedly improved this manuscript. This work was supported in part by a priming grant from the Cornell Center for Comparative and Population Genomics (to A.G.C., C.F.A., and N.D.S.) and by National Institutes of Health grant R01-GM36431 (to C.F.A.).
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Anderson L. K., Doyle G. G., Brigham B., Carter J., Hooker K. D., et al. , 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Auton A., Fledel-Alon A., Pfeifer S., Venn O., Segurel L., et al. , 2012. A fine-scale chimpanzee genetic map from population sequencing. Science 336: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E., Webster M. T., Ratnakumar A., Ponting C. P., Lindblad-Toh K., et al. , 2012. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 22: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T. L., and C. Elkan, 1994 Fitting a mixture model by expectation maximization to discover motifs in biopolymers, pp. 28–36 in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA. [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in Drosophila melanogaster. Nature 356: 519–520. [DOI] [PubMed] [Google Scholar]
- Begun D. J., Holloway A. K., Stevens K., Hillier L. W., Poh Y.-P., et al. , 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5: 2534–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Murray J. C., Sheffield V. C., White R. L., Weber J. L., 1998. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 63: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks L. D., Marks R. W., 1986. The organization of genetic variation for recombination in Drosophila melanogaster. Genetics 114: 525–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli E. T., Kliman R. M., Noor M. A. F., 2007. Fine-scale crossover rate heterogeneity in Drosophila pseudoobscura. J. Mol. Evol. 64: 129–135. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G., Wen X. Q., Ober C., Pritchard J. K., Przeworski M., 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319: 1395–1398. [DOI] [PubMed] [Google Scholar]
- Crawford A. M., Dodds K. G., Ede A. J., Pierson C. A., Montgomery G. W., et al. , 1995. An autosomal genetic linkage map of the sheep genome. Genetics 140: 703–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W. F., Miller J., Steen R., Merchant M. A., D. Damron-Boles et al, 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149–152. [DOI] [PubMed] [Google Scholar]
- Dumont B. L., Broman K. W., Payseur B. A., 2009. Variation in genomic recombination rates among heterogeneous stock mice. Genetics 182: 1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P., Smith N. G. C., 2005. A novel method with improved power to detect recombination hotspots from polymorphism data reveals multiple hotspots in human genes. Am. J. Hum. Genet. 77: 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 2000. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graffelman J., Balding D. J., Gonzalez-Neira A., Bertranpetit J., 2007. Variation in estimated recombination rates across human populations. Hum. Genet. 122: 301–310. [DOI] [PubMed] [Google Scholar]
- Hamblin M. T., Aquadro C. F., 1999. DNA sequence variation and the recombinational landscape in Drosophila pseudoobscura: a study of the second chromosome. Genetics 153: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y., Yano M., Shomura P., Sato M., Shimano T., et al. , 1998. A high-density rice genetic linkage map with 2275 markers using a single F-2 population. Genetics 148: 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt W., Fischer T. C., Winderl S., Fransz P., Torres-Ruiz R. A., 2001. The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. Plant J. 27: 285–296. [DOI] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2000. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics 155: 1415–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D., Wilson H. E., Mitchell A. R., Taylor B. A., Cooke H. J., 1994. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma 103: 46–55. [DOI] [PubMed] [Google Scholar]
- Kulathinal R. J., Bennettt S. M., Fitzpatrick C. L., Noor M. A. F., 2008. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc. Natl. Acad. Sci. USA 105: 10051–10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie E. J., Roeder G. S., 1988. A yeast centromere acts in cis to inhibit meiotic gene conversion of adjacent sequences. Cell 52: 863–873. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and E. H. Grell, 1967 Genetic Variations of Drosophila melanogaster. Pub. 627, Carnegie Institute, Washington, DC. [Google Scholar]
- Lynn A., Koehler K. E., Judis L., Chan E. R., Cherry J. P., et al. , 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296: 2222–2225. [DOI] [PubMed] [Google Scholar]
- Mahtani M. M., Willard H. F., 1998. Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res. 8: 100–110. [DOI] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Miller D. E., Takeo S., Nandanan K., Paulson A., Gogol M. M., et al. , 2012. A whole-chromosome analysis of meiotic recombination in Drosophila melanogaster. Genes Genomes Genet. 2: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H., 1912. Complete linkage in the second chromosome of the male of Drosophila. Science 36: 719–720. [Google Scholar]
- Muller H. J., 1916. The mechanism of crossing-over. Am. Nat. 50: 193–221. [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C., et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Adachi Y., Funahashi S., Niwa O., Yanagida M., 1986. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 5: 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M. W., Broman K. W., Mellersh C. S., Ray K., Acland G. M., et al. , 1999. A second-generation genetic linkage map of the domestic dog, Canis familiaris. Genetics 151: 803–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York. [Google Scholar]
- Neumann R., Jeffreys A. J., 2006. Polymorphism in the activity of human crossover hotspots independent of local DNA sequence variation. Hum. Mol. Genet. 15: 1401–1411. [DOI] [PubMed] [Google Scholar]
- Noor M. A. F., 2008. Connecting recombination, nucleotide diversity and species divergence in Drosophila. Fly 2: 255–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P. L., Goodstadt L., Bayes J. J., Birtle Z., Roach K. C., et al. , 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D., Audrey S. C., Noor M. A. F., 2006. A recombinational portrait of the Drosophila pseudoobscura genome. Genet. Res. 87: 23–31. [DOI] [PubMed] [Google Scholar]
- Parvanov E. D., Petkov P. M., Paigen K., 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Aquadro C. F., 2006. History and structure of sub-Saharan populations of Drosophila melanogaster. Genetics 174: 915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn M. I., Solari A. J., 1986. Recombination nodules in the oocytes of the chicken, Gallus domesticus. Cytogenet. Cell Genet. 43: 187–193. [DOI] [PubMed] [Google Scholar]
- Round E. K., Flowers S. K., Richards E. J., 1997. Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res. 7: 1045–1053. [DOI] [PubMed] [Google Scholar]
- Sackton T. B., Kulathinal R. J., Bergman C. M., Quinlan A. R., Dopman E. B., et al. , 2009. Population genomic inferences from sparse high-throughput sequencing of two populations of Drosophila melanogaster. Genome Biol. Evol. 1: 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E., Armstrong S. J., Santos J. L., Franklin F. C. H., Jones G. H., 2002. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C. H., Hubisz M. J., Lu J., et al. , 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104: 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman J. D., Stack S. M., 1995. Two-dimensional spreads of synaptonemal complexes from solanceous plants VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141: 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A., Perlman H., Yan Y. L., Walker C., Corley-Smith G., et al. , 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Davis J. C., Petrov D. A., 2005. Codon bias and noncoding GC content correlate negatively with recombination rate on the Drosophila X chromosome. J. Mol. Evol. 61: 315–324. [DOI] [PubMed] [Google Scholar]
- Singh N. D., Aquadro C. F., Clark A. G., 2009. Estimation of fine-scale recombination intensity variation in the white-echinus interval of D. melanogaster. J. Mol. Evol. 69: 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Jensen J. D., Clark A. G., Aquadro C. F., 2013. Inferences of demography and selection in an African population of D. melanogaster. Genetics 193: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Gregoretti I. V., Brick K., Khil P. J., Camerini-Otero R. D., et al. , 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley, and P. Green, 1996–2010 RepeatMasker Open-3.0. Available at http://www.repeatmasker.org.
- Stevison L. S., Noor M. A. F., 2010. Genetic and evolutionary correlates of fine-scale recombination rate variation in Drosophila persimilis. J. Mol. Evol. 71: 332–345. [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1913. A third group of linked genes in Drosophila ampelophila. Science 37: 990–992. [DOI] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D., 1990. The Z-DNA motif D(Tg)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human-cells in culture. Mol. Cell. Biol. 10: 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1975. Number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Zenger K. R., McKenzie L. M., Cooper D. W., 2002. The first comprehensive genetic linkage map of a marsupial: The tammar wallaby (Macropus eugenii). Genetics 162: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.