Abstract
In a field where structure has finally begun to have a real impact a series of new structures over the last two years have further extended our understanding of some of the critical regulatory events. Notably information has begun to flow from larger assemblies of components which allow insight into the often transient assemblies critical to complement regulation at the cell surface. This review will summarise the key structures determined since the last International Complement Workshop and the insights these have given us, before highlighting some questions that still require molecular frameworks to drive understanding.
Keywords: Complement, structure, regulation, protein complexes
Introduction
The last International Complement Workshop in 2010 in New York felt, to many of us there, to be the moment when the role of structure in defining and guiding the analyses of mechanisms came of age for complement. Thanks to the work of many groups over the years (reviewed in (Arlaud et al., 2007; Gal et al., 2009; Garlatti et al., 2010; Gros et al., 2008; Janssen and Gros, 2006; Janssen and Gros, 2007; Schmidt et al., 2008; Serruto et al., 2010; Springer, 2006)) the meeting saw a field where atomic level structures existed for the vast majority of molecules and where structural ideas were informing much of the biological discussion. In particular our understanding of the key activation steps associated with transition of C3 to C3b and regulation of this by regulators such as FH were being defined, as well as first insights into MAC assembly arising from structural studies of C8 and C9. Pathogen modes of escape from complement and molecular explanations for associations between polymorphisms and disease states were also being driven by structural insight. In the two years since, our picture has again leapt forwards with many new structures giving insight into these processes (Table 1 summarises key atomic structures determined in this time). This review cannot hope to discuss all of the structural studies published in the last two years and we apologise to anyone whose favourite structure is not discussed! But we will highlight structures which, to these authors’ minds, have answered or raised the most questions for the field as a whole in two broad areas; regulation at the level of C3 and assembly/regulation of the terminal pathway.
Table 1. Key Structures Published Since ICW 2010.
| Protein(s) | Accession code |
Method(s)/Resolution | Reference |
|---|---|---|---|
| Regulation at the level of C3 | |||
| C3b in complex with factors B and with Factor B & catalytically dead D |
2XWJ 2XWB |
X-ray 4.0 Å & 3.5 Å | (Forneris et al., 2010) |
| Factor I | 2XRC | X-ray 2.7 Å | (Roversi et al., 2011) |
| C3d in complex with fH19-20 | 3OXU 2XQW |
X-ray 2.1 Å & 2.3 Å | (Kajander et al., 2011; Morgan et al., 2011) |
| Extracellular portion of CD46 in complex with Adenovirus 11 fiber knob |
3O8E | X-ray 2.9 Å | (Persson et al., 2010) |
| SCIN in complex with C3b and C3c | 3OHX 3L5N 3L3O |
X-ray 7.5 Å - 3.4 Å | (Garcia et al., 2010) |
| iC3b | EMD-1908 | EM 24 Å | (Alcorlo et al., 2011) |
| Factor H domains 18-20 | 3SW0 | X-ray 1.8 Å | (Morgan et al., 2012) |
| Terminal Pathway | |||
| C6 | 3T5O | X-ray 2.9 Å | (Aleshin et al., 2012b) |
| C5b6 | 4E0S 4A5W |
X-ray 4.2 Å & 3.5 Å | (Aleshin et al., 2012a; Hadders et al., 2012) |
| sC5b9 | EMD-1991 | EM 24 Å | (Hadders et al., 2012) |
| C8 | EMD-1805 3OJY |
EM 25 Å & X-ray 2.5 Å | (Bubeck et al., 2011; Lovelace et al., 2011) |
| Other | |||
| Integrin interactions with iC3b | - | EM 20 Å | (Chen et al., 2012) |
Complement Regulation at the level of C3
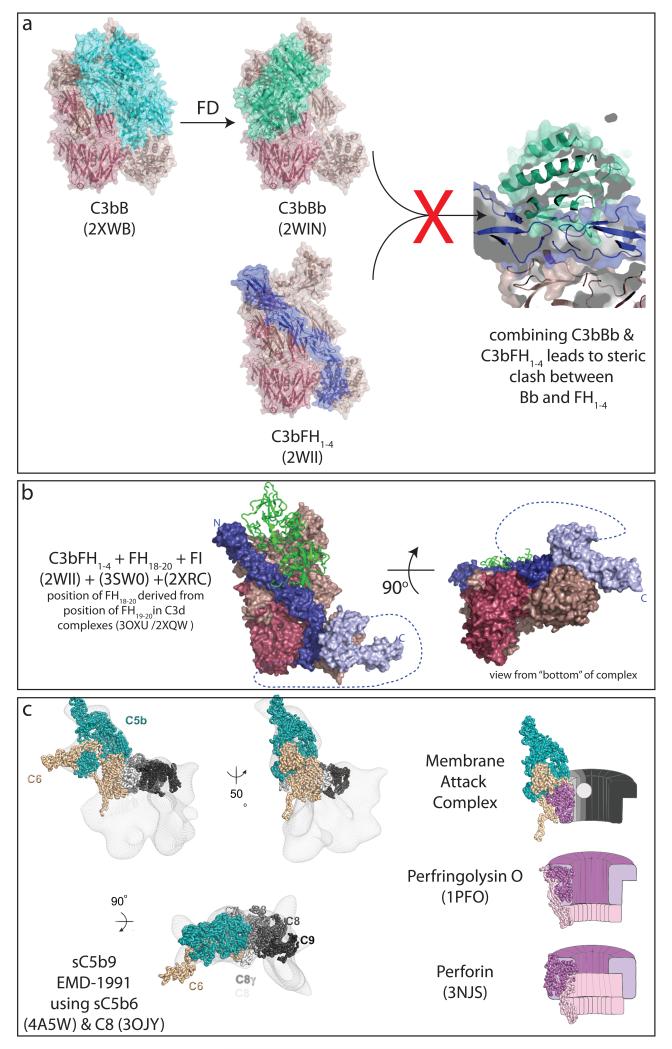
Much structural information in the last ten years has given insight into the molecular processes regulating complement at the level of C3 with structures of C3, C3a, C3d, C3b and C3b in complex with the regulatory region of FH being just some of the background to work in the last two years (Huber et al., 1980; Janssen et al., 2006; Janssen et al., 2005; Milder et al., 2007; Nagar et al., 1998; Torreira et al., 2009b; Wu et al., 2009). The most recent structures have built on the earlier work to give further insight the processes driving formation of the active convertase, C3bBb from C3b (Forneris et al., 2010) and into processes acting to limit activation by opposing this step either by cleavage of C3b by factor I (FI) (co-factor mediated regulation) or by dissociation of the convertase (so called decay acceleration) (Kajander et al., 2011; Morgan et al., 2012; Morgan et al., 2011; Roversi et al., 2011). The first structures giving insight into these processes were a pair of structures of C3b in complex with factor B (FB) with and without a catalytically inactive form of factor D (FD) (Forneris et al., 2010). These structures revealed that the pro-convertase (C3bB) contains FB in an “open” conformation compared either to earlier structures of FB in isolation (Milder et al., 2007) or FB in complex with cobra venom factor (Janssen et al., 2009; Krishnan et al., 2009). This opening exposes the FB scissile loop which is also seen to be mobile and disordered in the C3bB structure. Using a catalytically dead FD (active site Ser to Ala) allowed Forneris and co workers (Forneris et al., 2010) to visualise the C3bBD* complex and see that FD binds via its exosite to the VWA-SP linker of FB to induce further conformational change in FB. Binding of FD in the complex also leads to changes in FD to remodel the self-inhibitory loop previously seen occupying the substrate binding pocket (Jing et al., 1998; Narayana et al., 1994) generating the active catalytic triad ready for cleavage of FB. However, the changes at the active site did not seem to be driven by structural changes propagating from the exo-site which retains the same conformation in solution and in FD bound in the complex. The authors proposed that the paired structural rearrangements seen formed a “double-safety” catch with both co-factor dependent and substrate-induced proteolysis required to activate convertase. This new structure together with earlier models for the convertase (Rooijakkers et al., 2009; Torreira et al., 2009a) and for the regulatory domains of FH bound to C3b (Wu et al., 2009) reinforce the hypothesis that that regulators bound as seen in the fH/C3b complex cannot be bound to the C3bBb complex, but other than by the implication that steric clashes must somehow drive the process it still remains unclear precisely how decay acceleration occurs (Figure 1a).
Figure 1.
(a) The mechanism for decay acceleration of the C3bBb remains unclear despite the large structural advances since simple overlay of structures for C3bBb (Rooijakkers et al., 2009) and C3bFH1-4 (Wu et al., 2009) reveals the two structures are incompatible (due to clashes between FH1-4 and Bb) without revealing what conformational changes are required to promote dissociation of Bb from the C3bBb complex. (b) Recent structures allow construction of pseudo-atomic models for the complex of C3b/FH1-4/FH18-20/FI (Kajander et al., 2011; Morgan et al., 2012; Morgan et al., 2011; Roversi et al., 2011; Wu et al., 2009) showing the context in which the first cleavage event of co-factor regulation occurs. (c) Electron microscopy volumes for sC5b9 and atomic structures for C5b6 (Hadders et al., 2012), and C8 (Lovelace et al., 2011) suggest that the membrane attack complex pore assembles MACPF domains in an analogous fashion to bacterial CDC pores (e.g. perfringolysin O (Gilbert et al., 1998; Rossjohn et al., 1997)) rather than as proposed for perforin pores (Law et al., 2010). All pictures drawn using PyMol (www.pymol.org).
FH is amongst the most complex of the complement regulators to understand structurally being made up of a string of twenty CCP domains linked by variably flexible linkers. The conformational heterogeneity this induces, even in a pure sample of intact FH, makes it a challenging target for high resolution structural methods whilst the small size of the domains (~8kDa each) makes it a challenging target for visualisation using electron microscopy. Despite these challenges our structural understanding of FH has been growing from early studies of isolated domains and domain pairs (Barlow et al., 1992; Barlow et al., 1993; Norman et al., 1991) to more recent studies of portions of FH in complex with C3b (Wu et al., 2009) or mimics of cell-surface glycosaminoglycans (Prosser et al., 2007) (Blaum et al., 2010). Since the last meeting atomic structures of the C-terminal pair of domains (FH19-20) in complex with C3d (Kajander et al., 2011; Morgan et al., 2011) and a structure for the final three C-terminal domains (FH18-20) (Morgan et al., 2012) in combination with FRET and SAXS studies of larger fragments and intact FH mean that we have a much fuller picture of the way in which the complex network of interactions between the different portions of FH, C3b and cell-surface GAGs combine to specifically localise and orient FH on C3b for regulation (Blaum et al., 2010; Kajander et al., 2011; Morgan et al., 2011; Prosser et al., 2007; Wu et al., 2009). Particularly fascinating is the way in which both interactions with C3b are now seen to localise to the same face of the molecule meaning that the N- and C-termini of FH come much closer together than was previously thought (Figure 1b).
In the light of our increasing knowledge of the C3b/FH complex the only missing piece of structural information in our understanding of co-factor mediated regulation has been knowledge of the highly specific serine protease, factor I (FI). This too has been provided in the last year (Roversi et al., 2011) with the X-ray structure of FI purified from human serum being solved. This structure has revealed that FI circulates in serum with a highly disordered active site providing one mechanism by which specificity is achieved. Mapping of mutants onto the structure suggested that packing of the heavy chain against the base of the serine protease (light chain) acts allosterically to maintain inactivity solution. The structure of FI can be readily docked to the earlier structure of the complex of C3b and the regulatory N-terminus of FH (Wu et al., 2009) and this suggests that interactions between the heavy chain and the regulator are key to localising FI and presumably also key to releasing the heavy chain/light chain interactions that disorder the active site. This docking demonstrates that FI can pack against C3b and the co-factor with the active site of FI proximal to the first cleavage site in C3b implying that much of the specificity of FI is driven by both the co-factor driven release of heavy-chain allosteric inhibition and co-factor/C3b guided positioning of FI for cleavage. There remain however unanswered questions about how much rearrangement of FI is required to release allosteric inhibition and how the latter cleavages of C3b occur? Are these perhaps driven by rearrangement of pieces of C3b against a fixed co-factor/FI complex?
Assembly and Regulation of the Terminal Pathway of Complement
As of the last ICW meeting our knowledge of the structures involved in the terminal pathway was much more limited than of those involved earlier in the complement pathways. We had structural information about C5 (Fredslund et al., 2008), structures for portions of C8 (Hadders et al., 2007; Lovelace et al., 2008; Ortlund et al., 2002; Slade et al., 2008) (which also allowed construction of models for the MACPF domain of C9 (Hadders et al., 2007)) and structures of the FIM domains of C7 (Phelan et al., 2009). Taken together though these structures did not allow much insight into the process of assembly and regulation of the MAC. The last two years have seen many structures adding to our knowledge with information about intact C8 (both from EM (Bubeck et al., 2011) and then X-ray studies (Lovelace et al., 2011)), X-ray structures for C6 (Aleshin et al., 2012b) and the C5b6 complex (Aleshin et al., 2012a; Hadders et al., 2012) and EM reconstruction of the soluble, sC5b9 complex (Hadders et al., 2012). Taken together these structures now give us a framework on which to frame our questions about the terminal pathway. The structures of C5b6 were particularly exciting in that they show C6 adopting a highly extended conformation draped around C5b and trapping C5b in a conformation with the TED domain half-way between its location in C5 and the position it adopts in C3b. This observation seems to explain why C5b can only promote assembly of further components of the terminal pathway for a brief period of time following cleavage of C5 – presumably any C5b that does not encounter C6 during this time converts to a more C3b-like conformation and is then incompetent for further terminal pathway assembly. Once assembled to C5b6 the EM studies of the soluble sC5b9 complex suggest that further components dock sequentially onto this C5b6 platform without requiring further conformational changes. The MACPF domains of C9 appear to associate in a similar fashion to bacterial CDC pores, in contrast to the mode of assembly previously suggested for immune pores based on studies of perforin (Law et al., 2010) (Figure 1c). The EM studies also reveal that the soluble form of the complex is regulated by assembly of a large volume of protein (presumably protein S and vitronectin) below the arc made up of C6-C7-C8b-C8a-C9, presumably masking any exposed lipophilic portions of the MAC precursors and capping the ends of the arc preventing further assembly of C9. Further detail of the processes involved in assembly and regulation, particularly by CD59, will await studies of MAC inserted into membranes and higher resolution information about these complexes but by establishing the order of assembly and highlighting differences to the proposed mode of perforin assembly these structures have significantly advanced our understanding.
Concluding Remarks
Many other structural studies have also given insights into complement biology in the last two years including defining both the structural rearrangements that occur following FI cleavage to generate iC3b (Alcorlo et al., 2011) and then the domains involved in recognition of iC3b (Chen et al., 2012) by the integrin αXβ2 to promote recognition and phagocytosis of pathogens. However, despite this wealth of structural information, there are many questions for the complement-focussed structural biologist to try and answer. In particular the emerging structures of large complexes push us towards thinking about the architecture of the large complexes involved in regulation of complement such as the C1-complex/MBL/ficolins and C4bp. In some of these cases work is underway (e.g. (Dobo et al., 2009; Gaboriaud et al., 2011; Garlatti et al., 2010; Gout et al., 2011; Jenkins et al., 2006; Lang et al., 2010; Teillet et al., 2008) that is likely to give insight into the higher order assembly and perhaps its is this that will provide the structural focus at the meeting in 2014. Other questions remaining include generating a molecular understanding of how MAC pore formation is regulated by CD59, how the different convertases achieve specificity of activity and regulation and, particularly with the wealth of GWAS and other genetic studies, what are the molecular bases of the many genetic susceptibilities now associated with complement proteins? From the structural perspective – at least there seems to be plenty for us all to do!
Acknowledgements
Many thanks to all colleagues for conversations over many years that have informed opinions presented in this article and in particular to members of the Lea group. The authors apologise that limitations of space mean that invidious decisions have had to be made in terms of what structural work to discuss. We thank Doryen Bubeck for drawing Figure 1c. Work in the Lea group is supported by grants from the Wellcome Trust, The Medical Research Council UK and the Oxford Martin School.
Abbreviations
- MAC
membrane attack complex
- FH
factor H
- FI
factor I
- FD
factor D
References
- Alcorlo M, Martinez-Barricarte R, Fernandez FJ, Rodriguez-Gallego C, Round A, Vega MC, Harris CL, de Cordoba SR, Llorca O. Unique structure of iC3b resolved at a resolution of 24 A by 3D-electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13236–40. doi: 10.1073/pnas.1106746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleshin AE, Discipio RG, Stec B, Liddington RC. Crystal structure of c5b-6 suggests structural basis for priming assembly of the membrane attack complex. The Journal of biological chemistry. 2012a;287:19642–52. doi: 10.1074/jbc.M112.361121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleshin AE, Schraufstatter IU, Stec B, Bankston LA, Liddington RC, DiScipio RG. Structure of complement C6 suggests a mechanism for initiation and unidirectional, sequential assembly of membrane attack complex (MAC) The Journal of biological chemistry. 2012b;287:10210–22. doi: 10.1074/jbc.M111.327809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlaud GJ, Barlow PN, Gaboriaud C, Gros P, Narayana SV. Deciphering complement mechanisms: the contributions of structural biology. Molecular immunology. 2007;44:3809–22. doi: 10.1016/j.molimm.2007.06.147. [DOI] [PubMed] [Google Scholar]
- Barlow PN, Norman DG, Steinkasserer A, Horne TJ, Pearce J, Driscoll PC, Sim RB, Campbell ID. Solution structure of the fifth repeat of factor H: a second example of the complement control protein module. Biochemistry. 1992;31:3626–34. doi: 10.1021/bi00129a011. [DOI] [PubMed] [Google Scholar]
- Barlow PN, Steinkasserer A, Norman DG, Kieffer B, Wiles AP, Sim RB, Campbell ID. Solution structure of a pair of complement modules by nuclear magnetic resonance. Journal of molecular biology. 1993;232:268–84. doi: 10.1006/jmbi.1993.1381. [DOI] [PubMed] [Google Scholar]
- Blaum BS, Deakin JA, Johansson CM, Herbert AP, Barlow PN, Lyon M, Uhrin D. Lysine and arginine side chains in glycosaminoglycan-protein complexes investigated by NMR, cross-linking, and mass spectrometry: a case study of the factor H-heparin interaction. Journal of the American Chemical Society. 2010;132:6374–81. doi: 10.1021/ja1000517. [DOI] [PubMed] [Google Scholar]
- Bubeck D, Roversi P, Donev R, Morgan BP, Llorca O, Lea SM. Structure of human complement C8, a precursor to membrane attack. Journal of molecular biology. 2011;405:325–30. doi: 10.1016/j.jmb.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yu Y, Mi LZ, Walz T, Springer TA. Molecular basis for complement recognition by integrin alphaXbeta2. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4586–91. doi: 10.1073/pnas.1202051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobo J, Harmat V, Beinrohr L, Sebestyen E, Zavodszky P, Gal P. MASP-1, a promiscuous complement protease: structure of its catalytic region reveals the basis of its broad specificity. J Immunol. 2009;183:1207–14. doi: 10.4049/jimmunol.0901141. [DOI] [PubMed] [Google Scholar]
- Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–20. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredslund F, Laursen NS, Roversi P, Jenner L, Oliveira CL, Pedersen JS, Nunn MA, Lea SM, Discipio R, Sottrup-Jensen L, Andersen GR. Structure of and influence of a tick complement inhibitor on human complement component 5. Nature immunology. 2008;9:753–60. doi: 10.1038/ni.1625. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C, Frachet P, Thielens NM, Arlaud GJ. The human c1q globular domain: structure and recognition of non-immune self ligands. Frontiers in immunology. 2011;2:92. doi: 10.3389/fimmu.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal P, Dobo J, Zavodszky P, Sim RB. Early complement proteases: C1r, C1s and MASPs. A structural insight into activation and functions. Molecular immunology. 2009;46:2745–52. doi: 10.1016/j.molimm.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Garcia BL, Ramyar KX, Tzekou A, Ricklin D, McWhorter WJ, Lambris JD, Geisbrecht BV. Molecular basis for complement recognition and inhibition determined by crystallographic studies of the staphylococcal complement inhibitor (SCIN) bound to C3c and C3b. Journal of molecular biology. 2010;402:17–29. doi: 10.1016/j.jmb.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlatti V, Martin L, Lacroix M, Gout E, Arlaud GJ, Thielens NM, Gaboriaud C. Structural insights into the recognition properties of human ficolins. Journal of innate immunity. 2010;2:17–23. doi: 10.1159/000233475. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ, Rossjohn J, Parker MW, Tweten RK, Morgan PJ, Mitchell TJ, Errington N, Rowe AJ, Andrew PW, Byron O. Self-interaction of pneumolysin, the pore-forming protein toxin of Streptococcus pneumoniae. Journal of molecular biology. 1998;284:1223–37. doi: 10.1006/jmbi.1998.2258. [DOI] [PubMed] [Google Scholar]
- Gout E, Moriscot C, Doni A, Dumestre-Perard C, Lacroix M, Perard J, Schoehn G, Mantovani A, Arlaud GJ, Thielens NM. M-ficolin interacts with the long pentraxin PTX3: a novel case of cross-talk between soluble pattern-recognition molecules. J Immunol. 2011;186:5815–22. doi: 10.4049/jimmunol.1100180. [DOI] [PubMed] [Google Scholar]
- Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nature reviews. Immunology. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- Hadders MA, Beringer DX, Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–4. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- Hadders MA, Bubeck D, Roversi P, Hakobyan S, Forneris F, Morgan BP, Pangburn MK, Llorca O, Lea SM, Gros P. Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5b6 and sC5b9. Cell reports. 2012;1:1–8. doi: 10.1016/j.celrep.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Scholze H, Paques EP, Deisenhofer J. Crystal structure analysis and molecular model of human C3a anaphylatoxin. Hoppe-Seyler’s Zeitschrift fur physiologische Chemie. 1980;361:1389–99. doi: 10.1515/bchm2.1980.361.2.1389. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–6. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Gomes L, Koning RI, Svergun DI, Koster AJ, Fritzinger DC, Vogel CW, Gros P. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. The EMBO journal. 2009;28:2469–78. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJ, Gros P. Conformational complexity of complement component C3. Advances in experimental medicine and biology. 2006;586:291–312. doi: 10.1007/0-387-34134-X_20. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Gros P. Structural insights into the central complement component C3. Molecular immunology. 2007;44:3–10. doi: 10.1016/j.molimm.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–11. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- Jenkins HT, Mark L, Ball G, Persson J, Lindahl G, Uhrin D, Blom AM, Barlow PN. Human C4b-binding protein, structural basis for interaction with streptococcal M protein, a major bacterial virulence factor. The Journal of biological chemistry. 2006;281:3690–7. doi: 10.1074/jbc.M511563200. [DOI] [PubMed] [Google Scholar]
- Jing H, Babu YS, Moore D, Kilpatrick JM, Liu XY, Volanakis JE, Narayana SV. Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity. Journal of molecular biology. 1998;282:1061–81. doi: 10.1006/jmbi.1998.2089. [DOI] [PubMed] [Google Scholar]
- Kajander T, Lehtinen MJ, Hyvarinen S, Bhattacharjee A, Leung E, Isenman DE, Meri S, Goldman A, Jokiranta TS. Dual interaction of factor H with C3d and glycosaminoglycans in host-nonhost discrimination by complement. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2897–902. doi: 10.1073/pnas.1017087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Ponnuraj K, Xu Y, Macon K, Volanakis JE, Narayana SV. The crystal structure of cobra venom factor, a cofactor for C3- and C5-convertase CVFBb. Structure. 2009;17:611–9. doi: 10.1016/j.str.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A, Szilagyi K, Major B, Gal P, Zavodszky P, Perczel A. Intermodule cooperativity in the structure and dynamics of consecutive complement control modules in human C1r: structural biology. The FEBS journal. 2010;277:3986–98. doi: 10.1111/j.1742-4658.2010.07790.x. [DOI] [PubMed] [Google Scholar]
- Law RH, Lukoyanova N, Voskoboinik I, Caradoc-Davies TT, Baran K, Dunstone MA, D’Angelo ME, Orlova EV, Coulibaly F, Verschoor S, Browne KA, Ciccone A, Kuiper MJ, Bird PI, Trapani JA, Saibil HR, Whisstock JC. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468:447–51. doi: 10.1038/nature09518. [DOI] [PubMed] [Google Scholar]
- Lovelace LL, Chiswell B, Slade DJ, Sodetz JM, Lebioda L. Crystal structure of complement protein C8gamma in complex with a peptide containing the C8gamma binding site on C8alpha: implications for C8gamma ligand binding. Molecular immunology. 2008;45:750–6. doi: 10.1016/j.molimm.2007.06.359. [DOI] [PubMed] [Google Scholar]
- Lovelace LL, Cooper CL, Sodetz JM, Lebioda L. Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement. The Journal of biological chemistry. 2011;286:17585–92. doi: 10.1074/jbc.M111.219766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milder FJ, Gomes L, Schouten A, Janssen BJ, Huizinga EG, Romijn RA, Hemrika W, Roos A, Daha MR, Gros P. Factor B structure provides insights into activation of the central protease of the complement system. Nature structural & molecular biology. 2007;14:224–8. doi: 10.1038/nsmb1210. [DOI] [PubMed] [Google Scholar]
- Morgan HP, Mertens HD, Guariento M, Schmidt CQ, Soares DC, Svergun DI, Herbert AP, Barlow PN, Hannan JP. Structural analysis of the C-terminal region (modules 18-20) of complement regulator factor H (FH) PloS one. 2012;7:e32187. doi: 10.1371/journal.pone.0032187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HP, Schmidt CQ, Guariento M, Blaum BS, Gillespie D, Herbert AP, Kavanagh D, Mertens HD, Svergun DI, Johansson CM, Uhrin D, Barlow PN, Hannan JP. Structural basis for engagement by complement factor H of C3b on a self surface. Nature structural & molecular biology. 2011;18:463–70. doi: 10.1038/nsmb.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science. 1998;280:1277–81. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- Narayana SV, Carson M, el-Kabbani O, Kilpatrick JM, Moore D, Chen X, Bugg CE, Volanakis JE, DeLucas LJ. Structure of human factor D. A complement system protein at 2.0 A resolution. Journal of molecular biology. 1994;235:695–708. doi: 10.1006/jmbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- Norman DG, Barlow PN, Baron M, Day AJ, Sim RB, Campbell ID. Three-dimensional structure of a complement control protein module in solution. Journal of molecular biology. 1991;219:717–25. doi: 10.1016/0022-2836(91)90666-t. [DOI] [PubMed] [Google Scholar]
- Ortlund E, Parker CL, Schreck SF, Ginell S, Minor W, Sodetz JM, Lebioda L. Crystal structure of human complement protein C8gamma at 1. 2 A resolution reveals a lipocalin fold and a distinct ligand binding site. Biochemistry. 2002;41:7030–7. doi: 10.1021/bi025696i. [DOI] [PubMed] [Google Scholar]
- Persson BD, Schmitz NB, Santiago C, Zocher G, Larvie M, Scheu U, Casasnovas JM, Stehle T. Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS pathogens. 2010;6:e1001122. doi: 10.1371/journal.ppat.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan MM, Thai CT, Soares DC, Ogata RT, Barlow PN, Bramham J. Solution structure of factor I-like modules from complement C7 reveals a pair of follistatin domains in compact pseudosymmetric arrangement. The Journal of biological chemistry. 2009;284:19637–49. doi: 10.1074/jbc.M901993200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser BE, Johnson S, Roversi P, Herbert AP, Blaum BS, Tyrrell J, Jowitt TA, Clark SJ, Tarelli E, Uhrin D, Barlow PN, Sim RB, Day AJ, Lea SM. Structural basis for complement factor H linked age-related macular degeneration. The Journal of experimental medicine. 2007;204:2277–83. doi: 10.1084/jem.20071069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp JA, Gros P. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nature immunology. 2009;10:721–7. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–92. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- Roversi P, Johnson S, Caesar JJ, McLean F, Leath KJ, Tsiftsoglou SA, Morgan BP, Harris CL, Sim RB, Lea SM. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12839–44. doi: 10.1073/pnas.1102167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CQ, Herbert AP, Hocking HG, Uhrin D, Barlow PN. Translational mini-review series on complement factor H: structural and functional correlations for factor H. Clinical and experimental immunology. 2008;151:14–24. doi: 10.1111/j.1365-2249.2007.03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D, Rappuoli R, Scarselli M, Gros P, van Strijp JA. Molecular mechanisms of complement evasion: learning from staphylococci and meningococci. Nature reviews. Microbiology. 2010;8:393–9. doi: 10.1038/nrmicro2366. [DOI] [PubMed] [Google Scholar]
- Slade DJ, Lovelace LL, Chruszcz M, Minor W, Lebioda L, Sodetz JM. Crystal structure of the MACPF domain of human complement protein C8 alpha in complex with the C8 gamma subunit. Journal of molecular biology. 2008;379:331–42. doi: 10.1016/j.jmb.2008.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA. Complement and the multifaceted functions of VWA and integrin I domains. Structure. 2006;14:1611–6. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teillet F, Gaboriaud C, Lacroix M, Martin L, Arlaud GJ, Thielens NM. Crystal structure of the CUB1-EGF-CUB2 domain of human MASP-1/3 and identification of its interaction sites with mannan-binding lectin and ficolins. The Journal of biological chemistry. 2008;283:25715–24. doi: 10.1074/jbc.M803551200. [DOI] [PubMed] [Google Scholar]
- Torreira E, Tortajada A, Montes T, Rodriguez de Cordoba S, Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proceedings of the National Academy of Sciences of the United States of America. 2009a;106:882–7. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreira E, Tortajada A, Montes T, Rodriguez de Cordoba S, Llorca O. Coexistence of closed and open conformations of complement factor B in the alternative pathway C3bB(Mg2+) proconvertase. J Immunol. 2009b;183:7347–51. doi: 10.4049/jimmunol.0902310. [DOI] [PubMed] [Google Scholar]
- Wu J, Wu YQ, Ricklin D, Janssen BJ, Lambris JD, Gros P. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nature immunology. 2009;10:728–33. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]