Abstract
This paper explores the application of text mining to the problem of detecting protein functional sites in the biomedical literature, and specifically considers the task of identifying catalytic sites in that literature. We provide strong evidence for the need for text mining techniques that address residue-level protein function annotation through an analysis of two corpora in terms of their coverage of curated data sources. We also explore the viability of building a text-based classifier for identifying protein functional sites, identifying the low coverage of curated data sources and the potential ambiguity of information about protein functional sites as challenges that must be addressed. Nevertheless we produce a simple classifier that achieves a reasonable ∼69% F-score on our full text silver corpus on the first attempt to address this classification task. The work has application in computational prediction of the functional significance of protein sites as well as in curation workflows for databases that capture this information.
Keywords: text mining, information extraction, machine learning, catalytic site, biomedical literature, biomedical natural language processing, protein functional sites
1. Introduction
To facilitate progress in understanding and prediction of protein function, it is critical to populate databases with information about the physical aspects of protein function,1,2 including the location of functionally important residues on the protein and the biochemical properties of ligand-protein interactions. Drug discovery for treatment of diseases proceeds systematically from this information; drugs can be designed to target a specific functionally important site on the protein and can become the basis for large-scale drug screening experiments. However, such physical information is currently scarce compared to more qualitative information about protein function, such as pathway assignments or Gene Ontology annotations, despite its critical importance for characterization and eventual manipulation of protein behavior.
In previous work, we have shown that text mining can be integrated with protein structure-based methods for prediction of protein functional sites to identify high-quality predictions that are supported by evidence in the biomedical literature.3 The method we developed in that work is called Literature-Enhanced Automated Prediction of Functional Sites, or LEAP-FS. While that work showed that we were able to recover a good proportion of curated functional site annotations in existing databases, it did not attempt to classify the functional importance of each site more specifically, e.g., identifying catalytic sites. Automated identification of catalytic sites in the biomedical literature has application, for example, in genome annotation pipelines and in drug design. Such predictions provide fine-grained information regarding the biological significance of a specific functional site, influencing both the overall understanding of the role of a protein in a biological process, and how that protein might be modulated through drug intervention. The classification can also be employed within the curation pipeline for the development of resources such as the Catalytic Site Atlas4 to assist in identifying meaningful literature to be curated, and to highlight specific residue mentions within that literature that should be considered for inclusion in the resource.
Development of a functional site classifier would lay the foundation for generalizing the methods we previously developed in LEAP-FS. First, it supports the generalization of the methods to a broader set of the biomedical literature; we would like to be able to identify functionally important protein residues through analysis of literature that is not directly connected to proteins via curated links. The classifier would play a role in that generalization by assisting in the recognition of literature where residues are specifically discussed as being catalytically active. Second, classification of a residue mention as within a catalytic site gives us increased confidence that the residue mention is relevant for prediction of functional sites – it provides evidence that the residue is mentioned due to its functional importance.
In this work, we take a step towards finer-grained analysis of amino acid residues mentioned in text. We provide an analysis of the residues identified in publications linked to proteins in the Protein Data Bank in order to understand what relevant information is readily available in curated data sources. We further explore the development of a classifier which can classify amino acid residues as catalytic residues based on the textual context of the residue mention, e.g. for the positive cases below.
We propose a mechanism involving general base catalysis by the carboxy-terminal Trp270 carboxyl group [PMID 12356304]
it is possible that Arg 381 is one of the catalytic bases previously observed [PMID 9174368]
We find that while our classifier does a reasonable job in classifying positive instances, there is significant ambiguity around negative instances, both for the purpose of developing training material and during classification of held-out test data.
2. Related Work
BindingMOAD is a database of protein ligand-binding sites, which is updated through curation of approximately 2000 full text publications each year.5 To assist with this curation, a natural language processing system called BUDA, using the rule-based GATE system6 (gate.ac.uk) was developed to identify articles relevant to binding, and to extract protein-ligand interactions along with quantitative binding affinities. However, it has been noted that the curation of BindingMOAD cannot rely completely on this automated information extraction, due to ambiguities that persist in the extracted information.7 Furthermore, specific evaluation results of BindingMOAD have not been made available so we are unable to make detailed comparisons.
The Open Mutation Miner8 system extracts mutation information from full text publications and identifies mutation impact information including protein properties such as kinetic and stability data. The system also aims to capture protein function impacts, through detection of Gene Ontology9 molecular function terms via dictionary look-up with some morphological processing. A rule-based strategy is employed for association (grounding) of an impact to a mutation. We refer the reader to their work8 for a thorough review of other text mining systems that focus on extracting mutation information. The scope of these systems is different than our work, as we are interested in detecting all specific protein residue mentions, not only mutation sites, and we focus on catalytic and ligand binding sites.
Nagel et al10 address annotation of individual protein residues, focusing on binding and enzymatic activity. The goals of that work are closest to our own goals. They develop a mutual information-based classifier for detection and categorization of functional terms within a sentence, achieving F-scores of 0.57 for binding and 0.27 for enzymatic activity on a very small corpus of 100 manually annotated abstracts; association of residues to these terms is achieved using syntactic relationships. However, only 16 extracted functional annotations are produced by the system. The authors compared those annotations to information in UniProt and did find that that the text mining identified correct annotations, of which 11 were not already present in the resource. The evaluation of this work was very limited in scope; we will present a much larger-scale evaluation.
3. Methods
3.1. Amino Acid residue detection in text
To detect residues in biomedical publications, we employ a set of patterns that take advantage of regularities in how protein residues and mutations are expressed in text. These patterns are described in detail in previous work;3,11 that work showed that we were able to achieve F-scores of over 94% on a third-party gold standard.10 For the analysis reported here, we reused the previous results we generated, which included detection of single-letter abbreviations for mutations (e.g. D199S) but ignored single-letter abbreviations for individual residues. All residue and mutation mentions were normalised to a three-letter residue abbreviation with the specific position of the residue.
3.2. Corpus creation
We created two corpora for processing: a corpus of abstracts and a corpus of full text publications. The starting point for the creation of each of these corpora was the set of 17,595 PubMed identifiers referenced as the primary citation of records in the Protein Data Bank12 (PDB; www.pdb.org), using PDB data downloaded in May 2010 for the LEAP-FS system.
We were able to retrieve the full set of abstracts from a local Medline repository. We were also able to successfully retrieve 11,560 full text publications from this set. After running the residue and mutation detection step over these corpora, we identified 6,109 abstracts and 8,491 full text publications with residue mentions. We next applied a physical verification step for those residue mentions, in which each amino acid mention identified in the text must be matched to a physical residue in the corresponding PDB record, with both the position in the protein sequence and the specific amino acid matching (for mutations, either the wild type or the mutated amino acid was allowed to match). This step ensures that residues identified in the text are grounded to the appropriate protein sequence, and resulted in identifying 5,236 abstracts and 7,309 full text publications with physically verified text residues (PVTR) (in each case representing 86% of the original corpus).
3.3. Distant learning for training data creation
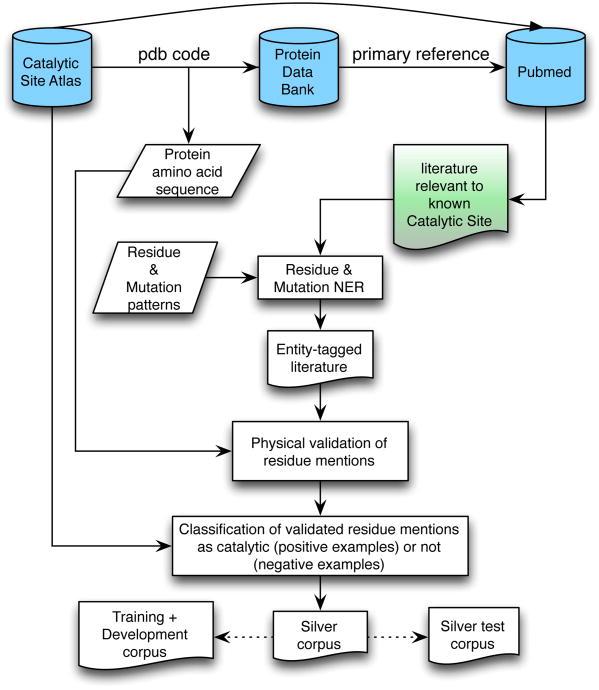
To avoid costly manual annotation of training data, we take advantage of high quality external knowledge to automatically generate appropriate training data. We have previously explored this strategy for creation of training data for extraction of protein-residue associations from text.11 We extend that approach here to create a “silver standard” data set – i.e. training data that we believe to be highly reliable, but which has not been manually verified. The architecture of the approach is outlined in Figure 1.
Fig. 1. Architecture for distant learning silver corpus creation.
The silver corpus creation starts with the abstract and full text corpora we collected, with each physically-verified text residue in the corpus serving as a potential training example. The annotation of the text residues in the corpus as well as the sub-selection of publications for the silver standard data set relies on external curated data. The external knowledge we rely on to build our training and test data is the curated links to the literature from the Protein Data Bank which form the basis of our corpus creation, coupled with literature-curated annotations of catalytic sites available in the Catalytic Site Atlas4 (CSA; www.ebi.ac.uk/thornton-srv/databases/CSA/). We refer to the subset of CSA annotations that are marked as coming from the literature as CSA-Lit. The annotated catalytic sites in CSA-Lit represent highly reliable positive information about the residues in PDB records that are catalytically active. Our training data focuses on the publications that have at least one physically-verified text residue that is annotated in CSA-Lit.
Because catalytic sites are also binding sites, and generally are a subset of functional sites, there is significant potential ambiguity about whether a given functional site is catalytic. To ensure that our training data cleanly captures specifically catalytic sites as positive instances but does not inadvertently include a catalytic site as a negative instance, we refer to other functional site resources to discard potentially ambiguous cases. Catalytic sites that are not in the CSA-Lit subset but are annotated in CSA (usually on the basis of sequence alignment with a known catalytic site) are discarded as they are not definitively catalytic, but very likely to be. We also consider any site identified in BindingMOAD as a binding site and any residue that is near a small molecule (NSM) in the corresponding PDB structure (see3 for details on how this is formally determined) as ambiguous. The logic we employ is formalized as follows:
For each PubMed article with at least one physically-verified text residue in CSA-Lit, for each physically-verified text residue in the article,
is it in CSA-Lit? (if yes, annotate as positive instance)
is it in CSA? (if yes, discard)
is it annotated in BindingMOAD? (if yes, discard)
is it a residue near a small molecule in the PDB structure? (if yes, discard)
otherwise, annotate the text residue as a negative instance.
Application of this strategy results in an imbalanced silver corpus for the abstracts, with 749 positive instances and 179 negative instances (in 259 abstracts), and a significantly larger and more balanced silver corpus for the full texts, with 5846 positive instances and 6095 negative instances (over 312 articles).
3.4. Applying basic machine-learning classification
The silver standard we created is designed to resemble the judgments which would be produced by a human without requiring an explicit annotation stage. The curators of CSA determined on the basis of a particular article whether a particular site was catalytic or not, which suggests that this information is available explicitly or implicitly in the text of the article. This in turn suggests that a machine-learning algorithm may be able to successfully classify some of the residue mentions on the basis of this textually-encoded information as catalytic.
In this section, we describe a fairly simplistic machine learning approach to this problem. This approach was designed to determine how readily the annotations could be determined using simple features based on the textual context surrounding the residue mention. In addition to this, it was desirable to have the features selected so the classifier could be trained on the relatively small abstracts-only portion of the corpus, since these are far more easily accessible than full text data. Because of the small size of this portion of the dataset, feature types which tend to suffer from data sparseness were not explored extensively. In particular, for word n-grams, from the few hundred instances in abstracts we have available there is unlikely to be enough information to meaningfully populate feature vectors for n > 1, so we only experimented with features based on word unigrams.
We report classification results with a model built using Zhang Le's Maxent Toolkita. In preliminary experiments, we achieved superior performance using this toolkit than with other tools. The corpora were preprocessed; sentences were identified using the Jena Sentence Boundary Detector tool13 and the text was tokenized and tagged with part-of-speech (POS) tags using the GENIA tagger.14 We experimented with the following feature sets:
TOKENS(b,e): the set of tokens in the range (b, e) relative to a physically-verified text residues (PVTR) token, not crossing sentence boundaries. For example, TOKENS(−2, −1) denotes the two tokens immediately preceding the PVTR. The value $ denotes the sentence boundary, so TOKENS($, −1) means all preceding tokens.
LEMMA(b,e) and BIOLEM(b,e): the same as TOKENS(b,e), but using lemmas of the tokens derived from the GENIA tagger and the BioLemmatizer,15 respectively.
MT: Match Type. Whether the PVTR was identified via mutation pattern such as Cys42Ala or a bare amino acid residue pattern such as Cys42.
Use of the full preceding sentential context, e.g. LEMMA($, −1), was found to be the most effective; smaller ranges tended to be detrimental. Features based on POS-tags were unhelpful. Unlemmatized tokens performed worse than lemmas (results not reported).
4. Results
4.1. Literature-based recovery of CSA annotations
To establish the context for the task of classifying functional sites as catalytic, we undertook an analysis of the data we had generated for our initial experiments with the LEAP-FS method. The aim of this analysis was to understand the proportion of the residues extracted from carefully selected biomedical publications that correspond to known catalytic sites. In our previous work,3 we established that a significant proportion of the residues identified in the publications we analysed corresponded to functionally active sites as recorded in both CSA and BindingMOAD and used this as evidence supporting the hypothesis that residue mentions in the literature have functional significance. Here, we specifically examine how well we are able to recover the functional site annotations in the CSA, essentially measuring the method's recall of curated annotations.
Table 1 summarises the results. Line 1 indicates the total amount of information in the Protein Data Bank that is linked to a PubMed ID. The last column (PMID,Site) indicates the number of unique combinations of PMIDs and residue locations in the Protein Data Bank. This represents an upper bound on the number of residues mentioned in text that we would expect to find. Line 2 represents the results of the analysis in our previous work; we identified 14,127 residue mentions in 5,236 PubMed abstracts; those residue mentions correspond to 44,701 physical residues in the PDB. Line 3 extends that to processing of full text publications. Lines 4-6 focus on the subset of PDB residues that are included in the Catalytic Site Atlas; we take these residues to be the set of known (or presumed) catalytic sites. We can see here that text mining of PubMed abstracts (Line 6) is only able to identify a small proportion (∼9%) of the catalytic residues that could be mentioned in some publication (Line 5). Restricting our analysis to those residues in the CSA that have been explicitly marked as having supporting evidence in the literature (the CSA-Lit subset; Lines 7-12) we find that we are able to recover a somewhat higher proportion, ∼17% for abstracts (476/2799). When we process full text as well, our coverage of CSA-Lit improves dramatically, to ∼38% (1052/2799) for all literature evidence we were able to access and detect.
Table 1. Statistics of PDB-PMID-Residue relationships in CSA.
PDB = Protein Data Bank. CSA = Catalytic Site Atlas. CSA-Lit = the subset of CSA annotations marked as based on literature. PMID = PubMed ID. A verified text residue is a residue that has been identified through text mining, and mapped to a physical residue in the corresponding PDB protein sequence. “Site” refers to a particular numbered location in a protein sequence.
| Source | Set | Residues | PDB | PMIDs | (PMID,Site) |
|---|---|---|---|---|---|
|
| |||||
| 1. PDB | PDB residues, with abstract | 17904740 | 30816 | 17595 | 4797110 |
| 2. PDB | PDB residues with verified text residues (abstracts) | 44701 | 9923 | 5236 | 14127 |
| 3. PDB | PDB residues with verified text residues (full text) | 7309 | 107153 | ||
| 4. CSA | PDB residues in CSA | 112031 | 17524 | ||
| 5. CSA | PDB residues in CSA, with abstract | 94327 | 14673 | 7587 | 29447 |
| 6. CSA | Verified text residues; match to CSA (abstracts) | 9059 | 3163 | 1630 | 2708 |
| 7. CSA-Lit | PDB residues in CSA-Lit | 6372 | 942 | ||
| 8. CSA-Lit | PDB residues in CSA-Lit, with abstract | 5586 | 831 | 823 | 2799 |
| 9. CSA-Lit | PDB residues in CSA-Lit, with abstract with at least one verified text residue | 2116 | 343 | 341 | 1139 |
| 10. CSA-Lit | Verified text residues; match to CSA-Lit (abstracts) | 878 | 259 | 259 | 476 |
| 11. CSA-Lit | Verified text residues; match to CSA-Lit (full text) | 312 | 805 | ||
| 12. CSA-Lit | Verified text residues; match to CSA-Lit (full text + abstract) | 444 | 1052 | ||
We further wish to understand the extent of the ambiguity we face in attempting the classification task. To assess this, we examined the annotation status of the physically verified text residues (PVTRs) in the full text data set. While the annotations of the CSA-Lit subset are clearly the most relevant to our classification task, representing literature-curated catalytic site annotations, all of the annotations in CSA are very likely to be valid catalytic sites, as they were derived through alignment with known catalytic sites in closely related structures. As we suggested above, many binding sites are also catalytic sites. Hence sites in PDB protein structures which are in close proximity with a small molecule (NSM = near small molecule), a characteristic strongly suggestive of a ligand binding site, as well as the curated subset of those sites represented in the BindingMOAD database,5 are also potentially catalytic. The overlap of the PVTRs with each of these sources is summarized in Table 2. The results show that a large proportion of the PVTRs overlap with some existing annotation for those sites (41.5%), despite only a small fraction having been formally curated as catalytic sites (0.8%). While this is a strong result for LEAP-FS – supporting the hypothesis that text residues are likely to be functionally important – it means that we have a large ambiguity set for our catalytic site classification task.
Table 2.
Analysis of the overlap of the physically verified text residues (PVTR) in our full text corpus with functional site annotations. Percentiles in parentheses are relative to the category.
| Category | Num PVTR | % PVTR | Num PMID | PMID % |
|---|---|---|---|---|
| All PVTR | 107153 | 100.0% | 7309 | 100.0% |
| in abstract | 6085 | (5.7%) | 2477 | (33.9%) |
| not in abstract | 101068 | (94.3%) | 4832 | (66.1%) |
|
| ||||
| PVTR CSA-Lit | 805 | 0.8% | 312 | 4.3% |
| in abstract | 237 | (29.4%) | 127 | (40.7%) |
| not in abstract | 568 | (70.6%) | 185 | (59.3%) |
|
| ||||
| PVTR any CSA | 5821 | 5.4% | 2413 | 33.0% |
| in abstract | 1252 | (21.5%) | 759 | (31.5%) |
| not in abstract | 4569 | (78.5%) | 1654 | (68.5%) |
|
| ||||
| PVTR BindingMOAD | 5652 | 5.3% | 698 | 9.5% |
| in abstract | 537 | (9.5%) | 239 | (34.2%) |
| not in abstract | 5115 | (90.5%) | 459 | (65.8%) |
|
| ||||
| PVTR NSM | 42603 | 39.8% | 5254 | 71.9% |
| in abstract | 3540 | (8.3%) | 1653 | (31.5%) |
| not in abstract | 39063 | (91.7%) | 3601 | (68.5%) |
|
| ||||
| PVTR any annotation | 44428 | 41.5% | 5566 | 76.2% |
| in abstract | 3900 | (8.8%) | 1804 | (32.4%) |
| not in abstract | 40528 | (91.2%) | 3762 | (67.6%) |
4.2. Classification results over silver corpus
The abstracts and full text corpora were split into a training subset and a test subset, with 80% of the articles in each corpus randomly selected for training and 20% reserved as a held-out test set. Table 3 provides a selection of the classification results over these subsets (test results in italics). Our experiments used 8-fold cross-validation. We include results for a baseline system, labelled BL, which is a majority-class classifier. Other lower-performing scenarios are not included. All results reported for LEMMA and BIOLEM are LEMMA($, −1) and BIOLEM($, −1), respectively. The column σ2 indicates the value for the σ2 Gaussian smoothing parameter to the MaxEnt learner (0.0, 1.0, 4.0 were tested).
Table 3.
Results using 8-fold cross-validation over the development and test sets. BL = Baseline; ME refers to the MaxEnt classification engine. A = abstracts; F = full text.
| Sections | Catalytic | Non-catalytic | F-score | ||||
|---|---|---|---|---|---|---|---|
| Eng. | σ2 | Features | Train | Test | P / R / F | P / R / F | Mic /Mac |
| ME | 0.0 | MT, LEMMA | A | A | 86.8/ 93.0 / 89.8 | 51.7 /34.6/41.4 | 81.5 /66.4 |
| ME | 1.0 | MT, LEMMA | A | A | 82.6 / 97.1 / 89.3 | 30.8 / 6.0 / 10.1 | 76.9 / 54.0 |
| ME | 4.0 | MT, LEMMA | A | A | 85.6 / 95.8 /90.4 | 56.7/ 25.6 / 35.2 | 81.8/ 65.5 |
| ME | 0.0 | MT, BIOLEM | A | A-test | 79.2 / 86.9 /82.9 | 17.4 /10.8 /13.3 | 69.0 /48.6 |
|
| |||||||
| BL | A, F | A | 82.2 /100.0/90.2 | 0.0 / 0.0 / 0.0 | 74.1 / 45.1 | ||
| ME | 1.0 | BIOLEM | A, F | A | 90.4/ 81.2 / 85.6 | 41.0/ 60.2 /48.8 | 79.5/68.1 |
| ME | 4.0 | MT, LEMMA | A, F | A | 90.0 / 74.6 / 81.5 | 34.5 /61.7/ 44.2 | 76.0 / 65.0 |
| ME | 0.0 | MT, BIOLEM | A, F | A-test | 89.2 / 80.0 /84.4 | 44.2 /62.2 /51.7 | 78.2 /68.8 |
|
| |||||||
| BL | A | F | 48.9 /100.0/ 65.7 | 0.0 / 0.0 / 0.0 | 32.1 / 32.8 | ||
| ME | 0.0 | LEMMA | A | F | 51.9 / 87.2 / 65.1 | 64.8 /22.5/ 33.4 | 56.2 / 56.5 |
| ME | 0.0 | MT, BIOLEM | A | F | 52.4/ 89.4 / 66.1 | 68.8 / 22.2 /33.6 | 57.8/58.1 |
| ME | 1.0 | MT, BIOLEM | A | F | 50.6 / 96.4 /66.3 | 73.9/ 9.7 / 17.2 | 56.8 / 57.3 |
| ME | 0.0 | MT, BIOLEM | A | F-test | 51.7 / 88.8 /65.3 | 64.8 /19.9 /30.5 | 56.0 /56.2 |
|
| |||||||
| BL | A, F | F | 48.9 /100.0/ 65.7 | 0.0 / 0.0 / 0.0 | 32.1 / 32.8 | ||
| ME | 1.0 | LEMMA | A, F | F | 63.5/ 72.6 /67.7 | 69.6 / 60.0 / 64.5 | 66.4/66.4 |
| ME | 0.0 | BIOLEM | A, F | F | 62.4 / 72.7 / 67.2 | 69.0 / 58.1 / 63.0 | 65.5 / 65.5 |
| ME | 1.0 | BIOLEM | A, F | F | 62.7 / 73.6 /67.7 | 69.7/ 58.1 / 63.3 | 66.0 / 66.0 |
| ME | 4.0 | MT, LEMMA | A, F | F | 63.5/ 60.9 / 62.2 | 64.0 /66.5/65.2 | 63.8 / 63.7 |
| ME | 0.0 | MT, BIOLEM | A, F | F-test | 69.2 / 68.4 /68.8 | 69.9 /70.7 /70.3 | 69.6 /69.5 |
The abstract development set contains 613 catalytic (positive) and 133 non-catalytic (negative) text instances, while the full text development set contains 4641 catalytic and 4846 non-catalytic text instances. The standard measurements of precision, recall, and f-score are calculated over these text instances. F-score is calculated through micro-averaging (Mic), i.e. across all text residues in the test set, and macro-averaging (Mac), i.e. averaging performance over the two categories catalytic vs. non-catalytic.
We note that the baseline system BL has non-zero values for the non-majority class (therefore, less than 100% majority class recall) in the cases of training on abstracts and full text together. This is because the majority class is calculated from the input data in the training fold. The folds are randomly determined at the document level rather than the text instance level. Coupled with more balanced instances in the full text data, this can result in the majority class in a given fold not being the same as the majority class in the entire data set.
5. Discussion
5.1. Full text versus abstracts
Examination of Table 1 shows a clear advantage when processing full publications as compared to abstracts, despite having access to a smaller proportion of the relevant literature (66% of relevant publications). This demonstrates that the increased level of detail that is available in full text publications16 is important for understanding of specific physical residues. Our results also indicate an advantage in processing both the abstracts and full text together.
Our results show some discrepancies between what is identified in abstracts as compared to the corresponding full text publications we were able to access. While it would be typical for a full text publication to contain the abstract, we found that our system was not able to identify (minimally) the same text residues as in the corresponding abstracts for 497 full text files. Further investigation revealed that more than half (250) of those full text publications were spurious – while there was text downloaded for a given PMID, it was not the actual publication context. This typically resulted from an error in the logic of our full text retrieval script or a subscription firewall. We found that in 157 publications we missed residue mentions due to conventions in the HTML to plain text conversion script that our residue detection patterns were not sensitive to. An additional 88 publications had no results in the full text data set because single letter mutations were not included in the full text processing.
5.2. Classifier performance
Examination of Table 3 shows several consistent patterns. First, the classifiers based on machine learning all easily outperform the baseline classifier; this effect is most pronounced on the more balanced full text test set. Second, the classifiers trained on more data (combining both abstracts and full text) outperform the classifiers trained on abstracts alone. The lack of complete subsumption of the abstract data in the full text data, as discussed above, likely contributes to this effect, but it also demonstrates the advantage of more training contexts to learn from. Third, the MT (match type) feature improves performance in most cases. Fourth, the results on the held-out test set are slightly lower than the corresponding results for the development set, except in the case of the final full text test run, trained on all the development data. This again shows the benefit of more data. However, the differences between the various feature sets we experimented with were small and not fully consistent across the system combinations we considered – sometimes BIOLEM gives an advantage over LEMMA and sometimes not, and various settings for the σ2 parameter affected P/R/F across the two categories inconsistently. We have experimented with a limited set of features in this work to test the viability of the approach; application of other features and other approaches to named entity recognition is warranted to achieve improved performance.
One complicating factor for the classifier arises from the distinction between a catalytic text mention of a given site in a protein, and a catalytic site. A catalytic site may be discussed in text for some reason that has nothing to do with its function and therefore a given text mention may not be appropriately categorized as “catalytic”, even if the corresponding protein site is a catalytic site. However, given our distance-based methodology for producing the training and test data for the classifier, we cannot discriminate between these cases. We annotate individual text mentions of PVTRs based on site-level information rather than considering whether the specific local textual context provides evidence of function.
An analysis of the classifier's performance at the level of a unique PVTR, rather than at the level of text mentions, is shown in Table 4. Here, we have aggregated over the classification of all text mention instances of a given (PMID, Site) pair. We have employed a simple majority vote of classifications over the instances – that is, if the majority of the individual text mentions are classified as catalytic, then the PVTR is classified as catalytic as well. In the case of a tie, we examine the scores of the classification. When the data is viewed this way, we see improved performance on the recall of catalytic sites, at a significant cost to precision. In contrast, the classification of non-catalytic sites has improved overall. These results therefore confirm the catalytic text mention/catalytic site difference, suggesting that many text mentions of catalytic sites are not clear references to its catalytic status, while it is possible to reliably rule a PVTR out as non-catalytic due to a lack of catalytic text mentions.
Table 4.
Results using 8-fold cross-validation over the development and test sets, aggregated by unique physically-verified text residue. See caption Table 3 for abbreviation definitions.
| Sections | Catalytic | Non-catalytic | F-score | ||||
|---|---|---|---|---|---|---|---|
| Eng. | σ2 | Features | Train | Test | P / R / F | P / R / F | Mic /Mac |
| BL | A | A | 78.9 /100.0/ 88.2 | 0.0 / 0.0 / 0.0 | 69.6 / 44.1 | ||
| ME | 0.0 | MT, LEMMA | A | A | 82.5/ 96.7 /89.1 | 65.8 /23.6/34.7 | 80.1/66.4 |
| ME | 4.0 | MT, LEMMA | A | A | 80.9 / 98.5 / 88.8 | 70.0/ 13.2 / 22.2 | 79.5 / 64.2 |
| ME | 1.0 | MT, BIOLEM | A | A | 79.7 / 99.0 / 88.3 | 60.0 / 5.7 / 10.3 | 77.4 / 59.8 |
| ME | 0.0 | MT, BIOLEM | A | A-test | 73.3 / 92.8 /81.9 | 33.3 / 9.7 /15.0 | 66.1 /52.3 |
|
| |||||||
| BL | A, F | A | 78.9 /100.0/88.2 | 0.0 / 0.0 / 0.0 | 69.6 / 44.1 | ||
| ME | 0.0 | LEMMA | A, F | A | 87.9 / 85.9 / 86.8 | 51.3 / 55.7 / 53.4 | 79.8 / 70.2 |
| ME | 4.0 | BIOLEM | A, F | A | 88.7/ 85.1 /86.9 | 51.6/ 59.4 /55.3 | 80.3/71.2 |
| ME | 0.0 | MT, LEMMA | A, F | A | 88.5 / 77.8 / 82.8 | 42.9 /62.3/ 50.8 | 76.6 / 67.8 |
| ME | 0.0 | MT, BIOLEM | A, F | A-test | 85.9 / 80.7 /83.2 | 55.6 /64.5 /59.7 | 77.0 /71.7 |
|
| |||||||
| BL | A | F | 19.3 /100.0/ 32.3 | 0.0 / 0.0 / 0.0 | 6.2 / 16.2 | ||
| ME | 0.0 | LEMMA | A | F | 22.6 / 94.5 / 36.5 | 94.6 /22.9/36.9 | 50.5 / 58.7 |
| ME | 0.0 | MT, BIOLEM | A | F | 22.8/ 95.8 /36.9 | 95.8 / 22.7 / 36.8 | 50.8/59.3 |
| ME | 1.0 | MT, BIOLEM | A | F | 20.8 / 99.7 / 34.5 | 99.2/ 9.6 / 17.5 | 40.8 / 57.2 |
| ME | 0.0 | MT, BIOLEM | A | F-test | 24.3 / 94.1 /38.6 | 92.1 /19.1 /31.6 | 48.5 /57.4 |
|
| |||||||
| BL | A, F | F | 19.3 /100.0/ 32.3 | 0.0 / 0.0 / 0.0 | 6.2 / 16.2 | ||
| ME | 1.0 | LEMMA | A, F | F | 36.7/ 78.6 /50.1 | 93.0 / 67.6 / 78.3 | 75.5/68.7 |
| ME | 1.0 | BIOLEM | A, F | F | 35.2 / 80.1 / 48.9 | 93.2/ 64.7 / 76.4 | 74.2 / 68.0 |
| ME | 4.0 | MT, LEMMA | A, F | F | 36.7/ 65.2 / 47.0 | 89.8 /73.2/80.6 | 75.4 / 66.1 |
| ME | 0.0 | MT, BIOLEM | A, F | F-test | 50.4 / 75.7 /60.5 | 92.2 /79.4 /85.3 | 80.8 /74.3 |
Addressing this problem could lead to overall improvement of the classifier, which we plan to explore in future work. We could build a classifier which aggregates information across text mentions to support classification of a unique PVTR rather than classification of each individual text mention. We could also explore a two-part solution, where the first part is to identify sentences that contain functional information about a site in a protein, and the second part is to classify that functional information more specifically. This would require development of a training corpus which provided more specific functional information. This could be done manually or by filtering text mentions in our existing corpus according to whether there is a detected Gene Ontology molecular function term within the same sentence, similar to the mutation grounding strategy of Naderi and Witte.8
5.3. Use of the classifier for improving curation of catalytic sites
Our data highlights (a) the low coverage of curated information about both catalytic sites and binding sites more generally, and (b) the significant ambiguity of functional sites.
The gap between curated information and the amount of inferred information in genomic databases is a well known problem,17 and we see clear evidence of that gap here. In comparing Lines 4 and 7 of Table 1, we see that the CSA-Lit curated subset represents less than 6% of the full CSA database. BindingMOAD is the curated subset of the PDB NSM data, focused on protein ligand binding sites. For the PVTRs in our corpus, only 13% of NSM sites we recover are also captured in BindingMOAD (PVTR NSM vs. PVTR BindingMOAD data in 2). While this difference in coverage could be because many of those NSM sites are not high-quality binding sites, it is more likely a reflection of the time and resources that manual curation requires. The fact that we are able to recover over forty thousand NSM sites in our corpus of 7,309 full text publications suggest that text mining can play a powerful role in closing this gap, by highlighting sites that have literature evidence of functional importance.
However, to close the annotation gap we must go one step further than identification, and perform finer-grained categorization of those PVTRs. The catalytic site classifier we have developed on the basis of our training data could be applied more broadly to the full set of 7,309 articles for which we have identified PVTRs. This would identify those PVTRs not in CSA-Lit that are most likely to be catalytic sites; those in turn can be prioritized for curation, and the specific document with the catalytic mention of the site can be provided to a database curator. These developments would enable higher-throughput in the curation process.
6. Conclusion
This paper has explored the applicability of text mining from the biomedical literature to the problem of detecting catalytic sites. We have presented two corpora in which protein residue mentions were annotated using reliable external knowledge about catalytic residues. Our analysis of these corpora according to their coverage of existing annotated resources showed that the literature is a good source of information about functionally significant protein sites, and furthermore that processing of full text publications is particularly important for achieving good recall of these sites from the literature. With respect to classification of these functional sites as catalytic, we observed that there is considerable ambiguity in assigning the functional role of a given site.
Nevertheless, we explored development of a classifier learned from our annotated silver corpora to enable automatic annotation of catalytic sites in the biomedical literature. Despite the ambiguity of catalytic sites, and the evaluation of the annotation at the level of individual text mentions of protein sites rather than aggregated over unique physical sites, the classifiers were able to achieve reasonably good performance with a simple set of features. Having established the viability of the approach, and having identified some of the challenges that arise in this task, we are confident that we in future work we will be able to develop new methods that improve upon the initial results presented here. This work represents an important step in the development of effective strategies for understanding functional characteristics of proteins at the level of specific residues, and for supporting curation of that information in databases, by exploiting the information available in the published literature.
Footnotes
Contributor Information
Karin Verspoor, Email: karin.verspoor@nicta.com.au, National ICT Australia, Victoria Research Lab, Parkville, VIC 3010 Australia.
Andrew Mackinlay, Email: andrew.mackinlay@nicta.com.au, National ICT Australia, Victoria Research Lab, Parkville, VIC 3010 Australia.
Judith D. Cohn, Email: jcohn@lanl.gov, Computer and Computational Sciences Division, Los Alamos National Laboratory, Los Alamos, NM 87545 USA.
Michael E. Wall, Email: mewall@lanl.gov, Computer and Computational Sciences Division, Los Alamos National Laboratory, Los Alamos, NM 87545 USA.
References
- 1.Marashi S-A. The necessity of extraction of ligand binding data from literature. EXCLI Journal. 2005;4:87–89. [Google Scholar]
- 2.Ofran Y, Punta M, Schneider R, Rost B. Beyond annotation transfer by homology: novel protein-function prediction methods to assist drug discovery. Drug Discov Today. 2005;10(21):1475–82. doi: 10.1016/S1359-6446(05)03621-4. [DOI] [PubMed] [Google Scholar]
- 3.Verspoor Karin M, Cohn Judith D, Ravikumar Komandur, Wall Michael E. Text mining improves prediction of protein functional sites. PLoS One. 2012;7(2):e32171. doi: 10.1371/journal.pone.0032171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter Craig T, Bartlett Gail J, Thornton Janet M. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Research. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson Mark L, et al. Updating Binding MOAD: Data Management and Information Workflow. New Mathematics and Natural Computation. 2010;06(01):49–56. [Google Scholar]
- 6.Cunningham Hamish, Maynard Diana, Bontcheva Kalina, et al. Text Processing with GATE (V. 6) 2011 [Google Scholar]
- 7.Benson Mark L, Smith Richard D, Khazanov Nickolay A, Dimcheff Brandon, Beaver John, Dresslar Peter, Nerothin Jason, Carlson Heather A. Binding MOAD, a high-quality protein-ligand database. Nucleic Acids Research. 2008;36(suppl 1):D674–D678. doi: 10.1093/nar/gkm911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naderi Nona, Witte Rene. Automated extraction and semantic analysis of mutation impacts from the biomedical literature. BMC Genomics. 2012;13(Suppl 4):S20. doi: 10.1186/1471-2164-13-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagel K, Jimeno-Yepes A, Rebholz-Schuhmann D. Annotation of protein residues based on a literature analysis: cross-validation against UniprotKB. BMC Bioinf. 2009;10 Suppl 8:S4. doi: 10.1186/1471-2105-10-S8-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravikumar Komandur, Liu Haibin, Cohn Judith D, Wall Michael E, Verspoor Karin M. Literature protein-residue associations with graph rules learned through distant supervision. J Biomed Sem. 2012;3(S3):S2. doi: 10.1186/2041-1480-3-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomanek Katrin, Wermter Joachim, Hahn Udo. POT 12th MEDINFO. IOS Press; 2007. A reappraisal of sentence and token splitting for life science documents; pp. 524–529. [PubMed] [Google Scholar]
- 14.Tsuruoka Yoshimasa, Tateishi Yuka, Kim Jin D, Ohta Tomoko, Mcnaught John, Ananiadou Sophia, Tsujii Jun’ichi. Developing a robust part-of-speech tagger for biomedical text. POT 10th Panhellenic Conf on Informatics. 2005:382–392. [Google Scholar]
- 15.Liu Haibin, Christiansen Tom, Baumgartner William, Verspoor Karin. Biolemmatizer: a lemmatization tool for morphological processing of biomedical text. J Biomed Sem. 2012;3:3. doi: 10.1186/2041-1480-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen K Bretonnel, Johnson Helen L, Verspoor Karin, Roeder Christophe, Hunter Lawrence. The structural and content aspects of abstracts versus bodies of full text journal articles are different. BMC Bioinf. 2010;11:1. doi: 10.1186/1471-2105-11-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumgartner William, Cohen K Bretonnel, Fox Lynne, Acquaah-Mensah George, Hunter Lawrence. Manual curation is not sufficient for annotation of genomic databases. Bioinf. 2007;23(13):i41–i48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]