Abstract
In mouse ear development, two bHLH genes, Atoh1 and Neurog1, are essential for hair cell and sensory neuron differentiation. Evolution converted the original simple atonal-dependent neurosensory cell formation program of diploblasts into the derived developmental program of vertebrates that generates two neurosensory cell types, the sensory neuron and the sensory hair cell. This transformation was achieved through gene multiplication in ancestral triploblasts resulting in the expansion of the atonal bHLH gene family. Novel genes of the Neurogenin and NeuroD families are upregulated prior to the expression of Atoh1. Recent data suggest that NeuroD and Neurogenin were lost or their function in neuronal specification reduced in flies, thus changing our perception of the evolution of these genes. This sequence of expression changes was accompanied by modification of the E-box binding sites of these genes to regulate different downstream genes and to form inhibitory loops among each other, thus fine-tuning expression transitions.
Keywords: bHLH genes, Neurosensory development, Neurosensory evolution, Ear development, Hair cells
Introduction: history and evolution of vertebrate mechanosensory cells and organs
The vertebrate ear arose in ancestral chordates and all living vertebrates have an ear with sensory hair cells for mechanoelectric transduction in a complicated three-dimensional system, the labyrinth. In addition, vertebrates have sensory neurons to conduct the information from the ear to the brain [1]. The mechanosensory hair cell of vertebrates has been hypothesized to be the transformation of ancestral mechanosensory cells that may go back to the unicellular ancestor of animals [2, 3]. The vertebrate ear develops from a placodal thickening of the ectoderm [4] through invagination, proliferation and cellular diversification (Fig. 1) into a sensory organ with a complex and intricate cytoarchitecture. The vertebrate inner ear contains fewer neuronal cell types than does the retina, whose development is governed by a complex interplay of multiple transcription factors [5] and may those allow an easier understanding. However, the vertebrate inner ear does have more cell types than the olfactory system, whose single principal neurosensory cell type already requires a sequential activation of several transcription factors for its development [6]. Additional subtypes of these different dominant cell types such as multiple amacrine cells in the retina, two types of hair cells in the cochlea and multiple receptor types in the olfactory epithelium, complicate matters, but these subtypes are not the subject of this review. Despite its complicated three-dimensional morphology, the vertebrate inner ear has only four principal cell types: sensory neurons that connect the ear with the brain [7], hair cells that convert mechanical stimuli into electric signals [8], supporting cells that provide cellular and mechanical support for hair cells, and otic epithelial cells that outline the labyrinth, some of which provide the K+-rich endolymph [9]. These principal cell types are found in and around the mammalian auditory end-organ, the cochlea and the vestibular labyrinth that consists of the other five inner ear sensory end-organs, the utricle, the saccule and the three canal cristae (Fig. 1).
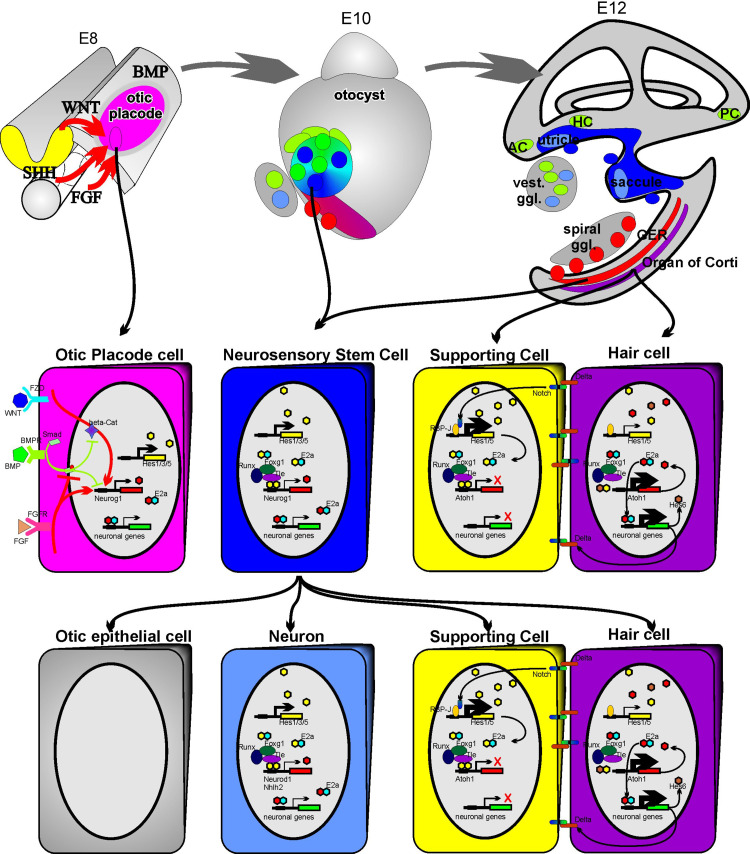
Fig. 1.
The morphogenesis and cellular interactions underlying mammalian inner ear development. Top panel The ear develops from the otic placode which in turn is induced to proliferate, invaginate to form the otocyst and undergo morphogenesis into the ear with the six sensory organs through multiple signals such as fibroblast growth factors (FGF), sonic hedgehog (SHH), wingless (WNT) and bone morphogenetic proteins (BMP). Neurons and hair cells arise from adjacent and partially overlapping areas (shown in color shades of each other) and may in certain cases share a clonal relationship, in particular for the utricle and saccule. Arrows indicate the topological origin of the cells shown at higher magnification in the middle panel. Middle panel: left The known and suspected signaling pathways to transform an ectodermal cell into an otocyst; left center the gene upregulation in the prosensory precursor stem cell; right the molecular interactions that, via lateral inhibition and the upregulation of Hes genes via the Delta/Notch pathway, help stabilize supporting cell differentiation. Neuron and hair cell differentiation in turn is driven by the upregulation of specific bHLH genes (Neurod1 for neurons and Atoh1 for hair cells). Bottom panel Single or adjacent cells from the saccular or utricular region may differentiate into otic epithelial cells (left), neurons (middle through upregulation of Neurod1) or generate hair cells and supporting cells (right). Cells of the greater epithelial ridge (GER) near the organ of Corti in the cochlea (top left) also have the capacity to differentiate into otic epithelial cells (spiral sulcus cells), spiral sensory neurons and, through application of Atoh1, into hair cells and supporting cells (modified from references [16, 20, 51])
Previously, we have proposed that the vertebrate sensory epithelia can be interpreted as a modification of the basic mechanosensory organ development of the fruit fly [9]. The present review investigates the current state of this hypothesis and illustrates the level of understanding of molecular processes achieved in the last 10 years with a focus on the bHLH genes and their function in ear neurosensory development and evolution. Most of the data we present here are supported through null mutant analyses of various genes, but some data on more recently discovered bHLH genes with inner ear function are not yet fully described in terms of null mutant analysis. Loss-of-function analyses provide good starting points to assess the importance of a given gene in certain developmental steps. The necessary mis- and overexpression studies to fully elucidate the function of a given gene with and without the normal context has barely begun in the vertebrate ear. We thus restrict our analysis to knockout or loss-of-function data supplemented by expression and available genomic sequence data.
Making neurosensory cells using conserved genes involved in differentiation regulation already in diploblastic animals
Three sets of knockout mutants have seemingly clarified the molecular basis for neurosensory development in the ear as being related to just three bHLH genes:
Neurogenin 1 (Neurog1) is necessary to induce sensory neurons [10].
Neuronal differentiation 1 (Neurod1) is necessary for sensory neuron survival and differentiation [11].
Atonal homolog 1 (Atoh1) is necessary for hair cell differentiation [8].
Over the last 10 years follow-up studies have verified and expanded upon these initial findings [12–19]. There is now a more complete understanding of the complexity of molecular interactions, and the necessary steps in development of the two major classes of cells, hair cells and neurons, underlying detection of mechanical stimuli in the mammalian inner ear and the conduction of this information to the brain are beginning to be refined. Most important are data that show the interaction of neurosensory specifying genes of the bHLH type with factors that regulate their topological and temporal expression patterns (upstream regulatory genes of bHLH genes). These genes define not only the place and time of bHLH gene up- and downregulation, but in doing so determine the number, anatomical location, and cell type of the neurosensory cells that will form [20]. To achieve this essential cellular specification in the inner ear requires multiple regulatory genes that cause these neurosensory cells to differentiate (downstream effects of bHLH genes), but also regulate the cellular interactions directing the fate of adjacent cells (Fig. 1). The Delta/Notch pathway functions in progenitor proliferation regulation and in supporting cell fate determination, and is well understood at the molecular level [18, 21]. However, in this review centered on the proneural bHLH gene we do not discuss nonneuronal genes as they play no direct role in the differentiation of neurosensory cells. We restrict our discussion to their role in cell fate determination of neurosensory progenitors by bHLH genes, which is the least understood aspect of the Delta/Notch system.
Evolution of molecular regulation of neurosensory cell development
The evolution of neurosensory cells is tightly interwoven with the evolution of neurons, and both can be viewed as a transformation of a generalized epidermal cell into a neuron-like cell with a novel sensory specialization [3, 22, 23]. This understanding was achieved through modifications of the general molecular schemes underlying proliferation and differentiation of epidermal cells as well as elucidation of the life cycle of single-cell organisms from vegetative cells to proliferation (mitosis) or sexual reproduction (meiosis). The molecular mechanisms underlying these events are tightly connected with bHLH genes, in particular the Myc/Max/Mad network for proliferation regulation. This network is already present in yeasts and the unicellular ancestor of animals [24–26]. Existing data on bHLH genes suggest that the evolution of multicellular organisms came about through multiplication of genes involved in cell-cycle regulation and diversification allowing the differentiation of novel cell types. For example, diploblastic coelenterates already have the same classes of bHLH genes found in triploblastic animals, and the principal bHLH gene families associated with neurosensory development are even found in demisponges [25]. In addition, it appears that in flies, long used as prototypical examples of invertebrate development, the bHLH genes may have been modified. Data suggest that in flies several bHLH genes have been lost secondarily or show reduced function, while the members of other bHLH gene families have been expanded or new families have been created [24]. Elucidating the neurosensory evolution of the ear can therefore be reduced to dissecting at the molecular level the evolution and developmental function of bHLH genes known to participate or suspected of participating in the neurosensory development of the ear (Fig. 2a).
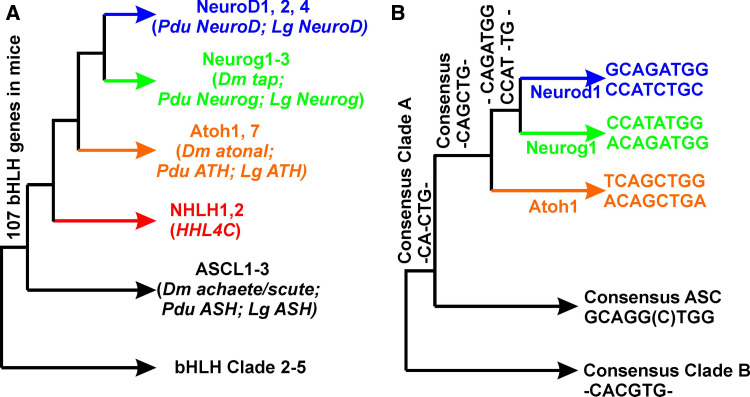
Fig. 2.
Interrelationship of bHLH genes relevant to the ear (a) and their E-box sequences (b). The achaete/scute sequence is used as a neuronal in-group, and other bHLH gene clades are used as outgroups. Note that Nhlh are in between the achaete/scute and the atonal family of genes. Most triploblasts have representatives of all three atonal gene family members, Atoh, Neurog and Neurod, except for the fly, (Drosophila melanogaster Dm) which lacks Neurod1. Since species belonging to both lophotrochozoans (Lottia gigantea Lg) and trochozoans (Platynereis dumerilii Pdu) have members of all three atonal associated families, the fly has secondarily lost Neurod and shows reduced function of Neurog. The consensus sequence of mammals shows that the central six nucleotides of the E-box are shared or differ by only one nucleotide. Likewise, the E-box motif of all atonal family members are closer to each other than to the outgroup sequences, suggesting that they coevolved with the changes in DNA binding motive in the atonal family members (compiled from references [24, 26–28])
In addition to the multiplication and functional diversification of bHLH genes there was also an evolution in their associated DNA binding domains as well as modification of the targeted E-box motifs themselves [27, 28]. Consistent with the expansion of the individual proneural bHLH gene families [25] is the modification of these E-box DNA elements through sequence alterations allowing modified bHLH genes to achieve a novel function through unique regulation of their downstream genes (Fig. 2b). Given that many downstream genes have more than one E-box site in their promoter(s), the increase in bHLH genes and in E-box motifs allowed the mixing of multiple genes in cell fate determination. This mixing greatly enhanced the diversity of cell types obtainable through topographically and temporally restricted overlapping proneural bHLH gene expression. Achieving this fine regulation became possible through the combined modification of the nucleotide sequence of the E-boxes in promoter regions, through the use of one or more of the multiple E-boxes to regulate the intensity of gene expression and through variation in the DNA binding affinities of the bHLH genes resulting from DNA binding domain alternations (Fig. 2). Superimposed on this binding affinity diversity resulting from bHLH and E-box sequence diversity, there is also diversity of transcription cofactor interactions near the binding sites [29, 30] as well as modification of the dimerizing sites of the proteins [31]. With this generalized evolutionary scheme in mind we next introduce the known details of ear-related bHLH genes as examples for the validity of these more global statements.
Evolution of bHLH genes for differentiation regulation of neurosensory development across phyla (proneural bHLH genes)
In the proneural bHLH gene family atonal, a single ancestral diploblast gene was multiplied independently in several lineages and vertebrates have two orthologs of atonal, Atoh1 and Atoh7 (formerly Math1 and Math5, respectively, in the mouse) [24, 26]. The zebrafish shows duplication of the Atoh1 gene and has evolved different functions for atoh1a and atoh1b [32]. In Drosophila as well, atonal has multiplied to form the additional genes absent multidendritic and olfactory sensilla (amos) [33] and cousin of atonal (cato) [34], both of which are more closely related to atonal than to the vertebrate Atoh1 and Atoh7 [26]. The amos gene is the proneural gene for a subset of multidendritic sensory neurons and two of three subtypes of olfactory sense organs, the third olfactory subtype being specified by atonal itself. The cato gene operates less as a proneural gene that confers neural competence on cells than as a neural differentiation factor, not unlike the vertebrate Neurod1. Strikingly, the functional equivalence of Drosophila atonal and mouse Atoh1 has been extensively tested through replacement studies both through expression of vertebrate Atoh1 instead of fly atonal [35], and of fly atonal instead of vertebrate Atoh1 [36]. Studies in the fly have shown significant functional differentiation between atonal and amos in chordotonal and olfactory organ specification, though amos is able to replace atonal in photoreceptor specification [37]. Interestingly, while the amos-specific functions are attributed to bHLH domain residues, the atonal-specific functions maps to the non-bHLH regions of atonal. These types of misexpression and replacement studies have not been as extensively performed for Atoh7, nor in a cross-species way for amos and cato. Indeed, whether Atoh7 is functionally competent to replace the fly atonal or the vertebrate Atoh1 needs to be tested through appropriate genetic manipulations. Likewise, the more recently found atonal-like genes in nonvertebrate triploblast species, Neurogenin and Neurod family members [24], need to be tested for functional characterization using approaches comparable to those outlined above for atonal/Atoh1. Specifically, given the similarities of E-boxes targeted by these bHLH proteins, it is possible that cross-activation will occur and likely contribute to differential levels of expression.
Despite these current limitations in analysis, it is clear that the promoter regions of Atoh1 and Atoh7 differ significantly [38]. Among other things, Atoh1 has a unique autoregulatory enhancer element containing an E-box in the 3′ region of the gene, which does not exist in Atoh7 genomic structure. Combined, these changes allow a spatially and temporally discrete regulation of both atonal paralogs in vertebrates thus tying them into very different functions (ear development for Atoh1 and eye development for Atoh7) that are served by the single atonal gene in flies (both ‘ears’ and eyes). Similar to Atoh1, fly atonal also has an autoregulatory enhancer [39, 40], but amos appears to lack such autoregulation [41]. The presence of such autoregulatory enhancers in fly atonal and mouse Atoh1 suggests that this may be the ancestral condition. Expression of other atonal-like genes in invertebrates other than flies needs to be investigated to elucidate the role of atonal/Atoh1 in neurosensory development and evolution and to be able to generalize data obtained thus far only in flies and vertebrates. Moreover, the E-boxes of those genes need to be characterized to be able to elucidate the cross-regulatory interaction between certain bHLH genes [16, 42, 43] and even in the same cell [44].
Obviously, selective differences in regulation of expression through promoter variations is necessary to acquire a new function but has to be accompanied by sequence modifications allowing grading to distinctly different regulation of downstream genes. These sequence differences must be tied into the evolution of novel E-box features in downstream genes to enhance the regulatory differences between the orthologs and paralogs of atonal. In the case of the vertebrate Atoh1/7 multiplication [24], we know that the high level of sequence similarity in the bHLH DNA binding domain is achieved with conservation of the atonal/Atoh1/Atoh7 E-boxes (Fig. 1b). In principle therefore, atonal, Atoh1 and Atoh7 should all be exchangeable as they all have a functionally identical DNA binding domain. The distinction between Atoh1 and Atoh7 is an example of evolution being not only on the cis-acting elements, but also on the trans effects. The cellular context of available genes expressed within those tissues leads to where the cellular context of available genes within those tissues whose expression leads to specific and unique cell type acquisitions. Similar trans effects have been proposed, and recently found to be important in the fly as well [29, 30]. This is certainly the case for the dichotomy between Atoh1 and Atoh7 with predominantly non-overlapping expressions of Atoh1 in the inner ear and of Atoh7 in the eye; in addition both genes have partially overlapping regions of expression in the brain as well as other unique expression areas in the digestive tract and Merkel cells.
Another scenario is the evolution of new or novel expression patterns in cells and tissues where these bHLH genes had not been previously found. For example, atonal is not expressed in the fly digestive system but Atoh1 plays a role in the development of secretory cells of the vertebrate intestine where it acts upstream of the bHLH gene Neurod1 which determines the fate of enteroendocrine cells [45]. Replacement studies have shown that this developmental function can also be served by the fly atonal gene [36]. It is likely that those downstream genes regulated by Atoh1 have evolved E-boxes with the common signaling sequence recognized by the atonal/Atoh1 DNA binding domain. This paradigm shows that within the atonal family of genes there is significant change in the promoter/enhancer regions to allow downstream genes to be differentially expressed, but the ability to regulate downstream genes seems to be highly conserved between family members within the same phylum as well as between phyla. Alterations in the binding affinity of these bHLH genes to the E-boxes could provide an alternative source for evolution. Thus, even slightly variable E-box sequences might be activated by atonal binding, provided there is sufficient binding affinity to permit expression of the targeted downstream gene. In addition, alterations in protein domains to form heterodimers can add to the complexity of bHLH gene evolution, but the latter part is the least understood aspect of bHLH gene evolution [31, 46].
In contrast to evolution of atonal/Atoh1 orthologs, evolution of other bHLH genes belonging to the NeuroD or Neurogenin families of genes indicates that these genes have been significantly altered, not only in their sequence, but in some lineages such as the fly, they have been lost altogether [26]. Furthermore, these transcription factors also bind to unique E-box motifs (Fig. 1b). A wider diversity in gene regulation is thus permitted through the activation of different downstream genes or modifying the action of atonal/Atoh1/Atoh7 on the same genes (wherever atonal and Neurogenin/NeuroD-associated E-boxes coexist). Working backward from the E-boxes therefore allows a set of bHLH genes to be functionally defined as family members with near-identical DNA binding capacity. Following the example of atonal/Atoh1 functional replacement, it is reasonable to assume that genes binding to the same E-box should be able to functionally replace each other. Several experiments [28, 47] have already demonstrated this ability to functionally replace certain bHLH genes by others with similar nucleotide binding specificity. In contrast, replacement by other bHLH genes was not successful likely because of incompatible binding to the defined or targeted E-box motif or because of protein interactions [46]. Knowing these limitations will allow future research to more directly test these predictions paying particular attention to the level of compensatory redundancy as different bHLH genes may bind to the same E-box, but may have different abilities to drive downstream genes via their differential binding affinities or interactions with different partners.
For example, the sequence-related bHLH genes Neurod1 and Neurog1 also entirely share one E-box binding motif (Fig. 1b) which in turn differs only by one nucleotide from Atoh1 binding motifs [28]. Thus, duplication of E-boxes and their sequence variations has not fully segregated all of the Neurod1 and Neurog1 signaling capacity, suggesting that function should at least partially be maintained on replacing Neurog1 by Neurod1. Beyond characterization of bHLH gene evolution, characterizing the evolution of E-boxes among phyla and verifying the functional equivalence of different bHLH genes through appropriate expression studies analogous to the work on atonal/Atoh1 [35, 36] are now needed to go beyond these tentative insights and establish both functional equivalence and consolidate the molecular basis of this equivalence or compensatory redundancy.
The developmental unit of neurosensory cell development: variations of a general theme?
Having now explored the evolution of proneural bHLH genes as transcription factors able to drive differential development of novel cell types, we need to evaluate how these cell types become organized into discrete organs. Clearly, this has to be achieved via upstream regulation that allows formation of single sensory cells in the epidermis, the most simple sensory organization (Fig. 3). This has been worked out in the cutaneous sensory organs of the fly and suggests that a single sensory mother cell will be selected which, through two rounds of proliferation, generates the necessary four cell types that constitute the minimal essential mechanosensory developmental unit (Fig. 3). This mechanism is molecularly best understood in insect single sensory cell development [48], but single cell formation or formation of aggregations of only a few sensory cells is also known in chordates [49, 50]. Unfortunately, the molecular basis for these single sensory cell formations in chordates is not yet elucidated and it remains to be shown that it parallels that in fly development. Likewise, patterning mechanisms specifying the topology of these single sensory cells are practically unknown in other phyla such as lophotrochozoans and trochozoans [24] and require additional investigations. Nevertheless, in theory a modified version of this cellular proliferation and decision making process could be the basis of inner ear neurosensory development, as we have previously proposed [9].
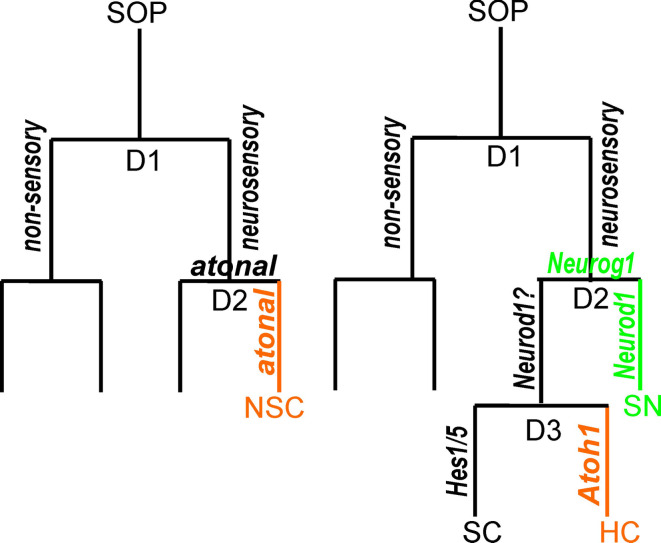
Fig. 3.
The known development of a fly cutaneous mechanosensor (left) and a hypothetical mammalian neurosensory development (right). A fly cutaneous mechanosensor forms through two divisions (D1, D2) out of a sense organ precursor (SOP). It is hypothesized that fly chordotonal organs follow the same principle with the exception that atonal is driving the differentiation of the neurosensory cell (NSC) and possibly also supports the second division. A similar generalized scheme seems to underlie mammalian ear development. A hypothetical module that starts with a sensory mother cell (referred to as neurosensory precursor cell in Fig. 1) undergoes a division to give rise to neurosensory and nonsensory precursors. The second division (D2) leads to upregulation of Neurog1 followed by upregulation of NeuroD1 to differentiate a sensory neuron. A third division of the sensory precursor cell will give rise to two cells that differentiate under the influence of the bHLH genes Atoh1 and Hes1/5 into hair cells (HC) and supporting cells (SC), respectively. As indicated in Fig. 1, this model may apply only to a restricted aspect of mammalian ear development (modified from references [9, 20])
Clearly, Neurog1 is the first bHLH gene upregulated in the mammalian inner ear (Fig. 4) and it activates the downstream genes Neurod1, Nhlh1 and Nhlh2, which govern neuronal development [10, 13, 14, 17]. Neurod1 primarily functions in regulating neuronal differentiation and survival [11, 14]. In addition, Neurog1 (and possibly Neurod1) appear to suppress Atoh1. First, in the absence of Neurog1 (and Neurod1) otic epithelial cells in the ductus reuniens, a nonsensory region known to give rise to sensory neurons [16, 51], can form hair cells [15]. Second, Neurog1 (and possibly Neurod1) have recently been shown to directly suppress Atoh1 upregulation in the inner ear [16], and suppression of Atoh1 by Neurod1 has recently been found in the cerebellum [44]. Since Neurod1, but not Neurog1, overlaps with Atoh1 in the sensory epithelia [11, 16] and Neurod1 is regulated by Neurog1 [10] it is likely that Neurod1 is mediating the effects of Neurog1 on hair cell differentiation. It has been noted before that certain similarities in Neurog1-null and Neurod1-null mice exist in cochlear hair cells on the apical turn [11, 13, 14]. In addition, Atoh1 inhibits Neurod1 expression in the utricle and saccule [16] resulting in an overexpression of Neurod1-LacZ in Neurod1-null mice [11].
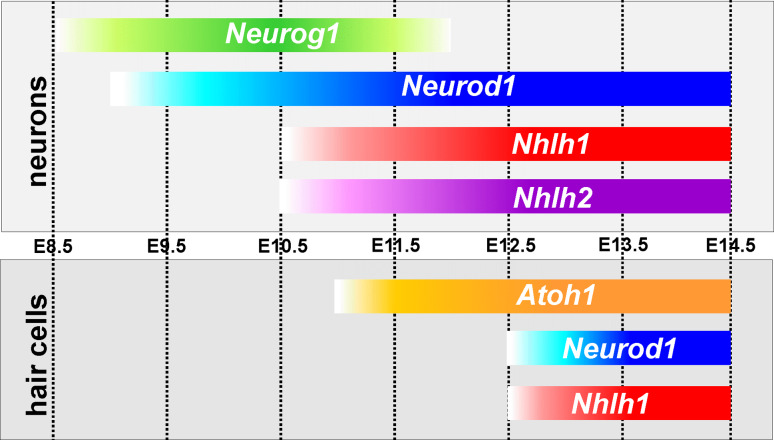
Fig. 4.
The expression profiles of mouse bHLH genes that play a role in neurosensory differentiation are shown for sensory neurons (top) and hair cells (bottom). The first bHLH gene upregulated in the ear is Neurog1 (around E8.75) followed immediately by NeuroD1 (around E9). Two additional bHLH genes are eventually upregulated in sensory neurons, Nhlh2 (in neurons only) and Nhlh1 (in neurons followed by hair cell expression). The exact time of expression of all four genes needs to be established using Q-PCR. bHLH gene expression in hair cells is much later, starting around E10.5 (as demonstrated by Q-PCR). Later, the bHLH genes Neurod1 and Nhlh1 are also expressed in hair cells as shown by in situ hybridization. Note that there is a 2-day delay between Neurog1 and Atoh1 upregulation in prosensory cells of the ear compared to the earliest expression of Atoh1 in postmitotic, differentiating hair cells. The upregulation of Neurod1 in sensory neurons is driven by Neurog1 whereas the upregulation of Neurod1 in hair cells is not, and appears in Neurog1-null mice. All start points of upregulation require verification with PCR to determine the exact dates (compiled from references [10, 15, 17, 19])
A further complication of these feed-forward and feedback loops of the three mammalian inner ear bHLH genes, outlined above, is the presence of additional bHLH genes. Nhlh2 (Nscl2) cooperates with Neurod1 in neuronal differentiation [17], but the function of Nhlh1 (Nscl1) in neuronal differentiation has not yet been fully explored. These bHLH genes are upregulated at E9 [10], whereas Atoh1 is upregulated approximately 2 days later [15] and after hair cells have exited the cell cycle (Fig. 4). After Atoh1 upregulation, upregulation of Nhlh1 and Neurod1 occurs in hair cells [14, 17], but both are expressed in areas of the otocyst that have been further specified much earlier in development [10, 11]. In addition, Neurod1 expression in hair cells is upregulated in the absence of Neurog1 [15] suggesting that either Atoh1 or an as yet unknown hair cell-specifying gene such as Sox2 [52] or Eya1 [53] can upregulate Neurod1 in the mammalian inner ear in addition to the well-established upregulation by Neurog1 [10].
What additional role Neurog1, Neurod1 and Nhlh1 play in hair cell development is unclear beyond the reported disruption of hair cell development in the apex of the cochlea [11, 13, 15] and near complete loss of all hair cells in the saccule [13]. Double-null mutations of Neurod1 and Nhlh1 have not yet been used to identify any effect on hair cells as shown for NeuroD1/Nhlh2 double-null mutants on neurons [17]. The fly NHLH ortholog (HLHC4 or CG3052) [26], is apparently expressed only in the embryonic CNS, making a role in development of the fly auditory system unlikely. However, functional characterization is lacking, and the role of Nhlh orthologs more broadly in neurosensory development, such as the olfactory system [54, 55], should be explored in invertebrates to allow a better understanding of the evolution of the interactions of these five bHLH genes that are now being recognized in vertebrate ear development (Figs. 3 and 4). Another gene recently identified in the developing ear is bHLHb5 [56]. However, this gene is not yet fully characterized in terms of its expression profile and its function in the existing null mutations [57]. How this bHLH gene interacts with the other thus far characterized bHLH genes in the developing ear remains unexplored.
Assuming that the basic principle of insect epidermal neurosensory development indeed reflects the same process across all triploblasts, it remains unclear how the changes needed to form the vertebrate developmental module were implemented. We previously based our assumption of the addition of another round of division on two arguments [9]. First, Neurod1 was known only for vertebrates and thus may have evolved to allow neuronal differentiation out of the prosensory domain. Second, most invertebrates with few exceptions have only neurosensory cells much like flies with only few showing segregation of sensory neurons specialized for information conduction and sensory cells specialized for sensory acquisition. In light of the recent suggestion that flies may have secondarily lost the NeuroD family of genes [24], it is more parsimonious to assume that the vertebrate developmental module, with the formation of both sensory neurons and sensory cells out of the prosensory anlage, has evolved early in triploblasts, and flies have selectively lost the molecular ability to form specialized sensory neurons that was variably retained in other lineages. A critical test of this hypothesis would be to study the molecular basis of neurosensory development in cephalopod statocysts. These organs show a mix of neurosensory cells next to distinct sensory cells and sensory neurons [9], and could show that NeuroD family members play an equivalent role to that in vertebrate ear development. Alternatively, recruiting Neurod1 to the neurosensory development of the vertebrate ear is unique to vertebrates, and NeuroD family members function very differently in the triploblast lineages that have those bHLH genes (Fig. 2a). Indeed the expression of NeuroD in trochozoans shows dramatic differences from that in vertebrates indicating an entirely different function. Unfortunately, this study [24] did not experimentally investigate the role of either Neurogenin or NeuroD in sensory organ development of the annelid Nereis, an important piece of information needed to evaluate the scenario outlined above. In light of these uncertainties it seems most parsimonious to assume that NeuroD evolution happened in ancestral triploblasts, but that evolution of the crucial incorporation of NeuroD1 downstream of Neurog1 to mediate neuronal differentiation of neuronal and suppress hair cell differentiation in neuronal precursors is a vertebrate novelty.
One of the predictions of our original hypothesis was that at least some hair cells and neurons should share a lineage and possibly have a clonal relationship (Figs. 1 and 3). Initially this idea was based on neurotrophin ligand expression in both developing sensory epithelia and delaminating sensory neurons that showed the topology of neuronal delamination near sensory epithelia [51]. This idea was further supported by findings that absence of Neurog1 results in loss of hair cells in the utricle, saccule and cochlea [12, 13] and the premature cell cycle exit of hair cells [15]. In addition, the cochlea is also truncated in Neurod1-null mice [11, 14], a gene downstream of Neurog1, and Neurod1 is coexpressed with Atoh1 in sensory hair cells [16]. Using an inducible Neurog1-Cre line for lineage tracing, this idea was further supported for the cochlea [58] and many vestibular hair cells [16], and a clonal relationship was proven for some neuron/hair cells in chicken development [59].
While all these data show an intimate relationship between neuronal and sensory development, including general otic epithelium or potentially prosensory domains such as the greater epithelial ridge next to the organ of Corti, the detailed relationships and whether the various feedback loops operate within lineage-related cells or are mediated by the Delta/Notch system of lateral inhibition remain to be demonstrated. Combined, these data give strong support to the idea that the evolution of neurosensory development can be reconstructed through molecular analysis of the ear neurosensory unit: the formation of a sensory neuron and a sensory hair cell via the molecular interactions of five distinct bHLH genes.
Recent data on zebrafish inner ear development have demonstrated yet another possible way that bHLH genes interact in ear development. The zebrafish atoh1a and atoh1b are used for the specification of hair cell precursors and their differentiation, and are expressed before neurog1 in the prosensory domain of the otic placode [32, 60]. Consistent with the complete independence of neurog1 and atoh1a/b expression is the fact that no alterations of hair cell development in the lateral line have been reported in zebrafish after knock-down of neurog1 [60]. Despite these apparent alterations, the regulation of neurod1 by neurog1 is maintained, and it is unclear if the duplication of atoh1 in zebrafish ancestors has resulted in these differences, or if the zebrafish inner ear development reflects the ancestral vertebrate state whereas mouse hair cells are derived. Data on lamprey are needed to establish an outgroup comparison to establish the timing of expression of these genes and to help understand if the inability of mammals to regenerate hair cells is related to the apparently delayed upregulation of Atoh1 in postmitotic hair cell precursors [15] instead of proliferating sensory precursors as in the zebrafish [32].
Aggregations of sensory precursors to form localized concentrations of prosensory precursor cells, the placodes in insects, cephalopods and vertebrates
Irrespective of these gaps in our knowledge, we are beginning to understand at least some factors that are important in coordinating the transformation of entire epithelial areas to become sensory arrays or a centralized nervous system. The assumption here is that the basic molecular mechanism transforming generalized epidermal cells into single sensory cells is orchestrated to happen in a larger, continuous set of cells. These epidermal cells, triggered simultaneously to clonally expand to generate the cellular material for large sensory arrays, are recognized in many animals and are uniformly referred to as placodes, which are thickenings of the epidermis, that ultimately leads to the formation of a large sensory organ [3]. Only in vertebrates is the molecular basis for such placode formation understood [4, 61], but many molecules that are known to be important in vertebrate placode specification are also found in nonvertebrates.
Many of the genes that are originally expressed in the otic placode function as protooncogenes and stimulate proliferation. For example, the protooncogene c-Myb is expressed in the developing chicken placode and it has been suggested that it is essential for the enhanced proliferation of placodal tissue [62]. Expression of these genes that are essential for upregulation of the proliferative capacity of ear progenitor cells need to be investigated in other animals with placodal formation such as flies and cephalopods to elucidate the molecular mechanisms.
Beyond upregulation of proliferation, the developing ear will be patterned by a number of diffusible and locally upregulated factors that combine to specify the axes of the developing ear and the areas of neurosensory formation (Fig. 1). The most important transcription factors for such placodal organization are factors that specify a unique area of epidermis and continue the enhanced proliferation of the otic placode to form the otocyst that grows into the ear. Chief among these factors are Eya1/Six1, Pax 2/8, Pou4f3, Sox2, Gata3, cMyb, Foxi1, Foxg1, Tbx1 and Fgf. Expression and mutational analysis has shown that these genes alone, and certainly in combination, are necessary for placodal formation with respect to both topology and initial upregulation of proliferation as well as neurosensory determination [3, 4, 53, 61]. How these genes interact, what their epistatic situation is, and how they activate the downstream bHLH genes in a possibly fairly restricted area (Fig. 1) are still under active investigation, and this interaction may not be conserved across vertebrates [20, 32].
Summary and conclusion
In this review we revisited a hypothesis that sought to explain the development and evolution of the vertebrate inner ear as an expansion of an ancestral neurosensory molecular developmental module. It is fair to say that the basic conclusion of our 10-year-old hypothesis has been extensively supported by some data but has also been modified by other data. Support comes from the molecular overlap of bHLH genes in sensory neurons and hair cells, the lineage relationship of some sensory neurons with hair cells and the coexpression of some bHLH genes in sensory neurons and hair cells. Unexpected differences have appeared between different vertebrates that require further investigation to establish the ancestral status. Future molecular dissection of this increasingly complex developmental module will require analysis of mechanosensory development in invertebrates with the basic complement of vertebrate bHLH gene families and expansion of the analysis of vertebrate ear development through selective misexpression to test the predictions of functional conservation across bHLH genes. Novel techniques and an expanded understanding of the comparative development of expression profiles will provide further progress toward a complete picture of molecular mechanisms of vertebrate mechanosensory development. Such understanding is needed for the most beneficial use of these insights to drive translational research into restoration of hearing loss in humans while minimizing risk associated with any genetic treatment.
Acknowledgments
This work was in part supported by a NIH-NIDCD grant (R01-DC005590).
References
- 1.Fritzsch B, Pauley S, Feng F, Matei V, Nichols DH. The evolution of the vertebrate auditory system: transformations of vestibular mechanosensory cells for sound processing is combined with newly generated central processing neurons. Int J Comp Psychol. 2006;19:1–24. [Google Scholar]
- 2.Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritzsch B, Beisel KW, Pauley S, Soukup G. Molecular evolution of the vertebrate mechanosensory cell and ear. Int J Dev Biol. 2007;51:663–678. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–472. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- 5.Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Kawauchi S, Beites CL, Crocker CE, Wu HH, Bonnin A, Murray R, Calof AL. Molecular signals regulating proliferation of stem and progenitor cells in mouse olfactory epithelium. Dev Neurosci. 2004;26:166–180. doi: 10.1159/000082135. [DOI] [PubMed] [Google Scholar]
- 7.Maklad A, Fritzsch B. Development of vestibular afferent projections into the hindbrain and their central targets. Brain Res Bull. 2003;60:497–510. doi: 10.1016/S0361-9230(03)00054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 9.Fritzsch B, Beisel KW, Bermingham NA. Developmental evolutionary biology of the vertebrate ear: conserving mechanoelectric transduction and developmental pathways in diverging morphologies. Neuroreport. 2000;11:R35–R44. doi: 10.1097/00001756-200011270-00013. [DOI] [PubMed] [Google Scholar]
- 10.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/S0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 11.Liu M, Pereira FA, Price SD, Chu MJ, Shope C, Himes D, Eatock RA, Brownell WE, Lysakowski A, Tsai MJ. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, Beisel KW, Wang VY. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, Johnson JE, Segil N, Groves AK. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- 17.Kruger M, Schmid T, Kruger S, Bober E, Braun T. Functional redundancy of NSCL-1 and NeuroD during development of the petrosal and vestibulocochlear ganglia. Eur J Neurosci. 2006;24:1581–1590. doi: 10.1111/j.1460-9568.2006.05051.x. [DOI] [PubMed] [Google Scholar]
- 18.Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, Kelley MW. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahan I, Kersigo J, Pan N, Fritzsch B (2010) Neurod1 regulates survival and formation of connections in the mouse ear and brain. Cell Tissue Res (in press) [DOI] [PMC free article] [PubMed]
- 20.Fritzsch B, Beisel KW, Hansen LA. The molecular basis of neurosensory cell formation in ear development: a blueprint for hair cell and sensory neuron regeneration? Bioessays. 2006;28:1181–1193. doi: 10.1002/bies.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 22.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 23.Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Res C Embryo Today. 2009;87:249–262. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simionato E, Kerner P, Dray N, Le Gouar M, Ledent V, Arendt D, Vervoort M. atonal- and achaete-scute-related genes in the annelid Platynereis dumerilii: insights into the evolution of neural basic-Helix-Loop-Helix genes. BMC Evol Biol. 2008;8:170. doi: 10.1186/1471-2148-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens JD, Roalson EH, Skinner MK. Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: genomic approach to cellular differentiation. Differentiation. 2008;76:1006–1022. doi: 10.1111/j.1432-0436.2008.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 29.Powell LM, Deaton AM, Wear MA, Jarman AP. Specificity of Atonal and Scute bHLH factors: analysis of cognate E box binding sites and the influence of senseless. Genes Cells. 2008;13:915–929. doi: 10.1111/j.1365-2443.2008.01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakada Y, Parab P, Simmons A, Omer-Abdalla A, Johnson JE. Separable enhancer sequences regulate the expression of the neural bHLH transcription factor neurogenin 1. Dev Biol. 2004;271:479–487. doi: 10.1016/j.ydbio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 32.Millimaki BB, Sweet EM, Dhason MS, Riley BB. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development. 2007;134:295–305. doi: 10.1242/dev.02734. [DOI] [PubMed] [Google Scholar]
- 33.Goulding SE, zur Lage P, Jarman AP. Amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/S0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 34.Goulding SE, White NM, Jarman AP. cato encodes a basic helix-loop-helix transcription factor implicated in the correct differentiation of Drosophila sense organs. Dev Biol. 2000;221:120–131. doi: 10.1006/dbio.2000.9677. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- 36.Wang VY, Hassan BA, Bellen HJ, Zoghbi HY. Drosophila atonal fully rescues the phenotype of Math1 null mice: new functions evolve in new cellular contexts. Curr Biol. 2002;12:1611–1616. doi: 10.1016/S0960-9822(02)01144-2. [DOI] [PubMed] [Google Scholar]
- 37.Maung SM, Jarman AP. Functional distinctness of closely related transcription factors: a comparison of the Atonal and Amos proneural factors. Mech Dev. 2007;124:647–656. doi: 10.1016/j.mod.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hufnagel RB, Riesenberg AN, Saul SM, Brown NL. Conserved regulation of Math5 and Math1 revealed by Math5-GFP transgenes. Mol Cell Neurosci. 2007;36:435–448. doi: 10.1016/j.mcn.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- 40.zur Lage PI, Powell LM, Prentice DR, McLaughlin P, Jarman AP. EGF receptor signaling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev Cell. 2004;7:687–696. doi: 10.1016/j.devcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Holohan EE, zur Lage PI, Jarman AP. Multiple enhancers contribute to spatial but not temporal complexity in the expression of the proneural gene, amos. BMC Dev Biol. 2006;6:53. doi: 10.1186/1471-213X-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/S0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 43.Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132:2709–2719. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan N, Jahan I, Lee JE, Fritzsch B. Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 2009;337:407–428. doi: 10.1007/s00441-009-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray SK, Leiter AB. The basic helix-loop-helix transcription factor NeuroD1 facilitates interaction of Sp1 with the secretin gene enhancer. Mol Cell Biol. 2007;27:7839–7847. doi: 10.1128/MCB.00438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- 47.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 48.Ghysen A, Dambly-Chaudiere C. A genetic programme for neuronal connectivity. Trends Genet. 2000;16:221–226. doi: 10.1016/S0168-9525(99)01969-1. [DOI] [PubMed] [Google Scholar]
- 49.Finger TE. Evolution of taste and solitary chemoreceptor cell systems. Brain Behav Evol. 1997;50:234–243. doi: 10.1159/000113337. [DOI] [PubMed] [Google Scholar]
- 50.Rasmussen SL, Holland LZ, Schubert M, Beaster-Jones L, Holland ND. Amphioxus AmphiDelta: evolution of Delta protein structure, segmentation, and neurogenesis. Genesis. 2007;45:113–122. doi: 10.1002/dvg.20278. [DOI] [PubMed] [Google Scholar]
- 51.Fritzsch B, Beisel KW, Jones K, Farinas I, Maklad A, Lee J, Reichardt LF. Development and evolution of inner ear sensory epithelia and their innervation. J Neurobiol. 2002;53:143–156. doi: 10.1002/neu.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 53.Zou D, Erickson C, Kim EH, Jin D, Fritzsch B, Xu PX. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum Mol Genet. 2008;17:3340–3356. doi: 10.1093/hmg/ddn229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki Y, Tsuruga E, Yajima T, Takeda M. Expression of bHLH transcription factors NSCL1 and NSCL2 in the mouse olfactory system. Chem Senses. 2003;28:603–608. doi: 10.1093/chemse/bjg051. [DOI] [PubMed] [Google Scholar]
- 55.Ruschke K, Ebelt H, Kloting N, Boettger T, Raum K, Bluher M, Braun T. Defective peripheral nerve development is linked to abnormal architecture and metabolic activity of adipose tissue in Nscl-2 mutant mice. PLoS One. 2009;4:e5516. doi: 10.1371/journal.pone.0005516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunelli S, Innocenzi A, Cossu G. Bhlhb5 is expressed in the CNS and sensory organs during mouse embryonic development. Gene Expr Patterns. 2003;3:755–759. doi: 10.1016/S1567-133X(03)00135-2. [DOI] [PubMed] [Google Scholar]
- 57.Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, Jung C, Hu L, Mok SI, Shah A, Savner EM, Tolias C, Corfas R, Chen S, Inquimbert P, Xu Y, McInnes RR, Rice FL, Corfas G, Ma Q, Woolf CJ, Greenberg ME. Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron. 2010;65:886–898. doi: 10.1016/j.neuron.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koundakjian EJ, Appler JL, Goodrich LV. Auditory neurons make stereotyped wiring decisions before maturation of their targets. J Neurosci. 2007;27:14078–14088. doi: 10.1523/JNEUROSCI.3765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satoh T, Fekete DM. Clonal analysis of the relationships between mechanosensory cells and the neurons that innervate them in the chicken ear. Development. 2005;132:1687–1697. doi: 10.1242/dev.01730. [DOI] [PubMed] [Google Scholar]
- 60.Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- 61.Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- 62.Leon Y, Miner C, Represa J, Giraldez F. Myb p75 oncoprotein is expressed in developing otic and epibranchial placodes. Dev Biol. 1992;153:407–410. doi: 10.1016/0012-1606(92)90126-2. [DOI] [PubMed] [Google Scholar]