Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is heterogeneous with regard to genic and allelic heterogeneity, as well as phenotypic variability. The genotype-phenotype relationship in ADPKD is not completely understood. Here, we studied 741 patients with ADPKD from 519 pedigrees in the Genkyst cohort and confirmed that renal survival associated with PKD2 mutations was approximately 20 years longer than that associated with PKD1 mutations. The median age at onset of ESRD was 58 years for PKD1 carriers and 79 years for PKD2 carriers. Regarding the allelic effect on phenotype, in contrast to previous studies, we found that the type of PKD1 mutation, but not its position, correlated strongly with renal survival. The median age at onset of ESRD was 55 years for carriers of a truncating mutation and 67 years for carriers of a nontruncating mutation. This observation allows the integration of genic and allelic effects into a single scheme, which may have prognostic value.
Autosomal dominant polycystic kidney disease (ADPKD) is the most common kidney disorder with a Mendelian inheritance pattern, with a prevalence ranging from 1/400 to 1/1000 worldwide.1 ADPKD shows both locus and allelic heterogeneity. Two causative genes—PKD1, located at 16p13.3,2 and PKD2, located at 4q213—have been identified, and the ADPKD mutation database (http://pkdb.mayo.edu/) describes >1000 pathogenic mutations (929 in PKD1 and 167 in PKD2 as of June 5, 2012), not including our most recent data.4
ADPKD also shows high phenotypic variability, as exemplified by the wide variation in the age at onset of ESRD,5 which is defined as the requirement of dialysis or transplantation. Genotype-phenotype correlation studies underscore two major issues. First, on average, ESRD occurs 20 years earlier in patients with PKD1 than those with PKD2,6,7 indicating a genic influence on the ADPKD phenotype. Second, the position of the PKD1 mutation is associated with the age at ESRD onset,8 suggesting an allelic influence on ADPKD phenotype. However, these observations were made >10 years ago, when mutational analysis of the PKD1 and PKD2 genes (particularly of the nonunique portion of the PKD1 gene2,9) was substantially less comprehensive and sophisticated than it is currently,4,10 the methods for predicting the potential pathogenicity of missense mutations were in their infancy, and the studied patient cohorts were relatively small. To confirm (or refute) these earlier observations, we performed a genotype and phenotype correlation study using the Genkyst cohort, which comprises patients with ADPKD recruited from all private and public nephrology centers in the Brittany region, namely, the western part of France.
Results
Description of the Cohort and Distribution of the PKD1 and PKD2 Mutations
This study included all 741 patients with ADPKD and molecular genetic analytic data (416 female and 325 male patients; 519 pedigrees) registered in Genkyst between early 2009 and July 2012. The mean age ± SD at inclusion was 53.4±14.8 years (range, 5.45–89.5 years). Pathogenic mutations in the PKD1 and PKD2 genes were found in 392 (75.5%) and 95 (18.3%) of the 519 pedigrees, respectively (Table 1). No mutation was identified in the remaining 32 pedigrees (6.2%); the overall detection rate was thus 93.8%. The phenotypic characteristics of each patient group, including age, sex, BP status with age at diagnosis of hypertension if available, and CKD stage in terms of estimated GFR (calculated using the Modification of Diet in Renal Disease formula), are provided in Table 1.
Table 1.
Patient characteristics
| Characteristic | PKD1 Mutation Carriers | PKD2 Mutation Carriers | Mutation-Negative Patients | Total |
|---|---|---|---|---|
| Patients (pedigrees), n (n) | 571 (392) | 133 (95) | 37 (32) | 741 (519) |
| Mean age (yr) | 51.96 | 58.91 | 56.01 | 53.4 |
| Sex (n) | ||||
| Male | 254 | 50 | 21 | 325 |
| Female | 317 | 83 | 16 | 416 |
| High BP | ||||
| Present, n (%) | 474 (83) | 107 (80.4) | 31 (83.7) | 612 |
| Age at diagnosis (yr) | 38.6 | 48.6 | 42.2 | 40.6 |
| Absent (n) | 92 | 26 | 6 | 124 |
| Unknown (n) | 5 | 0 | 0 | 5 |
| CKD stage according to estimated GFR (MDRD formula) (n) | ||||
| I | 74 | 21 | 4 | 90 (13.4) |
| II | 74 | 31 | 3 | 108 (14.6) |
| III | 91 | 38 | 8 | 137 (18.5) |
| IV | 61 | 12 | 4 | 77 (10.4) |
| V | ||||
| Total | 271 | 31 | 18 | 320 (43.2) |
| Requiring dialysis (n) | 51 | 15 | 4 | 70 (9.4) |
| With kidney transplant (n) | 192 | 11 | 7 | 210 (28.3) |
Unless otherwise noted, values in parentheses are percentages of patients. MDRD, Modification of Diet in Renal Disease.
A large number of the pathogenic mutations in the current study were confined to single pedigrees. Thus, a total of 342 different mutations were found in the 487 mutation-positive pedigrees (296 PKD1 mutations in 392 pedigrees and 46 PKD2 mutations in 95 pedigrees). Approximately two thirds of the 342 PKD1 mutation–positive pedigrees and almost 95% of the 95 PKD2 mutation–positive pedigrees carry truncating mutations (i.e., nonsense mutations, frameshift mutations, splicing mutations, and large rearrangements) (Table 2).
Table 2.
Distribution of pathogenic PKD1 and PKD2 mutations
| Gene/Mutation Type | Pedigrees, n (%) |
|---|---|
| PKD1 | 392 |
| Truncating mutations | |
| Total | 255 (65.1) |
| Frameshift | 130 (33.2) |
| Nonsense | 90 (22.9) |
| Splice | 32 (8.2) |
| Large rearrangements | 3 (0.8) |
| Nontruncating mutations | |
| Total | 137 (34.9) |
| Missense | 110 (28) |
| In-frame deletions or insertions | 27 (6.9) |
| PKD2 | 95 |
| Truncating mutations | |
| Total | 89 (93.7) |
| Frameshift | 13 (13.7) |
| Nonsense | 41 (43.1) |
| Splice | 20 (21.1) |
| Large rearrangements | 15 (15.8) |
| Nontruncating mutations | 6 (6.3) |
| Total (all mutation positive pedigrees) | 487 |
Genotype and Phenotype Correlations
Sex Influence
Using the Kaplan-Meier model, we found that male PKD1 mutation carriers presented with a poorer renal survival than female ones (median age at ESRD onset, 56.1 years for male patients versus 59.5 years for female patients; P<0.04). This result was confirmed using Cox multivariate analysis (P=0.03) (Table 3). However, we did not find any sex influence in the PKD2 mutation carriers. When the two datasets were combined, only a borderline significance was observed; the median age at ESRD onset was 62.8 years for male patients versus 65.4 years for female patients (P=0.05).
Table 3.
Details of univariate and multivariate Cox analysis for PKD1 mutation carriers
| Variable | Patients (n) | Univariate HR (95% CI) | P Value | Multivariate HR (95% CI) | P Value |
|---|---|---|---|---|---|
| Age | |||||
| <45 yr | 170 | 1 | 1 | ||
| 45–65 yr | 306 | 2.75 (1.91–3.96) | <0.0001 | 2.7 (1.88–3.93) | <0.0001 |
| >65 yr | 89 | 11.5 (5.42–24.5) | <0.0001 | 11.48 (5.34–24.5) | <0.0001 |
| Gender | |||||
| Female | 227 | 1 | 1 | ||
| Male | 236 | 1.31 (1.01–1.68) | 0.039 | 1.33 (1.03–1.72) | 0.029 |
| Mutation type | |||||
| Nontruncating | 142 | 1 | |||
| Truncating | 321 | 2.6 (1.9–3.56) | <0.0001 | 2.74 (2–3.76) | 0.0001 |
| High BP | |||||
| Presence | 474 | ||||
| Absence | 92 | 0.872 (0.53–1.43) | 0.40 | ||
| Smoking (active or stopped) | |||||
| Yes | 209 | ||||
| No | 311 | 1.107 (0.84–1.44) | 0.47 | ||
| Macroscopic hematuria | |||||
| Yes | 163 | ||||
| No | 358 | 1.195 (0.91–1.57) | 0.20 | ||
| Mutation position 5′ versus 3′ end | |||||
| 5′ end | 281 | ||||
| 3′ end | 289 | 1.113 (0.86–1.43) | 0.41 | ||
| Mutation position according to quartile of position | |||||
| 1st quartile | 147 | 1 | |||
| 2nd quartile | 138 | 1 (0.72–1.42) | 0.72 | ||
| 3rd quartile | 144 | 0.945 (0.67–1.34) | 0.75 | ||
| 4th quartile | 141 | 0.818 (0.57–1.17) | 0.67 |
Gene Influence
PKD1 mutation carriers had significantly poorer renal prognosis than their PKD2 counterparts, with respective median ages at ESRD onset of 58.1 (95% confidence interval [CI], 56.5–59.9) and 79.7 (95% CI, 76.8–82.6) years (P<0.001). The mean age at diagnosis of hypertension was 10 years earlier for PKD1 mutation carriers (n=315; 38.6±11.3 years) than for PKD2 mutation carriers (n=77; 48.6±11.5 years) (P<0.001) (Table 1).
Allelic Influence of the PKD1 Gene
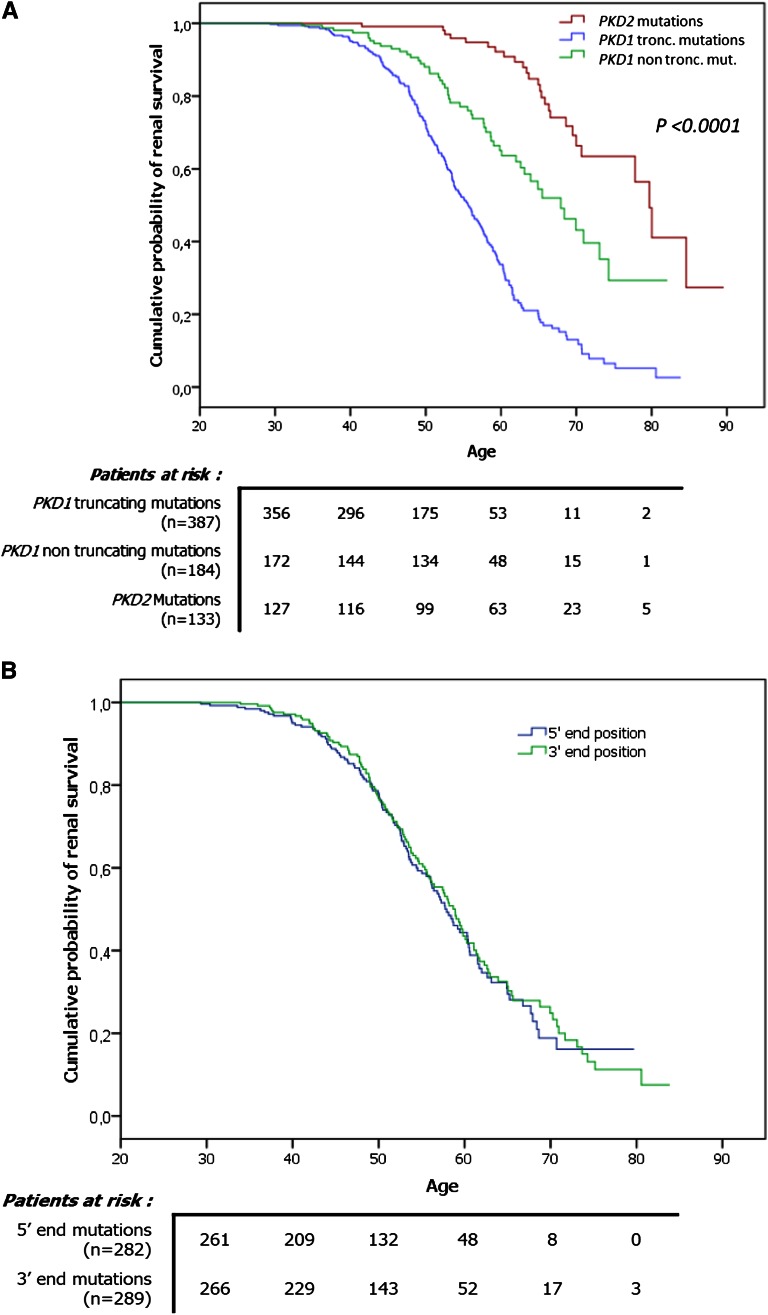
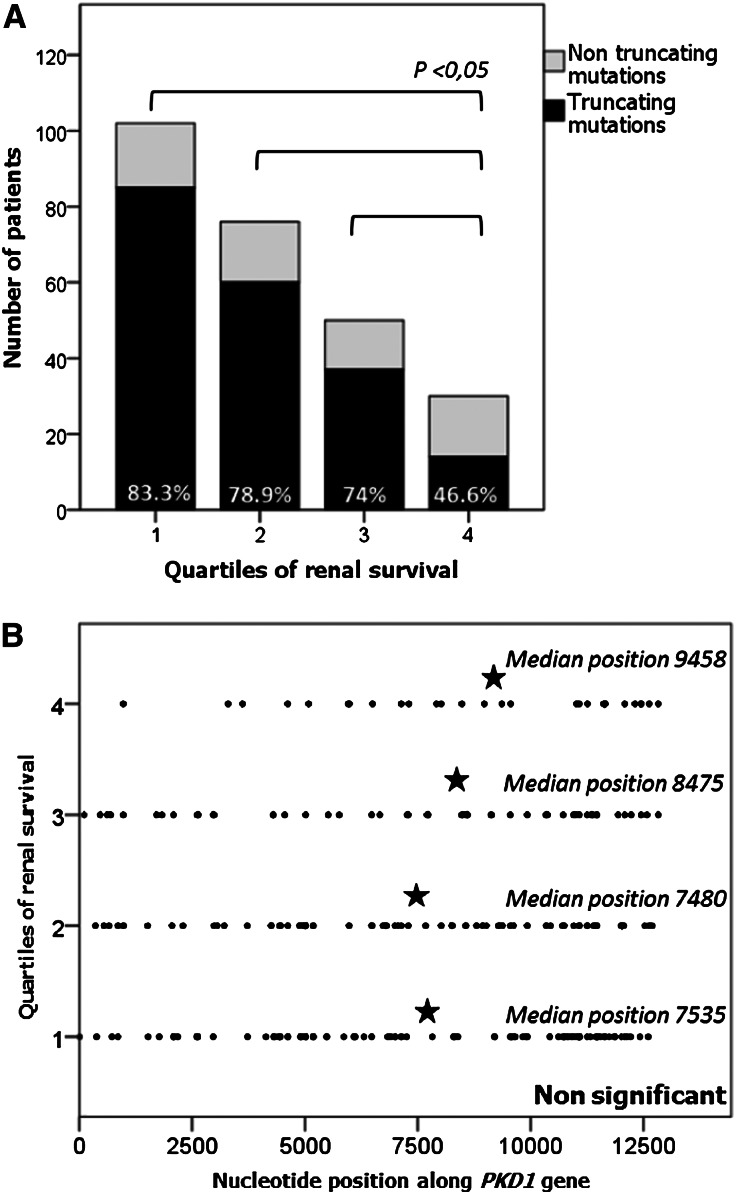
To address the issue of allelic influence on phenotype in ADPKD, we first investigated the relationship between the type of the mutation and renal survival. To this end, we studied renal survival in two main categories of PKD1 mutations, defined by their truncating effect on polycystin 1. As shown in Figure 1A, the median age at ESRD onset in the truncating mutation group (i.e., frameshift, nonsense, splice mutations, and large rearrangements) was 55.6 years (95% CI, 53.6–57.7 years), and mutations with no truncating effect (i.e., in-frame mutations and missense events) were associated with a 12-year delay in ESRD onset (median age, 67.9 years [95% CI, 62.4–73.4 years]) (P<0.0001). This difference was also reflected by the proportion of truncating mutations in four groups of patients, corresponding to the quartile of renal survival in patients with PKD1 (i.e., quartile 1, ESRD before 50.5 years; quartile 2, ESRD between 50.5 and 58.2 years; quartile 3, ESRD between 58.2 and 68.4 years; and quartile 4, patients >68.4 years without ESRD). In the mildest phenotype group (i.e., quartile 4), the proportion of truncating mutations (46.6%) was significantly lower than in each of the other three quartiles (quartile 1, 83.3%; quartile 2, 78.9%; and quartile 3, 74%; P<0.05) (Figure 2A). Moreover, we assessed the robustness of our survival analysis using a Cox multivariate model, taking into account age and sex. We found no effect of BP status, smoking, hematuria, or mutation position on renal survival in the univariate analysis (Table 3); therefore, these variables were not entered into the multivariate analysis. A strong effect of mutation type on renal survival was again observed; patients carrying a truncating PKD1 mutation were 2.74 times more likely to develop ESRD than those carrying a nontruncating PKD1 mutation (P<0.0001) (Table 3). Finally, the comparison of renal survival plots of male and female patients in the truncating mutation group showed that male patients with truncating mutations had significantly poorer prognoses than their female counterparts (respective median age at ESRD, 53.2 years [95% CI, 51.5–54.9 years] and 58.1 years [95% CI, 56.3–60 years]; P<0.001). However, the same comparison in the nontruncating mutation group did not show any significant difference.
Figure 1.
PKD1 mutation type, but not its location, influences renal survival. Renal survival plots. (A) Significant differences in renal survival between PKD1 truncating mutation carriers, PKD1 nontruncating mutation carriers and PKD2 mutation carriers. (B) The absence of a position effect of PKD1 mutation on renal survival.
Figure 2.
Patients with milder phenotypes are more likely to harbor non-truncating mutations. Distribution of the patients according to the type and location of PKD1 mutations in the four quartiles of renal survival. Quartile 1, ESRD before 50.5 years; quartile 2, ESRD between 50.5 and 58.2 years; quartile 3, ESRD between 58.1 and 68.4 years; quartile 4, patients >68.4 years without ESRD or reaching ESRD after 68.4 years. The 258 PKD1 mutation carriers presented in these figures have reached ESRD or are older than 68.4 years, corresponding to the lower limit of the last quartile. (A) Proportion of truncating mutations in patients with milder renal phenotypes, represented by quartile 4, is significantly lower than that in patients with more severe diseases (quartile 1, 2, or 3). (B) Median positions of mutations does not differ in the four groups of renal survival.
Second, we evaluated the influence of mutation location along the PKD1 gene. Patients with PKD1 were separated into two groups according to the mutation position around the median: nucleotide 7815 within exon 19. Renal survival plots comparing these two groups showed no significant difference between the 5′ and the 3′ end groups (P=0.69) (Figure 1B). In addition, we conducted the same analysis within four groups defined by the quartile of mutation position (quartile 1, 5′ end to nucleotide 4617; quartile 2, nucleotide 4617–7815; quartile 3, nucleotide 7815–10745; and quartile 4, nucleotide 10745–3′ end). Again, we found no significant differences in terms of renal survival. We further compared the median mutation positions in the four groups of patients corresponding to the quartile of renal survival in the PKD1 patients and found no significant differences (P=0.46) (Figure 2B). We also studied the position effect separately in the context of truncating and nontruncating mutations and, again, found no statistically significant differences (data not shown).
Discussion
We report here our genotypic and phenotypic analyses using data from Genkyst, one of the largest cohorts of patients with ADPKD worldwide. The Genkyst cohort is a valuable resource for investigating the role of genetic factors in renal disease evolution to ESRD because it aims to include all consenting patients followed in public and private nephrology centers of a unique geographic region, no matter the CKD stage. Indeed, the preferential inclusion of patients treated for ESRD (with dialysis or transplantation) could lead to the selection of only patients with severe phenotypes and may not take into account the milder forms of the disease.
Our overall mutation detection rate, 93.8%, was higher than the previously reported 86%–89%.11,12 This improvement is mainly due to the following two reasons: (1) the thorough analysis of the entire coding sequences and all the exon/intron boundaries of PKD1 and PKD2 for both conventional mutations (i.e., single nucleotide substitutions, small insertions, and small deletions) and gross genomic rearrangements4 and (2) the identification of a significant number of milder mutations. In addition, it may be possible that rare founder mutations have been considered as “recurrent” ones, thereby slightly increasing our mutation detection rate.
We describe here a high level of private mutations (70.2%), which is consistent with previous findings.10–12 We cannot exclude the possibility that some recurrent mutations might in fact be due to the presence of a common ancestor. However, to date, we have not been able to establish any genealogic link between these families despite careful reanalysis of the pedigrees. A consequence of this putative misclassification would be the creation of some artificial mutational hotspots. Nonetheless, this potential caveat is unlikely to have any undue influence on our main conclusions (see below) because our genotype and phenotype correlation was based on the number of included patients, irrespective of the number of families involved.
Analysis of renal survival (Figure 1A) confirmed the previous observation that PKD2 mutations are associated with renal survival that is approximately 20 years longer than that seen with PKD1 mutations. However, our observed median ages at onset of ESRD for PKD1 and PKD2 mutation carriers (58.1 years versus 79.7 years) are 3.8 and 5.7 years older than the previously reported 54.3 and 74 years,7 respectively. This discrepancy may be attributed to the following reasons. First, the previous study7 was performed 13 years ago. Improvements in nephroprotective strategies over this period may have contributed to the increased renal survival. Second, all our patients were recruited from the same region in France and thus form a relatively homogeneous population. The presence of common protective genetic or environmental factors in these patients cannot be excluded. Third, and perhaps most important, earlier studies typically analyzed traditional, large families, which are often affected by severe mutations. In contrast, our study identified a significant number of mild or “hypomorphic” alleles.
The sex effect reported in our study for the PKD1 gene, with an earlier onset of ESRD in male patients, has previously been described,13,14 but not unanimously.7,8 Nonetheless, this sex influence is moderate, both in our study and in previous ones, which may explain the conflicting results.
To our knowledge, this is the first study to show a strong effect of PKD1 mutation type on renal phenotype, but we found no effect of mutation location at the population level. Indeed, we showed a 12-year delay in ESRD onset in patients harboring a truncating mutation of PKD1 compared with patients with nontruncating mutation of PKD1. The risk of developing ESRD was 2.74 times higher in patients with a truncating mutation than in patients with a nontruncating mutation of PKD1. Within the PKD1 population, this aspect is, to our knowledge, the most effective predictive factor of the evolution of ESRD ever reported.
In contrast to our conclusions, Rossetti et al. reported that the position but not the type of PKD1 mutation correlates with the severity of renal disease in ADPKD.8 Specifically, on the basis of the analysis of 324 patients with PKD1 from 80 different families, in whom the age at onset of ESRD was known for 152 patients, the authors concluded that “patients with mutations in the 5′ region [i.e., 5′ to the median position at nucleotide 7812; the full-length PKD1 mRNA sequence is ∼14,000 bp] had significantly more severe disease than the 3′ group; median time to ESRD was 53 and 56 years, respectively (P = 0.025).” In contrast, the authors found no significant differences among patients with truncating mutations, in-frame changes, and missense mutations in PKD1. Thus, in terms of allelic influence, our findings are in direct opposition to previous ones.8 The three caveats mentioned in the introduction (i.e., the improvements in both molecular analyses and the scoring of missense events in the last 10 years and the smaller size of the former study cohort) may certainly explain this discrepancy. Moreover, our findings are indirectly supported by the literature. Although no similar observations have been reported in autosomal dominant diseases, two studies performed in the context of autosomal or X-linked recessive diseases have shown that truncating mutations are significantly more severe than nontruncating mutations (predominantly missense mutations).15,16 Most of the nontruncating PKD1 mutations in our study are also missense mutations (Table 2). It is tempting to speculate that some of these missense mutations may represent “hypomorphic” alleles.17 In fact, this hypothesis is well supported by recent publications. Cases of milder ADPKD phenotypes have been reported in members of pedigrees harboring missense mutations compared with relatives, co-inheriting the missense mutation with another mutation in PKD1 or PKD2.18–20 Moreover, the dosage dependence of cytogenesis has recently been highlighted by a new PKD1 murine model.21
As illustrated in Figure 1A, our analysis led to a refinement of the genotypic and phenotypic relationship in ADPKD, with nontruncating PKD1 mutations identified as being associated with an intermediate severity in terms of age at ESRD onset (67.9 years) between truncating PKD1 mutations (55.6 years) and PKD2 mutations (79.7 years). This new information, which integrates the genic and allelic influences on phenotype into a single scheme, may be of significant prognostic value. Moreover, the costs of sequencing reactions and analyses are expected to decrease steadily in the coming years. Many factors have contributed to this decrease, such as the automation of reaction preparations (i.e., for PCR reactions) and the development of next-generation sequencing technologies, as recently described for PKD1 and PKD2.10 In that setting, the molecular analysis of PKD1 and PKD2 could be more readily conducted for determining prognosis. In the context of emerging targeted therapies, this new prognostic tool may help identify patients who could benefit from treatment at a presymptomatic stage of the disease. However, it is important to keep in mind that for each of the two mutation type categories, extreme ESRD ages exist. Thus, taking into account the mutation type alone to predict renal outcome in PKD1 individual patients could be misleading.
Beyond the demonstration of this allelic influence upon renal survival for PKD1, the identification of modifier genes is an important goal, but studies to this end have remained inconclusive to date.5 Nevertheless, case reports of early and severe ADPKD have recently highlighted the role of mutations in additional PKD genes (HNF1β, PKD1, PKD2, or PKHD1) in aggravating the phenotype.19 In the future, integrating the different components of genetic heterogeneity in ADPKD, such as the gene involved, the type of PKD1 mutation, and the coexistence of eventual modifier genes, will be complex but may improve our ability to predict the severity of renal disease.
Concise Methods
Patients
Genkyst is a regional cohort involving up to 73 nephrologists working in all 17 private and public nephrology centers in the Brittany region (Bretagne), France. It registers clinical and molecular genetic data of all consenting patients with ADPKD from this area. The ADPKD diagnosis was based on renal ultrasonographic findings in accordance with previously described criteria.22 All 741 participating patients (from 519 pedigrees) with molecular genetic data were recruited between early 2009 and July 2012, through nephrology consultation, from dialysis centers, or during consultations or hospitalizations for monitoring of kidney transplants. Repartition of the 741 patients according to estimated GFR using the MDRD formula is available in Table 1. All participants provided informed consent, and the local ethics committee approved the study.
Mutation Analysis
Most of the 741 participating patients had been included in our recent mutation report, which analyzed patients from both the Brittany region and other regions in France.4 For the newly recruited patients, mutation detection and pathogenicity prediction for missense mutations were performed as previously described.4 Briefly, to assess the pathogenicity of missense variants, four different methods were combined: Grantham matrix scoring system, Align Grantham Variation Grantham Deviation;23 PolyPhen (http://genetics.bwh.harvard.edu/pph/);24,25 Sorting Intolerant from Tolerant;26 and Mutation Taster.27 All missense mutations predicted to be pathogenic (together with their predicted scores by the different algorithms and the number of affected and unaffected members) are provided in Supplemental Table 1.
Statistical Analyses
All statistical analyses were performed using SPSS software, version 19 (SPSS, Inc., Armonk, NY). Renal survival was analyzed using the Kaplan-Meier method. The differences between the survival curves of the tested groups were assessed by means of a log-rank test with a 0.05 significance level. The effects of age (<45 years, 45–65 years, >65 years), sex, and truncating versus nontruncating mutations on renal survival were analyzed using the Cox proportional hazard model. A variable was entered into the multivariate analysis when it was found to be associated with the occurrence of ESRD in univariate analysis at a conservative threshold of 20%. The presence of high BP (defined as systolic BP >140 mmHg, diastolic BP >90 mmHg, or the need for antihypertensive medication), positive history of macroscopic hematuria, smoking status (former or active), and mutation position were excluded from the multivariate analysis because they failed to meet this criterion. The final model was chosen by performing a backward selection based on the maximum likelihood estimation. Adjusted hazard ratios with their 95% CIs are presented. In addition, the means were compared using the t test and medians using a nonparametric Mann-Whitney test or a Kruskal-Wallis test depending on the number of groups.
Disclosures
None.
Acknowledgments
The authors would like to thank all the patients and the Genkyst Group investigators: Dr. Moal, Dr. Tanquerel, Dr. Hanrotel, Dr. Hemon, Dr. Mesguen, Dr. Kersale, Dr. Treguer (Centre Hospitalier Régional Universitaire Brest), Professor Dantal, Dr. Dufay, Professor Giral, Dr. Meurette, Dr. Couvrat, Dr. Garandeau, Professor Fakhouri, Dr. Lino, Dr. Touzot, Dr. Ristea, Professor Blancho, Dr. Hodemon-Corne (Centre Hospitalier Universitaire, Nantes), Professor Le Pogamp, Dr. Frouget, Professor Vigneau, Dr. Laruelle, Dr. Gie, Dr. Rivalan, Dr. Dolley-Hitze, Dr. Richer, Dr. Gosselin (Centre Hospitalier Universitaire, Rennes), Professor Michez, Dr. Mandart, Dr. Menoyo, Dr. Pincon, Dr. Coulibaly (Centre Hospitalier de Bretagne Atlantique, Vannes), Dr. Potier, Dr. Stanescu, Dr. Ang, Dr. Le Cacheux, Dr. Leonetti, Dr. Baluta (Centre Hospitalier de Saint Brieuc), Dr. Siohan, Dr. Metes (Centre Hospitalier de Cornouailles, Quimper), Dr. Latif, Dr. Massad (Centre Hospitalier du centre Bretagne, Pontivy), Dr. Strullu, Dr. Depraetre, Dr. Le Mee, Dr. Chaffara (Association des Urémiques de Bretagne, Brest), Dr. Terki, Dr. Goulesque (Société Brestoise du Rein Artificiel, Brest), Dr. Sawadogo, Dr. Chamontin, Dr. Georgescu (Centre Hospitalier de Bretagne Sud, Lorient), Dr. Benarbia, Dr. Dimulescu, Dr. Durand (Association des Urémiques de Bretagne, Quimper), Dr. Besnier, Dr. Blanpain, Dr. Durault, Dr. Regnier-Le Coz (Centre Hospitalier de Saint-Nazaire), Dr. Legrand (Association des Urémiques de Bretagne, Lorient), Dr. Ferrier, Dr. Menoyo, Dr. Rifaat (ECHO, Vannes), Dr. Savoiu (ECHO, Saint Herblain), Dr. Bertheleme (Centre Héliomarin, Roscoff).
The authors would also like to acknowledge the clinical research team (Stéphanie Bouvier, Christelle Ratajczack, and Christelle Guillerm-Regost) and the molecular genetic laboratory (Joelle Creff, Isabelle Quéré, Sandrine Maestri, Caroline Benech, and Sylvia Redon).
This study was conducted with the support of the National Plan for Clinical Research (PHRC regional 2010), the Société Française de Néphrologie, and the Institut National de la Santé et de la Recherche Médicale (INSERM).
Part of this work was presented as an oral communication at the annual meeting of the American Society of Nephrology, November 2012, San Diego, California.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “The Mutation, a Key Determinant of Phenotype in ADPKD,” on pages 868–870.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2012070650/-/DCSupplemental.
References
- 1.Dalgaard OZ, Nørby S: Autosomal dominant polycystic kidney disease in the 1980’s. Clin Genet 36: 320–325, 1989 [DOI] [PubMed] [Google Scholar]
- 2.The European Polycystic Kidney Disease Consortium : The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 77: 881–894, 1994 [DOI] [PubMed] [Google Scholar]
- 3.Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, Reynolds DM, Cai Y, Gabow PA, Pierides A, Kimberling WJ, Breuning MH, Deltas CC, Peters DJ, Somlo S: PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272: 1339–1342, 1996 [DOI] [PubMed] [Google Scholar]
- 4.Audrézet M-P, Cornec-Le Gall E, Chen J-M, Redon S, Quéré I, Creff J, Bénech C, Maestri S, Le Meur Y, Férec C: Autosomal dominant polycystic kidney disease: Comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat 33: 1239–1250, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Harris PC, Rossetti S: Determinants of renal disease variability in ADPKD. Adv Chronic Kidney Dis 17: 131–139, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torra R, Badenas C, Darnell A, Nicolau C, Volpini V, Revert L, Estivill X: Linkage, clinical features, and prognosis of autosomal dominant polycystic kidney disease types 1 and 2. J Am Soc Nephrol 7: 2142–2151, 1996 [DOI] [PubMed] [Google Scholar]
- 7.Hateboer N, v Dijk MA, Bogdanova N, Coto E, Saggar-Malik AK, San Millan JL, Torra R, Breuning M, Ravine D: Comparison of phenotypes of polycystic kidney disease types 1 and 2. European PKD1-PKD2 Study Group. Lancet 353: 103–107, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Rossetti S, Burton S, Strmecki L, Pond GR, San Millán JL, Zerres K, Barratt TM, Ozen S, Torres VE, Bergstralh EJ, Winearls CG, Harris PC: The position of the polycystic kidney disease 1 (PKD1) gene mutation correlates with the severity of renal disease. J Am Soc Nephrol 13: 1230–1237, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Bogdanova N, Markoff A, Gerke V, McCluskey M, Horst J, Dworniczak B: Homologues to the first gene for autosomal dominant polycystic kidney disease are pseudogenes. Genomics 74: 333–341, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Rossetti S, Hopp K, Sikkink RA, Sundsbak JL, Lee YK, Kubly V, Eckloff BW, Ward CJ, Winearls CG, Torres VE, Harris PC: Identification of gene mutations in autosomal dominant polycystic kidney disease through targeted resequencing. J Am Soc Nephrol 23: 915–933, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossetti S, Consugar MB, Chapman AB, Torres VE, Guay-Woodford LM, Grantham JJ, Bennett WM, Meyers CM, Walker DL, Bae K, Zhang QJ, Thompson PA, Miller JP, Harris PC, CRISP Consortium : Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 18: 2143–2160, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Tan Y-C, Blumenfeld JD, Anghel R, Donahue S, Belenkaya R, Balina M, Parker T, Levine D, Leonard DGB, Rennert H: Novel method for genomic analysis of PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease. Hum Mutat 30: 264–273, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Gabow PA, Johnson AM, Kaehny WD, Kimberling WJ, Lezotte DC, Duley IT, Jones RH: Factors affecting the progression of renal disease in autosomal-dominant polycystic kidney disease. Kidney Int 41: 1311–1319, 1992 [DOI] [PubMed] [Google Scholar]
- 14.Orskov B, Christensen KB, Feldt-Rasmussen B, Strandgaard S: Low birth weight is associated with earlier onset of end-stage renal disease in Danish patients with autosomal dominant polycystic kidney disease. Kidney Int 81: 919–924, 2012 [DOI] [PubMed] [Google Scholar]
- 15.McEntagart M, Parsons G, Buj-Bello A, Biancalana V, Fenton I, Little M, Krawczak M, Thomas N, Herman G, Clarke A, Wallgren-Pettersson C: Genotype-phenotype correlations in X-linked myotubular myopathy. Neuromuscul Disord 12: 939–946, 2002 [DOI] [PubMed] [Google Scholar]
- 16.Snoeckx RL, Huygen PLM, Feldmann D, Marlin S, Denoyelle F, Waligora J, Mueller-Malesinska M, Pollak A, Ploski R, Murgia A, Orzan E, Castorina P, Ambrosetti U, Nowakowska-Szyrwinska E, Bal J, Wiszniewski W, Janecke AR, Nekahm-Heis D, Seeman P, Bendova O, Kenna MA, Frangulov A, Rehm HL, Tekin M, Incesulu A, Dahl H-HM, du Sart D, Jenkins L, Lucas D, Bitner-Glindzicz M, Avraham KB, Brownstein Z, del Castillo I, Moreno F, Blin N, Pfister M, Sziklai I, Toth T, Kelley PM, Cohn ES, Van Maldergem L, Hilbert P, Roux A-F, Mondain M, Hoefsloot LH, Cremers CWRJ, Löppönen T, Löppönen H, Parving A, Gronskov K, Schrijver I, Roberson J, Gualandi F, Martini A, Lina-Granade G, Pallares-Ruiz N, Correia C, Fialho G, Cryns K, Hilgert N, Van de Heyning P, Nishimura CJ, Smith RJH, Van Camp G: GJB2 mutations and degree of hearing loss: A multicenter study. Am J Hum Genet 77: 945–957, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vujic M, Heyer CM, Ars E, Hopp K, Markoff A, Orndal C, Rudenhed B, Nasr SH, Torres VE, Torra R, Bogdanova N, Harris PC: Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 21: 1097–1102, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei Y, Lan Z, Wang K, Garcia-Gonzalez M, He N, Dicks E, Parfrey P, Germino G, Watnick T: A missense mutation in PKD1 attenuates the severity of renal disease. Kidney Int 81: 412–417, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergmann C, von Bothmer J, Ortiz Brüchle N, Venghaus A, Frank V, Fehrenbach H, Hampel T, Pape L, Buske A, Jonsson J, Sarioglu N, Santos A, Ferreira JC, Becker JU, Cremer R, Hoefele J, Benz MR, Weber LT, Buettner R, Zerres K: Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J Am Soc Nephrol 22: 2047–2056, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, Chauveau D, Rees L, Barratt TM, van’t Hoff WG, Niaudet P, Torres VE, Harris PC, Harris PC: Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int 75: 848–855, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopp K, Ward CJ, Hommerding CJ, Nasr SH, Tuan H-F, Gainullin VG, Rossetti S, Torres VE, Harris PC: Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J Clin Invest 122: 4257–4273, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei Y, Obaji J, Dupuis A, Paterson AD, Magistroni R, Dicks E, Parfrey P, Cramer B, Coto E, Torra R, San Millan JL, Gibson R, Breuning M, Peters D, Ravine D: Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol 20: 205–212, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavtigian SV, Byrnes GB, Goldgar DE, Thomas A: Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum Mutat 29: 1342–1354, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramensky V, Bork P, Sunyaev S: Human non-synonymous SNPs: Server and survey. Nucleic Acids Res 30: 3894–3900, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunyaev S, Hanke J, Aydin A, Wirkner U, Zastrow I, Reich J, Bork P: Prediction of nonsynonymous single nucleotide polymorphisms in human disease-associated genes. J Mol Med (Berl) 77: 754–760, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC: SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40[Web Server issue]: W452–457, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D: MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7: 575–576, 2010 [DOI] [PubMed] [Google Scholar]