Abstract
The human body is colonized by a vast array of microbes, which form communities of bacteria, viruses and microbial eukaryotes that are specific to each anatomical environment. Every community must be studied as a whole because many organisms have never been cultured independently, and this poses formidable challenges. The advent of next-generation DNA sequencing has allowed more sophisticated analysis and sampling of these complex systems by culture-independent methods. These methods are revealing differences in community structure between anatomical sites, between individuals, and between healthy and diseased states, and are transforming our view of human biology.
The microbes that exist in the human body are collectively known as the human microbiota. This amazingly complex and poorly understood group of communities has an enormous impact on humans. An increasing number of conditions are being examined for correlative and causative associations with the microbiome — which, in this Review, is used to refer to the microbiota and the habitat it colonizes (Box 1). Each one of the many microbial communities has its own structure and ecosystem, depending on the body environment it exists in. The fundamental goal of human microbiome research is to measure the structure and dynamics of microbial communities, the relationships between their members, what substances are produced and consumed, the interaction with the host, and differences between healthy hosts and those with disease.
BOX 1. Terminology.
Biodiversity is a measure of the complexity of a community. It is affected by the number of taxa (richness) and their range of abundance (evenness). High biodiversity occurs when many taxa (high richness) are present at similar abundances (an even distribution).
Commensals are organisms that benefit from another organism but that have no harm or benefit themselves. Microbes of the microbiome were thought to be commensals that benefited from the human host but did no harm. Many of these organisms provide benefits to the human host and so have a mutualistic relationship.
Contig is a stretch of contiguous sequence in a genome assembly.
Coverage is the number of times a genome or gene is sequenced. In a genome sequenced to coverage, each nucleotide in the sequence appears, on average, in 100 reads.
Genome assembly is the process of constructing a genome sequence from short subsequences by sequencing many random fragments from a sheared genome. The random short sequences are compared, and overlapping common sequences are used to determine their orientation and order with respect to each other. A consensus sequence is constructed from this layout. Usually there are gaps, but when contigs can be arranged in the correct order and orientation, these longer stretches are called scaffolds.
Metagenomics was defined83 as a process for identifying genes specifically by their function by cloning them directly from the environment and expressing genes in a surrogate host84. Therefore, gene function was known even if the sequence was not sufficient for functional inference, such as when it encoded a protein of previously unknown function. This definition, also known as functional metagenomics, is widely used. More recently, metagenomics refers to general analyses of microbial communities by culture-independent methods, which do not necessarily focus on function. The combined genomes of the microbes in a community are thought of as the community metagenome. Another type of metagenomic analysis focuses on the structure of these aggregate genomes in a community.
Microbiome in this Review refers to the microbiota and the habitat it colonizes and is analogous to the term biome in ecology. Microbiome is also used to refer to the collective genomes of the microbes — what is now the metagenome, and may have originally been coined by Joshua Lederberg (cited by Hooper and Gordon85). However, it is also used for the more ecologically consistent meaning. A microbiome can be a specific body site, such as the gut microbiome, but the human microbiome is often used to refer to the collection of microbiomes of the human body.
Mutualism is a type of symbiosis in which both organisms benefit. This is one type of relationship seen in the human microbiome.
Operational taxonomic unit in microbiome research is a group of organisms with 16S ribosomal RNA gene sequences that show a certain level of identity. This group is often used as a surrogate for a species when the 16S rRNA sequences are at least 97% identical.
Pathogenic microbe is one with the potential to cause disease.
Read is the primary output of DNA sequencing, consisting of a short stretch of DNA sequence that is produced from sequencing a region of a single DNA fragment.
Shotgun sequencing is the process of randomly breaking (often by shearing) a long DNA molecule (for example, a complete chromosome) and then sequencing the resultant DNA fragments, which each come from a different location in the original long DNA molecule.
Virome is the collection of viruses in the microbiota.
Despite an explosion in human-microbiome research, these communities are still the dark matter of the body. The microbiome has been called another organ1–4 because of its products, its responsiveness to the environment and its integration with other systems. Sometimes referred to as our second genome5, the genes of microbes that make up the microbiome outnumber human genes by more than 100-fold, with over 3 million bacterial genes in the gut alone6,7. These extensive microbial ecosystems are not limited to the human body. Microbes and their communities dominate the environment and occupy a vast range of niches. Environmental metagenomics was developed extensively before being applied to the human body8,9, and methods from other disciplines have had a significant effect on human-microbiome research. Defining complicated microbial ecosystems and developing tools to probe their workings is an important research enterprise of twenty-first century microbiology.
The complexity of microbial communities makes studying them challenging. There may be hundreds of different species, and enumerating what organisms are present with standard microbiological techniques is not possible because many organisms have never been grown in culture and may require special, as yet unknown, growth conditions. In addition, the abundance of some microbes can range over orders of magnitude, so deep sampling is required to detect the less-abundant members. Culture-independent methods of taking a microbial census began about 25 years ago and were based on targeted sequencing of 5S and 16S ribosomal RNA genes10, which differ for each species and are a convenient identifier. As this became a tractable research area, next-generation sequencing (NGS) technologies (Table 1) were developed and allowed more extensive analyses, both targeted 16S rRNA gene sequencing and whole-genome shotgun sequencing of microbes in communities en masse. The number of culture-independent metagenomic investigations of the human microbiome has mushroomed, and it is one of the most studied areas of microbiology with significant potential to benefit clinical practice. This culture-independent methodology is broadly applied outside human-microbiome research and is expanding our knowledge of the environment. This Review describes how NGS approaches are transforming human-microbiome studies, and posing questions and challenges for the future.
Table 1.
DNA sequencing platforms used for microbiome analysis
| Platform | Method | Characteristics | 16S rRNA | Shotgun | Comments |
|---|---|---|---|---|---|
| Established
| |||||
| Sanger-based or capillary-based instrument | Fluorescent, dideoxy terminator | 750-base reads High accuracy |
Full length sequenced with 2–3 reads | Long reads help with database comparisons | Most costly method Relatively low throughput, so low coverage of 16S or shotgun |
| Roche–454 | Pyrosequencing light emission | 400-base reads | Up to 3 variable regions per read | Long reads help with database comparisons | Cost limits shotgun coverage but 16S coverage is good |
| Illumina | Fluorescent, stepwise sequencing | 100–150-base reads | Only 1 variable region per read | Short reads do not seem to limit analysis | Very high coverage owing to high instrument output and very low cost |
|
| |||||
| Not yet widely used
| |||||
| Ion Torrent | Proton detection | More than 200-base reads | Like other NGS | Like Illumina | Expect high coverage, but longer reads than Illumina |
| PacBio | Fluorescent, single-molecule sequencing | Up to 10- kilobase reads Low accuracy |
Accuracy an issue for correct taxon identification | Long reads could help assembly | Attractive for long reads, but lower accuracy limits applications |
| Oxford Nanopore* | Electronic signal as DNA passes through pore Single-molecule sequencing |
Long reads | Unknown | Long reads could help assembly | Not yet available |
At the time of publication, the Oxford Nanopore system was not available, and information provided is based on company presentations. Ion Torrent and PacBio are both available but have not been widely used for microbiome analysis. The Illumina MiSeq instrument is expected to provide 250-base reads in the near future.
Single organisms and microbial communities
In the past, research on microbial interactions with humans has focused on single pathogenic organisms. Studies of communities of non-pathogenic microbes in the body were limited because the organisms were thought to be benign, with minor effects on human health compared with pathogens. Microbiome research has led to new interest in the communities of non-pathogenic microbes that inhabit the human body, and the need to describe the genomes of these organisms to understand the human microbiome has been recognized.
Every community of the microbiome has its own characteristics (Table 2). For the gut community, for example, high biodiversity is associated with a healthy state and reduced biodiversity occurs in patients with conditions such as Crohn’s disease11, whereas for tissues of the vagina, a lower biodiversity exists in healthy individuals and a bloom of organisms occurs in patients with vaginosis12. To understand why different sites have different properties, the mechanisms that lead to the disruption of ecosystems and to disease, and exceptions to generalities about a tissue, researchers require knowledge of the structure and behaviour of microbial communities.
Table 2.
Characteristics of bacteria, microbial eukaryotes and viruses in the human microbiome
| Characteristic | Bacteria | Viruses | Eukaryotic microbes |
|---|---|---|---|
| Genome size | 0.5–10 megabases | 1–1,000 kilobases | 10–50 megabases |
| Number of taxa in the human microbiome | At least thousands | Unknown, but could be as many as bacteria | Unknown, but may be fewer than bacteria |
| Relative abundances | Highly variable | Highly variable | Unknown |
| Targeted detection methods | Sequencing of genes such as 5S and 16S rRNA | No universal method for genes, but virus-specific polymerase chain reaction assays for some | Sequencing of 18S rRNA gene Spacer region in rRNA |
| Shotgun approach to analyses | Alignment to reference genomes or database comparison | Database comparison | Alignment to reference genomes or database comparison |
| Subspecies or strain diversity | Modest sequence variation Horizontal gene transfer also contributes |
High sequence variation | Unknown |
Microbial communities benefit the host by providing functions such as digestion of nutrients13 or protection against infection14. Antibiotic treatment perturbs the microbiome15,16 by reducing its size and altering its composition. This disturbance can lead to infection17–19, and antibiotic-resistant organisms such as Clostridium difficile — normally controlled by the microbiome — can overgrow and create problems20. More complex community contributions also exist, such as interactions with host immune and inflammatory systems21,22 or production of metabolites involving hybrid pathways from multiple organisms, including host–microbe pathways23. Understanding these phenomena will ultimately allow the microbiome to be manipulated so that, for example, transplants of microbial communities could treat C. difficile infections24,25.
Whether the microbial ecology of the human body can be simplified to the properties of single organisms is unknown. Many organisms have never been cultured and may be adapted to life in a community environment rather than a pure culture. For organisms for which growth requirements are understood, there is a dependence on secreted products from other community members. For example, secreted siderophores26 are small molecules that help microbes to scavenge iron, which is a limiting factor for growth in the body. So even the study of individual organisms can be dependent on studying the community.
Dissecting a microbiome
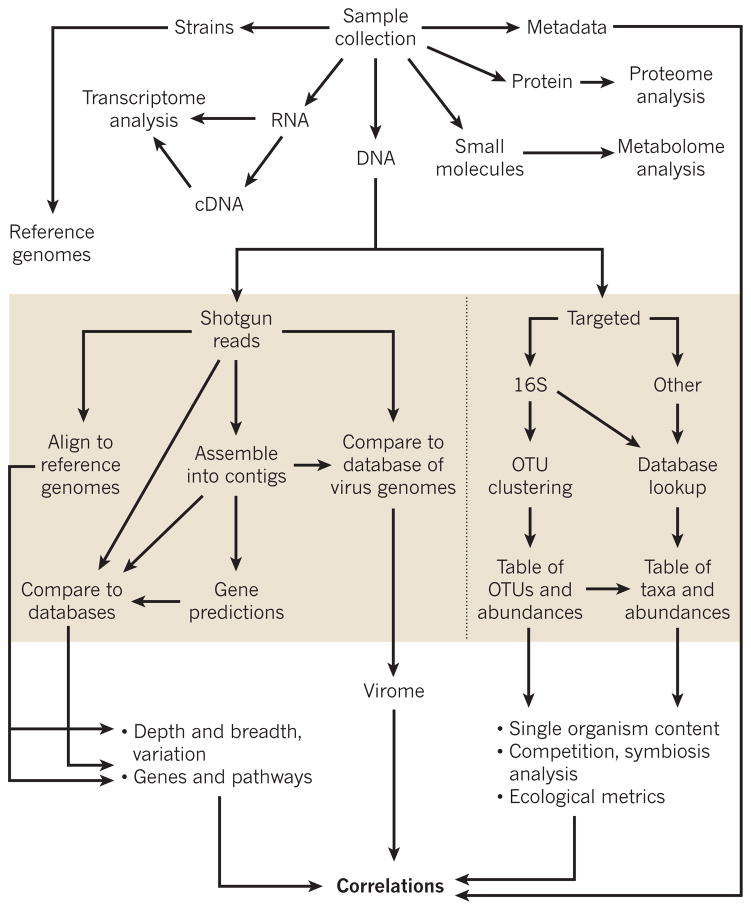
Analysis of community structure (Fig. 1) focuses on either targeted regions (such as the 16S rRNA gene) or shotgun sequencing to catalogue the genes that are present. Additional analysis involves sequencing genomes of individual organisms to produce a catalogue of reference genomes27, and analysing RNA to describe the transcriptome and identify RNA viruses. Non-genomic analyses include proteomic and metabolomic studies, but these are not discussed here. Every sample should be well-annotated with clinical metadata, so that, ultimately, the microbiome’s genetic and community structures can be correlated with the individual’s phenotype.
Figure 1. Data and analysis workflow for microbiome analysis.
From a microbiota sample, DNA, RNA and protein can be extracted, and metadata and strains of bacteria obtained. Data from DNA can be supplemented with proteome and transcriptome analysis. During primary analysis, shotgun techniques can produce reads from DNA, which are then aligned to reference genomes to identify variants and community population genetics, assembled into contigs to make gene predictions or compared with databases. Alternatively, targeted sequencing such as 16S rRNA gene sequencing can be used to take a community census, and these data are then compared with databases to create tables of taxa and abundance, or analysed with software programs to cluster the reads into OTUs to create tables of abundance. The derivative data is used in secondary analysis for ecological metrics or competition and symbiosis analysis. In addition, shotgun reads and comparisons with reference genomes and databases can be used to build pathways and reconstruct the capabilities of a community. The combination of these analyses will contribute to understanding the differences within and between individuals.
Census of organisms
Modern metagenomic analyses of microbial communities were developed from culture-independent methods for taking a census of organisms present in a community and their abundances. Although DNA reassociation kinetics provides information on community diversity and structure28, there is no accounting for organisms that may be tracked between samples. Methods more useful for providing information on the entire structure often focus on signature sequences that distinguish taxa (detected by hybridization to arrays of diagnostic oligonucleotides29), various methods for fingerprinting polymerase chain reaction (PCR) products (such as single-strand conformation polymorphisms or terminal restriction fragment length polymorphisms) or DNA sequencing of targeted PCR products. Sequencing of 16S rRNA genes is the main method of taking a community census because fingerprinting methods do not adequately measure low-abundance organisms30.
16S rRNA differs for each bacterial species. A bacterial species is hard to define, but is often taken as organisms with 16S rRNA gene sequences having at least 97% identity — an operational taxonomic unit (OTU). A 16S rRNA gene sequence of about 1.5 kilobases has nine short hypervariable regions that distinguish bacterial taxa; the sequences of one or more of these regions are targeted in a community census.
Before the introduction of NGS methods, the prevailing approach was to clone full-length 16S rRNA genes after PCR with primers that would amplify genes from a wide range of organisms. Cloned 16S rRNA genes were sequenced by the Sanger method, which required two or three reads to cover the entire gene. Accuracy was crucial because sequencing errors led to misclassification. The cost and effort required for the Sanger method limited the depth of sampling, and studies often produced about 100 sequences per specimen. This method identified the dominant organisms in a community, but analysis of less abundant organisms was limited.
Introducing NGS to 16S rRNA gene analysis led to marked improvements in cost and depth of sampling. The Roche–454 platform has dominated microbial community analysis31. As the read length for 454 pyrosequencing is about 400 bases, only a portion of the 16S rRNA gene can be sampled, and many different studies have targeted between one and three of the hypervariable regions, with different hypervariable regions targeted in different studies. Using a portion of the 16S rRNA gene led to a loss of sensitivity (some taxa cannot be reliably defined at the species level, although high confidence identification of higher taxonomic ranks is possible), nevertheless gains in depth of sampling and cost savings outweigh this caveat. The US Human Microbiome Project (HMP)32 has sequenced more than 10,000 specimens from healthy adults on the 454 platform by targeting V3 to V5 regions in the 16S rRNA gene and producing, on average, 7,000 sequences per specimen33, which is a vast expansion on the Sanger method of sequencing analysis. The results of the HMP, which sampled 18 body sites, provide an in-depth definition of the human microbiome. Another study16 that focused on the effects of the antibiotic ciprofloxacin reported the ‘rare biosphere’ in the gut. This study documented perturbation of taxa and recovery from antibiotic treatment, as well as minor constituents that did not recover after antibiotic treatment. Such analyses will be important in identifying individuals who are at risk of side effects from antibiotic treatment, for example overgrowth of pathogens such as C. difficile or life-threatening antibiotic-associated diarrhoea.
When using 16S rRNA gene sequencing to compare individuals it is not necessary to know which organisms are present, only whether the spectra of 16S rRNA gene sequences are similar and the degree of difference between samples. Projects that compare healthy cohorts and those with disease to determine whether there is a difference in the microbiome, or examine the effects of diet, antibiotic treatment or environmental factors on the microbiome, all focus on detecting differences in communities, rather than identifying actual taxa. A loss of sensitivity for organism identification can be tolerated, and NGS allows cost-effective deep sampling of large cohorts, which is needed to reach statistically significant conclusions. The Illumina sequencing platform has been applied to metagenomics projects34–36, but because this sequencing platform currently produces reads of 100 bases (HiSeq system) to 150 bases (MiSeq system), only a single hypervariable region can be sequenced. However, this further loss of sensitivity does not preclude the use of the Illumina platform for the comparative projects already described in this Review. An early application of this platform was its use in a study of vaginal microbiomes in patients with HIV, for which comparisons of patients with conditions such as vaginosis before and after antibiotic therapy were examined37. As a result of the exceptional increases in numbers of reads and the lower cost associated with the Illumina platform, it is becoming more widely used for 16S rRNA gene-sequence profiling and continues the microbiome-analysis trend of deeper sampling at lower costs.
Shotgun sequencing for cataloguing organisms
Targeted sequencing is a powerful tool for assessing the organisms that are present in microbial communities, but it is limited in terms of the functional and genetic information produced. Organisms for which the genome sequences are known (currently there are several thousand sequenced bacterial genomes) can be used to infer the genes and functional capabilities of the community (Fig. 1). However, many organisms have no reference sequence. Furthermore, a reference sequence does not completely describe the genes that are contributed by an organism. There is considerable variation in the genomes between strains of the same species. Two strains of Escherichia coli, O157:H7 and K-12, both have 16S rRNA gene sequences of E. coli, but differ in hundreds of genes. There are limits to what can be learned about the genetic content of communities from 16S rRNA gene sequences alone.
Moving beyond this level of functional inference requires a gene-based census. This catalogue of genes can be provided by shotgun sequencing of DNA that has been extracted from the community as a whole and samples the mixture of genomes that make up the metagenome (Fig. 1). In a community in excess of hundreds of species with varying abundance, deep sequencing is needed to sample minor constituents that are not necessarily unimportant. The bacterial concentration in the gut can be 1011 cells ml−1 (refs. 38, 39), so for an organism that is present at a concentration of 1 per 106 there are 105 cells ml−1, which is sufficient for the organism’s products, such as metabolites and toxins, to have an effect on the community and the host.
Illumina sequencing of faecal samples produced 4 gigabases per sample and 10 Gb per sample in the Metagenomics of the Human Intestinal Tract (MetaHIT)6 and HMP33 projects, respectively, which corresponded to tens of millions of reads per sample. At this depth of sequencing, the genomes of minor constituents such as E. coli (with an abundance of about 1% or lower) are sampled almost completely, and organisms with an even lower abundance have some of their genome represented. This extraordinary sampling of complex microbial communities is made possible by producing large amounts of data and by the low cost of NGS methods.
Shotgun sequence data, in addition to 16S rRNA gene analysis, provide information on the organisms that make up communities. Extracting 16S rRNA gene sequences from shotgun reads to determine the organisms present is possible; however, targeted 16S rRNA gene sequencing tends to introduce biases (owing to the broad-range PCR used to amplify 16S rRNA gene sequences or the choice of region within the 16S rRNA gene), which shotgun sequencing does not. Shotgun sequencing is less sensitive than targeted rRNA sequencing because a small fraction of the sequences are from 16S rRNA genes. Another approach is to align shotgun sequences to bacterial reference genomes33,40,41, allowing the relative abundance of species to be determined on the basis of the number of reads that align to each reference genome (also useful for the comparative studies already described). The MetaHIT project has used this approach to classify individuals into different groups, called enterotypes, on the basis of the community structure in their faecal samples40. The same enterotypes have been found in 16S rRNA gene-based analysis42. The vaginal microbiome has also been classified into five groups43. These observations suggest the human microbiome may exist in distinct states in different people, although correlation with environmental, genetic or health status is not yet clear. Stratifying future studies depending on which community class an individual belongs to may be important for identifying correlations with phenotypic data.
The need for reference genome sequences is clear both to infer genetic content of organisms identified by 16S rRNA genes and to identify sources of shotgun reads by aligning to reference genomes, and so determining organismal content of communities from shotgun data. NGS techniques have reduced the cost of bacterial sequences to less than US$1,000 per genome and led to an increase in the production of ‘complete’ genome sequences. Current methodology relies mainly on Illumina shotgun sequencing and a variety of methods to assemble the reads into a genome. The product is not a true complete genome, but a high-quality draft that covers almost all of the genome and results in a high-quality base sequence27. Programmes such as the HMP32,44 and the Genomic Encyclopedia of Bacteria and Archaea (GEBA)45 are producing reference genomes by the thousands.
Although bacteria are the main components of the human microbiome, eukaryotic microbes and viruses (both human viruses and bacteriophages) are also present (Table 2). The study of eukaryotic microbes is not as advanced as that of bacteria46, but the organisms are identified by signature sequences (such as fingerprinting and 18S rRNA) and shotgun sequencing analogous to bacteria. The number of reference genomes for eukaryotic microbes is smaller than that for bacteria, and progress will depend on addressing this shortfall.
By contrast, considerable effort is being given to characterizing the genomes of human viruses47 and bacteriophages48, known as the virome (Box 1). This work is based on shotgun sequencing (Fig. 1), although oligonucleotides microarrays for virus detection are also used49,50. Viral sequences can be detected in shotgun data from different body sites, and viruses can also be enriched by processing samples before DNA extraction51. Virome analysis by shotgun sequencing of microbial communities (discussed later) has led to the identification of human viruses52–54, as well as the detection of known viruses in healthy subjects and diseases of unknown aetiology55. Likewise, bacteriophages are found to be highly diverse at different body sites56–58, with differences between individuals as a result of diet59 or disease states60,61.
Sequencing for gene catalogues and functional inference
Metagenomic shotgun data also sample community gene content, which is useful to define community capabilities and identify particular members. Deep sequencing, such as that used in the MetaHIT and the HMP, broadly samples the genomes of even minor constituents, facilitating the identification of genes present within a given community (Fig. 1). By using the sequence reads themselves, or by first assembling them into contigs (Box 1), sequence data can be compared with databases such as the National Institutes of Health’s GenBank to identify which genes are present. De novo prediction of genes from metagenomic data is also possible33, which provides motifs for functional inference even if the sequence does not find a match in a database. Finally, alignment of reads or contigs to reference genomes identifies which organisms are present, along with their known gene content. These methods convert metagenomic sequence data into catalogues of genes that can be further analysed.
Gene catalogues can be compared with databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG)62, which sorts gene products into pathways and processes. Such analyses provides lists of pathways, identify which pathway genes are in the community and quantify the abundances of genes and pathways63. Comparing gene catalogues to specialized metabolic databases, such as the Carbohydrate-Active Enzymes database64, is also useful. Carbohydrate-degrading capabilities of communities differ between body sites, suggesting the carbohydrate spectrum of each body site has determined which organisms and pathways are present65.
In addition to pathway analysis, determining the presence and abundance of genes, such as antibiotic-resistance genes or virulence factors, in a community is possible using similar methods to those already described, and can shed light on pathogen burden in an individual and consequences of antibiotic treatment. The importance of functional analyses cannot be overemphasized, and functional properties of communities are thought to be more important than their taxonomic composition66.
Computational tools and strategies
Metagenomic data are a rich source of information for the sequencing and analysis methods already discussed67,68. The data analysis workflow has three phases. In the first phase, primary data are processed and filtered depending on the application. For 16S rRNA gene sequencing, the quality of analysis is important so that organisms are not misclassified. Initial processing addresses read quality, chimaerism (a read formed from different 16S rRNA genes), read length after removing low-quality bases and related issues69–73. For shotgun sequence data6,33 — in addition to sequence quality — artefacts such as duplicate reads must also be addressed, as well as computationally removing contamination from human sequences. Removal of human and bacterial sequences is important in read processing for virome analysis47,55 (Fig. 1).
Following production of processed reads, the second phase involves generating various derivative data sets. For 16S rRNA gene analysis, tables of taxa and abundance are produced by comparisons with 16S rRNA sequence databases or by using software packages to cluster the reads into OTUs74,75. Comparing shotgun reads to gene databases, such as GenBank or KEGG, by using the Basic Local Alignment Search Tool (BLAST), for example, produces lists of genes and the number of matched reads7,33,63. Alignment of reads to reference genomes produces tables of breadth and depth of coverage, by reads of each genome41. In each of these data sets, there is more biological information to be gleaned and added through further analysis. Not all reads match sequences in databases because not all organisms have a reference genome sequenced. In addition, reads may match genes whose function has not been elucidated. These sequences of unknown origin or function can be a sizeable fraction and the effect of this uninformative portion of data on analyses and conclusions is not clear.
The third phase of analysis uses these derivative data to produce trees or other representations of the similarity of communities, abundance curves, biodiversity plots, and other ecological and statistical descriptors of community structure74,75 (Fig. 1). A list of hits from BLAST is used to build metabolic pathways for reconstruction of community capabilities63. Alignments to reference genomes are further analysed for variants and population genetics of communities. Computational analysis can also be used to determine which organisms co-occur or rarely co-occur as evidence for symbiosis or competition, respectively, or to follow the dynamics of community structure in longitudinal time series76.
Some analyses pose significant computational challenges. Comparisons to gene databases at the protein level are particularly demanding because shotgun sequences must be translated into polypeptides in all six reading frames, and each must be compared with a gene database represented at the protein level. Using conventional BLASTx programs for this comparison in large data sets, such as the HMP, could take decades, so supercomputers, accelerated BLAST programs or both must be used33. A lack of efficient software and large enough computer clusters are often bottlenecks for metagenomic analysis, because sequencing and data production are not limiting factors. Management of large data sets and computing resources are receiving more attention, with cloud-computing services seeming to be a viable alternative77.
Future directions and challenges
The rapid rise in metagenomic studies has solved many problems but, as the field has grown, other questions have been raised. Existing methodology is becoming more sophisticated, and sequencing technology is making exponential advances (Table 1). The Illumina platform introduced instruments that were more appropriate for sequencing smaller genomes, with faster run times and longer read lengths, offering more flexibility for metagenomic applications. The long read length of the PacBio platform has the potential to help distinguish the reads from different organisms, which is a challenge for metagenomic shotgun sequencing. The technology produced by Oxford Nanopore promises long reads and short run times in a scalable system, and is therefore a good match for microbial applications. Reducing the amount of DNA needed for shotgun sequencing will allow communities in smaller anatomical regions, such as within the gastrointestinal tract, to be studied separately rather than together with other regions as is the case with the current methodology. Short run-time instruments and reductions in sample size will also hasten the introduction of microbiome analysis to the clinic, where analyses of patient samples must be quick and able to deal with limited amounts of material. Ultimately, the aim of human-microbiome research is its application as a diagnostic, therapeutic and preventive tool in the clinic.
The main limitation of using shotgun data is the large number of organisms that have not been cultured, let alone sequenced. These organisms are therefore under-represented in databases, and their shotgun reads are anonymous. When community shotgun data are assembled into genomes to obtain genome sequences for new organisms, contig sizes are typically small as a result of lower organism abundance and the challenges associated with assembly of a complex mixture. The long read lengths of PacBio and Oxford Nanopore instruments should help with these challenges, as will the development of assembly algorithms for metagenomic data. Expanding the catalogue of reference genomes by producing reference sequences for individual uncultured organisms is an active area. Methods that use cell sorting to isolate organisms, coupled with sequencing and assembly techniques for single-cell DNA preparations, are producing new genome sequences78,79 and, in high-throughput mode, could complement shotgun metagenomics for analysing communities.
One problem associated with genomic data is that it does not address whether an organism is alive or has succumbed to host defences or antibiotic treatment. However, the data can be complemented with transcriptome analysis, or proteomic and metabolomic data sets, which analyse gene expression and metabolic data that are more likely to be derived specifically from living cells.
The simultaneous advances in human genetics and genomics offer opportunities for combining studies of host genotype with microbiome phenotype. Methods for viewing the microbiome as a quantitative trait and relating this to host genotype are being developed80. Advances in host–microbiome studies are also coming from combining immunology and human-microbiome research81,82. Moreover, continued development of statistical methods in microbiome research, such as advances in power analysis, will aid experimental design and future analysis.
Acknowledgments
The author gratefully acknowledges generous support from the National Institutes of Health.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The author declares no competing financial interests.
Readers are welcome to comment on the online version of this article at go.nature.com/1oqsjw.
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B, Goldberg D, Murdock C, Xi C, Gilsdorf JR. Conceptualizing human microbiota: from multicelled organ to ecological community. Interdiscip Perspect Infect Dis. 2008;2008:613979. doi: 10.1155/2008/613979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Possemiers S, Bolca S, Verstraete W, Heyerick A. The intestinal microbiome: a separate organ inside the body with the metabolic potential to influence the bioactivity of botanicals. Fitoterapia. 2011;82:53–66. doi: 10.1016/j.fitote.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Shanahan F. The host–microbe interface within the gut. Best Pract Res Clin Gastroenterol. 2002;16:915–931. doi: 10.1053/bega.2002.0342. [DOI] [PubMed] [Google Scholar]
- 5.Bruls T, Weissenbach J. The human metagenome: our other genome? Hum Mol Genet. 2011;20:R142–R148. doi: 10.1093/hmg/ddr353. [DOI] [PubMed] [Google Scholar]
- 6.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. This paper presents initial findings on the gut microbiome from the MetaHIT project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. This paper presents analysis of data from the HMP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein JL, Marsh TL, Wu KY, Shizuya H, DeLong EF. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergin KL, et al. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl Environ Microbiol. 1998;64:3075–3078. doi: 10.1128/aem.64.8.3075-3078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 11.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 13.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 14.Srikanth CV, McCormick BA. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: a three-way crosstalk. Interdiscip Perspect Infect Dis. 2008;2008:626827. doi: 10.1155/2008/626827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobsson HE, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller CP, Bohnhoff M, Rifkind D. The effect of an antibiotic on the susceptibility of the mouse’s intestinal tract to Salmonella infection. Trans Am Clin Climatol Assoc. 1956;68:51–55. [PMC free article] [PubMed] [Google Scholar]
- 18.Sekirov I, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulligan ME. Epidemiology of Clostridium difficile-induced intestinal disease. Rev Infect Dis. 1984;6:S222–S228. doi: 10.1093/clinids/6.supplement_1.s222. [DOI] [PubMed] [Google Scholar]
- 21.Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol. 2011;23:353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsland BJ. Regulation of inflammatory responses by the commensal microbiota. Thorax. 2012;67:93–94. doi: 10.1136/thoraxjnl-2011-200750. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 25.Brandt LJ, Reddy SS. Fecal microbiota transplantation for recurrent clostridium difficile infection. J Clin Gastroenterol. 2011;45:S159–S167. doi: 10.1097/MCG.0b013e318222e603. [DOI] [PubMed] [Google Scholar]
- 26.D’Onofrio A, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. This paper presents methods and analysis for large-scale production of reference genome sequences from human-microbiome organisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 29.Nelson TA, et al. PhyloChip microarray analysis reveals altered gastrointestinal microbial communities in a rat model of colonic hypersensitivity. Neurogastroenterol Motil. 2011;23:169–177. doi: 10.1111/j.1365-2982.2010.01637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bent SJ, et al. Measuring species richness based on microbial community fingerprints: the emperor has no clothes. Appl Environ Microbiol. 2007;73:2399–2401. doi: 10.1128/AEM.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The NIH HMP Working Group et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. This paper describes the data sets and resources of the HMP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazarevic V, et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009;79:266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claesson MJ, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gloor GB, et al. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS ONE. 2010;5:e15406. doi: 10.1371/journal.pone.0015406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hummelen R, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS ONE. 2010;5:e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zubrzycki L, Spaulding EH. Studies on the stability of the normal human fecal flora. J Bacteriol. 1962;83:968–974. doi: 10.1128/jb.83.5.968-974.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luckey TD. Introduction to intestinal microecology. Am J Clin Nutr. 1972;25:1292–1294. doi: 10.1093/ajcn/25.12.1292. [DOI] [PubMed] [Google Scholar]
- 40.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin J, et al. Optimizing read mapping to reference genomes to determine composition and species prevalence in microbial communities. PloS ONE. 2012;7:e36427. doi: 10.1371/journal.pone.0036427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108:S4680–S4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10:287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 45.DOE Joint Genome Institute. A Genomic Encyclopedia of Bacteria and Archaea. US Department of Energy; 2012. http://www.jgi.doe.gov/programs/GEBA/ [Google Scholar]
- 46.Parfrey LW, Walters WA, Knight R. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front Microbiol. 2011;2:153. doi: 10.3389/fmicb.2011.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wylie KM, Weinstock GM, Storch GA. Emerging view of the human virome. Transl Res. 2012 Apr 24; doi: 10.1016/j.trsl.2012.03.006. http://dx.doi.org/10.1016/j.trsl.2012.03.006. [DOI] [PMC free article] [PubMed]
- 48.Breitbart M, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palacios G, et al. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casas V, Rohwer F. Phage metagenomics. Methods Enzymol. 2007;421:259–268. doi: 10.1016/S0076-6879(06)21020-6. [DOI] [PubMed] [Google Scholar]
- 52.Allander T, et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkbeiner SR, et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathogens. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breitbart M, Rohwer F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques. 2005;39:729–736. doi: 10.2144/000112019. [DOI] [PubMed] [Google Scholar]
- 55.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. Sequence analysis of the human virome in febrile and afebrile children. PLoS ONE. 2012;7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2011;6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proc Natl Acad Sci USA. 2012;109:3962–3966. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Breitbart M, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 59.Minot S, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willner D, Furlan M. Deciphering the role of phage in the cystic fibrosis airway. Virulence. 2010;1:309–313. doi: 10.4161/viru.1.4.12071. [DOI] [PubMed] [Google Scholar]
- 61.Lepage P, et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 2008;57:424–425. doi: 10.1136/gut.2007.134668. [DOI] [PubMed] [Google Scholar]
- 62.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE. 2012;7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol (Lond ) 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raes J, Foerstner KU, Bork P. Get the most out of your metagenome: computational analysis of environmental sequence data. Curr Opin Microbiol. 2007;10:490–498. doi: 10.1016/j.mib.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 68.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS ONE. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wright ES, Yilmaz LS, Noguera DR. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol. 2012;78:717–725. doi: 10.1128/AEM.06516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Angiuoli SV, White JR, Matalka M, White O, Fricke WF. Resources and costs for microbial sequence analysis evaluated using virtual machines and cloud computing. PLoS ONE. 2011;6:e26624. doi: 10.1371/journal.pone.0026624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chitsaz H, et al. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nature Biotechnol. 2011;29:915–921. doi: 10.1038/nbt.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dichosa AE, et al. Artificial polyploidy improves bacterial single cell genome recovery. PLoS ONE. 2012;7:e37387. doi: 10.1371/journal.pone.0037387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 84.Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 85.Hooper LV, Gordon JI. Commensal host–bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]