Abstract
Recently, small peptides have been shown to modulate aggregation and toxicity of the amyloid-β protein (Aβ). As such, these new scaffolds may help discover a new class of biotherapeutics useful in the treatment of Alzheimer's disease. Many of these inhibitory peptide sequences have been derived from natural sources or from Aβ itself (e.g., C-terminal Aβ fragments). In addition, much earlier work indicates that tachykinins, a broad class of neuropeptides, display neurotrophic properties, presumably through direct interactions with either Aβ or its receptors. Based on this work, we undertook a limited screen of neuropeptides using ion mobility-mass spectrometry to search for similar such peptides with direct Aβ binding properties. Our results reveal that the neuropeptides leucine enkephalin (LE) and galanin interact with both the monomeric and small oligomeric forms of Aβ1-40 to create a range of complexes having diverse stoichiometries, while some tachyknins (i.e., substance P) do not. LE interacts with Aβ more strongly than galanin, and we utilized ion mobility-mass spectrometry, molecular dynamics simulations, gel electrophoresis/Western blot, and transmission electron microscopy to study the influence of this peptide on the structure of Aβ monomer, small Aβ oligomers, as well as the eventual formation of Aβ fibrils. We find that LE binds selectively within a region of Aβ between its N-terminal tail and hydrophobic core. Furthermore, our data indicate that LE modulates fibril generation, producing shorter fibrillar aggregates when added in stoichiometric excess relative to Aβ.
Introduction
Alzheimer's disease (AD) is currently the sixth leading cause of death worldwide, directly affecting over 24 million people globally.1 Critically, of the top six causes of death, AD is the only cause for which the mortality rate has not decreased over the past 5 years.1, 2 There are several competing hypotheses describing the onset and etiology of AD.3 One of the most prominent of these theorems centers on the uncontrolled aggregation of Aβ peptides varying from 36-43 amino acids in length, produced from proteolytic cleavage of the amyloid precursor protein (APP) in vivo; these Aβ species proceed to form amyloid fibrils or plaques in the brain that are comprised primarily of Aβ1-40 and Aβ1-42.4, 5 While the eventual formation of amyloid fibrils serves to demarcate the phenomenology of the disease, and fibrils are found prominently in the brains of AD patients, current data suggests that the causative Aβ-related agent, if any, takes the form of small oligomers that may range from dimers to dodecamers.6-10 Because Aβ peptides aggregate quickly both in vivo and in vitro, likely occupying a number of structural and oligomeric states simultaneously, the study of Aβ aggregation and its prevention has proven challenging for classical structural biology and biophysics tools.11
A number of small peptides and small peptide fragments have been found to inhibit the formation of Aβ fibrils and, in some cases, possess protective or restorative properties with respect to the neuronal degeneration that accompanies AD.12-17 Such peptides are sought after as potential biotherapeutics for AD primarily due to their natural ability to cross membranes and the blood-brain barrier.18, 19 For example, fragment sequences derived from the C-terminal section of Aβ1-42 (C-terminal fragments, CTFs) have been found to directly interact with full-length Aβ peptides and inhibit fibril formation and toxicity.14 Similarly, proline-rich hydrophobic peptides have been found to alter Aβ1-42 folding and fibril formation.20. Seminal work in AD indicated that tachykinin neuropeptides possess the ability to ameliorate the neurotoxic effects of Aβ peptides.21 Specifically, substance P and physalaemin were found to be inhibitors of Aβ-induced neurotoxicity in hippocampal neurons at μM concentrations, whereas other related peptides did not display the same abilities. Whether the observed neurotrophic activity of these tachykinin peptides is related to their specific roles as neuronal agonists, or if they interact directly with soluble forms of Aβ monomers or oligomers is currently unknown.
Relatively recently, ion mobility-mass spectrometry (IM-MS) has been used to investigate a number of amyloid forming peptides and proteins, providing details on the structures and identities of the oligomers formed prior to amyloid fibril formation.22, 23 IM separates peptides, proteins, and protein complexes according to differences in ion collision cross-section (CCS) and charge.24 When combined with MS, this technology is a powerful approach to both complex mixture analysis25 and for structural studies of heterogeneous biological complexes.26 IM-MS has been used to identify the oligomers produced by β-microglobulin and transthyretin tetramer23, 27, 28, in the case of the former providing detailed structural and composition information on the soluble oligomers formed. Aβ has also been analyzed in detail using IM-MS, revealing much regarding the structure of soluble oligomers and the action of potential inhibitors on the fibril formation process in vitro. Specifically, IM-MS has been employed to assess the influence of the CTF Aβ39-42 on the oligomerization of full length Aβ14 While the addition of CTF to full length Aβ did not alter fibril formation, IM-MS showed discrete differences in the oligomerization pathway of Aβ when bound to the CTF in a manner linked to decreases in Aβ-induced neurotoxicity.
Inspired both by this previous IM-MS work on CTF-mediated Aβ oligomerization 14, and also the earlier work described above relating tachykinin-type neuropeptides to neurotrophic effects in neurons doped with excess Aβ,21 we screened a limited panel of neuropeptides for their direct interactions with Aβ monomers and small oligomers using IM-MS. While we find no evidence of direct interaction between substance P and monomeric/dimeric Aβ1-40, we do detect Aβ interactions with leucine enkephalin (LE) and galanin. Of the two neuropeptides, LE displays a stronger noncovalent bond (Aβ:LE dissociation constant (Kd) equivalent to Aβ1-40 dimer formation), and thus in this work we characterized this complex in detail using IM-MS, molecular dynamics (MD) simulations, gel electrophoresis, and transmission electron microscopy (TEM). Concentration-dependent doping of Aβ1-40 with LE leads to the formation of peptide oligomers having a broad range of stoichiometries, and we find that subsequent LE additions to monomeric Aβ are less favored than multiple LE additions to the Aβ dimer. Furthermore, our MD data, filtered by CCS values, indicate that the most likely binding site for LE is within the N-terminus of the peptide, a region similar to other natural products that have been shown to inhibit Aβ fibril formation and neurotoxicity.29 Finally, our gel data demonstrate that LE-doped Aβ1-40 samples exhibits the increased amount of Aβ species with MW ≤ 25 kDa, compared to that observed under LE-untreated conditions. From TEM analysis, mainly truncated fibrillar Aβ are observed in contrast to the long amyloid fibrils typically formed by the peptide in isolation. We discuss these results in the context of biotherapeutic development for AD, as well as Aβ fibril formation in general.
Experimental
General
Reagents were purchased from commercial sources and used without further purification unless specified otherwise. Aβ1-40 was purchased from Anaspec (Fremont, CA, USA). Aβ1-40 was dissolved in the supplied 1% (v/v) ammonium hydroxide and diluted with pH 6.9 100 mM ammonium acetate (Sigma-Aldrich, St. Louis, MO, USA). Peptide concentration for the stock solution was calculated from absorbance at 280 nm (ε = 1450 M-1 cm-1) using a Thermo Scientific Genesys 10UV spectrometer (Vernon Hills, IL, USA) or Agilent 8453 spectrophotometer (Santa Clara, CA, USA). LE acetate hydrate (YGGFL) was purchased from Sigma-Aldrich (St. Louis, MO, USA) and prepared in pH 6.9 100 mM ammonium acetate. Somatostatin 14, substance P, galanin, and neurotensin were purchased from Anaspec and prepared in 100 mM ammonium acetate. Sequence identity comparisons between different neuropeptides studied here were performed with the LALNVIEW tool, accessed through the ExPASy bioinformatics resource.30
IM-MS
Mass spectra were collected on a quadrupole-ion mobility-time-of-flight (ToF) mass spectrometer (Synapt G2 HDMS, Waters, Milford, MA, USA) with a nano-electrospray ionization (nESI) source. Protein ions were generated using a nESI source and optimized to allow transmission of noncovalent protein complexes using nanoflow electrospray capillaries prepared as described previously.31 Protein complex ions were generated using an aliquot of the sample (ca. 7 μL) sprayed from the nESI emitter at a capillary voltage of 1.7 kV. The source was operated in positive ion mode with the sample cone at 50 V. The bias voltage was 45 V, with backing pressure at 5.42 mbar and ToF pressure at 7.94 × 10-7 mbar. The traveling-wave IM separator was operated at a pressure of approximately 3.0 mbar of nitrogen and helium. Mass spectra were calibrated externally using a solution of cesium iodide (100 mg/mL) and analyzed using MassLynx 4.1 and Driftscope 2.0 software (Waters, Milford, MA, USA). CCS (Ω) measurements were externally calibrated using a database of known values in helium, using values for peptides and proteins that bracket the likely CCS and ion mobility values of the unknown ions.27, 32 We report the standard deviations (σ) from triplicate measurements of CCS, but real errors for these values must incorporate the errors involved in the calibration process (an additional ±3%). Samples were prepared by mixing stock solutions (as prepared above) of neuropeptide and Aβ1-40 at pH 6.9, to generate a final Aβ1-40 concentration of 20 μM. Samples were incubated on ice for 1 h prior to IM-MS analysis.
Docking Studies
Flexible ligand docking studies were performed using AutoDock Vina.33 LE was created using PyMOL and studies were conducted against the Aβ1-40 monomer (aqueous solution NMR structure, PDB 2LFM).34 Twenty docking studies, one with each conformation within the PDB file, were performed. The structures for LE and Aβ1-40 were prepared for use with AutoDock Vina using AutoDock Tools.33 Hydrogens were added to Aβ1-40 and the peptide was contained within a search space sized to contain the whole monomer. No modifications were made to the LE, and torsions were kept as the default selected in AutoDock Tools. The exhaustiveness for the docking studies was set at 8, resulting in 9 output models for each Aβ1-40 conformation. Docked models of LE were visualized with Aβ1-40 using Pymol.
MD Simulations
Simulations were started using the minimized Aβ1-40 solution NMR structure (PDB 2LFM)34 The simulations were performed using periodic boundary conditions in a dodecahedron with the minimum distance between the simulated molecules and the box wall being 1.0 nm. GROMACS ligand topology was prepared using the GlycoBioChem PRODRG2 server.35 The MD simulations were carried out using the GROMACS software package36 and GROMOS96 force field.37 To constrain the bond lengths in the Aβ1-40 and LE, the LINCS algorithm was used, allowing an integration time step of 2 fs. Long-range electrostatic interactions were treated with the particle mesh Ewald method. Temperature was maintained using the method of Berendsen et al.38 The LE and the Aβ1-40 were separately coupled to external temperature bath with a temperature-coupling constant of 0.1 ps.
The system was energy-minimized by steepest decent for 500 steps. After equilibration, simulated annealing was performed for the Aβ1-40:LE complex in the gas-phase. The three most basic the Aβ1-40 side chains (R5, K16, and K28) were charged. The system was heated from 300 K to 500 K over 100 ps, then cooled down to 300 K over the next 100 ps. The cycle was repeated over 20 ns in order to allow for escape from local minima and enhance equilibration. For the Aβ1-40:LE complex, 20 independent simulated annealing runs, each running for a total of 20 ns, were performed from the lowest energy complexes generated by AutoDock Vina. From the MD trajectory generated, 100 structures were sampled at 300 K and the CCS was calculated with Mobcal using the trajectory method algorithm.39, 40 Models of the Aβ1-40:LE complexes were visualized in Pymol. In total 2000 structures were generated. Of the 1090 structures which were within ±3% of the experimentally determined CCS, the 201 structures with lowest energy were analyzed to determine the Aβ1-40 residues within 4 Å of LE. Standardized values (Z-scores) were calculated for each residue of Aβ1-40 and plotted in standard deviation (σ) space in order to determine the relative likelihood of LE binding within a given region of Aβ.
Kd Measurements by MS
Dissociation constant (Kd) values for the neuropeptides with Aβ1-40 were calculated using the relative intensity of each species from the mass spectra, as described previously.41 We modified this method to accommodate multiple ligand binding events with the following assumptions: 1) the spray and detection efficiency of all species are similar, 2) the ligand concentration is sufficiently high so that [L]eq remains constant and 3) the ligand binds to the complex one at a time in a stepwise fashion.
For the equilibrium binding of A (unbound protein) and B (a ligand/binding partner):
| (1) |
| (2) |
Where Rx is an equilibrium quotient between the bound form of the protein (Cx, having x ligands attached) when interacting with ligand B, and its unbound form (A).
| (3) |
Eqn. 3 above defines Rx for all bound forms of the protein (Ci) and:
| (4) |
allows for the determination of a Kd for any given step in the sequential equilibrium described in Eqn. 1, where C0 = A. Standard deviation values (σ) for the Kd measurements reported here are shown from three replicate measurements.
Aβ Aggregation Experiments
Aβ experiments were performed according to the previously published methods.42-49 Prior to experiments, Aβ1-40 was dissolved in ammonium hydroxide (NH4OH, 1% v/v, aqueous), aliquoted, and lyophilized overnight, and stored at –80 °C. A stock solution of fresh Aβ was prepared by dissolving the peptide in 1% NH4OH (10 μL) and diluting with ddH2O. The Aβ stock solution was diluted to a final concentration of 25 μM in a buffered solution containing ammonium acetate (100 mM, pH 6.9). The Aβ samples were incubated with 0, 1, 3, or 5 equiv LE (1.2 mM stock solution in same buffered solution) at 37 °C with constant agitation for 24 h.
Samples from the experiment were analyzed by gel electrophoresis and visualized by Western blot using an anti-Aβ antibody (6E10).42-49 Each sample was separated on a 10-20% Tris-tricine gel (Invitrogen, Grand Island, NY, USA) and transferred onto a nitrocellulose membrane. The nitrocellulose was blocked with bovine serum albumin (BSA, 3% w/v, Sigma, St. Louis, MO, USA) in Tris-buffered saline (TBS, Fisher, Pittsburgh, PA, USA) containing 0.1% Tween-20 (Sigma: TBS-T) for 2 h at room temperature. Afterward, the membrane was incubated with the anti-Aβ antibody 6E10 (1:2000, Covance, Princeton, NJ) in a solution of 2% BSA (w/v in TBS-T) overnight at 4 °C. The horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:5000, Cayman Chemical, Ann Arbor, MI, USA) in 2% BSA was added for 1 h at room temperature. The ThermoScientific SuperSignal West Pico Chemiluminescent Substrate was used to visualize the protein bands.
TEM
TEM images were taken using Phillips CM-100 transmission electron microscope (Microscopy and Image Analysis Laboratory, University of Michigan, MI, USA) using a magnification factor of 25,000. Samples for TEM were prepared according to the previously reported methods.42-47, 49 Glow-discharged grids (Formar/Carbon 300-mesh, Electron Microscopy Sciences, Hatfield, PA, USA) were treated with Aβ samples from the in vitro inhibition experiments (5 μL) for 2 min at room temperature. Excess sample was removed using filter paper followed by washing with ddH2O five times. Each grid was stained with uranyl acetate (1% w/v ddH2O, 5 μL, 1 min). Upon removal of excess uranyl acetate, the grids were dried for 15 min at room temperature.
Results
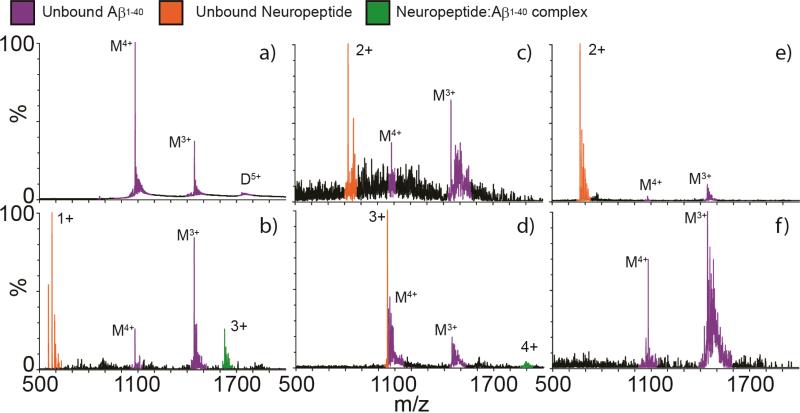
A limited neuropeptide screen for binding with Aβ1-40 was conducted with five neuropeptides, varying in mass and structure. Masses of the neuropeptides in this study ranged from 555.62 Da (LE) to 3158.5 Da (galanin). Aβ1-40 prepared for nESI-IM-MS analysis in 100 mM ammonium acetate results in a mass spectrum containing both 3+ and 4+ monomer and 5+ dimer ions in high relative abundances (Fig. 1). IM-MS analysis of this same dataset (data not shown) reveals evidence of additional oligomeric forms of the peptide, as well as additional conformational forms of the monomeric 4+ peptide (vide infra). Aβ1-40: neuropeptide binding was only detected between Aβ1-40 and two neuropeptides, LE and galanin, under our experimental conditions (Fig. 1). Complex formation between Aβ1-40 and substance P, somatostatin, or neurotensin is not evident in our dataset. The Aβ1-40 binding strength is greater for LE than for galanin, as reported in the relative intensity differences detected by MS between the free Aβ1-40 monomer peaks and the respective complex ion signals. Due to the apparently enhanced strength of the Aβ:LE interaction under the conditions of our screen relative to all other neuropeptides studied, we focused on characterizing the structure of this complex further.
Figure 1.
Analyses of Aβ1-40 incubated with one equivalent of each neuropeptide by nESI-IM-MS. (A) Mass spectrum of Aβ1-40 only, with signals corresponding to monomeric and dimeric peptides marked with ‘M’ and ‘D’, respectively. Satellite peaks observed correspond to alkali metal adducts commonly observed in nESI-MS. Aβ1-40 was then mixed with equivalent amounts of (B) LE (free [M+H]+ at m/z = 556.6), (C) somatostatin (free [M+2H]2+ at m/z = 820.5), (D) galanin (free [M+3H]3+ at m/z = 1053.8), (E) substance P (free [M+2H]2+ at m/z = 675.5) and (F) neurotensin (no free signal detected, peptide mass = 1674.0). LE and galanin are the two neuropeptides in this set where we detect complexes with Aβ1-40 (complexes signals in green), while the other neuropeptides screened result in signals for unbound Aβ1-40 (purple) and unbound neuropeptide (orange). Poorer signal intensities are recorded in panels C and F due to signal suppression surrounding the addition of the neuropeptides indicated.
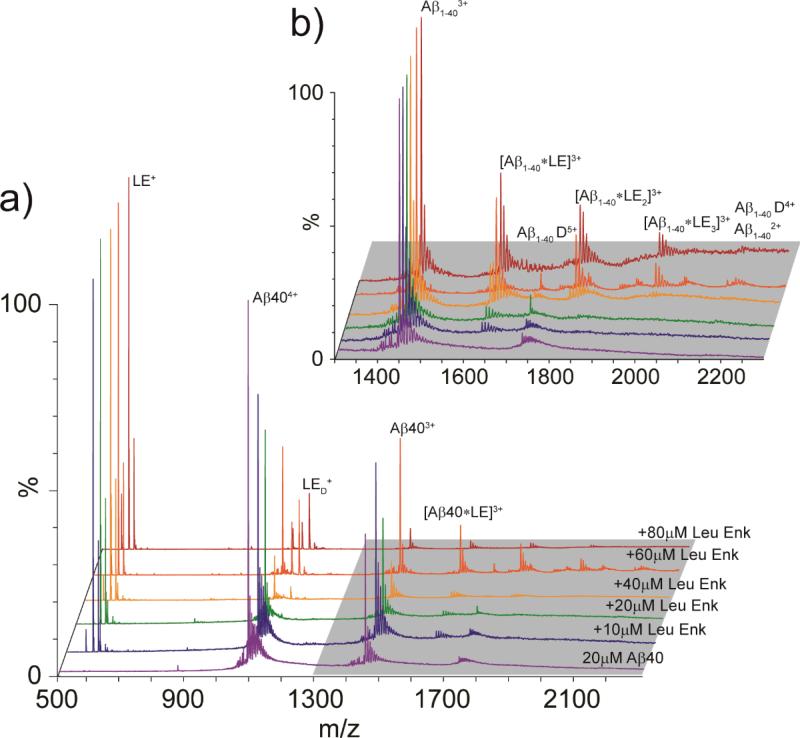
Up to 80 μM LE was titrated into a 20 μM Aβ solution, and the resultant complexes were detected using IM-MS (Fig. 2). The 1:1 complex of Aβ1-40:LE is first observed in our IM-MS data when 10 μM LE is added. At a 1:2 ratio of Aβ1-40:LE, the 1:2 and 1:3 complexes of Aβ1-40:LE is detected, both of which occupy a 3+ charge state (Fig. 2B). Complexes of Aβ1-40 dimer with LE are also observed with lower intensity in the Aβ1-40:LE mixture (20/60 μM, respectively) (vide infra). Dissociation constants (Kd) for each of the Aβ1-40:LE complexes identified were calculated as an average across the concentration ramp (Table 1). Measured Kd values for Aβ:LE complexes are in the μM range, with similar Kd values for both the monomer and dimer complexes. While the 1:1 complex exhibits a Kd of 61.7 μM (similar to the Kd we measure for the LE with the Aβ dimer, at 56 μM), the 1:2 and 1:3 complexes possess Kd values of 99.2 and 55.7 μM respectively, indicating a slight thermodynamic barrier in the formation of Aβ:2LE. Aβ dimer, however, does not exhibit signs of a similar barrier, as LE-related Kd values are recorded to be 76.7 and 49.8 μM for the formation of 2:1 and 2:2 complexes, respectively.
Figure 2.
(A) MS spectra for Aβ1-40 acquired at 20 μM (purple). Aβ1-40 was then incubated with increasing concentrations of LE, ranging from 10-80 μM (stoichiometric ratios from 0.5 to 4). At sufficiently high LE concentrations, Aβ1-40 is seen in complex with LE, producing Aβ:LE complexes ranging from 1:1: to 1:3. (B) A magnified region of the spectrum shown in A (grey highlight), where signals corresponding to Aβ:LE complexes are labeled.
Table 1.
Kd values for LE:Aβ1-40 Complexes Measured by MS
| Complex | Kd (μM) | σ |
|---|---|---|
| Aβ1-40 Dimer | 56.0 | 44.7 |
| [Aβ1-40 + LE]3+ | 61.7 | 27.2 |
| [Aβ1-40 + LE2]3+ | 99.2 | 30.6 |
| [Aβ1-40 + LE3]3+ | 55.7 | 21.7 |
| [Aβ1-40 Dimer + LE]5+ | 76.7 | 15.6 |
| [Aβ1-40 Dimer + LE2]5+ | 49.8 | 6.5 |
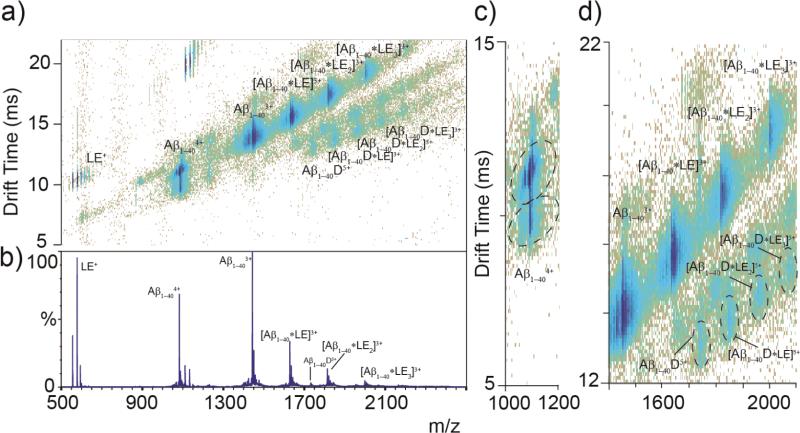
Using IM-MS, it is possible to measure the CCSs of both free Aβ1-40 and Aβ:LE complexes simultaneously (Fig. 3). While the 3+ charge state of Aβ1-40 and the Aβ1-40 monomer:LE complexes all occupy a single closely related family of structures, as evidenced by the drift time profiles recorded for these ions, the 4+ Aβ1-40 monomer has at least two main conformational families, along with a third minor structure for which we did not record data here.29 The more compact form of Aβ 4+ has a CCS of 622 Å2 and the more extended, 678 Å2. These values are similar to previous reports.29 The measured cross section of LE is 165 Å2; however, the change in CCS of Aβ1-40 monomer when in complex with LE is only 44-47 Å2 indicating a closely packed interaction. The changes in size for the dimer complexes are similar in magnitude to those of the monomer (27-51 Å2), also likely representing a tightly packed complex. The intensities that we observe for the Aβ dimer-based complexes are lower than those of the Aβ monomer-related complexes (Fig. 3D), likely due to the lower concentration of free Aβ dimer in solution.
Figure 3.
(A) IM-MS data for Aβ1-40 (20 μM) incubated with LE (60 μM) for 1 h on ice, where Aβ:LE complexes are labeled, along with free peptide. IM separation allows for the identification of Aβ dimer complexes previously undetected by MS alone. Aβ1-40 is seen in complex with LE at stoichiometric ratios up to 1:3 Aβ1-40:LE and 2:3 Aβ1-40:LE. (B) MS dataset for the IM-MS plot shown in A. (C) Two main conformations of Aβ1-404+ are identified, with a third minor conformer, as observed previously.29 (D) A magnified region of the IM-MS data shown in A, showing detail on Aβ1-404+: LE complexes. Monomer complexes are observed in greater relative abundance than those related to dimeric Aβ.
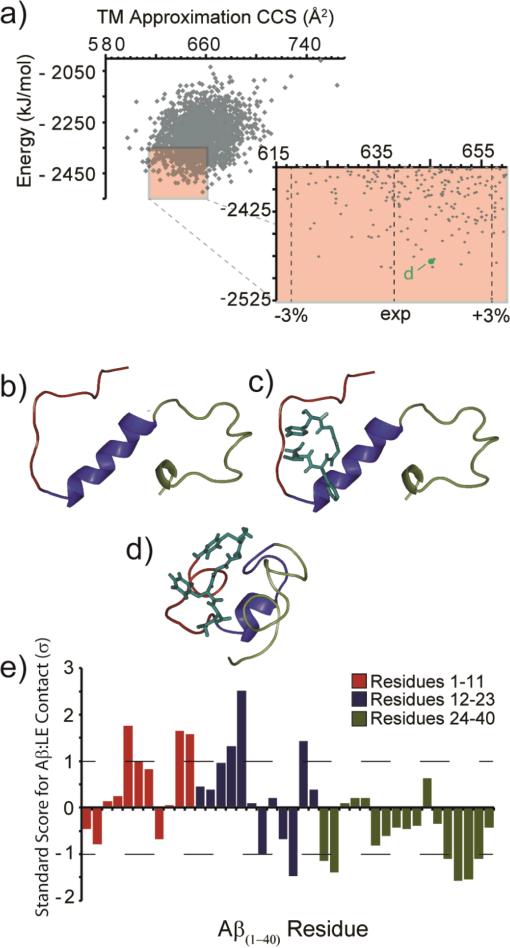
To visualize the most likely configurations of the 1:1 Aβ1-40: LE complex, rigid peptide-flexible ligand docking followed by simulated annealing MD was performed. Of the 2000 resultant structures after annealing simulations, 1090 had CCS values within 3% of the experimentally measured value for the complex (638 Å2), as calculated by the trajectory method. These structures were additionally filtered by the energy axis from MD data, resulting in 201 sample structures that are most-likely to represent the structure of the gas-phase ions in our expeirments. From this population, a lowest-energy sample configuration was compared to both the starting NMR (PDB ID: 2LFM) and LE docked structures in Figure 4 (Fig. 4B-D). We further analyzed the 201 low energy structures to determine the LE side-chain groups within 4 Å of any Aβ1-40 residue and counted these as potential interactions. As expected, the hydrophobic residues Phe and Tyr in LE interacted more frequently with Aβ1-40 than the Leu and in much greater frequency than the two Gly residues. The Aβ1-40 region with most frequent LE interactions in our MD dataset lies in the region between residues Glu-3 and Lys-16, with the most frequent LE interactions occurring with the Arg-5, Tyr-10, Glu-11, Lys-16, and Glu-22 side chains. Generally, our MD results, filtered according to our experimental CCS data, indicate that LE interacts preferentially with the N-terminus and hydrophobic core of the Aβ1-40 monomer.
Figure 4.
(A) Output from all molecular dynamics simulations. The lowest energy 201 structures with CCS values within ±3% of experiment are highlighted (red box). (B) The structure of Aβ monomer (PDB 2LFM). (C) A docked structure of Aβ (PDB 2LFM) with LE using AutoDock Vina. (D) A representative low energy model (indicated in A) from the main structural family identified from our MD simulations, in agreement with experimental CCS values. Colors represent the N-terminus (red), core/helix region (blue), and the C-terminus (green). (E) A plot of the standard score (Z-score) for Aβ residues within 4 Å of the bound LE. Larger values denote contacts of greater significance on the standard deviation (σ) scale. Negative values denote contacts of reduced significance.
In order to assess whether the interaction of LE with Aβ1-40 could influence aggregation of the peptide, in vitro aggregation studies were conducted and samples of the resulting Aβ species were analyzed by gel electrophoresis/Western blot and TEM (Fig. 5). The gel electrophoresis/Western blot results present the distribution of Aβ species, including aggregates that are able to penetrate the gel, based on their molecular weight (MW).29, 43-46, 49 Upon incubation of Aβ species with LE (1, 3, or 5 equiv), the amount of Aβ species with MW ≤ 25 kDa, visualized in the gel, is relatively increased compared to that observed under LE-untreated conditions (Fig. 5A). For a qualitative comparison against these gel results, TEM was used to identify whether LE could restructure gross Aβ aggregate morphology (Fig. 5B). In the absence of LE, large Aβ aggregates with mainly fibrillar morphologies are indicated. On the other hand, from the samples containing Aβ and LE (3 or 5 equiv), shorter fibrillar species are shown as the main Aβ species. Taken together, LE demonstrates an ability to moderately alter Aβ aggregate formation in vitro.
Figure 5.
Influence of LE on Aβ1-40 aggregation in vitro. (A) Visualization of Aβ species generated in the absence and presence of LE by gel electrophoresis and Western blotting (6E10). Experimental conditions: [Aβ1-40] = 25 μM; [LE] = 0, 25, 75, or 125 μM; 100 mM ammonium acetate, pH 6.9; 37 °C; 24 h; agitation. (B) TEM images of Aβ species in the absence and presence of LE (3 and 5 equiv) from samples in A. The scale bar depicts 500 nm.
Discussion
Neuropeptide screening results shown in Figure 1 not only indicate Aβ:LE and Aβ:galanin complex formation, but also detect no direct complexes between Aβ and substance P, a neuropeptide known to possess neurotrophic properties with respect to Aβ-induced neurotoxicity.50 This result suggests that either substance P interacts with larger toxic oligomers that are not detected in our IM-MS datasets, or that the action of substance P is related to its role as a neuronal agonist, where it may act to block Aβ interactions with critical cell surface receptors. The original work identifying the neurotrophic effects of tachykinin neuropeptides in the context of Aβ localized the critical amino acid sequence involved in the putative interaction to GSNKGAIIGLM, which shares broad sequence homology within the tachykinin family and corresponds to residues 25-35 of the Aβ peptide.21 This sequence bears little resemblance to the amino acid sequence of either LE (YGGFL) or galanin (GWTLNSAGYLLGPHAVGNHRSFSDKNGLTS). For example, the strongest identity between galanin and Aβ25-35 exist in a six amino acid sequence between residues 23 and 28 in galanin and residues 2 through 7 in the Aβ fragment, resulting in only a 33% sequence identity within that region. Since early reports for Aβ:tachykinin interactions suggested that the C-terminus of Aβ played a role ,21 the fact that our results for peptides that contain little sequence identity to tachykinins and target the N-terminus is not surprising. Interestingly, galanin has also been implicated in the etiology of AD, as the peptide has been found within fibrils innervating surviving cholinergic neurons.51 While the known physiological concentrations of both neuropeptides is insufficient to drive the formation of the relatively weak interactions discovered in this report in vivo, the local concentrations of these peptides, which generally co-localize with Aβ,52 may be sufficient to interact with Aβ and influence complex equilibria towards the complex formation.
IM-MS structure and oligomer population data for peptides and proteins must be interpreted carefully due to the relatively unique environment employed during the analysis. First, during the nESI process, many peptides may be trapped in rapidly evaporating droplets and are thus forced to generate artificial complexes due to solvent evaporation.53 While this situation is typically avoided by simply lowering the overall sample concentration,26 the concentration accessed in this report is in excess of those that typically limit the production of ESI-artifact oligomers. The data in Figure 1, however, serve as control experiments in this regard, validating the specificity of the interactions identified for LE and galanin against a panel of either larger or similarly sized peptides. No Aβ complexes were detected for the other peptides in our panel (i.e., substance P, somatostatin, or neurotensin), even when added in large excess in solution, indicating that oligomers observed in our IM-MS data are both specific to LE and galanin and likely formed in solution. Since the structural measurements generated by IM-MS take place in the gas-phase, a certain amount of local structural rearrangement is expected for peptide complex ions. Such rearrangements can be observed in Figure 4D. The key to utilizing gas-phase IM-MS data effectively in assessing the structure of biomolecules is to seek evidence of correlated structural elements, rather than a wholesale identity between solvent-free and native-state structures.54 However, we find that the binding region likely accessed by LE in solution is apparently retained in our lowest energy gas-phase structures and, indeed, throughout our simulation results (Fig. 4E) despite the rearrangements observed for the Aβ peptide backbone. We also note that while the Aβ data presented here focuses on positive ions, most reports of IM-MS data for this peptide contain data for negative ions in order to exploit its charge obtained in solution and to simplify data interpretation and MD simulations.55 While we are actively pursuing negative ion mode data for Aβ bound to neuropeptides, the data in this report focused on positive ions primarily to avoid the signal intensity and instrumental noise limitations associated with acquiring such data. We deemed ion signal intensity of primary importance in our studies due to the relatively weak Aβ:neuropeptide interactions we wished to probe.
Once we overcame the challenges associated with the collection and interpretation of IM-MS data on these complexes, we noted several advantages of the method for studying Aβ:neuropeptide complexes in comparison to other structural biology tools. The concentration range accessed by the IM-MS approach is comparatively low relative to other approaches that are capable of recording complex size and shape information.56-61 In addition, due to the heterogeneous nature of the Aβ:LE complexes interrogated, most spectroscopic probes would report structure and Kd values averaged over many co-existing complexes and assemblies. We found IM analysis to be especially important in the detection of dimer-related complexes, which may have gone completely overlooked if detected by MS alone. The plots shown in Figure 3 group these signals together to form a trend line easily distinguished from other signals associated with Aβ and LE.62 Trends observed for the CCS associated with the addition of LE to Aβ are especially informative (Table 2) and indicate that highly compact forms are favored for this complex in the context of both Aβ monomers and dimers. Interestingly, the compactness of the resulting complex recorded by IM-MS has no correlation with the resultant binding constant of that complex (Kd, Table 1), perhaps providing additional evidence of local structural rearrangements in these Aβ:LE assemblies upon introduction to the gas phase. Finally, the ability to evaluate individually the structure and stability of Aβ:LE complexes is a feat that few other techniques can accomplish, and as such the values reported here, while in broad agreement with other studies of Aβ stability29 form a unique resource on Aβ:neuropeptide interactions.
Table 2.
CCS values for LE:Aβ1-40 Complexes Measured by IM-MS
| Complex | m/z | CCS (Å2) | σ * |
|---|---|---|---|
| LE+ | 557.0 | 165 | 0.22 |
| Aβ1-404+ | 1083.5 | 622 | 13.2 |
| Aβ1-404+ | 1083.5 | 678 | 0.29 |
| Aβ1-403+ | 1444.3 | 594 | 0.38 |
| [Aβ1-40 + LE]3+ | 1629.6 | 638 | 0.84 |
| [Aβ1-40 + LE2]3+ | 1815.0 | 683 | 0.43 |
| [Aβ1-40 + LE3]3+ | 2000.3 | 731 | 0.18 |
| Aβ1-40 Dimer5+ | 1733.0 | 946 | 12.7 |
| [Aβ1-40 Dimer + LE]5+ | 1844.2 | 982 | 0.21 |
| [Aβ1-40 Dimer + LE2]5+ | 1955.4 | 1032 | 1.54 |
| [Aβ1-40 Dimer + LE3]5+ | 2066.6 | 1060 | 22.6 |
Standard deviations from measurements in triplicate. For real errors, including those from IM calibration, an additional ~3% must be added to these values.
The ability of endogenous or exogenous molecules to modulate Aβ aggregate formation has been of interest, primarily in an effort to redirect aggregation from producing toxic intermediates. Altering the thermodynamic and/or kinetic parameters of this process using molecules like LE could offer insight into this problem; however, traditional methods to assess the degree of fibril formation, such as the fluorescence-based assay using the amyloid-specific dye thioflavin-T (ThT), do not always adequately represent these changes.29, 42, 63, 64 In the case of LE, its inherent interactions with both ThT and Aβ aggregates would have interfered with data interpretation (data not shown). Thus, gel electrophoresis/Western blot studies in conjunction with TEM could offer a more complete picture of the ability of LE to influence Aβ1-40 aggregation. Upon incubation of Aβ with LE, there are slightly different distributions of peptide sizes detected by gel electrophoresis/Western blot, compared to that from the LE-untreated Aβ sample, indicating that LE could not completely block or alter the fibril formation trajectory. To complement these results, TEM images show differences in aggregate morphology that occur during aggregation when LE is present, particularly on larger aggregates. While the aggregate populations could not be completely characterized or quantified by TEM, primarily larger sized aggregates can be visualized. The TEM images display that samples containing excess LE (i.e., 3 or 5 equiv) indicate altered aggregates of large sizes (e.g., mainly shorter fibrils), which is consistent with the gel data that showed minimal changes to low MW populations. Thus, weak ligand binding may not preclude it from influencing downstream fibrillization provided that it favors interactions near the N-terminus of Aβ, similar to LE; however, adjustment of additional parameters in such compounds might be required to fully alter structural properties of Aβ aggregates at earlier stages.
The Aβ:LE binding region identified in Figure 4 possesses some features similar the Aβ binding region previously identified in our work with (–)-epigallocatechin-3-gallate (EGCG), a natural product found in green tea.29 Many previous reports have identified EGCG as a potent inhibitor of Aβ fibril formation and toxicity.65, 66 Our very recent work has further indicated that the main role of EGCG in inhibiting Aβ fibril formation likely relies on critical interactions with bound metal ions, such as Cu(II) and Zn(II).29 Like LE, we found that EGCG binds preferentially to the cleft between the N-terminus and central helix regions of metal-free Aβ1-40.29 EGCG and LE bind to this region of Aβ1-40 with similar affinities (Kd relative to Aβ monomer for both LE and EGCG is ~60 μM) in the absence of metal ions; thus, both produce relatively similar downstream effects in Aβ fibril formation once bound (Fig. 5). EGCG has been found to recover neuronal activity in cell culture, thus making the natural product a key target in AD drug design efforts. 29, 65 While the data presented in this report is too preliminary to make similar claims related to the potential of LE as a general scaffold or optimization target for future AD therapies, it is clear from the data presented here that the nature of the binder, as well as the binding site accessed, both play significant roles in tuning the ability of small molecules to modulate Aβ fibril formation and ameliorate disease phenotypes.
Conclusions
Here, we describe studies employing IM-MS, MD simulations, gel electrophoresis/Western blot, and TEM which determine both the presence and structure of Aβ:LE complexes, as well as their influence on Aβ oligomerization and fibril formation. In addition to LE, our IM-MS data on a limited panel of neuropeptides detected Aβ:galanin interactions, but no complexes were detected between Aβ and somatostain, neurotensin, or substance P. The latter result is especially informative, as it indicates that previously identified neurotrophic effects for substance P relative to Aβ are likely the result of the neuropeptide interacting with larger Aβ oligomers or due to its agonist activity relative to neuronal receptor sites. Aβ:LE complexes ranging in stoichiometry from 1:1 to 2:3 are detected in our dataset, and CCS values indicate that the complexes favor a compact configuration. A detailed IM-MS analysis of Aβ:LE complexes indicates that the small neuropeptide is likely bound in a cleft between the Aβ N-terminus and its hydrophobic core region in a manner similar to EGCG.29 The gel/Western blot data suggest minimal change in the size distribution of the Aβ species incubated with LE, but TEM data for Aβ samples doped with LE display mainly shorter fibrils in lieu of potentially non-toxic amorphous aggregates that were observed in previous work using metal-containing Aβ samples doped with EGCG.29 Overall, the dataset presented here generates intriguing correlations between binding affinity, binding site, and resultant fibril morphology that will likely aid in the pursuit of both small molecules and biotherapeutics for AD. Future work in our group will continue to pursue neuropeptides, including the interaction with galanin we observed in these data, as a potential Aβ binders, fibril formation inhibitors, and neurotrophic agents.
Acknowledgments
The authors thank Russell Bornschein (UM) for his aid in developing the Kd relationships shown in this work. In addition, we thank Richard D. Smith (UM) and Jordan J. Clark (UM) for their aid in the MD simulations presented here. This work was supported by funding from the NIH GM-095832 (to B.T.R.), the Alfred P. Sloan Foundation and the Ruth K. Broad Biomedical Research Foundation (to M.H.L.). M.T.S is funded by an NIH Training Grant (T32 CA140044) and A.S.D. acknowledges the National Science Foundation for a Graduate Research Fellowship.
REFERENCES
- 1.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Alzheimers Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer's Association Alzehimers Dement. 2012;8:131–168. doi: 10.1016/j.jalz.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe DJ. Phys. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien RJ, Wong PC. Annu. Rev. Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper JD, Lansbury PT., Jr Annu. Rev. Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 6.Kirkitadze MD, Bitan G, Teplow DB. J. Neuro. Sci. Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 7.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 8.Necula M, Kayed R, Milton S, Glabe CG. J. Biol. Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 9.Ono K, Condron MM, Teplow DB. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, Bowers MT. Nature Chemistry. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitan G, Fradinger EA, Spring SM, Teplow DB. Amyloid. 2005;12:88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- 12.Yamin G, Ruchala P, Teplow DB. Biochemistry. 2009;48:11329–11331. doi: 10.1021/bi901325g. [DOI] [PubMed] [Google Scholar]
- 13.Murray MM, Bernstein SL, Nyugen V, Condron MM, Teplow DB, Bowers MT. J. Am. Chem. Soc. 2009;131:6316–6317. doi: 10.1021/ja8092604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gessel MM, Wu C, Li H, Bitan G, Shea J-E, Bowers MT. Biochemistry. 2012;51:108–117. doi: 10.1021/bi201520b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bohrmann B, Tjernberg L, Kuner P, Poli S, Leve-Trafit B, Näslund J, Richards G, Huber W, Döbeli H, Nordstedt C. J. Biol. Chem. 1999;274:15990–15995. doi: 10.1074/jbc.274.23.15990. [DOI] [PubMed] [Google Scholar]
- 16.Janusz M, Woszczyna M, Lisowski M, Kubis A, Macala J, Gotszalk T, Lisowski J. FEBS Lett. 2009;583:190–196. doi: 10.1016/j.febslet.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 17.Orner BP, Liu L, Murphy RM, Kiessling LL. J. Am. Chem. Soc. 2006;128:11882–11889. doi: 10.1021/ja0619861. [DOI] [PubMed] [Google Scholar]
- 18.Boado RJ. Drug News Perspect. 2008;21:489–503. doi: 10.1358/dnp.2008.21.9.1290820. [DOI] [PubMed] [Google Scholar]
- 19.Egleton RD, Davis TP. Peptides. 1997;18:1431–1439. doi: 10.1016/s0196-9781(97)00242-8. [DOI] [PubMed] [Google Scholar]
- 20.Bharadwaj P, Head R, Martins R, Raussens V, Sarroukh R, Jegasothy H, Waddington L, Bennett L. Food Funct. 2013;4:92–103. doi: 10.1039/c2fo30111c. [DOI] [PubMed] [Google Scholar]
- 21.Yanker BA, Duffy LK, Kirschner DA. Science. 1990;240:279. doi: 10.1126/science.2218531. [DOI] [PubMed] [Google Scholar]
- 22.Baumketner A, Bernstein SL, Wyttenbach T, Bitan G, Teplow DB, Bowers MT, Shea J-E. Protein Sci. 2006;15:420–428. doi: 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith DP, Radford SE, Ashcroft AE. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6794–6798. doi: 10.1073/pnas.0913046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemmer DE, Jarrold MF. J. Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 25.Bohrer BC, Mererbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Annu. Rev. Anal. Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 27.Ruotolo BT, Hyung S-J, Robinson PM, Giles K, Bateman RH, Robinson CV. Angew. Chem. Int. Ed. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 28.Borysik AJH, Read P, Little DR, Bateman RH, Radford SE, Ashcroft AE. Rapid Commun. Mass Spectrom. 2004;18:2229–2234. doi: 10.1002/rcm.1613. [DOI] [PubMed] [Google Scholar]
- 29.Hyung S-J, DeToma AS, Brender JR, Lee S, Vivekanadan S, Kochi A, Choi J-S, Rammamoorthy A, Ruotolo BT, Lim MH. Proc. Natl. Acad. Sci. U. S. A. 2013 doi: 10.1073/pnas.1220326110. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duret L, Gasteiger E, Perriere G. CABIOS. 1996;12:507–510. doi: 10.1093/bioinformatics/12.6.507. [DOI] [PubMed] [Google Scholar]
- 31.Hernández H, Robinson CV. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 32.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal. Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 33.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. Biochem. Biophys. Res. Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schüttelkoph AW, van Aalten DMF. Acta Crystallogr. D. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 36.Lindahl E, Hess B, Spoel D. v. d. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 37.van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG. GROMOS96 manual and users guide. 1996 [Google Scholar]
- 38.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 39.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J. Phys. Chem. 1996;100:16082–16086. [Google Scholar]
- 40.Shvartsburg AA, Jarrold MF. Chem. Phys. Lett. 1996;261:86–91. [Google Scholar]
- 41.Wang W, Kitova EN, Klassen JS. Anal. Chem. 2003;75:4945–4955. doi: 10.1021/ac034300l. [DOI] [PubMed] [Google Scholar]
- 42.Mancino AM, Hindo SS, Kochi A, Lim MH. Inorg. Chem. 2009;48:9596–9598. doi: 10.1021/ic9014256. [DOI] [PubMed] [Google Scholar]
- 43.Hindo SS, Mancino AM, Braymer JJ, Liu Y, Vivekanandan S, Ramamoorthy A, Lim MH. J. Am. Chem. Soc. 2009;131:16663–16665. doi: 10.1021/ja907045h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi J-S, Braymer JJ, Nanga RPR, Ramamoorthy A, Lim MH. Proc. Natl. Acad. Sci. U. S. A. 2012;107:21990–21995. doi: 10.1073/pnas.1006091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi J-S, Braymer JJ, Park SK, Mustafa S, Chae J, Lim MH. Metallomics. 2011;3:284–291. doi: 10.1039/c0mt00077a. [DOI] [PubMed] [Google Scholar]
- 46.DeToma AS, Choi J-S, Braymer JJ, Lim MH. ChemBioChem. 2011;12:1198–1201. doi: 10.1002/cbic.201000790. [DOI] [PubMed] [Google Scholar]
- 47.Braymer JJ, Choi J-S, DeToma AS, Wang C, Nam K, Kamph JW, Ramamoorthy A, Lim MH. Inorg. Chem. 2011;50:10724–10734. doi: 10.1021/ic2012205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He X, Park HM, Hyung S-J, DeToma AS, Kim C, Ruotolo BT, Lim MH. Dalton Trans. 2012;41:6558–6566. doi: 10.1039/c2dt12207c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pithadia AS, Kochi A, Soper MT, Beck MW, Liu Y, Lee S, DeToma AS, Ruotolo BT, Lim MH. Inorg. Chem. 2012;51:12959–12967. doi: 10.1021/ic302084g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kowall NW, Beal MF, Busciglio J, Duffy LK, Yanker BA. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7247–7251. doi: 10.1073/pnas.88.16.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Counts SE, Perez SE, Mufson EJ. Cell. Mol. Life Sci. 2008;65:1842–1853. doi: 10.1007/s00018-008-8159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swaab DF. Prog. Brain Res. 1982;55:97–122. doi: 10.1016/S0079-6123(08)64192-8. [DOI] [PubMed] [Google Scholar]
- 53.Kitova EN, El-Hawiet A, Schnier PD, Klassen JS. J. Am. Soc. Mass Spectrom. 2012;23:431–441. doi: 10.1007/s13361-011-0311-9. [DOI] [PubMed] [Google Scholar]
- 54.Zhong YY, Hyung SJ, Ruotolo BT. Expert Rev. Proteomics. 2012;9:47–58. doi: 10.1586/epr.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teplow DB, Lazo ND, Bitan G, Bernstein S, Wyttenbach T, Bowers MT, Baumketner A, Shea JE, Urbanc B, Cruz L, Borreguero J, Stanley HE. Acc. Chem. Res. 2006;39:635–645. doi: 10.1021/ar050063s. [DOI] [PubMed] [Google Scholar]
- 56.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 57.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Int. J. Mass. Spectrom. 2005;240:301–315. [Google Scholar]
- 58.Mertens HDT, Svergun DI. J. Struct. Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 59.Irvine GB. Anal. Chim. Acta. 1997;352:387–397. [Google Scholar]
- 60.Lebowitz J, Lewis MS, Schuck P. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonvin AM, Boelens R, Kaptein R. Curr. Opin. Chem. Biol. 2005;9:501–508. doi: 10.1016/j.cbpa.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 62.Ruotolo BT, Gillig KJ, Stone EG, Russell DH. J. Chromatogr. B. 2002;782:385–392. doi: 10.1016/s1570-0232(02)00566-4. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki Y, Brender JR, Hartman K, Ramamoorthy A, Marsh ENG. Biochemistry. 2012;51:8154. doi: 10.1021/bi3012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hudson SA, Ecroyd H, Kee TW, Carver JA. FEBS J. 2009;276:5960–5972. doi: 10.1111/j.1742-4658.2009.07307.x. [DOI] [PubMed] [Google Scholar]
- 65.Choi YT, Jung CH, Lee SR, Bae JH, Baek WK, Suh MH, Park J, Park CW, Suh SI. Life Sci. 2001;70:603–614. doi: 10.1016/s0024-3205(01)01438-2. [DOI] [PubMed] [Google Scholar]
- 66.Bieschke J, Russ J, Friedrich RP, Ehrnhoefer DE, Wobst H, Neugebauer K, Wanker EE. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7710–7715. doi: 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]