Abstract
Regulated trafficking of neurotransmitter receptors is critical to normal neurodevelopment and neuronal signaling. Group I mGluRs (mGluR1/5 and their splice variants) are G protein-coupled receptors enriched at excitatory synapses, where they serve to modulate glutamatergic transmission. The mGluR1 splice variants mGluR1a and mGluR1b are broadly expressed in the central nervous system and differ in their signaling and trafficking properties. Several proteins have been identified that selectively interact with mGluR1a and participate in receptor trafficking but no proteins interacting with mGluR1b have thus far been reported. We have used a proteomic strategy to isolate and identify proteins that co-purify with mGluR1b in Madin-Darby Canine Kidney (MDCK) cells, an established model system for trafficking studies. Here, we report the identification of 10 novel candidate mGluR1b-interacting proteins. Several of the identified proteins are structural components of the cell cytoskeleton, while others serve as cytoskeleton-associated adaptors and motors or endoplasmic reticulum-associated chaperones. Findings from this work will help unravel the complex cellular mechanisms underlying mGluR trafficking under physiological and pathological conditions.
Keywords: G protein-coupled receptors, interacting proteins, metabotropic glutamate receptors, mGluR1b, proteomics, trafficking
Trafficking of neurotransmitter receptors to synaptic sites in dendrites and axons is critical for the establishment of neuronal connections during development and for neuronal signaling. Dysregulated trafficking of membrane receptors, whether because of aberrant localization or failed insertion/removal in the plasma membrane, may lead to abnormal signaling with pathological consequences (Conn et al. 2007).
Metabotropic glutamate receptors (mGluRs) are G protein-coupled receptors enriched at excitatory synapses, where they serve to regulate glutamatergic neurotransmission. Group I mGluRs (mGluR1/5) activate the phospholipase C/inositol trisphosphate and the ERK-MAPK pathways and regulate the opening of ion channels (Hermans and Challiss 2001; Kim et al. 2003). mGluR1/5 control neurotransmission both post-synaptically, by regulating neuronal excitation, and pre-synaptically, by regulating neurotransmitter release (Conn and Pin 1997). Signaling by mGluR1/5 is implicated in cortical development, activity-dependent synaptic plasticity and neuropsychiatric disorders including schizophrenia, Parkinson’s disease, addiction and mental retardation. Importantly, abnormal mGluR1/5 signaling appears to underlie some of the cognitive deficits that characterize fragile X syndrome, the most common inherited cause of mental retardation (Dolen et al. 2007).
The mGluR1 receptor exists in at least four alternatively spliced variants (mGluR1a–d), which differ in the composition and length of their intracellular tails (Conn and Pin 1997). mGluR1a and mGluR1b are broadly expressed in the central nervous system and show differences in their regional distribution (Ferraguti et al. 1998; Mateos et al. 1998, 2000). Within neurons, the receptor variants target to different subcellular compartments. In the brain, mGluR1a is broadly distributed throughout dendritic arborization whereas mGluR1b is mostly restricted to soma and proximal dendritic regions (Ferraguti et al. 1998; Mateos et al. 2000); moreover, in spinal cord neurons mGluR1b is also found in the axonal hillock and fibers (Alvarez et al. 2000). The differential localization of mGluR1a/b within neurons suggests that they may play a different role in the regulation of neurotransmission. At present, little is known about the molecular mechanisms underlying transport of neurotransmitter receptors to different neuronal compartments.
Epithelial MDCK cells are an established model system for the study of protein trafficking. In these cells, membrane proteins can be selectively targeted to basolateral versus apical compartments by mechanisms that are similar to those employed by neurons to regulate trafficking to dendrites versus axons, respectively (Horton and Ehlers 2003). In MDCK cells, mGluR1a localizes to basolateral membranes whereas mGluR1b is targeted to the apical domain; targeting signals identified in the intracellular tails of the receptors are critical for their differential trafficking (Francesconi and Duvoisin 2002). Several proteins have been identified that interact with mGluR1a, but not mGluR1b, and are involved in receptor trafficking. Homer proteins, which bind to a motif present in the carboxy-terminal tail of mGluR1a and mGluR5 (Tu et al. 1998; Xiao et al. 1998), regulate receptor exit from the endoplasmic reticulum (ER; Roche et al. 1999), clustering (Das and Banker 2006) as well as lateral movement (Serge et al. 2002) in the neuronal membrane. Tamalin, a scaffold protein that binds to the distal portion of the tail of mGluR1a is also involved in intracellular trafficking of mGluR1a (Kitano et al. 2002). However, little is known about mGluR1b interacting proteins that might participate in its intracellular trafficking and/or receptor signaling.
Here, we describe the use of a proteomic strategy to identify proteins interacting with mGluR1b. Using this strategy, we have isolated and identified 10 proteins that co-purify with mGluR1b in MDCK cells. We further show that several of the identified proteins associate with native mGluR1b in the brain, where they may play an important role in regulating synaptic trafficking of the receptor under physiological and pathological conditions.
Materials and methods
Recombinant constructs
A modified form of the TAP tag (Rigaut et al. 1999) was generated by standard cloning techniques. A tandem repeat of the Z domain of protein A from Staphylococcus Aureus was derived by polymerase chain reaction (PCR) from the pEZZ18 vector. The Tobacco Etch Virus (TEV) recognition site was derived from the pProExHTa vector (Invitrogen, Carlsbad, CA, USA). A six-histidine tag was added in frame after the TEV site by PCR. The complete TAP tag was inserted into cDNA constructs encoding myc-tagged mGluR1a/b (Francesconi and Duvoisin 2002) after residue Leu-28 and upstream of the myc tag. The TAP-tagged receptors were subcloned in the pUHD10.3 vector (Gossen and Bujard 1992).
Cell culture and transfection
MDCK (T23 parental clone) and HEK293 cells were cultured as previously described (Francesconi and Duvoisin 1998, 2002). HEK293 cells were transiently transfected with Lipofectamine 2000 as recommended by the manufacturer. T23 cells, expressing a tetracycline-repressible transactivator (encoded by the vector pUHD15.1), were co-transfected with tagged receptors and the pCB7 plasmid (encoding hygromycin resistance) as described (Francesconi and Duvoisin 2002). Selection and maintenance medium contained 200 μg/mL hygromycin and 20 ng/mL doxycycline. To induce receptor expression, cells were rinsed with fresh medium and incubated in the absence of doxycycline for 1–3 days. For protein purification, cells (8 × 109 total) were plated in 245 mm dishes and grown in serum-free UltraMDCK medium in the presence of doxycycline until fully polarized. After removing doxycycline, cells were maintained in culture for three more days.
Tandem affinity purification
Tandem affinity purification (TAP) is a method to isolate putative receptor-interacting proteins at different stages of intracellular trafficking and affords purification of near-pure macromolecular complexes under native conditions via sequential rounds of enrichment of the tagged receptor. All purification steps were carried out at 4°C unless otherwise indicated. Cells expressing tagged-mGluR1a/b were scraped from plates and centrifuged at 4000 g for 5 min. The cell pellets were washed twice with 100 mL phosphate-buffered saline and once with 100 mL hypotonic buffer (10 mM Tris–HCl, 1.5 mM MgCl2, pH 8.0). Pellets were suspended in 1 mL/g (wet weight) NP-40 buffer (Roche Applied Science, Indianapolis, IN, USA) (6 mM NaH2PO4, 4 mM Na2HPO4, 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40) with 0.1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mg/mL each antipain, aprotinin, leupeptin, pepstatin. Cells were ruptured with a Dounce homogenizer with three sets of 10 strokes and incubated for 1 h with gentle mixing. After centrifugation at 15 000 g for 30 min, the soluble extracts were incubated with 0.8 mL of IgG-sepharose beads (GE Healthcare, Piscataway, NJ, USA) for 2 h with constant mixing and transferred to columns; unbound material was collected by gravity flow. The packed beads were washed at 21°C with 30 mL of IPP150 buffer (50 mM Tris–HCl, 150 mM NaCl, 0.1% NP-40, pH 8.0) and 10 mL of TEV cleavage buffer (10 mM Tris–HCl, 150 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 0.1% NP-40, pH 8.0). The beads were incubated for 2 h at 30°C with 1 mL fresh TEV cleavage buffer and 350 Units of recombinant TEV protease (Invitrogen). The eluate was recovered by gravity flow and 1 mL of fresh TEV cleavage buffer added to the column; the combined fractions were dialyzed (molecular weight cut off 10 kDa) for 16 h against PB150 buffer (50 mM NaH2PO4, 150 mM NaCl, 0.05% Tween 20, pH 8.0). The purified dialyzed proteins were incubated for 1 h with 0.2 mL pre-equilibrated Ni-NTA resin (Qiagen, Valencia, CA, USA). The beads were transferred to a column and washed three times with 10 mL wash buffer (50 mM Na phosphate, 150 mM NaCl, 2 mM 2-mercaptoethanol, 10 mM imidazole, 0.05% NP-40, pH 6.0); bound proteins were eluted with 6 × 0.2 mL elution buffer (50 mM Na phosphate, 150 mM NaCl, 2 mM 2-mercaptoethanol, 250 mM imidazole, 0.05% NP-40, pH 6.0). Eluted fractions were pooled and precipitated with TCA. Protein pellets were suspended in sample buffer, heated at 60°C for 3 min and fractionated by SDS-PAGE on 4–20% gradient gels; thioglycollate (11 mg/mL) was added to the cathode running buffer. Proteins were visualized by staining with either Silver or Coomassie R250, according to standard procedures. Identification of proteins by mass spectrometry was carried out by the Laboratory for Macromolecular Analysis and Proteomics (Albert Einstein College of Medicine, Bronx, NY, USA). Briefly, Coomassie-stained gel slabs corresponding to individual or multiple visible bands were excised, destained, reduced and digested with trypsin for ≥ 16 h at 30°C. The digested products were analyzed by tandem mass spectrometry in combination with nano-LC separation. The acquired MS/MS spectra were used to search an annotated database using the Pro-ID algorithm. MS/MS spectra were obtained from samples derived from two independent purifications; each sample was analyzed independently two times. Only sequence matching peptides with a cross-correlation (XCorr) value > 2, a delta correlation (ΔCn) value > 0.1 and ion percentage > 30 are presented and were used to search an annotated database.
Pull-down assays
A full-length cDNA encoding human α-actinin-4 (Open Biosystems, Huntsville, AL, USA) was subcloned in pGEX4T-2 and transformed in BL21 (D3) bacterial cells (Stratagene, La Jolla, CA, USA). Protein expression was induced with 1 mM IPTG for 16 h at 20°C. Cells were recovered by centrifugation (4000 g) and suspended in cold phosphate buffer (PB) with 1 mM PMSF and 1 μg/mL each aprotinin and leupeptin. After centrifugation, the bacterial pellet was suspended in extraction buffer (50 mM Tri–HCl, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 8.5) and incubated on ice for 1 h with lysozyme (100 μg/mL) and bacterial protease inhibitors (Sigma-Aldrich, St. Louis, MO, USA). After adding sarcosyl (1%), the suspension was sonicated and incubated with 1% Triton X-100 for 30 min. The soluble extract was recovered by centrifugation at 10 000 g for 30 min and incubated with glutathione-agarose beads for 30 min at 21°C. The beads were washed with 10 mL extraction buffer containing 0.5% Triton X-100 and 1 mM DTT. Bound proteins were recovered by incubation for 10 min with 2 ml of 10 mM reduced glutathione. Eluted proteins were dialyzed overnight against PB and quantified. For pull-down assays, transfected HEK293 cells were suspended in binding buffer (20 mM HEPES, 100 mM NaCl, pH 7.4), sonicated and incubated with 1% NP-40 and 0.5% Triton X-100 for 30 min. After centrifugation, the soluble extract was diluted (1:1) with binding buffer and the protein content quantified. Three milligrams of protein were incubated overnight with 365 pmol each of GST or GST-Actn4 immobilized onto glutathione-agarose beads. The beads were washed three times with binding buffer containing 0.1% Tween-20, once with buffer without detergent and eluted with sample buffer.
Immunoprecipitation
HEK293 cells were suspended in lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 0.25% Na deoxycholate, 1% NP-40, pH 7.4) with 1 mM Na3VO4 and protease inhibitors (1 mM PMSF, 1 mg/mL each aprotinin, leupeptin and pepstatin) and incubated on ice for 15 min. Cell lysates were centrifuged at 20 800 g for 30 min and pre-cleared by incubation with Protein A-sepharose (GE Healthcare) for 60 min. Pre-cleared lysates (0.5–1 mg at 1 mg/mL) were incubated with primary antibodies for 16 h; immunoprecipitates with mouse monoclonal antibodies were incubated with 2.4 μg rabbit anti-mouse IgG for 1 h. The immunocomplexes were incubated with Protein A beads for 3–4 h with continuous mixing, washed three times with lysis buffer, once with PB and eluted with sample buffer. For immunoprecipitation of native mGluR1b, brain cortices or cerebella from adult Wistar rats (Charles River Laboratories, Wilmington, MA, USA) were homogenized in 10 mM Tris–HCl (pH 7.4), 5 mM EDTA, 320 mM sucrose and protease inhibitors. The homogenate was spun at 800 g for 10 min and the recovered supernatant (S1) centrifuged at 10 000 g for 15 min; the solubilized pellet (P2) or the S2 fraction was used for immunoprecipitation. The immunocomplex was captured with anti-rabbit IgG conjugated to agarose or Protein A beads and analyzed by immunoblot with primary antibodies and the TrueBlot detection system (eBioscience, San Diego, CA, USA) or horseradish peroxidase-conjugated secondary antibodies.
Immunofluorescence
HEK293 cells were plated on glass coverslips and MDCK cells on Transwells polycarbonate filters. Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline supplemented with 1 mM MgCl2, 0.1 mM CaCl2 and permeabilized with 0.3% Triton X-100 for 10 (MDCK) or 5 min (HEK293) at 21°C. After incubation with blocking buffer (10% normal goat serum, 2% bovine serum albumin) primary antibodies were applied for 18 h at 4°C. Cells were washed with PB and incubated with secondary antibodies conjugated to Alexa Fluor 488 (Invitrogen) or Cy3 (Jackson Immunoresearch Labs, West Grove, PA, USA) for 1 h, rinsed and mounted on glass slides with the antifade Prolong (Invitrogen). The following antibodies were used for these studies: rabbit anti-Actn4 (Axxora, San Diego, CA, USA), rabbit anti-GFP Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse anti-myc (StressGen, Victoria, BC, Canada), mouse anti-α-tubulin (clone DM 1A; Sigma-Aldrich) and mouse anti-vimentin (Lab Vision, Fremont, CA, USA). Mouse anti-E-cadherin (clone rr1) was obtained from Developmental Studies Hybridoma Bank (supported by NICHD and maintained by the University of Iowa, Department of Biological Sciences). The anti-mGluR1b antibody was raised in rabbit against peptide KKRQPEFSPSSQCPSAHAQL corresponding to the distal portion of the mGluR1b carboxyl-tail. Laser scanning confocal microscopy was carried out with Zeiss LSM 510 (Zeiss, Thornwood, NY, USA) and 5 DUO V2.
Results
Generation of TAP-tagged mGluR1
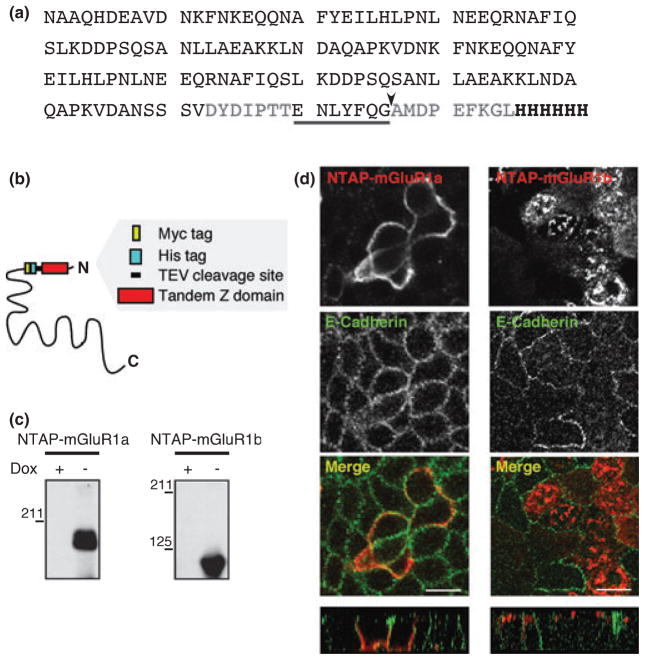
The goal of this study was to implement a biochemical strategy that would enable the identification of mGluR1b-interacting proteins potentially involved in receptor trafficking. The purification of significant amounts of highly hydrophobic transmembrane receptors under native conditions (i.e., conditions preserving receptor native conformation and interaction with binding partners) is particularly challenging because of the low-yield of proteins extracted from membranes by mild, non-ionic detergents. The Tandem Affinity Purification (TAP) method, originally developed in yeast, allows purification of near-pure protein complexes from relatively small amounts of material (Rigaut et al. 1999; Gavin et al. 2002). Because of these advantages, we undertook to adapt the TAP method to mammalian cells to identify proteins that are present in a complex with the native mGluR1b receptor. We first generated a modified form of the original TAP tag (Rigaut et al. 1999). Like the original TAP, the modified tag (NTAP) is composed of two IgG-binding units (Z domain) of protein A, followed by a TEV protease recognition site; however, the modified TAP contains a six-histidine tag instead of a calmodulin-binding peptide downstream of the TEV cleavage site (Fig. 1a and b). The intracellular tails of both mGluR1a and -1b contain motifs involved in trafficking and protein interaction; we reasoned that these motifs could be masked or occluded by insertion of the relatively bulky NTAP tag at the carboxyl terminus of the receptors. We therefore inserted the NTAP tag in the extracellular domain of rat mGluR1a/b, after residue Leu-28 and upstream of a myc tag inserted in the receptors (Fig. 1b; Francesconi and Duvoisin 2002); the additional myc tag affords specific and easy detection of the recombinant receptors with anti-myc antibodies.
Fig. 1.
Generation of mGluR1 receptors tagged with NTAP. (a) Amino acid sequence of the NTAP tag. NTAP is composed of two IgG-binding domains (Z domain), a TEV protease recognition sequence and a six-histidine tag. The consensus TEV cleavage site is underlined; the site where cleavage occurs is indicated by an arrow. Spacer sequences appear in gray and the six-histidine tag in bold characters. (b) Schematic representation of the NTAP tag inserted after the leader peptide of the receptors and upstream of a myc epitope. (c) Inducible expression of NTAP-mGluR1a/b. Western blot analysis with a myc antibody of extracts from representative stable MDCK clones in the presence (+) or absence (−) of doxycycline. (d) Localization of NTAP-mGluR1a and NTAP-mGluR1b in polarized MDCK cells. The receptors are labeled with TRITC-IgG (red) and the basolateral membrane with anti-E-cadherin (green). Shown are XY and XZ (bottom panels) views of optical slices; scale bars, 10 μm.
We have used a heterologous system, MDCK cells, to isolate and identify proteins that may be involved in trafficking of mGluR1. MDCK cells are a well-established model for the study of polarized trafficking; cellular mechanisms underlying vectorial transport in these cells are believed to share some common properties with mechanisms employed in neurons to regulate protein transport to dendrites versus axons (Horton and Ehlers 2003). Thus, uncovering the molecular machinery that participates in trafficking of mGluRs in MDCK cells may provide valuable insights towards understanding the mechanisms that regulate their trafficking in neurons. We first generated stable MDCK cell lines expressing the tagged receptors (NTAP-mGluR1a and NTAP-mGluR1b) under the control of a tetracycline-repressible promoter. Receptor expression in stable clones was inhibited by presence of doxycycline in the culture medium but rapidly induced upon removal of the drug (Fig. 1c), demonstrating tight transcriptional regulation of the exogenous constructs. To examine whether the tagged receptors were correctly targeted to the cell membrane, we labeled polarized cells expressing NTAP-mGluR1a or NTAP-mGluR1b with anti-myc and anti-E-cadherin antibodies. We found that, similarly to the corresponding wild-type receptors (Francesconi and Duvoisin 2002), NTAP-mGluR1a localized to the basolateral domain positive for E-cadherin, a marker of basolateral membranes, whereas NTAP-mGluR1b localized to the apical compartment (Fig. 1d). These results indicate that presence of the NTAP tag in the extracellular N-terminus of mGluR1a/b does not alter receptor trafficking.
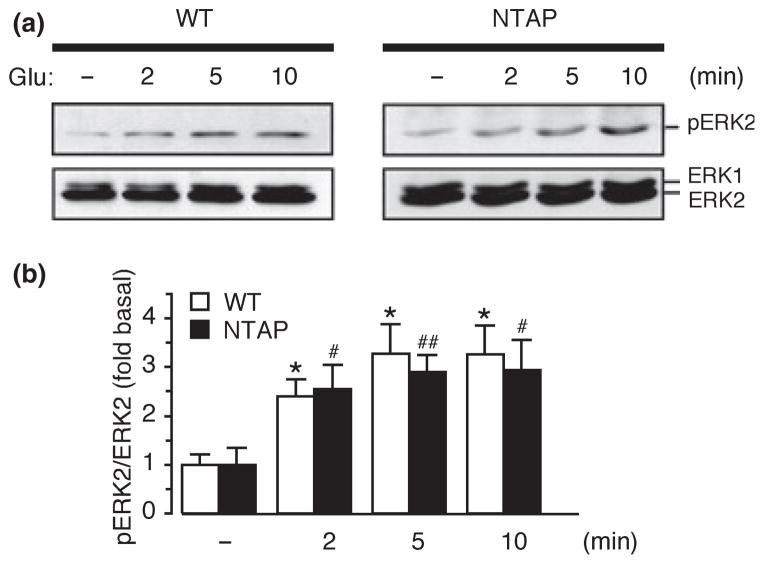
The presence of a relatively large tag in the extracellular domain of the receptors may potentially affect their function by inducing conformational changes and/or by interfering with ligand binding. To test this possibility, we assessed the impact of the NTAP tag on mGluR1a-dependent activation of the ERK-MAPK pathway. The mGluR1a receptor was chosen for these functional studies because its signaling properties have been extensively characterized in both heterologous cells and neurons. We co-transfected wild-type or tagged receptor together with the tetracycline transactivator in HEK293 cells (which do not express endogenous glutamate receptors) in the absence of doxycycline and measured glutamate-stimulated, mGluR1-dependent ERK1/2 phosphorylation. In cells expressing the wild-type receptor, stimulation with glutamate (1 mM) induced an approximately three-fold increase in ERK2 phosphorylation at 5 min, as determined by western blots probed with anti-phospho-ERK1/2 (Thr202/Tyr204) and anti-ERK1/2 antibodies (Fig. 2). In cells expressing NTAP-mGluR1a, glutamate induced an increase in ERK2 phosphorylation comparable to that induced by wild-type receptors (Fig. 2), indicating that the presence of the NTAP tag in the extracellular domain of the receptors does not significantly affect their function and transport to the plasma membrane.
Fig. 2.
The NTAP tag does not affect receptor signaling to ERK-MAPK. (a) Representative immunoblots for pERK1/2 and ERK1/2 of extracts from HEK293 cells expressing wild-type or NTAP-tagged mGluR1a and stimulated with glutamate for indicated times. (b) Quantitative analysis of experiments like those represented in (a); glutamate-induced ERK2 activation is calculated as ratio of the band densities of pERK2/ERK2 normalized to basal level. Means ± sem, n = 4; *p < 0.05 (relative to wild-type) and #p < 0.05, ##p < 0.01 (relative to NTAP).
Purification of the mGluR1b complex
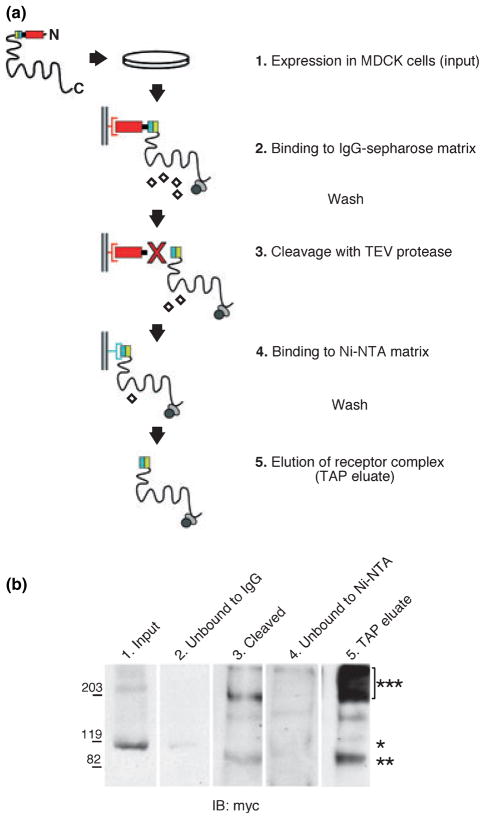
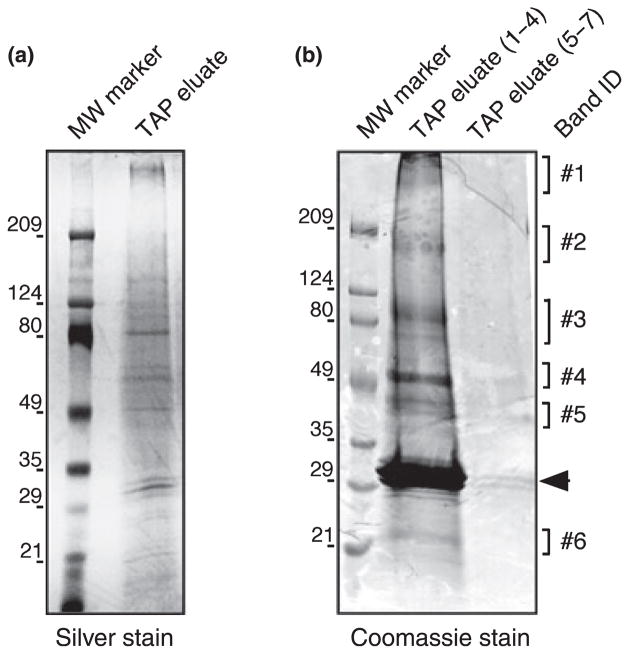
To purify NTAP-mGluR1b in a complex with potential interacting proteins, we implemented a dual affinity purification procedure that consists of two sequential rounds of enrichment of the tagged ‘bait’ protein (Fig. 3a; Rigaut et al. 1999). First, tagged receptors in the soluble cell extract were bound to IgG-sepharose beads, followed by extensive wash to eliminate unbound contaminant proteins. Immobilized receptors were released from the sepharose matrix by cleavage with TEV protease, which specifically recognizes a site embedded in the tag. Recovered cleaved receptors were bound to a Ni-NTA matrix via the remaining six-histidine tag, and further purified by extensive wash to remove residual contaminant proteins. We first examined whether the different components of the NTAP tag retained their functional integrity in the context of the native receptor. Extraction of MDCK cells with the non-ionic detergent NP-40 allowed significant (although partial) recovery of NTAP-mGluR1b in the soluble fraction (Fig. 3b, lane 1). After incubation with IgG-sepharose, a small amount of receptor was found in the unbound fraction (Fig. 3b, lane 2), indicating that affinity purification via the Z domain was effective. Incubation with TEV protease yielded cleaved receptors of the expected molecular mass that were recovered in the column flow-through (Fig. 3b, lane 3), indicating efficient and specific cleavage of the tag by the protease. After incubation with Ni-NTA matrix, only traces of receptor were observed in the unbound fraction (Fig. 3b, lane 4) whereas subsequent incubation with elution buffer resulted in efficient recovery of bound receptors (Fig. 3b, lane 5), indicating that the exposed six-histidine tag could mediate efficient binding to the matrix. Together, these results show that the modified NTAP tag affords efficient recovery of ‘native’ receptors. To identify proteins potentially involved in trafficking of mGluR1b, we plated NTAP-mGluR1b transfected MDCK cells at high density and allowed them to grow for several days in the presence of doxycycline. After the cells had reached confluency and formed fully polarized monolayers, as determined by visual inspection, we induced NTAP-mGluR1b expression by removing doxycycline from the culture medium. Tagged receptors in a complex with bound proteins were recovered from soluble MDCK cell extracts by dual affinity purification. Protein samples were collected at each step of the purification procedure and the final eluates were combined into two separate pools; proteins in the collected samples were analyzed by SDS-PAGE and visualized by Silver- (Fig. 4a) or Coomassie-staining (Fig. 4b). Analysis of the protein content in the IgG-sepharose and Ni-NTA unbound fractions showed that most of the ‘input’ proteins were eliminated during the purification (data not illustrated), indicating that two sequential rounds of enrichment greatly reduced the abundance of non-specific proteins. Analysis of the final eluates from different preparations consistently showed discrete protein bands distributed over a range of molecular mass (Fig. 4), indicating that the affinity purification procedure was efficient and reproducible.
Fig. 3.
Affinity purification of NTAP-mGluR1b. (a) Schematic representation of the dual affinity purification protocol adapted from Rigaut et al. (1999). (b) Western blot analysis with anti-myc of samples collected during the purification procedure. Lane 1, input receptor; lane 2, receptor in IgG-sepharose column flow-through; lane 3, TEV-cleaved receptor; lane 4, receptor unbound to Ni-NTA; lane 5, receptor in final eluate. Predicted MW is ~ 120 kDa for NTAP-mGluR1b (*), 99 kDa for the cleaved monomer (**) and 198 kDa for the cleaved dimer (***). As for all mGluRs, the most abundant form of the receptor is oligomeric (***).
Fig. 4.
Purified NTAP-mGluR1b associated complex. (a) Representative Silver-stained gel. (b) Representative Coomassie-blue stained gel; fractions 1–4 and 5–7 containing the TAP eluate were pooled. Band ID indicates the regions of the gel that were excised and analyzed by mass spectrometry. MW in kDa is at left; the arrowhead indicates bands corresponding to residual TEV protease.
Identification of proteins co-purified with mGluR1b
We used mass spectrometry to determine the identity of proteins that co-purify with NTAP-mGluR1b. Proteins were extracted from slabs of gel corresponding to individual or groups of bands stained by Coomassie and subjected to tryptic digestion. Peptide sequence information obtained by the MS/MS spectra (supporting information Table S1) was used to search an annotated protein database. Using this strategy, we identified 10 candidate interacting proteins (Tables S1 and 1) that could be broadly categorized as ‘cytoskeleton’ (actin, α-tubulin, β-tubulin, vimentin), ‘cytoskeleton-associated’ (α-actinin-4), ‘motor’ (myosin-Ic), ‘chaperone’ (hDj9) and ‘metabolic’ (ATP synthase beta chain, glutamate dehydrogenase) proteins. An additional protein, stromal cell derived factor 2, identified by this method has no known function. Peptides corresponding to mGluR1b were recovered from two different bands (Fig. 4b, bands 1 and 3) of >200 and ~ 100 kDa corresponding, respectively, to the oligomeric and monomeric forms of the receptor. A group of three bands of ~ 30 kDa that was present with variable abundance in different preparations (Fig. 4) was readily identified as the TEV protease complex. Contamination with recombinant protease has been previously reported (Knuesel et al. 2003) and is likely due to its high abundance in relation to the abundance of ‘bait’ and co-purified proteins. No peptides were obtained from a band (or group of bands) of ~200 kDa (Fig. 4b, band 2).
Table 1.
Proteins co-purified with mGluR1b in MDCK cells
| Protein ID | Gene ID | * Accession | Peptides | † MW | Coverage (%) | Band ID |
|---|---|---|---|---|---|---|
| mGluR1 | Grm1 | gi:2495074 | 4 | 101/204 | 6.4 | 1, 3 |
| Myosin-Ic (Myosin-I beta) | MyoIc | gi:13431721 | 2 | 118 | 1.7 | 3 |
| Non-muscle alpha-actinin4 | ACTN4 | gi:13123943 | 11 | 102 | 17.8 | 3 |
| Glutamate dehydrogenase | GLUD1 | gi:4885281 | 2 | 61 | 7.8 | 4 |
| ATP synthase beta chain | ATP5B | gi:114549 | 3 | 56 | 8.4 | 4 |
| Vimentin | VIM | gi:167887751 | 2 | 53 | 4.3 | 4 |
| Alpha-tubulin | Tuba | gi:5174733 | 5 | 50 | 15.2 | 5 |
| Beta-tubulin | Tubb | gi:5174739 | 12 | 50 | 45.1 | 5 |
| Alpha actin | Actin | gi:4501883 | 4 | 42 | 13.5 | 5 |
| hDj9 | hDj9 | gi:6567166 | 2 | 40 | 7.5 | 5 |
| Stromal-cell-derived factor-2 | SDF2 | gi:6677893 | 1 | 23 | 11.3 | 6 |
NCBI protein database.
Approximate MW.
Validation of putative mGluR1b-interacting proteins
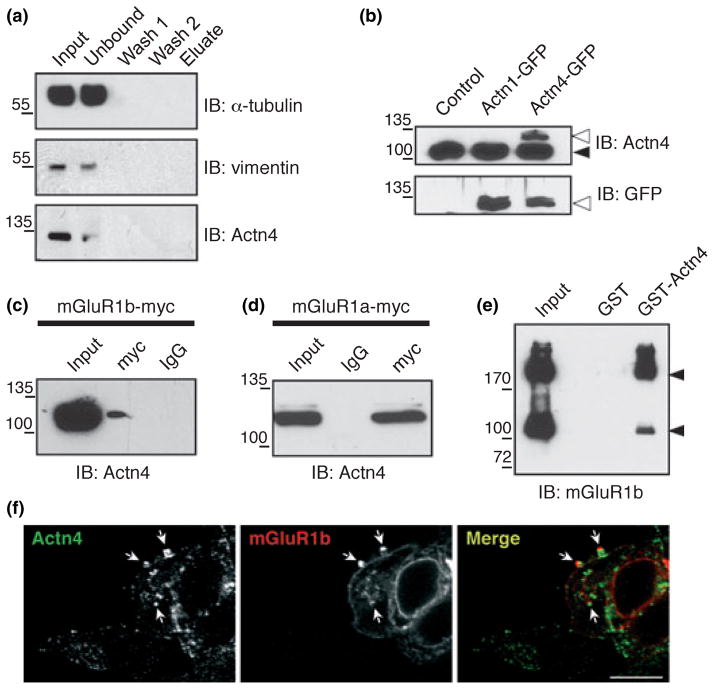
The sensitivity of Tandem Affinity Purification allows the co-purification of proteins that interact directly with the tagged ‘bait’ as well as of proteins that associate with the ‘bait’ indirectly, via intermediate binding-proteins (Gavin et al. 2002). To rule out the possibility that endogenous cellular proteins expressed at high levels could spuriously bind to the IgG-Sepharose and/or Ni-NTA matrix during the purification procedure, we performed Tandem Affinity Purification from control extracts prepared from cells that do not express mGluR1a/b. Input lysate, proteins recovered from intermediate purification steps and the final eluate were analyzed by western blot with antibodies to selected candidate interacting proteins (α-tubulin, vimentin and α-actinin-4; Fig. 5a) that are integral to or associated with the cytoskeleton. Proteins recovered in the first unbound fraction amounted to a large proportion of input lysate whereas no immunoreactive bands were detected in the final eluate, indicating that the TAP procedure is highly selective and affords efficient removal of non-specific ‘contaminant’ proteins. To assess whether proteins co-purified with mGluR1b are indeed receptor-interacting proteins in vivo, we focused on proteins likely to be involved in receptor trafficking. We first assessed a potential interaction of mGluR1b with α-actinin-4 (Actn4). Actn4 is a member of a small gene family of four homologous actin-binding proteins (Actn1–4) known to mediate bundling of actin filaments (Honda et al. 1998). To examine whether mGluR1b and Actn4 interact, we undertook three different approaches. First, we co-transfected myc-tagged mGluR1b and Actn4 in HEK293 cells and used an antibody to the myc epitope to immunoprecipitate mGluR1b. The anti-myc antibody, but not control IgG, precipitated Actn4 as shown by immunoblot with an anti-Actn4 antibody (Fig. 5c). The anti-Actn4 antibody is selective for Actn4 and does not recognize its closest homologue, Actn1 (Honda et al. 1998), as shown by its lack of reactivity with transfected Actn1-GFP (Fig. 5b). To examine whether Actn4 also interacts with the mGluR1a splice variant, we co-transfected HEK293 cells with Actn4 and myc-tagged mGluR1a and immunoprecipitated the receptor with the anti-myc antibody. Actn4 was efficiently precipitated by anti-myc but not by a control IgG (Fig. 5d), indicating that Actn4 interacts with both mGluR1b and mGluR1a. Second, we tested whether the interaction between mGluR1b and Actn4 could occur in vitro. We performed pull-down assays in which lysates from HEK293 cells expressing mGluR1b-myc were applied to immobilized GST-Actn4 or control GST, washed and collected the eluate containing bound proteins. GST-Actn4, but not GST, precipitated mGluR1b-myc (Fig. 5e), indicating that mGluR1b can interact with Actn4 in vitro. Next, we immunolabeled HEK293 cells transfected with mGluR1b-myc and examined the receptor cellular localization in relation to endogenous Actn4. Labeling with anti-Actn4 antibodies showed the presence of Actn4 in intracellular vesicular structures and in protrusion of the plasma membrane (Fig. 5f). Immunolabeling with anti-myc antibody revealed the presence of mGluR1b at the plasma membrane but also enrichment in an intracellular compartment (Fig. 5f); close apposition and overlap of mGluR1b-myc immunoreactivity to Actn4-positive structures was readily observed both at the plasma membrane and in intracellular vesicles (Fig. 5f). Together, these findings indicate that Actn4 is a novel binding partner of mGluR1a/b.
Fig. 5.
Actn4 interacts with mGluR1a/b. (a) Protein extracts (15 mg) from HEK293 cells that do not express mGluR1a/b were subjected to Tandem Affinity Purification. Equal volumes of input lysate (containing 25 μg of protein, corresponding to 0.0016% of total lysate), unbound fraction recovered from the IgG-Sepharose column, first (wash 1) and second (wash 2) wash fractions and 33% of the final eluate were analyzed by immunoblot with antibodies to α-tubulin (top panel), vimentin (middle panel) and α-actinin-4 (bottom panel). (b) The anti-Actn4 antibody recognizes Actn4-GFP but not Actn1-GFP in transfected HEK293 cells. The black arrowhead points to endogenous Actn4; white arrowheads point to Actn1/4-GFP. (c and d) Actn4 co-precipitates with mGluR1a/b-myc. The myc immunocomplex from HEK293 cells transfected with mGluR1b-myc (c) or mGluR1a-myc (d) was probed with anti-Actn4 antibody. (e) GST-Actn4 pulls down mGluR1b-myc from HEK293 cell extracts. Purified GST-Actn4 and GST bound to beads were incubated with extracts from HEK293 cells transfected with mGluR1b-myc. Proteins bound to GST-Actn4 and GST were probed with anti-mGluR1b antibody. Arrowheads indicate the monomeric and dimeric form of the receptor. (f) Representative images of HEK293 cells transfected with mGluR1b-myc and labeled with anti-Actn4 (green) and anti-myc (red). Arrows point to areas of immunoreactivity apposition and/or overlap. Shown are XY optical slices; scale bar, 10 μm.
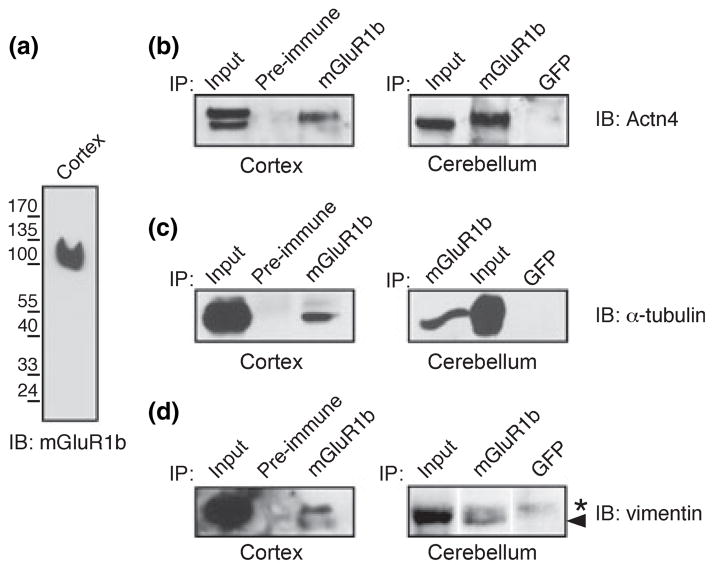
Next, we assessed whether proteins co-purified in a complex with NTAP-mGluR1b in MDCK cells associate with the native receptor in the brain. Toward this end we raised an antibody against the last 20 residues of the mGluR1b carboxyl-tail; this peptide is specific for mGluR1b and is not conserved in mGluR1a. The anti-mGluR1b antibody reacts specifically with recombinant mGluR1b transfected in HEK293 cells (Fig. 5e, lane 1) and recognizes a single band of ~ 100 kDa, the predicted molecular weight of mGluR1b, in extracts from rat brain cortex (Fig. 6a). Next, we used the mGluR1b antibody to immunoprecipitate the endogenous receptor from rat brain cortex and cerebellum and assessed its interaction with candidate proteins identified by proteomics by probing the immunocomplexes with antibodies specific to Actn4 (Fig. 6b), α-tubulin (Fig. 6c) and vimentin (Fig. 6d). These proteins were chosen for validation of interaction with the receptor in native tissue because they are expressed in the brain and specific reagents are readily available. Immunoblots probed with antibodies selective for Actn4 showed that anti-mGluR1b, but not control pre-immune serum or anti-GFP, precipitated endogenous Actn4 (Fig. 6b). Similarly, immunoblots probed with anti-α-tubulin (Fig. 6c) or anti-vimentin (Fig. 6d) demonstrated specific immunoprecipitation of the endogenous proteins by anti-mGluR1b, but not by control pre-immune serum or anti-GFP, in the brain. Together these findings indicate that at least a subset of proteins identified in a complex with NTAP-mGluR1b in MDCK associate with native mGluR1b in the brain.
Fig. 6.
Association of putative interacting proteins with native mGluR1b in the brain. (a) The anti-mGluR1b antibody raised against a peptide in the carboxyl-tail of mGluR1b reacts with the native receptor in extracts from rat brain cortex. (b–d) Actn4 (b), α-tubulin (c) and vimentin (d) co-precipitate with native mGluR1b in extracts from rat brain cortex and cerebellum. Arrowhead points to the vimentin band.
Discussion
Regulated trafficking of neurotransmitter receptors plays an important role in neurotransmission. Signaling by mGluR1 is critical to the regulation of excitatory neurotransmission, synaptic plasticity and to the establishment of synaptic circuitry during development. Genetic deletion of mGluR1 in mice results in decreased hippocampal long-term potentiation, abnormal associative learning and fear conditioning (Aiba et al. 1994a), impaired cerebellar long-term depression and motor coordination and defects in synapse formation in the cerebellum (Aiba et al. 1994b; Ichise et al. 2000). Dysregulation of group I mGluR signaling is implicated in a number of neuropsychiatric disorders including schizophrenia (Pietraszek et al. 2007), addiction (Kenny and Markou 2004) and Fragile X syndrome (Dolen et al. 2007). The prominent mGluR1 splice variants in the brain, mGluR1a and 1b, exhibit differing spatial patterns of expression and differential targeting within neurons (Ferraguti et al. 1998; Mateos et al. 1998, 2000). These observations suggest that different intracellular proteins might interact with mGluR1a and 1b and target the receptor isoforms to different neuronal microdomains. Here we report the identification of 10 proteins that co-purify with mGluR1b in MDCK cells and validate the interaction of three of these proteins with native mGluR1b in the brain. At least six of the identified proteins are known to play a role in intracellular trafficking and two proteins among them, Actn4 and myosin Ic, are enriched at the apical membrane of native polarized cells and participate in trafficking at the luminal domain (Boyd-White et al. 2001; Kim et al. 2002). Findings in the present study demonstrate that the TAP method is a powerful method for the identification of novel mGluR1 interacting partners implicated in receptor trafficking.
Actn4 is an actin bundling protein involved in both cell motility and endocytic traffic. Actn4 is expressed in the brain and forms a ternary complex with densin-180 and CaMKIIα, that are enriched at the post-synaptic density of excitatory synapses (Walikonis et al. 2001). In fibroblasts, Actn4 is involved in the recycling of internalized membrane receptors (Yan et al. 2005) and therefore it is likely to contribute to the regulation of mGluR1b endocytic trafficking and recycling. Interestingly, mGluR5 has been shown to interact with Actn1, over-expression of which increases mGluR5 surface expression (Cabello et al. 2007). Group I mGluRs are constitutively internalized from the plasma membrane (Fourgeaud et al. 2003; Bhattacharya et al. 2004); thus, recycling mechanisms are likely to play an important function in regulating the abundance of receptors expressed at the cell surface. Myosin Ic, another protein present in the NTAP-mGluR1b complex, also associates with endosomes and may participate to receptor trafficking by mediating movement of mGluR1b-containing recycling vesicles via the actin cytoskeleton.
Four proteins – actin, α-tubulin, β-tubulin, vimentin –purified in a complex with mGluR1b are part of the cell cytoskeleton. The observation that α- and β-tubulin are part of the mGluR1b complex points to a role of microtubules in trafficking of the receptor. In MDCK cells, microtubules are involved in transport from the trans-Golgi network to the apical domain and in transcytosis from the basolateral to the apical domain. Previous reports have shown that mGluR1a (Ciruela et al. 1999) and mGluR5 (Serge et al. 2003) interact with β-tubulin, whereas mGluR7 was found to interact with α-tubulin (Saugstad et al. 2002). Interestingly, the interaction between mGluR1a and β-tubulin is mediated by the last 86 residues of the receptor carboxyl tail that are not conserved in mGluR1b. These observations suggest that other domains in mGluR1 may also participate in the interaction with tubulin, or that alternative splicing generates a tubulin-binding region in the mGluR1b carboxyl tail. An alternative possibility is that the interaction of β-tubulin with mGluR1b is not direct but mediated through other proteins present in the mGluR1b-associated complex. Collectively, these observations suggest that microtubules may play an important role in mGluR trafficking in neurons.
Vimentin is a phosphorylated intermediate filament protein involved in multiple cellular functions including adhesion, membrane traffic and scaffolding of protein complexes at the cell membrane (Ivaska et al. 2007). Vimentin associates with the Golgi complex and autophagosomes and is important for endo-lysosomal traffic (Styers et al. 2005). In addition, integrity of the intermediate filament cytoskeleton is important for establishment of polarity in epithelial cells, possibly by a mechanism involving the regulation of cholesterol content in lipid rafts that are involved in apical transport (Styers et al. 2005). In the brain, vimentin is expressed in glia but not in post-mitotic neurons. However, vimentin expression can be re-induced in neurons by cell injury and participate in retrograde axonal transport of some proteins (Perlson et al. 2005).
In neurons and in heterologous cells, a substantial pool of mGluR1b resides intracellularly, likely in a sub-compartment of the ER (Remelli et al. 2008). Regulated export from the ER could constitute a means to control receptor expression level at the cell surface and interaction with signaling proteins. One of the proteins identified in a complex with mGluR1b is hDj9 (also known as ERdj3 or DnaJ homolog subfamily B member 11), an ER-associated Hsp40 co-chaperone. HDj9 is part of a chaperone complex that binds to nascent proteins and presumably assists their folding (Meunier et al. 2002). Interestingly, SDF2-L1, a homolog of the SDF-2 protein (Hamada et al. 1996) that co-purifies with mGluR1b, is also a component of this complex. These observations suggest that hDj9 and SDF-2 itself may associate with mGluR1b in the ER and participate in receptor folding and/or transport in the secretory pathway.
Two additional proteins that co-purify with mGluR1b, ATP synthase beta chain (ATP5B) and glutamate dehydrogenase, are metabolic enzymes present in the inner mitochondria matrix. The presence of mitochondrial proteins in the mGluR1b complex may possibly be due to non-specific interactions formed during the purification process and not to interactions formed in intact cells. However, both ATP synthase beta chain (Chi and Pizzo 2006) and glutamate dehydrogenase are also present in subcellular compartments outside of mitochondria. ATP5B localizes to the cell membrane where it binds apolipoprotein A–I (Martinez et al. 2003). In addition, ATP5B binds to caveolin-1 and translocates to plasma membrane caveolae in response to cholesterol (Wang et al. 2006). Membrane-bound glutamate dehydrogenase has been shown to be involved in the association of lysosomes to microtubules, to which it binds in an ATP-dependent manner (Rajas et al. 1996). At present, the specific function/s of ATP5B and glutamate dehydrogenase outside of mitochondria are not well understood.
Because of its capacity of affording the recovery of highly purified protein complexes, Tandem Affinity Purification allows the isolation of proteins that interact directly or indirectly with the tagged ‘bait’ and of proteins that may interact only transiently. In our study, the use of a full-length receptor in combination with purification under ‘native’ conditions is likely to yield physiologically relevant candidate interactors. Consistent with this, we have shown that proteins found in a complex with mGluR1b in MDCK cells associate with the native receptor in the brain. Since mGluR1b differs from other mGluR1 variants only in the composition of the intracellular tail, it is likely that at least some of the identified proteins may also interact with other mGluR1 isoforms; future studies are warranted to address the selectivity and the functional impact of the identified interactions. Novel mGluR1 interacting partners identified in this study may represent novel targets for pharmacologic intervention in neuropsychiatric disorders.
Supplementary Material
Acknowledgments
We thank Drs Robert M. Duvoisin and Michael V. L. Bennett for support and encouragement, and Dr Anne Müsch for advice. We thank the Laboratory for Macromolecular Analysis and Proteomics of AECOM (Dr Ruth Hogue Angeletti, Director) for MS/MS analysis and the High Resolution Optical Imaging Facility (Dr Kostantin Dobrenis, Director) of AECOM for assistance with confocal microscopy. This work was supported by grants from NARSAD, FRAXA and NIH NS47684 (to A.F.) and NIH NS20752 (to RSZ). RSZ is the F.M. Kirby Professor of Neural Repair and Protection.
Abbreviations used
- DTT
dithiothreitol
- MDCK
Madin-Darby Canine Kidney cells
- PB
phosphate buffer
- PMSF
phenylmethylsulfonyl fluoride
- TEV
tobacco etch virus
Footnotes
Additional Supporting information may be found in the online version of this article:
Table S1 Peptide sequences identified by mass spectrometry.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aiba A, Chen C, Herrup K, Rosenmund C, Stevens CF, Tonegawa S. Reduced hippocampal long-term potentiation and context-specific deficit in associative learning in mGluR1 mutant mice. Cell. 1994a;79:365–375. doi: 10.1016/0092-8674(94)90204-6. [DOI] [PubMed] [Google Scholar]
- Aiba A, Kano M, Chen C, Stanton ME, Fox GD, Herrup K, Zwingman TA, Tonegawa S. Deficient cerebellar long-term depression and impaired motor learning in mGluR1 mutant mice. Cell. 1994b;79:377–388. [PubMed] [Google Scholar]
- Alvarez FJ, Villalba RM, Carr PA, Grandes P, Somohano PM. Differential distribution of metabotropic glutamate receptors 1a, 1b, and 5 in the rat spinal cord. J Comp Neurol. 2000;422:464–487. doi: 10.1002/1096-9861(20000703)422:3<464::aid-cne11>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M, Babwah AV, Godin C, Anborgh PH, Dale LB, Poulter MO, Ferguson SSG. Ral and phospholipase D2-dependent pathway for constitutive metabotropic glutamate receptor endocytosis. J Neurosci. 2004;24:8752–8761. doi: 10.1523/JNEUROSCI.3155-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd-White J, Srirangam A, Goheen MP, Wagner MC. Ischemia disrupts myosin I beta in renal tubules. Am J Physiol Cell Physiol. 2001;281:C1326–1335. doi: 10.1152/ajpcell.2001.281.4.C1326. [DOI] [PubMed] [Google Scholar]
- Cabello N, Remelli R, Canela L, et al. Actin-binding protein alpha-actinin-1 interacts with the metabotropic glutamate receptor type 5b and modulates the cell surface expression and function of the receptor. J Biol Chem. 2007;282:12143–12153. doi: 10.1074/jbc.M608880200. [DOI] [PubMed] [Google Scholar]
- Chi AL, Pizzo SV. Cell surface F1F0 ATP synthase: a new paradigm? Ann Med. 2006;38:429–438. doi: 10.1080/07853890600928698. [DOI] [PubMed] [Google Scholar]
- Ciruela F, Robbins MJ, Willis AC, McIlhinney RAJ. Interactions of the C terminus of metabotropic glutamate receptor type 1a with rat brain proteins: evidence for a direct interaction with tubulin. J Neurochem. 1999;72:346–354. doi: 10.1046/j.1471-4159.1999.0720346.x. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- Conn PM, Ulloa-Aguirre A, Ito J, Janovick JA. G protein-coupled receptor trafficking in health and disease: lessons learned to prepare for therapeutic mutant rescue in vivo. Pharmacol Rev. 2007;59:225–250. doi: 10.1124/pr.59.3.2. [DOI] [PubMed] [Google Scholar]
- Das SS, Banker GA. The role of protein interaction motifs in regulating the polarity and clustering of metabotropic glutamate receptor mGluR1a. J Neurosci. 2006;26:8115–8125. doi: 10.1523/JNEUROSCI.1015-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji BD, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraguti F, Conquet F, Corti C, Grandes P, Kuhn R, Knopfel T. Immunohistochemical localization of the mGluR1b metabotropic glutamate receptor in the adult rodent forebrain: evidence for a differential distribution of mGluR1 splice variants. J Comp Neurol. 1998;400:391–407. [PubMed] [Google Scholar]
- Fourgeaud L, Bessis AS, Rossignol F, Pin JP, Olivo-Marin JC, Hemar A. The metabotropic glutamate receptor mGluR5 is endocytosed by a clathrin-independent pathway. J Biol Chem. 2003;278:12222–12230. doi: 10.1074/jbc.M205663200. [DOI] [PubMed] [Google Scholar]
- Francesconi A, Duvoisin RM. Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J Biol Chem. 1998;273:5615–5624. doi: 10.1074/jbc.273.10.5615. [DOI] [PubMed] [Google Scholar]
- Francesconi A, Duvoisin RM. Alternative splicing unmasks dendritic and axonal targeting signals in metabotropic glutamate receptor 1. J Neurosci. 2002;22:2196–2205. doi: 10.1523/JNEUROSCI.22-06-02196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bösche M, Krause R, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Tashiro K, Tada H, et al. Isolation and characterization of a novel secretory protein, stromal cell-derived factor-2 (SDF-2) using the signal sequence trap method. Gene. 1996;176:211–214. doi: 10.1016/0378-1119(96)00251-x. [DOI] [PubMed] [Google Scholar]
- Hermans E, Challiss RA. Structural, signaling and regulatory properties of the group I metabotropic glutamate receptors: prototypic family C G-protein-coupled receptors. Biochem J. 2001;359:465–484. doi: 10.1042/0264-6021:3590465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Neuronal polarity and trafficking. Neuron. 2003;40:277–295. doi: 10.1016/s0896-6273(03)00629-9. [DOI] [PubMed] [Google Scholar]
- Ichise T, Kano M, Hashimoto K, Yanagihara D, Nakao K, Shigemoto R, Katsuki M, Aiba A. mGluR1 in cerebellar Purkinje cells essential for long-term depression, synapse elimination, and motor coordination. Science. 2000;288:1832–1835. doi: 10.1126/science.288.5472.1832. [DOI] [PubMed] [Google Scholar]
- Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Kenny PJ, Markou A. The ups and downs of addiction: role of metabotropic glutamate receptors. Trends Pharmacol Sci. 2004;25:265–272. doi: 10.1016/j.tips.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee-Kwon W, Park JB, Ryu SH, Yun CH, Donowitz M. Ca(2 +)-dependent inhibition of Na+/H+ exchanger 3 (NHE3) requires an NHE3-E3KARP-alpha-actinin-4 complex for oligomerization and endocytosis. J Biol Chem. 2002;277:23714–23724. doi: 10.1074/jbc.M200835200. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim YS, Yuan JP, Petralia RS, Worley PF, Linden DJ. Activation of the TRPC1 cation channel by metabotropic glutamate receptor mGluR1. Nature. 2003;426:285–291. doi: 10.1038/nature02162. [DOI] [PubMed] [Google Scholar]
- Kitano J, Kimura K, Yamazaki Y, Soda T, Shigemoto R, Nakajima Y, Nakanishi S. Tamalin, a PDZ domain-containing protein, links a protein complex formation of group 1 metabotropic glutamate receptors and the guanine nucleotide exchange factor cytohesins. J Neurosci. 2002;22:1280–1289. doi: 10.1523/JNEUROSCI.22-04-01280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel M, Wan Y, Xiao Z, Holinger E, Lowe N, Wang W, Liu X. Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol Cell Proteomics. 2003;2:1225–1233. doi: 10.1074/mcp.T300007-MCP200. [DOI] [PubMed] [Google Scholar]
- Martinez LO, Jacquet S, Esteve JP, et al. Ectopic b-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nature. 2003;421:75–78. doi: 10.1038/nature01250. [DOI] [PubMed] [Google Scholar]
- Mateos JM, Azkue J, Benitez R, et al. Immunocytochemical localization of the mGluR1b metabotropic glutamate receptor in the rat hippocampus. J Comp Neurol. 1998;390:225–233. doi: 10.1002/(sici)1096-9861(19980112)390:2<225::aid-cne5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mateos JM, Benitez R, Elezgarai I, et al. Immunolocalization of the mGluR1b splice variant of the metabotropic glutamate receptor 1 at parallel fiber-Purkinje cell synapses in the rat cerebellar cortex. J Neurochem. 2000;74:1301–1309. doi: 10.1046/j.1471-4159.2000.741301.x. [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multi-protein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Pietraszek M, Nagel J, Gravius A, Schäfer D, Danysz W. The role of group I metabotropic glutamate receptors in schizophrenia. Amino Acids. 2007;32:173–178. doi: 10.1007/s00726-006-0319-9. [DOI] [PubMed] [Google Scholar]
- Rajas F, Gire V, Rousset B. Involvement of a membrane-bound form of glutamate dehydrogenase in the association of lysosomes to microtubules. J Biol Chem. 1996;271:29882–29890. doi: 10.1074/jbc.271.47.29882. [DOI] [PubMed] [Google Scholar]
- Remelli R, Robbins MJ, McIlhinney RA. The C-terminus of the metabotropic glutamate receptor 1b regulates dimerization of the receptor. J Neurochem. 2008;104:1020–1031. doi: 10.1111/j.1471-4159.2007.05034.x. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Roche KW, Tu JC, Petralia RS, Xiao B, Wenthold RJ, Worley PF. Homer 1b regulates the trafficking of group I metabotropic glutamate receptors. J Biol Chem. 1999;274:25953–25957. doi: 10.1074/jbc.274.36.25953. [DOI] [PubMed] [Google Scholar]
- Saugstad JA, Yang S, Pohl J, Hall RA, Conn PJ. Interaction between metabotropic glutamate receptor 7 and alpha tubulin. J Neurochem. 2002;80:980–988. doi: 10.1046/j.0022-3042.2002.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serge A, Fourgeaud L, Hemar A, Choquet D. Receptor activation and Homer differentially control the lateral mobility of metabotropic glutamate receptor 5 in the neuronal membrane. J Neurosci. 2002;22:3910–3920. doi: 10.1523/JNEUROSCI.22-10-03910.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serge A, Fourgeaud L, Hemar A, Choquet D. Active surface transport of metabotropic glutamate receptors through binding to microtubules and actin flow. J Cell Sci. 2003;116:5015–5022. doi: 10.1242/jcs.00822. [DOI] [PubMed] [Google Scholar]
- Styers ML, Kowalczyk AP, Faundez V. Intermediate filaments and vesicular membrane traffic: the odd couple’s first dance? Traffic. 2005;6:359–365. doi: 10.1111/j.1600-0854.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Walikonis RS, Oguni A, Khorosheva EM, Jeng CJ, Asuncion FJ, Kennedy MB. Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2 +/calmodulin-dependent protein kinase II and (alpha)-actinin. J Neurosci. 2001;21:423–433. doi: 10.1523/JNEUROSCI.21-02-00423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chen Z, Wang X, Shyy JY, Zhu Y. Cholesterol loading increases the translocation of ATP synthase b chain into membrane caveolae in vascular endothelial cells. Biochim Biophys Acta. 2006;1761:1182–1190. doi: 10.1016/j.bbalip.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Petralia RS, et al. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Yan Q, Sun W, Kujala P, Lotfi Y, Vida TA, Bean AJ. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol Biol Cell. 2005;16:2470–2482. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.