Abstract
BACKGROUND & AIMS
Early embryogenesis involves cell fate decisions that define the body axes and establish pools of progenitor cells. Development does not stop once lineages are specified; cells continue to undergo specific maturation events, and changes in gene expression patterns lead to their unique physiological functions. Secretory pancreatic acinar cells mature postnatally to synthesize large amounts of protein, polarize, and communicate with other cells. The transcription factor MIST1 is expressed by only secretory cells and regulates maturation events. MIST1-deficient acinar cells in mice do not establish apical-basal polarity, properly position zymogen granules, or communicate with adjacent cells, disrupting pancreatic function. We investigated whether MIST1 directly induces and maintains the mature phenotype of acinar cells.
METHODS
We analyzed the effects of Cre-mediated expression of Mist1 in adult Mist1– deficient (Mist1KO) mice. Pancreatic tissues were collected and analyzed by light and electron microscopy, immunohistochemistry, real-time polymerase chain reaction analysis, and chromatin immunoprecipitation. Primary acini were isolated from mice and analyzed in amylase secretion assays.
RESULTS
Induced expression of Mist1 in adult Mist1KO mice restored wild-type gene expression patterns in acinar cells. The acinar cells changed phenotypes, establishing apical-basal polarity, increasing the size of zymogen granules, reorganizing the cytoskeletal network, communicating intercellularly (by synthesizing gap junctions), and undergoing exocytosis.
CONCLUSIONS
The exocrine pancreas of adult mice can be remodeled by re-expression of the transcription factor MIST1. MIST1 regulates acinar cell maturation and might be used to repair damaged pancreata in patients with pancreatic disorders.
Keywords: DIMM, Exocrine Pancreas Disease, Secretion, Transcription
Although vertebrate embryogenesis is responsible for the initial establishment of individual cell fates, most cells require additional maturation events to ensure their appropriate specialization in the adult organism. These events often include creation of intracellular polarity, setting up cell– cell communication, and expanding (qualitative and quantitative) gene expression patterns so that cells are able to perform their unique functions. Despite significant progress in defining transcriptional networks that control early cell fate decisions, little is known about how key transcription factors regulate maturation events. This is a particularly important area of investigation because genetic alterations that disrupt the specialization properties of differentiated cells often lead to disease states. Thus, defining the transcriptional circuits that maintain the adult cell phenotype is critical for devising intervention strategies for different diseases.
The exocrine pancreas serves as an excellent model to study cell maturation events. A hallmark of the adult exocrine organ is formation of well-organized acini units consisting of individual acinar cells that show an apical-basal cellular polarity, clustering of zymogen granules, and extensive intercellular signaling networks. The initial specification of the cells begins early in mouse pancreas development (~embryonic day 12.5), but final maturation, including accumulation of abundant rough endoplasmic reticulum, maximum expression of digestive enzyme transcripts, apically localized zymogen granules, and the ability to regulate exocytosis events, occurs during late embryonic and postnatal development.1–3 A key protein involved in this maturation process is MIST1 (also known as BHLHA15), a basic helix-loop-helix transcription factor whose expression in the pancreas is restricted to acinar cells.4,5 Embryonic Mist1 null (Mist1KO) acinar cells fail to establish an apical-basal organization, fail to position zymogen granules, and fail to generate intercellular communication.4–6 The block in the final differentiation steps leads to a damaged exocrine pancreas that shows defective exocytosis properties,4,7,8 altered cell proliferation,9 and a lack of appropriate cell– cell junctions,6 making the exocrine organ highly susceptible to pancreatic diseases including pancreatitis and pancreatic cancer.6,7,10
The Mist1 gene is unique in that it is expressed exclusively in secretory lineages including salivary gland acinar cells, zymogenic cells of the stomach, lactating mammary epithelial cells, Paneth cells of the intestine, and in antibody- secreting plasma cells.4,5,11–13 DIMM, the Drosophila ortholog of MIST1, is expressed similarly in secretory peptidergic neurons that produce neuropeptides.14–16 The MIST1/DIMM expression patterns suggest that these proteins have a central role in controlling specific secretory properties associated with each of these differentiated cell types. Indeed, studies by Tian et al17 have shown that MIST1 controls expression of the small guanosine triphosphatase genes Rab26 and Rab3d, both of which are critical to forming appropriate zymogen granules in stomach zymogenic cells. Similarly, DIMM activates several genes involved in regulated secretion of neuropeptides.14 In all cases, activation or repression of MIST1/DIMM target genes occurs via binding to specific E-box regulatory elements.14,17–19
Studying maturation events in adult pancreatic cells has been hampered by a lack of a tractable genetic system in which changes in cell organization can be followed within the context of the intact organ. To address this question, we generated a novel system in which expression of a Mist1 transgene was induced in disorganized Mist1KO acinar cells within the context of the adult mouse pancreas. Our studies show the remarkable ability of the Mist1 transcriptional network to restore wild-type gene expression patterns to Mist1KO acinar cells. The altered gene expression produced dramatic changes in cellular phenotypes, including generation of apical-basal polarity, increased zymogen granule size, reorganization of the actin cytoskeletal network, and restoration of intercellular communication. In addition, regulated exocytosis function was fully restored upon MIST1 induction. Each of these changes occurred in adult acinar cells in the absence of cell regeneration. These results show that the injured adult exocrine pancreas can be remodeled via expression of a single transcription factor, offering new research paradigms in repairing damaged organs.
Materials and Methods
Mouse Strains and Genotyping
Mist1CreER/+ and Mist1CreER/CreER mice have been described previously.10,20 The LSL-Mist1myc transgene was generated by replacing the LacZ gene with a myc-tagged Mist1 coding sequence in the pCAG-CAT-LacZ plasmid that contains a Lox-STOP-Lox cassette.21 Pronuclear injections and generation of LSL-Mist1myc mice were performed by the Purdue University Transgenic Mouse Core Facility. Genotyping and recombination primer sets are listed in the Supplementary Materials and Methods section. Induction of Cre-ERT2 activity was accomplished by single administration of tamoxifen (200 µL of 20 mg/mL) via oral gavage to adult mice (8–9 wk). All experiments were performed with mice on a C57BL/6 background, and all animal studies were conducted in compliance with the National Institutes of Health and the Purdue University IACUC guidelines.
Histology and Immunohistochemistry
Mouse pancreata were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned, and stained using conventional histologic techniques. Tissue sections were deparaffinized and retrieved using the 2100-Retriever (Pick-Cell Laboratories, Amsterdam, The Netherlands) and antigen unmasking solution (Vector Laboratories, Burlingame, CA). Blocking was performed by incubating sections with 3% H2O2 for 10 minutes and using the M.O.M. blocking kit (Vector Laboratories). Primary antibodies were applied for 1 hour at room temperature or 4°C overnight with biotinylated secondary antibodies added for 10 minutes at room temperature. Visualization was accomplished via diaminobenzidine peroxidase staining (Vector Laboratories) or tertiary, avidin-conjugated Alexa Fluor 594 (Invitrogen, Camarillo, CA). Primary antibodies and conditions are provided in the Supplementary Materials and Methods section.
Electron Microscopy
Pancreata were fragmented into 1-mm cubes and fixed in 3% paraformaldehyde/0.5% glutaraldehyde in phosphate-buffered saline, gradually dehydrated, and embedded in Epon resin (Electron Microscopy Sciences, Hatfield, PA). Electron micrographs were obtained from ultra-thin sections using a Philips CM-100 transmission electron microscope (Philips, Andover, MA).
RNA Expression Analysis
Pancreatic RNA was isolated using the RNeasy system (Qiagen, Valencia, CA). RNA processing, hybridization, and initial analysis of the Affymetrix Mouse Gene 1.0 ST expression arrays (Affymetrix, Santa Clara, CA) was performed by the Indiana University Center for Medical Genomics according to the manufacturer’s protocols. The complete set of .CEL files is available at the National Center for Biotechnology Information Gene Expression Omnibus (GEO accession number GSE34232). For real-time polymerase chain reaction (PCR) analysis, RNA was converted to complementary DNA using the iScript complementary DNA synthesis kit (Bio-Rad, Hercules, CA) and amplified with FastStart Universal SYBR Green (Roche Applied Science, Indianapolis, IN) using an ABI 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). Real-time PCR primers are listed in the Supplementary Materials and Methods section.
Chromatin Immunoprecipitation
Mouse pancreata were disrupted using a dounce homogenizer and fixed in 1% formaldehyde for 10 minutes. Fixation was quenched in 125 mmol/L glycine for 5 minutes, and nuclei were disrupted with 10 mL lysis buffer (10 mmol/L Tris-HCl, pH 7.5, 10 mmol/L NaCl, 3 mmol/L MgCl2, 0.5% octylphenoxypoly-ethoxyethanol, and 1 mmol/L phenylmethylsulfonyl fluoride). Samples then were sonicated 5 times and precleared with Protein A sepharose beads (GE Healthcare, Latham, NY). Incubation with 2 µg anti-MIST1 or control IgG was performed overnight at 4°C with rocking. Bound protein/DNA complexes were resuspended in TE buffer supplemented with 195 mmol/L NaCl. Cross-linking was reversed by incubation at 65°C overnight and digested with proteinase K at 56°C for 4 hours. DNA was purified using a Qiaquick PCR purification kit (Qiagen) and used for real-time PCR.
Isolated Acini
Primary acini were isolated from wild-type (WT), Mist1KO, and Mist1KO/LSL-Mist1myc mice as described previously.10 Single-cell injections of 6-carboxyfluorescein (Invitrogen) were imaged 3 minutes after injection. Amylase secretion assays were performed using published protocols.22 Briefly, whole pancreata were isolated, washed in Hank’s Balanced Salt Solution, and injected with 0.1 mg/mL collagenase P (Roche Applied Science) in serum-free media supplemented with bovine serum albumin. Pancreata then were rocked at 37°C for 40 minutes in fresh collagenase P solution, sheared via passage through narrow-orifice pipettes, and filtered through 150-µm nylon cloth. Isolated acini were washed, given a 30-minute rest period at 37°C, and then stimulated with 30 µmol/L cholecystokinin. Supernatants and cell pellets were collected for quantification of percentage amylase release via the Infinity Amylase Detection Kit (ThermoFisher Scientific, Waltham, MA).
Results
Establishing an Inducible Mist1 Mouse Model
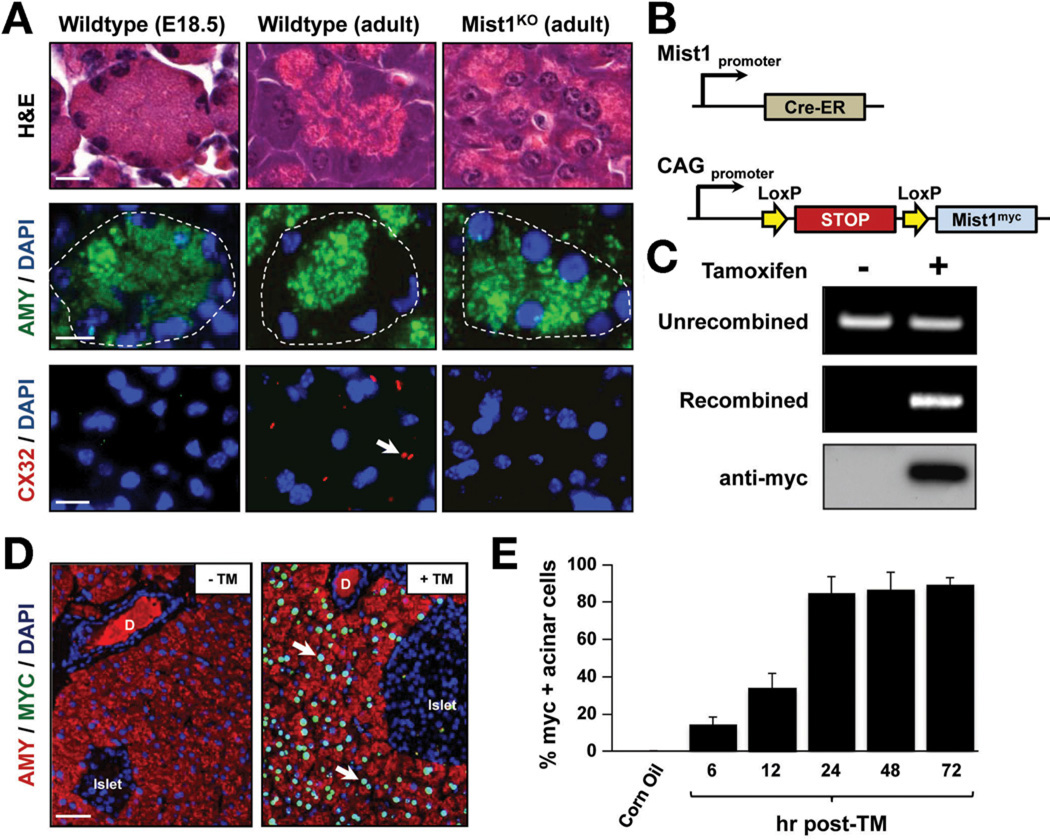
Mouse pancreatic acinar cells are specified during early pancreatic development but final maturation of the cells occurs postnatally and is dependent on an intact MIST1 transcriptional network (Figure 1A).1–3,5,6 Given the importance of maturation events to a normal exocrine pancreas, we asked if activation of a single transcription factor in damaged adult acinar cells could re-establish maturation properties and cell function. To this end, we developed an inducible transgenic mouse line that expressed a myc-tagged MIST1 protein upon Cre-mediated recombination of an upstream Lox-STOP-Lox cassette (LSL-Mist1myc) (Figure 1B). Crossing LSL-Mist1myc mice to Mist1CreER/+ mice10,20 generated Mist1CreER/CreER/LSL-Mist1myc offspring that lacked the endogenous Mist1 allele (Mist1KO/LSL-Mist1myc). Upon exposure to tamoxifen (TM), the transcriptional LSL stop cassette was excised and expression of Mist1myc was driven by the constitutive CAG promoter exclusively in acinar cells (Figure 1C and D).21 Mist1myc-expressing cells were observed as early as 6 hours after TM, with peak response (~90% acinar cells) achieved by 24 hours (Figure 1E). Importantly, no overt phenotype was observed in the acinar, ductal, or islet cell compartments of Mist1CreER/+ (Mist1HET)/LSL-Mist1myc mice that maintained constitutive Mist1myc expression for more than 3 months (Supplementary Figure 1A and B).
Figure 1.
Characterization of the LSL-Mist1myc transgenic line. (A) Standard H&E, amylase (AMY), and Cx32 immunofluorescence depicts adult Mist1KO acini showing similarities to embryonic cells, in contrast to adult, fully developed WT acinar cells. Arrows, Cx32 gap junctions. E, embryonic day. (B) Schematic of the Mist1Cre-ER genomic locus and LSL-Mist1myc transgene. (C) Genomic PCR and immunoblots reveal the TM-restricted recombination and expression of the LSL-Mist1myc transgene. (D) Upon TM, only acinar cells express nuclear MIST1myc.protein (arrows). D, duct. (E) Time-course of MIST1myc expression after TM treatment. Scale bars: (A) 10 µm; (D) 50 µm.
Genome-Wide Expression Changes Upon Mist1myc Induction
The Mist1KO phenotype suggested that Mist1 operates postnatally to maintain acinar cell organization and function in the adult organ. To test this hypothesis, we asked if induced MIST1myc could shift acinar gene expression profiles and reverse the disorganized exocrine pancreas of adult Mist1KO organs. We chose 36 hours after TM as a time point when approximately 90% of Mist1KO acinar cells expressed the LSL-Mist1myc transgene and reasoned that this sampling would allow us to identify early changes in adult gene expression patterns owing to induced MIST1myc.
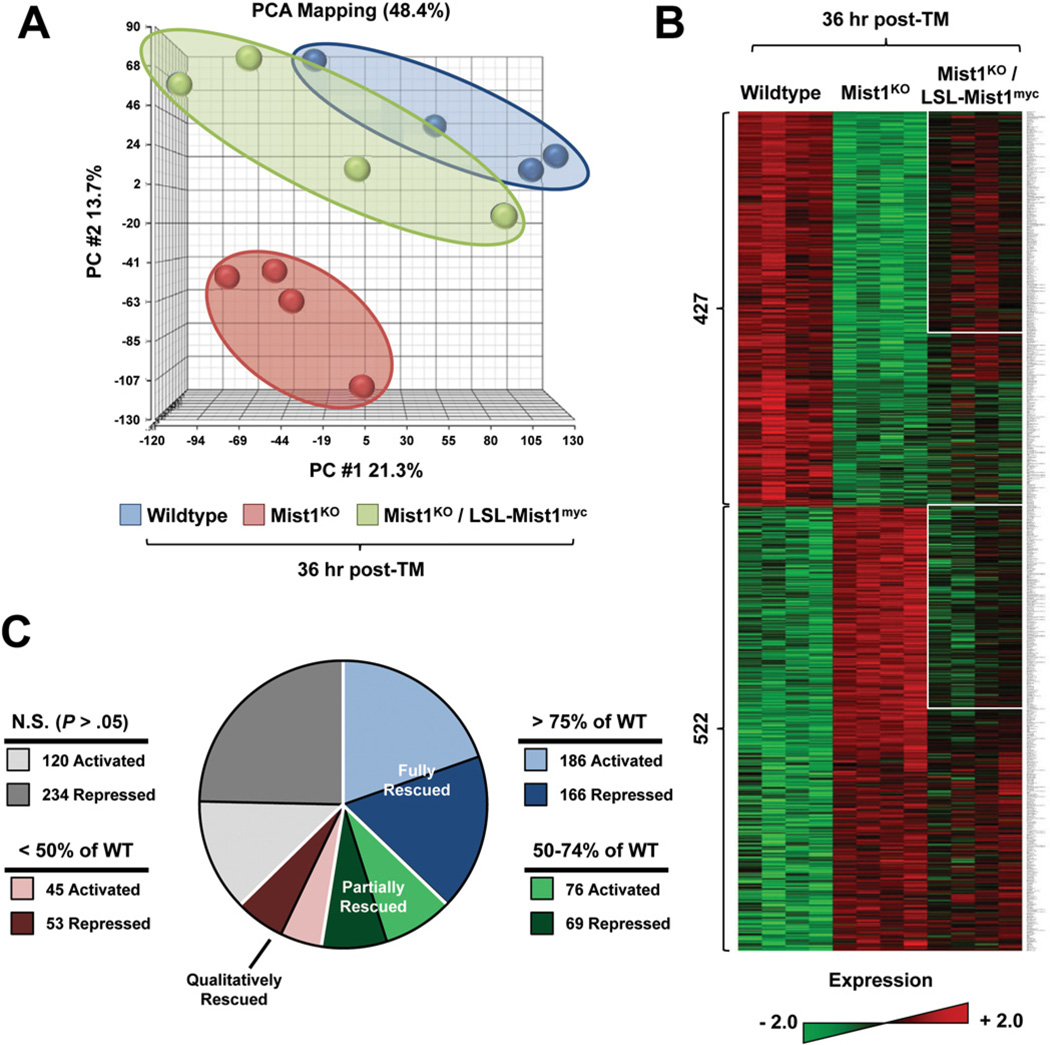
WT, Mist1KO, and Mist1KO/LSL-Mist1myc littermates (4 per group) were given a single dose of TM, after which pancreatic RNA was harvested 36 hours after TM, converted to complementary DNA, and hybridized to Affymetrix Gene 1.0 ST expression arrays. Principal component analysis revealed a clear separation in gene expression profiles between Mist1KO and WT pancreata (Figure 2A). Interestingly, within 36 hours of inducing LSL-Mist1myc, the gene expression signature of Mist1KO/LSL-Mist1myc pancreata was remodeled with an overall shift in gene expression profiles approaching WT signatures (Figure 2A and B). Analysis of the array data revealed a cohort of 949 genes (P < .05; fold-change, >1.5) that were expressed differentially between WT and Mist1KO pancreata (Figure 2B and C). Of these, 353 (37%) were classified as fully rescued in Mist1KO/LSL-Mist1myc samples, with 186 genes up-regulated and 166 genes down-regulated within this short time frame. A gene was considered fully rescued if its relative array expression value was within 25% of WT levels. For example, MIST1myc-activated genes that had ≥75% of WT levels and MIST1myc-repressed genes that were ≤125% of WT levels were considered fully rescued. Similarly, 145 genes (15%) had a partial rescue profile, reaching 50%–74% (activated genes) or 126%–150% (repressed genes) of WT expression levels by 36 hours (Figure 2B and C). Another 98 genes had significant qualitative differences but only modest quantitative changes toward WT levels (<50% for activated genes and >150% for repressed genes). Interestingly, 354 genes (37%) that showed significant differences in expression between WT and Mist1KO pancreata remained unchanged in the Mist1KO/LSL-Mist1myc samples, suggesting that these were target genes that responded slowly to MIST1myc or were genes with altered expression patterns in adult Mist1KO pancreata but were not direct MIST1 gene targets (Figure 2B and C).
Figure 2.
Expression of LSL-Mist1myc for 36 hours induces extensive changes in gene expression. (A) Principle component analysis (PCA) of WT, Mist1KO, and Mist1KO/LSLMist1myc mice after TM. Each small circle represents an individual animal. See the Supplementary Materials and Methods section for details. (B) Representative heat map of 4 individual WT, Mist1KO, and Mist1KO/LSL-Mist1myc pancreas RNA samples. The 949 genes that showed a significant difference in expression (P < .05; fold-change, >1.5) between WT and Mist1KO are indicated. White boxes represent fully and partially rescued genes. (C) Pie chart illustrating the degree of recovery for the 949 identified genes in Mist1KO/LSL-Mist1myc mice after TM. See the text for details.
Specific Mist1 Gene Targets
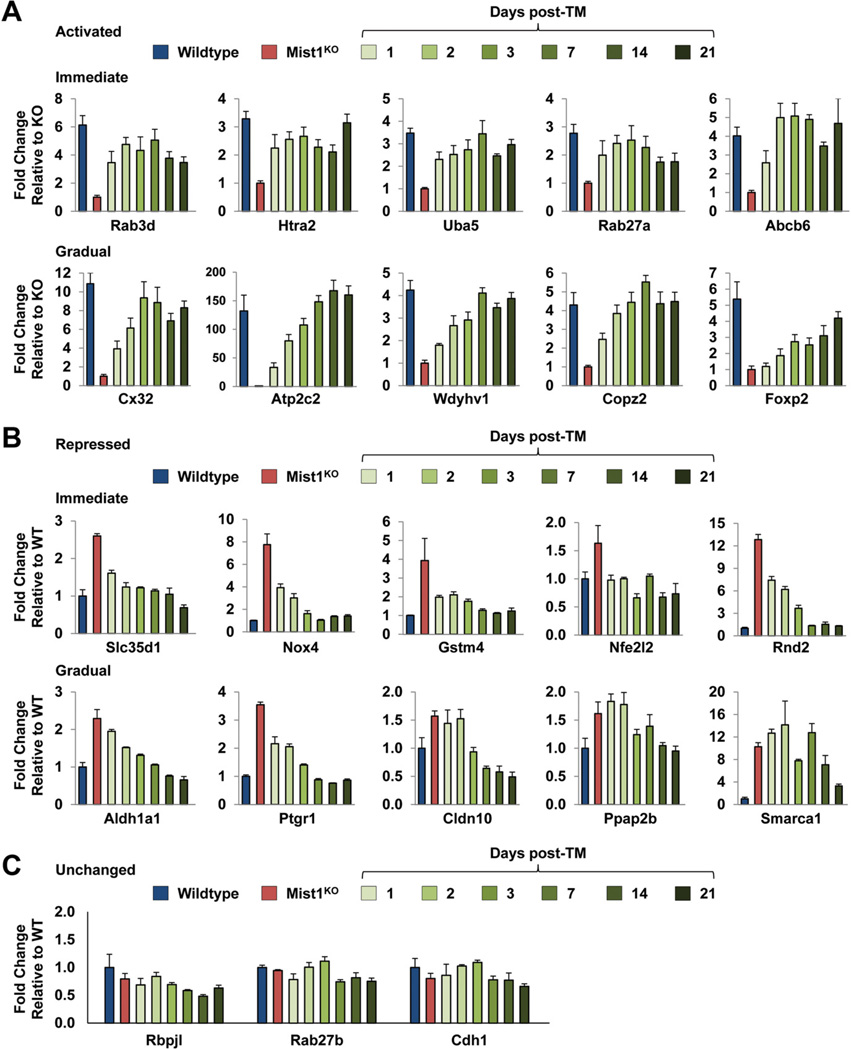
The gene array analyses identified a cassette of acinar genes that rapidly responded to induced MIST1myc expression in the adult organ. To understand the functional significance of gene expression changes, we focused on 20 genes for the following reasons: (1) they were significantly up-regulated or down-regulated upon MIST1myc induction, (2) they were identified previously as potential MIST1 targets (connexin32 [Cx32], Atp2c2, and Rab3d),6,17,23 (3) they contained potential MIST1 binding sites, and (4) their gene products likely were critical to normal acinar cell function (Supplementary Figure 2). Pancreatic RNA was isolated from adult Mist1KO/LSL-Mist1myc mice 1–21 days after TM, and the transcript levels of individual genes were determined by reverse-transcription quantitative PCR and compared with WT and Mist1KO pancreata RNA expression levels. As shown in Figure 3A and B, a number of genes (Rab3d, Htra2, Uba5, Rab27a, Abcb6, Slc35d1, Nox4, Gstm4, Nfe2l2, and Rnd2) showed rapid transcript changes (up and down) that approached WT levels by 24 hours after TM and that were maintained at near WT levels over the 3-week time course. In contrast, other genes (Cx32, Atp2c2, Wdyhv1, Copz2, Foxp2, Aldh1a1, Ptgr1, Cldn10, Ppap2b, and Smarca1) showed a gradual change in transcript levels that nonetheless achieved WT levels over the 21-day period. Regardless of the differences in rates of expression, the MIST1myc-induced up-regulated and down-regulated changes were specific to this gene set because the expression profiles for a number of other genes were not significantly different between WT and Mist1KO samples and remained unresponsive to MIST1myc expression (Figure 3C). Likewise, induction of MIST1myc in Mist1CreER/+ mice had no effect on the expression of putative MIST1 target genes, including Cx32, Atp2c2, and Rnd2 (Supplementary Figure 1C). These results confirm that increased Mist1 expression per se does not lead to misregulation of the acinar cell transcriptome. Instead, it is the induction of MIST1myc in a damaged Mist1KO background that generates specific gene expression changes.
Figure 3.
Long-term expression of LSL-Mist1myc further influences gene expression patterns in pancreatic acinar cells. (A and B) Mist1KO/LSL-Mist1myc mice (3 mice per time point) were given a single dose of TM and then RNA was harvested at the indicated times. Reverse-transcription quantitative PCR was used to compare the expression profiles of genes from WT, Mist1KO, and Mist1KO/LSL-Mist1myc pancreata. (C) Control genes Rbpjl, Rab27b, and Cdh1 show no significant changes in gene expression over this time-course.
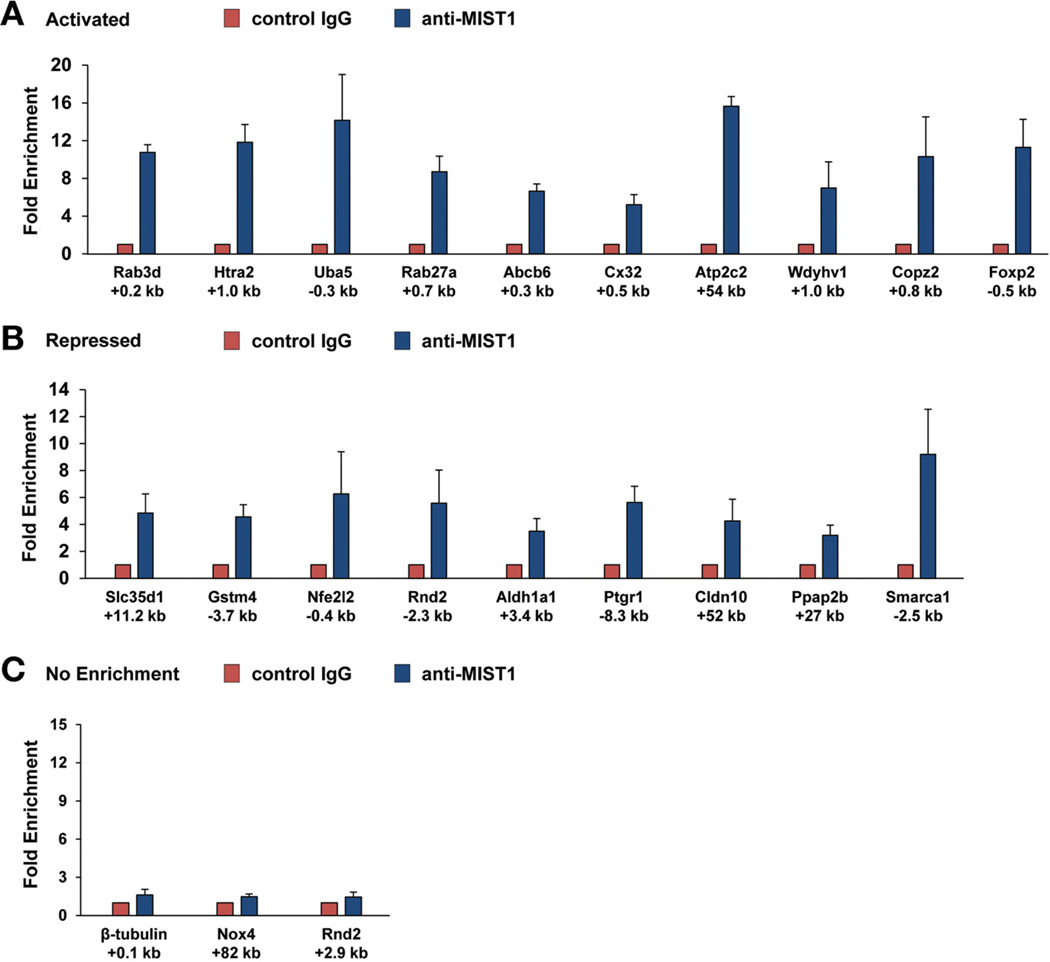
The robust responses of the 20 candidate genes to MIST1myc suggested that they are direct MIST1 targets. To test this hypothesis, chromatin immunoprecipitation (ChIP) assays were performed with anti-MIST1 antibody on WT pancreata to identify MIST1 binding sites. Initial PCR primer pairs were chosen based on 2 criteria: a focus on the 5’ promoter and first intronic regions of each gene and on ChIP-seq results, which tentatively identified MIST1 binding sites within the mouse genome. As predicted, most genes showed MIST1-specific ChIP enrichment with the exception of Nox4, which showed no enrichment with several different primer sets (Figure 4). Sequence analysis revealed a significant concentration of GC or TA E-box target sites situated at or near the ChIP amplicon regions for most of the putative target genes (Supplementary Figure 3), suggesting that MIST1 activates or represses gene expression through direct binding to E-box regulatory elements located within the promoter and first intronic regions.18 Several exceptions were found that lacked proximal regulatory sites including the Slc35d1, Atp2c2, Cldn10, and Ppap2b genes, in which the predicted (by ChIP-seq) and confirmed MIST1 binding sites were considerably distal (+11 kb → +54 kb) to the transcription start sites. The combined gene expression and ChIP analyses support the idea that induced Mist1myc affects gene expression by binding to promoters, distant enhancers, or introns of its target genes to promote or repress gene transcription.
Figure 4.
Chromatin immunoprecipitation assays confirm MIST1 interaction with target genes. (A and B) Pancreas chromatin from 3 WT mice was processed for ChIP assays using anti-IgG (control) or anti-MIST1. In most cases, MIST1 binding was enriched significantly in the anti-Mist1 groups. (C) Notably, no enrichment was observed with control genes (β-tubulin) or at alternative sites within target genes (eg, Rnd2 +2.9 kb). In addition, Nox4 showed no MIST1 binding at the predicted MIST1 binding site. The positions of the amplicon regions with respect to each gene are indicated below the graphs.
Adult Remodeling of the Exocrine Pancreas Upon Mist1myc Expression
Although identification of individual MIST1 target genes provides a roadmap to understanding the molecular mechanisms by which MIST1 regulates gene expression, the focus of this study was to investigate if induced MIST1myc could reverse exocrine defects in an adult intact organ. To examine this possibility, we administered TM to WT, Mist1KO, and Mist1KO/LSL-Mist1myc mice, and then harvested pancreata at 1, 3, 7, and 21 days after treatment. Multiple tissue samples from each time point were examined for known defects associated with the Mist1KO exocrine pancreas, including alterations in cellular organization, zymogen granule dynamics, and ductal lumen size.
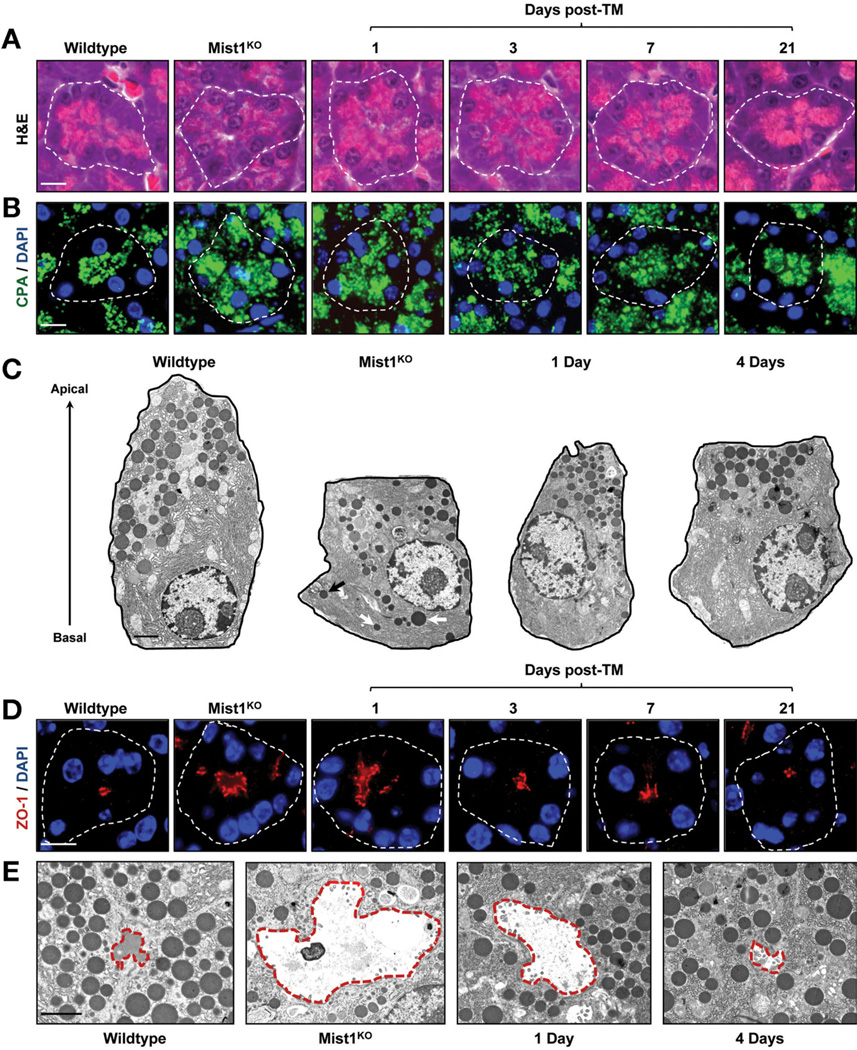
As previously reported,5 Mist1KO acinar cells lacked an apical-basal polarity in which nuclei and rough endoplasmic reticulum normally accumulate at the basal aspect and zymogen granules cluster at the apical pole (Figure 5A and B). However, expression of the LSL-Mist1myc transgene re-established the characteristic cellular polarity observed in WT pancreata. Within 24 hours, nuclei began to accumulate basally, with near-complete cellular reorganization by 7 days after TM (Figure 5A and B). As part of this reorganization, aberrant zymogen granule localization also was rescued, with enhanced zymogen granule clustering evident by 1 day after TM (Figure 5B and C; Supplementary Figure 4A). Similarly, the smaller-sized zymogen granules found in Mist1KO acinar cells rapidly achieved WT diameters within 4 days after TM (Figure 5C; Supplementary Figure 4B). Interestingly, acinar reorganization was entirely owing to changes in the current adult cell population. There was no evidence of altered acinar cell regeneration during the 3-week time course (unpublished results).
Figure 5.
Induced MIST1myc rescues the Mist1KO phenotype in adult acinar cells. (A and B) H&E and carboxypeptidase (CPA) immunofluorescence reveals the organization of zymogen granules within individual acini units of WT, Mist1KO, and Mist1KO/LSL-Mist1myc pancreata 1, 3, 7, and 21 days after TM. (C) Transmission electron microscopy views of individual acinar cells showing the mislocalized zymogen granules (arrows) in Mist1KO samples. (D and E) Analysis of lumen size in WT, Mist1KO, and Mist1KO/LSL-Mist1myc acini by zonula occludens 1 (ZO-1) immunofluorescence and transmission electron microscopy. White outlines, individual acini; red outlines, individual lumens. DAPI, 4′,6-diamidino-2-phenylindole. Scale bars: (A, B, and D) 10 µm; (C and E) 2 µm.
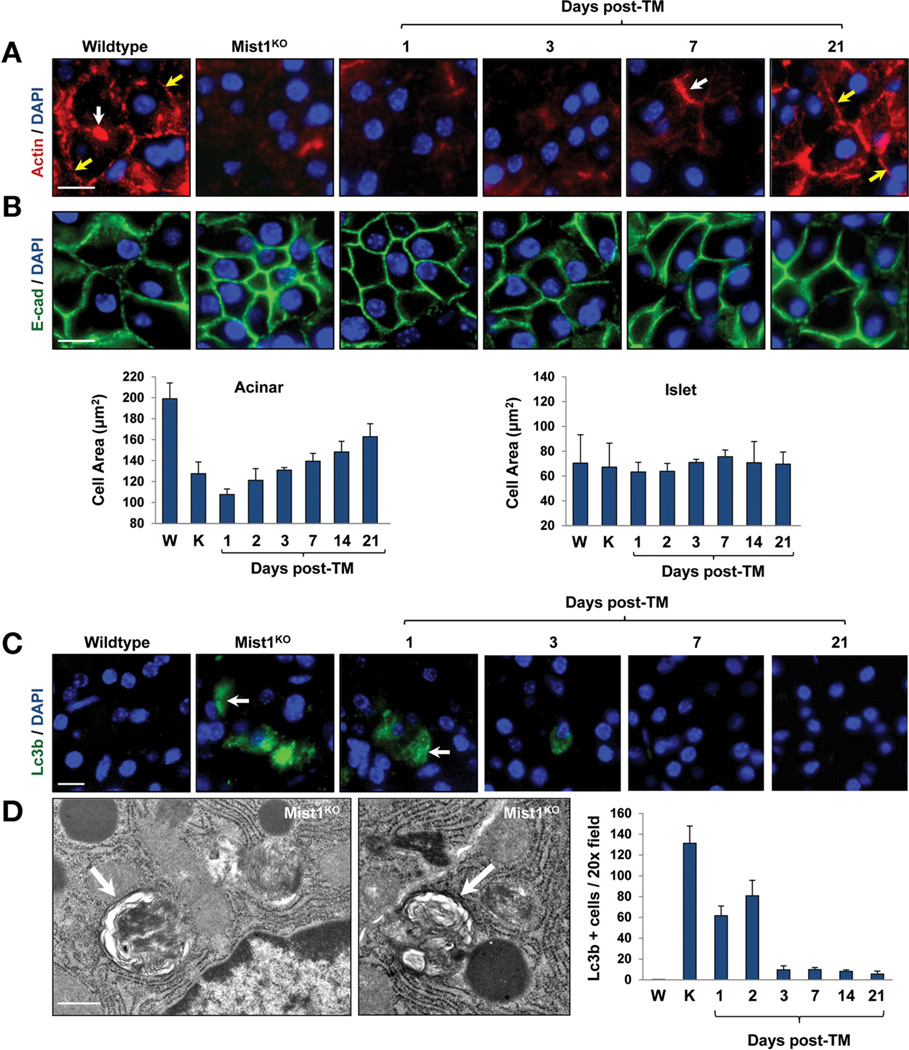
Another key characteristic of pancreatic acini were the apical, tight junction– dependent lumens where secreted zymogens enter the ductal system. Typical WT lumens were focal and small whereas Mist1KO acini lumens were highly dilated, with reduced secretion content (Figure 5D and E). Induction of MIST1myc in Mist1KO/LSL-Mist1myc mice rapidly led to closure of the lumen openings (Figure 5D and E). Changes in lumen sizes also were accompanied by alterations in the actin cytoskeleton and in the overall size of MIST1myc-expressing acinar cells. Although Mist1KO acinar cells were small and lacked a significant cortical and terminal actin web,5 induction of MIST1myc re-established the actin cytoskeleton (Figure 6A). Likewise, MIST1myc expression led to increases in the size of Mist1KO acinar cells but had no affect on islet cell size within the same tissue (Figure 6B). The basal level of autophagy that previously was reported for Mist1KO acinar cells5 also was completely eliminated within 3 days of MIST1myc induction (Figure 6C and D). Together, these studies place MIST1 within a critical transcriptional network that controls the organization of the acinar cell cytoskeleton to maintain proper apical-basal polarity and zymogen granule dynamics. Similarly, the constant state of physiological stress in the absence of MIST1 is rescued by induced expression of LSL-Mist1myc in adult cells.
Figure 6.
Induced MIST1myc rescues the actin cytoskeleton and cell size defects of Mist1KO acinar cells. (A) Anti–β-actin immunofluorescence showing restoration of the terminal (white arrows) and cortical (yellow arrows) actin webs after TM-induced MIST1myc expression. (B) E-cadherin immunofluorescence and quantification of acinar and islet cell areas over the TM time course. (C and D) LC3B immunofluorescence and TEM reveal that Mist1KO acinar cells show high levels of autophagic vacuoles, whereas induced MIST1myc expression reduces their presence toWT levels. (C) LC3B-positive cells (arrows); (D) individual autophagic vesicles (arrows). Scale bars: (A, B, and C) 10 µm; (D) 0.5 µm. DAPI, 4′,6-diamidino-2-phenylindole; E-cad, E-cadherin.
Mist1 Regulates Acinar Cell Communication and Secretion
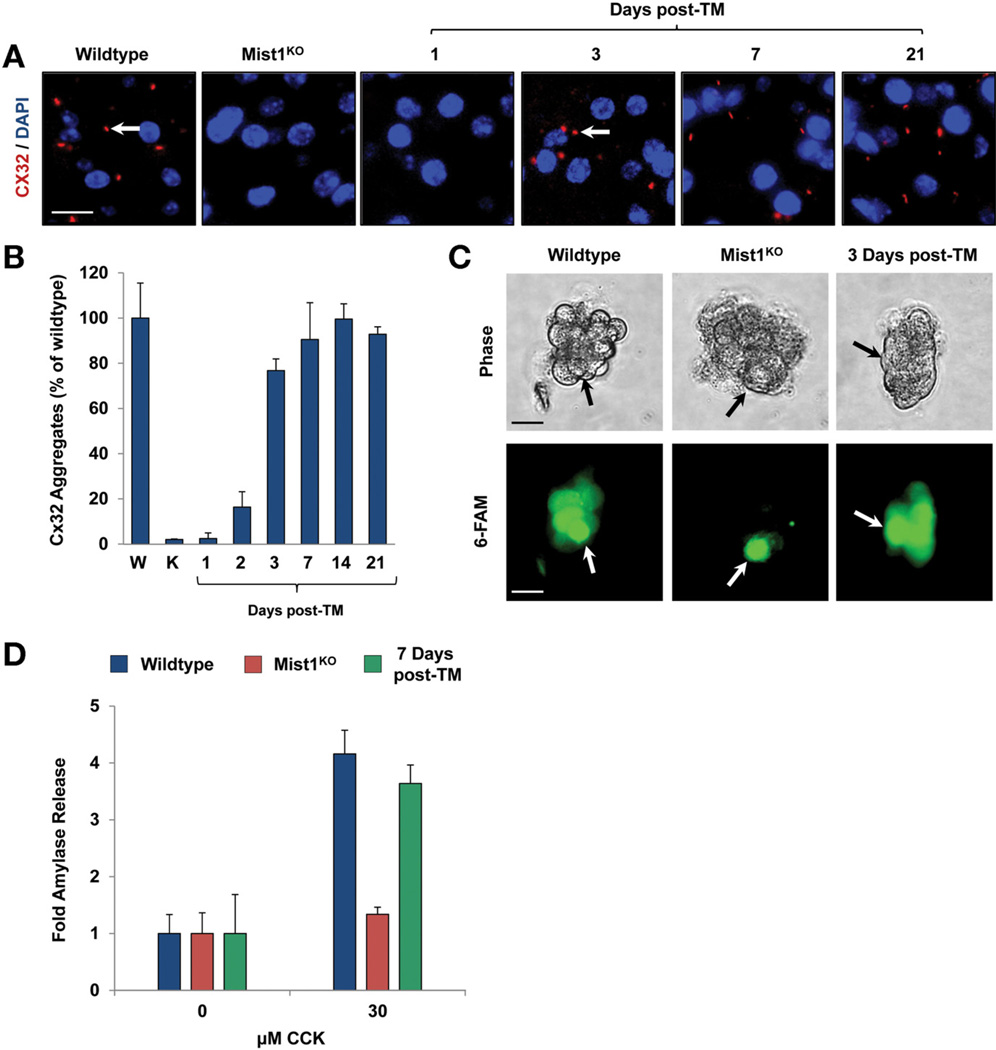
MIST1 is essential for efficient cell–cell communication owing, at least in part, to up-regulation of the gap-junction protein Cx32.6,8 Indeed, Mist1KO acinar cells were completely devoid of Cx32-containing gap junctions (Figure 7A). To determine if induced MIST1myc could re-establish gap junction complexes, Mist1KO/LSL-Mist1myc pancreata were examined for restoration of Cx32 gap junctions over a 3-week post-TM period. As predicted from the Cx32 transcript analysis (Figure 3A), Cx32 gap junctions assembled on adult Mist1KO/LSL-Mist1myc acinar cells by 3 days after TM (Figure 7A and B). In addition, the newly synthesized gap junctions were fully functional. Single-cell injections of 6-carboxyfluorescein into isolated acini confirmed that the transport of 6-carboxyfluorescein into neighboring cells was restored within 3 days of MIST1myc expression (Figure 7C), showing that adult acinar cell intercellular communication can be remodeled by ectopic MIST1 protein.
Figure 7.
Cx32-containing gap junctions, intercellular communication, and regulated exocytosis are rescued by MIST1myc expression. (A and B) Cx32-containing gap junctions are produced upon Mist1myc expression. Arrows show gap junction complexes. (C) Single-cell injections of 6-carboxyfluorescein (6-FAM) into isolated acini reveal that intercellular transfer of 6-FAM into neighboring acinar cells occurs after MIST1myc induction. Arrows show the injected cell. (D) Quantification of amylase secretion from WT, Mist1KO, and Mist1KO/LSL-Mist1myc acini after cholecystokinin (CCK) stimulation. The defect in secretion observed with Mist1KO acinar cells is reversed completely by induced LSL-Mist1myc expression. Scale bars: (A) 10 µm; (C) 20 µm. DAPI, 4′,6-diamidino-2-phenylindole.
Finally, to examine if induced MIST1myc could rescue functional exocytosis in Mist1KO acinar cells, we performed amylase secretion assays on adult WT, Mist1KO, and Mist1KO/LSL-Mist1myc mice 7 days after TM. As previously reported,8 Mist1KO acini were completely unresponsive to cholecystokinin induction of regulated exocytosis (Figure 7D). However, Mist1KO mice expressing MIST1myc showed restored amylase secretion that reached WT acinar cell efficiencies 7 days after TM (Figure 7D). Together, these studies show that the defects observed in Mist1KO granule secretion can be recovered robustly and quickly by expression of exogenous MIST1myc in quiescent, adult pancreatic acinar cells within the context of the intact organ.
Discussion
Embryonic cell fate decisions define the body axes and establish progenitor cell pools that direct organogenesis. Much has been learned about the transcriptional networks that are instrumental in permitting cell lineages to develop where loss of key transcription factors can have devastating consequences to this initial body plan. Although the importance of cell fate choices is obvious, development does not cease after cells commit to a specific lineage. Instead, most cells undergo a further type–specific maturation in which changes in the cellular transcriptome are needed to ensure proper gene products are available for cells to perform their unique physiological functions.24
Understanding the molecular pathways instrumental in cell maturation has been hampered by a lack of inducible systems in which the course of specialization can be followed in the intact organ. In this study, we asked if induced MIST1 expression could remodel the adult exocrine pancreas because Mist1KO-associated defects leave the organ susceptible to bouts of pancreatitis and pancreatic cancer.7,10 We found that within 36 hours of MIST1 induction, the expression of 352 genes was qualitatively and quantitatively rescued, with 186 up-regulated and 166 down-regulated. Detailed analysis of a cohort of genes confirmed the presence of MIST1-bound regulatory elements that contained CATATG and CAGCTG E-boxes, the preferred binding sites of the vertebrate MIST1 and Drosophila DIMM proteins.14,17,23,25 The ability of MIST1 to activate and repress different target genes was expected because previous studies have shown that MIST1/DIMM serve as transcriptional activators or repressors depending on the gene context.6,9,17–19,23,25
Investigation of 20 target genes revealed that their functions were involved intimately in the specialization of acinar cells, namely generating correct cellular polarity and establishing a proper secretory pathway. The expression profiles of genes encoding a tight junction protein (Cldn10) involved in cell polarity26 and a Rho guanosine triphosphatase (Rnd2) that regulates actin cytoskeletal dynamics27 were restored to WT levels upon MIST1myc induction. Similarly, a group of genes (Rab3d, Rab27a, Atp2c2, Gjb1, and Copz2) known to be critical for exocytosis were mis-expressed in Mist1KO acinar cells and rescued by MIST1myc. Gjb1, which encodes Cx32, is critical to gap junction formation, allowing cells to use an intercellular Ca2+ signaling cascade for efficient secretion.6 Mist1KO acinar cells failed to express Gjb1, but this defect readily was reversed by MIST1myc expression, which restored Cx32 gap junctions and intercellular communication. The genes encoding RAB3D and RAB27A, small guanosine triphosphatases that are known to be important in vesicle trafficking,17,28 and COPZ2, a subunit of the coatomer protein complex involved in vesicle formation,29 also were up-regulated upon MIST1myc expression, leading to proper zymogen granule trafficking and increased vesicle size. These results were in agreement with Tian et al,17 who showed that forced expression of MIST1 in gastric cell lines led to up-regulation of Rab3d and increased vesicle formation. Similarly, the mis-expression of DIMM in Drosophila peptidergic neurons induces genes encoding dense-core vesicle proteins.25 MIST1myc also rapidly induced the expression of Atp2c2, the gene encoding the acinar secretory pathway Ca2+-adenosine triphosphatase23 and rescued the Ca2+-dependent exocytosis defects associated with loss of Mist1.
We also identified additional genes (Htra2, Uba5, Abcb6, Nox4, Gstm4, Nfe2I2, and Aldh1a1) that regulate cellular stress pathways. Htra2 encodes a serine protease that plays a protective role against cellular stress.30 Similarly, Nfe2I2, encoding a basic leucine zipper transcription factor, activates antioxidant and cellular detoxification programs by targeting specific downstream modulators.31 Another MIST1 target gene was Uba5, encoding an E1-like, ubiquitin-activating enzyme that activates the post-translational modifier protein ubiquitin fold modifier 1.32 Ubiquitin fold modifier 1 is expressed to high levels in acinar cells and protects pancreatic cells from ER stress–induced apoptosis.33 In agreement with these altered gene expression profiles, Mist1KO acinar cells have shown deficient ER-stress pathway responses7 and increased levels of autophagy5 that completely were ameliorated by expression of MIST1myc. Of the identified stress-related genes that came out of our rescue screens, only one (Nox4) was found to not bind MIST1 protein. Interestingly, Nox4 transcription is regulated by Nfe2I2,34 which itself is a MIST1 gene target. Future studies will focus on mapping the entire regulatory network by which MIST1 controls aspects of acinar cell function.
MIST1 is a unique transcription factor in that it functions primarily to ensure the correct qualitative and quantitative expression levels of specific secretory cell genes to establish proper, regulated, high-capacity exocytosis. Recently, Mills and Taghert35 proposed that MIST1/DIMM be classified as “scaling factors” that are tasked with controlling entire subcellular domains associated with a mature cell type. In support of this idea, our studies revealed that MIST1 indeed controls aspects of acinar cell organization that are involved in regulated exocytosis events. The importance of MIST1 to maintaining cellular organization also is apparent in a number of pancreatic diseases in which loss of Mist1 exacerbates pancreatitis and pancreatic cancer formation.7,10 Similarly, loss of MIST1 is a prognostic indicator of gastric carcinomas.36,37 Indeed, alteration of intracellular organization is likely critical to disease formation in many different organ contexts. The ability of Mist1 to alter the phenotype of an adult acinar cell offers new therapeutic opportunities for treating a number of diseases, including pancreatitis and pancreatic cancer.
Supplementary Material
Acknowledgments
The authors thank the Purdue Center for Cancer Research Transgenic Mouse Core Facility for generating the transgenic mouse strains used in this study and David Miley for generating tissue sections.
Funding
This work was supported by grants from the National Institutes of Health (DK55489 and CA124586 to S.F.K., and DK61220 to R.J.M.), a grant from Phi Beta Psi Sorority (S.F.K.), and a Clinical and Translational Sciences Institute Predoctoral Fellowship (D.D.).
Abbreviations used in this paper
- ChIP
chromatin immunoprecipitation
- Cre-ER
Cre recombinase-estrogen receptor
- Cx32
connexin32
- PCR
polymerase chain reaction
- TM
tamoxifen
- WT
wild type
Footnotes
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2012.04.011.
Conflicts of interest
The authors disclose no conflicts.
References
- 1.de Assis GF, Cestari TM, Sesso A, et al. Post-natal maturation of acinar cells of the guinea pig pancreas: an ultrastructural morphometric study. Anat Histol Embryol. 2003;32:36–41. doi: 10.1046/j.1439-0264.2003.00435.x. [DOI] [PubMed] [Google Scholar]
- 2.Jamieson JD, Gorelick FS, Chang A. Development of secretagogue responsiveness in the pancreas. Scand J Gastroenterol Suppl. 1988;151:98–103. doi: 10.3109/00365528809095920. [DOI] [PubMed] [Google Scholar]
- 3.Rovira M, Delaspre F, Massumi M, et al. Murine embryonic stem cell-derived pancreatic acinar cells recapitulate features of early pancreatic differentiation. Gastroenterology. 2008;135:1301–1310. 1310, e1–e5. doi: 10.1053/j.gastro.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson CL, Kowalik AS, Rajakumar N, et al. Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech Dev. 2004;121:261–272. doi: 10.1016/j.mod.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Pin CL, Rukstalis JM, Johnson C, et al. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol. 2001;155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rukstalis JM, Kowalik A, Zhu L, et al. Exocrine specific expression of Connexin32 is dependent on the basic helix-loop-helix transcription factor Mist1. J Cell Sci. 2003;116:3315–3325. doi: 10.1242/jcs.00631. [DOI] [PubMed] [Google Scholar]
- 7.Kowalik AS, Johnson CL, Chadi SA, et al. Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1123–G1132. doi: 10.1152/ajpgi.00512.2006. [DOI] [PubMed] [Google Scholar]
- 8.Luo X, Shin DM, Wang X, et al. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J Biol Chem. 2005;280:12668–12675. doi: 10.1074/jbc.M411973200. [DOI] [PubMed] [Google Scholar]
- 9.Jia D, Sun Y, Konieczny SF. Mist1 regulates pancreatic acinar cell proliferation through p21 CIP1/WAF1. Gastroenterology. 2008;135:1687–1697. doi: 10.1053/j.gastro.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi G, Zhu L, Sun Y, et al. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology. 2009;136:1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsey VG, Doherty JM, Chen CC, et al. The maturation of mucussecreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 12.Bredemeyer AJ, Geahlen JH, Weis VG, et al. The gastric epithelial progenitor cell niche and differentiation of the zymogenic (chief) cell lineage. Dev Biol. 2009;325:211–224. doi: 10.1016/j.ydbio.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capoccia BJ, Lennerz JK, Bredemeyer AJ, et al. Transcription factor MIST1 in terminal differentiation of mouse and human plasma cells. Physiol Genomics. 2011;43:174–186. doi: 10.1152/physiolgenomics.00084.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park D, Shafer OT, Shepherd SP, et al. The Drosophila basic helix-loop-helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide-amidating enzyme. Mol Cell Biol. 2008;28:410–421. doi: 10.1128/MCB.01104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamanaka Y, Park D, Yin P, et al. Transcriptional orchestration of the regulated secretory pathway in neurons by the bHLH protein DIMM. Curr Biol. 2010;20:9–18. doi: 10.1016/j.cub.2009.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park D, Taghert PH. Peptidergic neurosecretory cells in insects: organization and control by the bHLH protein DIMMED. Gen Comp Endocrinol. 2009;162:2–7. doi: 10.1016/j.ygcen.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Tian X, Jin RU, Bredemeyer AJ, et al. RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol. 2010;30:1269–1284. doi: 10.1128/MCB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran T, Jia D, Sun Y, et al. The bHLH domain of Mistl is sufficient to activate gene transcription. Gene Expr. 2007;13:241–253. doi: 10.3727/000000006780666975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemercier C, To RQ, Carrasco RA, et al. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J. 1998;17:1412–1422. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habbe N, Shi G, Meguid RA, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araki K, Araki M, Miyazaki J, et al. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc Natl Acad Sci U S A. 1995;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams JA. Isolation of rodent pancreatic acinar cells and acini by collagenase digestion. [Accessed November 13, 2010];The Pancreapedia: Exocrine Knowledge Base. 2010 Available at: http://dx.doi.org/10.3998/panc.2010.18.
- 23.Garside VC, Kowalik AS, Johnson CL, et al. MIST1 regulates the pancreatic acinar cell expression of Atp2c2, the gene encoding secretory pathway calcium ATPase 2. Exp Cell Res. 2010;316:2859–2870. doi: 10.1016/j.yexcr.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald RJ, Swift GH, Real FX. Transcriptional control of acinar development and homeostasis. Prog Mol Biol Transl Sci. 2010;97:1–40. doi: 10.1016/B978-0-12-385233-5.00001-5. [DOI] [PubMed] [Google Scholar]
- 25.Park D, Hadzic T, Yin P, et al. Molecular organization of Drosophila neuroendocrine cells by dimmed. Curr Biol. 2011;21:1515–1524. doi: 10.1016/j.cub.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inai T, Sengoku A, Guan X, et al. Heterogeneity in expression and subcellular localization of tight junction proteins, claudin-10 and -15, examined by RT-PCR and immunofluorescence microscopy. Arch Histol Cytol. 2005;68:349–360. doi: 10.1679/aohc.68.349. [DOI] [PubMed] [Google Scholar]
- 27.Pacary E, Heng J, Azzarelli R, et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron. 2011;69:1069–1084. doi: 10.1016/j.neuron.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasai K, Ohara-Imaizumi M, Takahashi N, et al. Rab27a mediates the tight docking of insulin granules onto the plasma membrane during glucose stimulation. J Clin Invest. 2005;115:388–396. doi: 10.1172/JCI22955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moelleken J, Malsam J, Betts MJ, et al. Differential localization of coatomer complex isoforms within the Golgi apparatus. Proc Natl Acad Sci U S A. 2007;104:4425–4430. doi: 10.1073/pnas.0611360104. [DOI] [PubMed] [Google Scholar]
- 30.Moisoi N, Klupsch K, Fedele V, et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brainspecific transcriptional stress response. Cell Death Differ. 2009;16:449–464. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- 31.Chanas SA, Jiang Q, McMahon M, et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng M, Gu X, Zheng D, et al. UBE1DC1, an ubiquitin-activating enzyme, activates two different ubiquitin-like proteins. J Cell Biochem. 2008;104:2324–2334. doi: 10.1002/jcb.21791. [DOI] [PubMed] [Google Scholar]
- 33.Lemaire K, Moura RF, Granvik M, et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS One. 2011;6:e18517. doi: 10.1371/journal.pone.0018517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pendyala S, Moitra J, Kalari S, et al. Nrf2 regulates hyperoxiainduced Nox4 expression in human lung endothelium: identification of functional antioxidant response elements on the Nox4 promoter. Free Radic Biol Med. 2011;50:1749–1759. doi: 10.1016/j.freeradbiomed.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills JC, Taghert PH. Scaling factors: transcription factors regulating subcellular domains. Bioessays. 2012:3410–3416. doi: 10.1002/bies.201100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lennerz JK, Kim SH, Oates EL, et al. The transcription factor MIST1 is a novel human gastric chief cell marker whose expression is lost in metaplasia, dysplasia, and carcinoma. Am J Pathol. 2010;177:1514–1533. doi: 10.2353/ajpath.2010.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam KT, Lee HJ, Sousa JF, et al. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology. 2010;139:2028 e9–2037 e9. doi: 10.1053/j.gastro.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.