Abstract
MHC class I–related chain A (MICA) antigens are surface glycoproteins strongly implicated in innate immunity, and the MICA gene is highly polymorphic. Clinical observations suggest a role for donor MICA antigens expressed on transplant endothelial cells in the alloimmune response, but the effect of MICA genotype is not well understood. Here, we investigated the immunologic effect of the A5.1 mutation, related to the common MICA*008 allele. Compared with wild-type endothelial cells (ECs), homozygosity for MICA A5.1 associated with an endothelial phenotype characterized by 7- to 10-fold higher levels of MICA mRNA and MICA proteins at the cell surface, as well as exclusive release in exosomes instead of enzymatic cleavage. Mechanistically, we did not detect quantitative changes in regulatory microRNAs. Functionally, A5.1 ECs enhanced NKG2D interaction and natural killer cell activation, promoting NKG2D-dependent lysis of ECs. In kidney transplant recipients, polyreactive anti-MICA sera bound preferentially to ECs from MICA A5.1 donors, suggesting that MICA*008(A5.1) molecules are the preferential antigenic determinants on ECs of grafts. Furthermore, the incidence of MICA A5.1 mismatch revealed a statistically significant association between donor MICA A5.1 and both anti-MICA sensitization and increased proteinuria in kidney recipients. Taken together, these results identify the A5.1 mutation as an immunodominant factor and a potential risk factor for transplant survival.
MHC class I–related chain A (MICA) antigens are surface glycoproteins strongly implicated in innate immunity.1,2 MICA is a ligand for the activating immunoreceptor NKG2D, a highly conserved C-type lectin-like membrane glycoprotein expressed on essentially all natural killer (NK) cells, as well as on γδ and αβ CD8(+) T cells.2–4
MICA proteins are physiologically expressed at the cell surface of a restricted number of cell types, including endothelial cells (ECs), epithelial cells, fibroblasts, dendritic cells, and activated TCD4+ and B lymphocytes.5 MICAs are stress-induced proteins regulated at the cell surface by infection (i.e., viruses and some intracellular bacteria), heat shock, DNA damage response,6 and oncogenic transformation.3 Dysregulation of MICA is associated with tumor escape7 but also causes autoreactive T cell stimulation, thus promoting autoimmune diseases.8
The MICA gene is highly polymorphic, and >70 alleles have been reported so far (http://hla.alleles.org).9,10 MICA also has a triplet repeat microsatellite polymorphism (GCT) within exon 5 encoding for the transmembrane region. Seven GCT (alanine) repeats have been described, corresponding to 4(A4), 5(A5), 6(A6), 7(A7), 8(A8), 9(A9), or 10(A10) alanine repetitions within the transmembrane region. Additionally, a mutation has been associated with certain A5 repeat alleles. This mutation consists of a guanine insertion after the second of five trinucleotide repeats (A5.1) that causes a frameshift mutation leading to a premature intradomain stop codon.11,12 Previous studies suggest that MICA A5.1 encodes a truncated protein with possible aberrant protein expression and cellular localization.13
Although genetic matching of the classic HLA antigens is clearly a major determinant of successful organ transplant outcome, clinical studies demonstrate that MICA is another polymorphic genetic factor involved. Initial studies reported on specific antibodies against MICA in the serum of patients who had rejected kidney allografts, suggesting a role for these molecules in transplant immunopathology.14,15 Expression of MICA in transplanted organs has been demonstrated, and anti-MICA antibodies have been associated with both acute and chronic rejection in renal,16,17 pancreatic,18 and heart transplants.19 In renal transplantation, anti-MICA antibodies after transplantation have been reported in 5%–9% of recipients and cause a 10% decrease in graft survival at 1 year.20 Together, these findings suggest a role for donor MICA antigens expressed on transplant ECs in the alloimmune response. However, MICA genotyping is not routinely achieved, and a possible correlation between MICA polymorphism, MICA expression, and function on the ECs of the graft is still unknown. Moreover, the molecular bases for MICA allospecific immunization are not well understood.
The aim of this study was to evaluate the functional effect of MICA A5.1 mutation on MICA expression by ECs and its clinical relevance in organ transplantation. Here, we examined the frequency of MICA A5.1 mutation among a cohort of kidney transplant donors. We demonstrate that MICA A5.1 mutation leads to abnormal expression of both surface MICA expression and release of soluble and exosomal MICA antigens by ECs. We show here that endothelial MICA A5.1 expression enhances NKG2D engagement on NK cells and is a major antigenic determinant of the allele-specific anti-MICA humoral response in kidney transplant recipients.
Results
Predominant MICA A5.1 Mutation in Kidney Transplants Is Associated with MICA Protein Alteration in Donor ECs
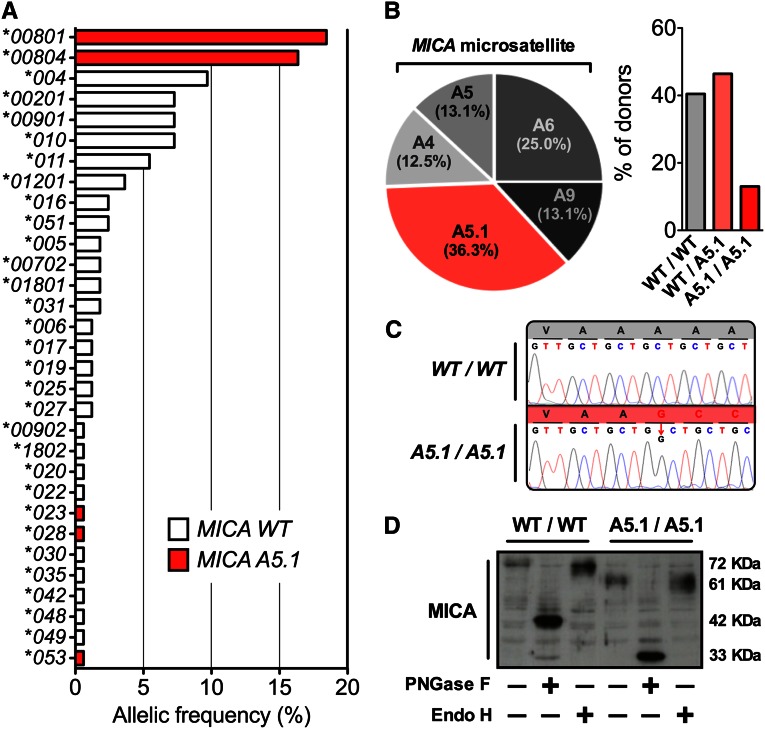
Cultures of donor ECs corresponding to renal transplantations performed in our institute (ITUN, Nantes, France) between 1999 and June 2012 and prospectively isolated and stored have been used to determine the effect of MICA A5.1 mutation on endothelial phenotype and immune functions. Consistent with our previous study,21 MICA genotyping of transplant donors (n=84) indicated that A5.1 mutation was associated with four alleles: *023, *028, *053, and *008, the last being the most represented allele in our cohort (34.5%, including MICA*0801 [18.4%] and MICA*0804 [16.1%]) (Figure 1A). Overall, the frequency of the MICA A5.1 mutation was 59.5% in our cohort, including 13.1% (n=11) of A5.1 homozygous and 46.4% (n=39) of heterozygous carriers (Figure 1B). MICA A5.1 variant contains 5 GCT repeats plus a nucleotide insertion (GGCT) (Figure 1C). This insertion causes a frameshift mutation leading to a premature intradomain stop codon within the transmembrane region, which deletes the MICA cytoplasmic tail. As a consequence of this mutation, expression of shortened MICA proteins in EC cultures from A5.1 homozygous donors compared with wild-type (WT) donors (33 kD versus 42 kD, respectively) (Figure 1D) was observed by Western blotting after protein deglycosylation.
Figure 1.
MICA A5.1 mutation is predominant in kidney transplants and leads to a truncated protein in graft ECs. (A) MICA allele distribution in a cohort (n=84) of kidney donors. MICA genotyping was performed as reported in the Concise Methods section. MICA alleles associated with MICA A5.1 mutation are shown in red. (B) Distribution of exon5 microsatellite polymorphism in our cohort of transplant donors (left panel) and distribution of MICA WT and A5.1 genotype (right panel). Data are expressed as percentages of the total population. (C) Representative electrophoregrams of MICA exon5 microsatellite sequences, homozygous WT/WT and A5.1/A5.1 donors. (D) A representative Western blot showing MICA proteins expressed in vascular ECs isolated from transplant donors. EC lysates were obtained from cultures issued from WT (MICA A9) or MICA A5.1 homozygous individuals. Cell lysates (20 µg) were pretreated overnight with or without endoglycanase F (PNGase F) or endoglycosidase H (Endo H) for deglycosation before electrophoresis. Immunoblotting was performed after protein transfer using an anti-MICA monoclonal antibody (AMO1). Experiment is representative of three separate experiments.
Elevated MICA Expression on Graft ECs Features MICA A5.1 Donors and Enhances NKG2D Activation
To determine whether MICA A5.1 mutation may change EC phenotype, MICA expression was compared on primary EC cultures issued from MICA A5.1 homozygous (A5.1/A5.1; n=4), heterozygous (WT/A5.1; n=4), and control (WT/WT, n=4) transplant donors. First, FACS analysis (Figure 2A) shows that MICA surface expression was significantly higher on ECs from A5.1/A5.1 donors than those from controls (mean fluorescence intensity [MFI] ± SD, 1085.7±190.5 for A5.1/A5.1 versus 155.7±40.3 for WT/WT; P=0.0073). In contrast, all groups expressed equal levels of HLA-A, -B, and -C and other NKG2D ligands (NKG2DL): MICB and UL-16–binding protein (ULBP)-2 and -3 (Figure 2, B–E). ECs with intermediate MICA expression were heterozygous for the A5.1 mutation.
Figure 2.
MICA A5.1 proteins are overexpressed on graft's EC surface and trigger NKG2D activation. (A–E) Endothelial expression of MICA (A), MICB (B), ULBP2 (C), ULPB3 (D), and MHC class I (E) at cell surface was established by FACS analysis performed on ECs from homozygous WT/WT (n=4), MICA A5.1/A5.1 (n=4), and heterozygous WT/A5.1 (n=4) donors. Upper panel shows representative histograms of fluorescence intensity, and lower panel shows a quantitative analysis from four individual EC cultures (**P<0.01). (F) Exacerbated expression of MICA A5.1 proteins on ECs increases NKG2D internalization on NK cells. NKG2D expression on NKL was analyzed by FACS 24 hours after co-culture with EC monolayers from MICA WT/WT (n=4) or A5.1/A5.1 (n=4) individuals. Representative histograms of fluorescence intensity (left) and a quantitative analysis (right) from four MICA WT/WT and four A5.1/A5.1 individual EC cultures are shown (*P<0.05). (G) Exacerbated expression of MICA A5.1 proteins on ECs increases NKG2D-dependent NK cytotoxic activity. NKLs were used as effector cells and ECs with MICA WT/WT (n=3) or MICA A5.1/A5.1 (n=5) genotype were used as targets at various effector-to-target (E:T) ratios. Cytotoxicity was measured using a 4-hour 51Cr release assay. Results are expressed as mean ± SEM of specific lysis (*P<0.05). Results shown are representative of three independent experiments.
The NKG2D receptor stimulates activating signals for cytotoxicity by binding NKG2DL (MICA/B or ULBP1/2/3) on target cells.22 Here, two models were used to study the functional effect of MICA A5.1 EC phenotype. First, coculture of NKL cells with EC monolayers was performed and showed (Figure 2F) that elevated MICA cell surface expression on A5.1/A5.1 ECs significantly decreases NKG2D expression on NK cells compared with WT/WT ECs (32.40%±6.67% versus 50.80%±4.93%; P=0.03), confirming increased ligand/receptor interaction. Second, NK activation and cytotoxic activity toward allogeneic ECs was measured. Enhanced MICA expression at EC target provokes a significant increase in lysis (51.20%±11.11% versus 32.96%±2.85% of lysis at 100:1 effector cell–to–target cell ratio, for A5.1/A5.1 and WT/WT, respectively; P=0.04) (Figure 2G). Together, these findings provide evidence that transplant donors carrying the A5.1 mutation have an elevated expression of MICA protein on the ECs of grafts that triggers an enhanced NKG2D-dependent EC lysis by effector cells.
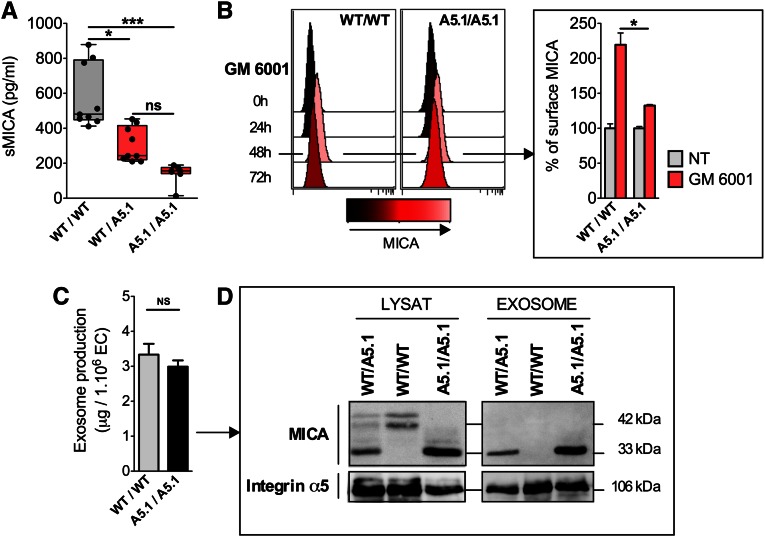
MICA A5.1 Mutation Changes the Release of Soluble and Exosomal MICA by ECs
A feature of MICA proteins is their ability to be released as soluble form by proteolytic cleavage.7 To define whether a higher expression of cell-bound MICA may also result in an increased level of soluble MICA (sMICA), EC culture supernatants were collected and sMICA was quantified by ELISA. Unexpectedly, release of sMICA was strongly reduced for MICA A5.1 heterozygous cells (1.82-fold decrease compared with the wild-type [WT] group; P<0.05), whereas almost no soluble form was produced by A5.1 homozygous ECs (Figure 3A). Consistent with an impaired shedding of MICA from A5.1 cells, inhibition of metalloproteinases was significantly less efficient to maintain MICA surface expression on A5.1 carriers than on WT ECs (32.5%±3.2% versus 119.5%±33.8% of basal MICA increase; P=0.03) (Figure 3B). A previous study reported the shedding of MICA A5.1 proteins into exosomes23 as a mechanism of immune escape used by some tumor cell lines. Here, the release of exosomes was similar in quantity from both WT and A5.1 homozygous ECs (3–4 µg/106 ECs per 48 hours) (Figure 3C). However, consistent with high cell surface expression, immunoblotting indicated that exosomes released from A5.1 ECs expressed higher levels of MICA mutated proteins, with a dose-response observed for heterozygous and homozygous cells, whereas exosomes from WT ECs were not associated with MICA molecules (Figure 3D). Thus, the genetic variant MICA A5.1 strongly affects MICA expression by altering cell surface MICA and the release of soluble and exosomal forms of MICA from ECs.
Figure 3.
MICA A5.1 mutated proteins are preferentaly released in exosomes rather than by proteolytic cleavage. Conditioned media from WT/WT (n=9), WT/A5.1 (n=9), and A5.1/A5.1 (n=7) MICA EC cultures were collected at 120 hours and analyzed by ELISA (*P<0.05, ***P<0.001). (B) Time course analysis of membrane-bound MICA after treatment of ECs with or without (NT) an inhibitor of metalloproteinases (GM6001) measured by flow cytometry. Representative histograms (left) and a relative quantification obtained from four EC WT/WT or A5.1/A5.1 are shown (right). Data are expressed as relative percentages of MICA expression (*P<0.05). (C) Quantification of exosomes from 48-hour conditioned media from MICA WT/WT (n=3) and A5.1/A5.1 (n=3) EC cultures. (D) Exosomes were purified by successive ultracentrifugations. Total EC lysates and lysates from purified EC-derived exosomes (20 µg) were pretreated overnight with PNGase F before Western blot analysis. Immunoblotting was performed using anti-MICA/B antibodies (BAMO1). Blots were rehybridized using anti-integrin α5 antibodies. Results shown are representative of three independent experiments.
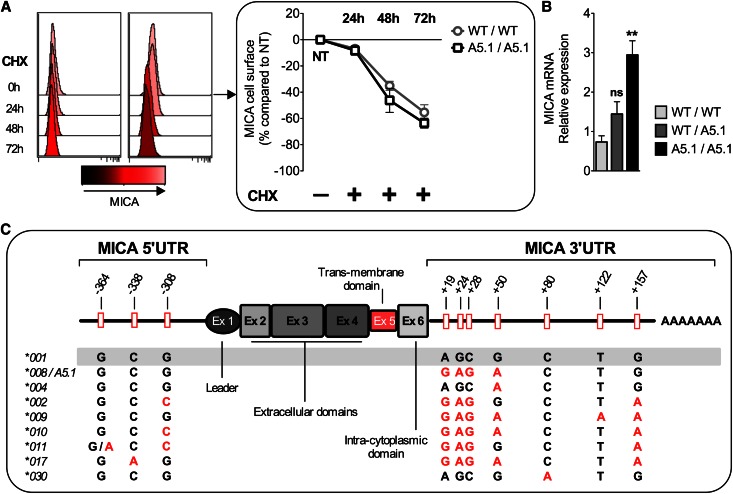
Transcriptional and Post-transcriptional Control of MICA A5.1 Expression
Next, we investigated the regulatory mechanisms that may trigger elevated MICA expression on MICA A5.1 ECs. First, time courses of membrane-bound MICA decay were similar for WT and A5.1 ECs in the presence of cycloheximide, indicating that protein stability was not involved (Figure 4A). In contrast, quantitative PCR analyses indicated that A5.1 phenotype correlates with significantly higher mRNA steady state (1.96±0.83- and 4.00±0.96-fold increase for A5.1 heterozygous and homozygous, respectively, versus WT MICA; P=0.0085) (Figure 4B).
Figure 4.
MICA A5.1 overexpression is not due to increased protein stability but results from a transcriptional control. (A) ECs with WT/WT (n=3) or A5.1/A5.1 (n=6) MICA genotypes were treated by cycloheximide (CHX), and MICA expression was analyzed by flow cytometry at different time points. Representative histograms from individual donors (left) and a quantitative analysis (right) are shown. Decrease in parallel kinetics of membrane-bound MICA was observed by FACS. (B) MICA transcripts in ECs were quantified by quantitative PCR, and results from WT/WT (n=6), WT/A5.1 (n=4) and MICA A5.1/A5.1 (n=4) are expressed as relative expression calculated by the 2−ΔΔCt method (**P<0.01). (C) Location and allele-specificity of the SNPs identified by sequencing the 5′UTR and 3′UTR regions from 34 EC cultures, including alleles MICA*008(A5.1), *004, *002, *009, *010, *011, *017, and *030.
Among transcriptional mechanisms, regulatory microRNAs (miRNAs) have been recently shown to control basal MICA expression in cells.24 We found no quantitative change in the miRNAs previously reported to control MICA expression (miR-20a, -106b, -373, -520b) or in miRNAs that we found predicted to target MICA 3′UTR (http://www.targetscan.org) (miR-105, -520f, -636) (data not shown). We further speculated that change in MICA A5.1 transcription rate could alternatively result from an associated polymorphism in the promoter region or in the 3′UTR region that is a target for miRNA regulatory activity.25 To test this hypothesis, genomic DNA, in proximal 5′ and 3′UTRs, from a set of MICA WT (n=24) and A5.1 (n=10) transplant donors was sequenced. As a result, three novel single-nucleotide polymorphisms (SNPs) were identified in the proximal 5′ untranslated region (UTR) (at −364, −338, and −308 from start codon), but none were specifically associated with MICA A5.1 (Figure 4C). Similarly, seven novel SNPs were found in the MICA 3′UTR region (at positions +19, +24, +28, +50, +80, +122, and +157 from the stop codon). Although none were specifically associated with MICA*008(A5.1), we cannot exclude their contribution to regulatory mechanisms.
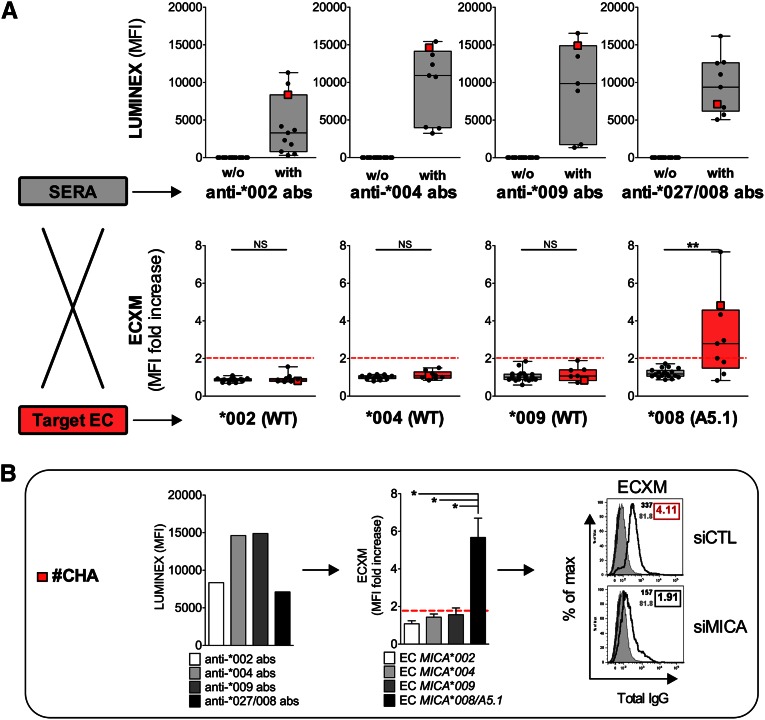
Graft ECs from MICA A5.1 Donors Are Predominant Targets of Anti-MICA Responses in Kidney Allograft Recipients
The prevalence of anti-MICA antibodies after transplantation is around 5%–9%, and the presence of antibodies decreases graft survival by 10% at 1 year after transplantation.20 To investigate the clinical effect of high MICA expression on graft ECs (i.e., transplant from a MICA A5.1 donor) we analyzed the specificity of anti-MICA responses in a cohort of kidney allograft recipients. Sera from 28 recipients with anti-MICA antibodies in the absence of anti-HLA antibodies were selected for the study. First, antibody specificity for the various MICA alleles of the sera was determined by Luminex assays. Then, we developed an EC cross-match assay (ECXM) to test the MICA-based reactivity of the sera on a cellular relevant target. To this end, sera were incubated with four EC cultures issued from donors homozygous for the four major MICA alleles represented in our donor cohort: MICA*002(WT), MICA*004(WT), MICA*009(WT), and MICA*008(A5.1). A representative example of ECXM positivity is shown in Supplemental Figure 1. Anti-MICA antibody binding to the various ECs was detected by FACS, as we previously described.26 Luminex analysis showed that when present in the sera, intensities of anti-MICA*008/*027 antibodies in the sera were not higher than intensities observed for other anti-MICA antibodies as determined by MFI. When tested on EC cultures expressing physiologic levels of membrane-bound MICA, sera bind only to ECs from MICA*008(A5.1) donors (Figure 5A). Surprisingly, similar or even higher antibody levels defined on beads coated with MICA*002, *004, or *009 proteins did not react with EC cultures from MICA*002(WT), *004(WT), or *009(WT) donors. These findings are illustrated in Figure 5B and Supplemental Figure 2, which compare the reactivity of a set of recipient’s sera on beads and ECs. Together, these data show, for the first time to our knowledge, that anti-MICA antibodies bind to target graft ECs in an allele-specific manner and also suggest that the level of MICA protein on A5.1 mutated EC is a key and limiting parameter. To test this hypothesis, ECXM were repeated on MICA*008(A5.1) ECs that previously been MICA-silenced using specific small interfering RNA (siRNA) (Figure 5B) (efficacy of MICA silencing is shown in Supplemental Figure 3). Here, we demonstrate that reducing MICA level on MICA*008(A5.1) ECs abrogates recipient’s sera binding to donor ECs.
Figure 5.
Donor ECs overexpressing MICA A5.1 antigens are predominant targets of anti-MICA antibodies in kidney allograft recipients. (A) Quantitative analysis of anti-MICA antibodies in 28 sera from transplant recipients by Luminex MICA single antigens and ECXM. The four major specificities are shown (MICA*002, *004, *009, *027/*008). Data from Luminex analysis (upper panel) are expressed as MFI. (A) ECXM (lower panel) was performed by incubating ECs homozygous for MICA*002(WT), *004(WT), *009(WT), and *008(A5.1) with the sera previously characterized by Luminex assay (upper panel). ECs issued from a MICA*010(null) donor and expressing no MICA at cell surface was used as a control. Results are expressed as a ratio between MFI obtained on indicated EC and MFI obtained on EC MICA*010(null) target. (B) For demonstration, data obtained with one serum (CHA) are presented (in red in part A). Serum reactivity determined by Luminex (left panel) and found by ECXM performed on four homozygous MICA ECs (medium panel) are shown. Positive ECXM was abolished after MICA silencing using MICA-specific siRNAs (right panel). Isotype-matched IgG controls are presented (dark-gray shading). ECXM on MICA*010(null) (light-gray shading) and MICA*008/A5.1 EC targets (black lines) are shown. Positive (red) and negative (black) ECXM scores are indicated.
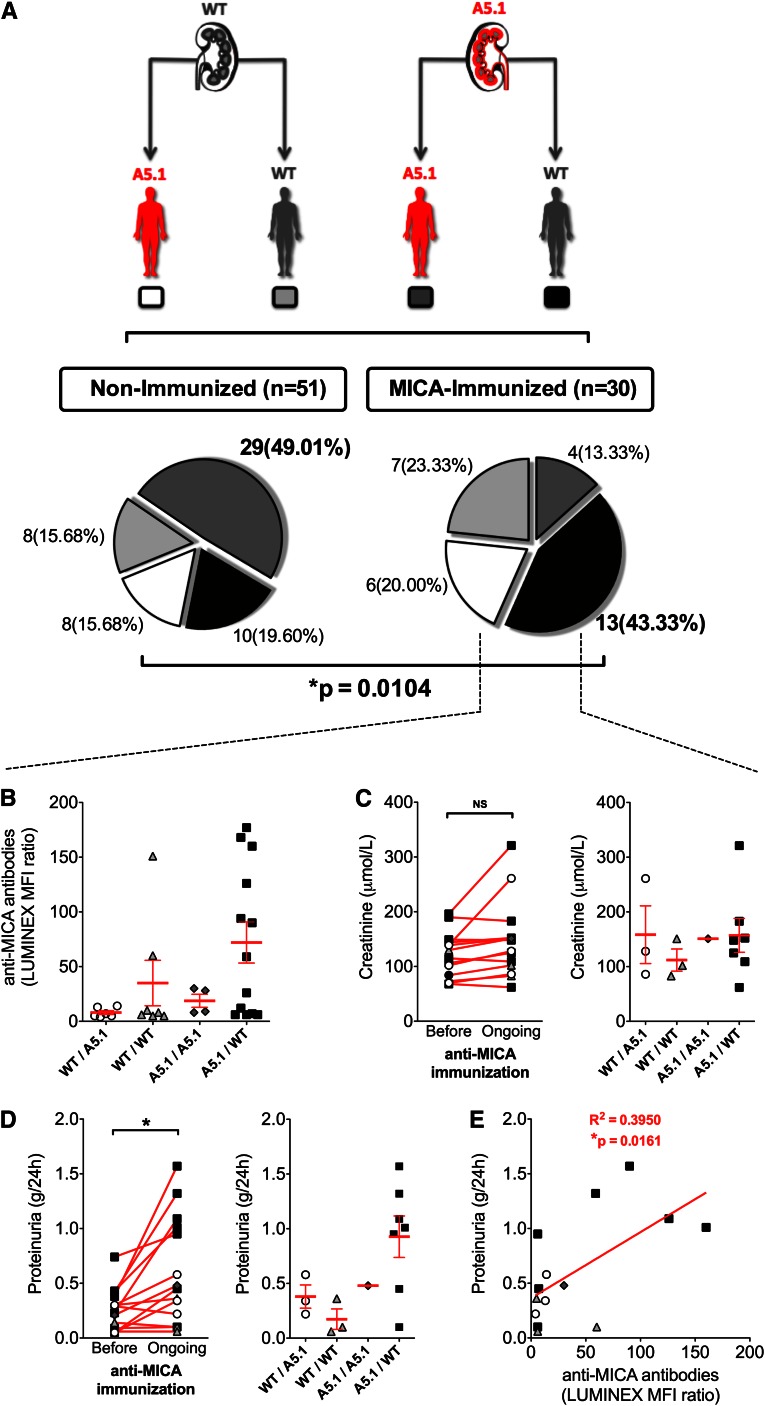
MICA A5.1 Mismatch between Transplant Donor and Recipient Is a Major Determinant for Anti-MICA Immunization and a Risk Factor for Transplant Outcome
As a consequence of the preceding findings attributing to MICA A5.1 changes in graft EC phenotype and function, in circulating forms of MICA released by ECs and the immunodominance of anti-MICA antibodies, we hypothesized that a mismatch in MICA A5.1 between donor (D) and recipient (R), in particular the combination D(A5.1)/R(WT), could promote MICA sensitization. To test this hypothesis, the clinical effect of a MICA A5.1 mismatch was investigated in a cohort of D/R pairs (n=81) with or without post-transplant MICA sensitization, as we previously determined by Luminex single antigen assays. Our threshold for positive MICA sensitization was an MFI ratio ≥5. Demographic data for both groups are presented in Table 1. MICA-immunized and MICA-nonimmunized patients in our cohort were selected among transplant recipients with at least 2 years of postgraft follow-up. Follow-up time and time to anti-MICA antibody detection after transplant in our cohort are reported in Table 1. Genotyping for A5.1 mutation was performed for both donors and recipients. D/R pairs were classified according to their MICA genotypes, and four MICA D/R combinations were defined. The distribution of the D/R combinations was then statistically analyzed in the two groups of patients according to MICA sensitization (Figure 6). Our results show that the combination D A5.1/R WT was overrepresented in the group of MICA-sensitized patients (n=13; 43.3%) compared with the group of nonimmunized recipients. Of note, the D/R full matching for A5.1 was predominant (49.01%) in the nonimmunized patients and low within the MICA-sensitized group (13.3%). Statistical analysis of MICA A5.1 and WT distribution among donors and recipients in both groups demonstrates a significant association between D/R MICA A5.1 mismatch and anti-MICA alloimmunization, particularly when donors carry the A5.1 mutation (P=0.0104).
Table 1.
Demographic data
| Patients | Nonimmunized | MICA-Immunized |
|---|---|---|
| Patients (n) | 51 | 30 |
| Donors | ||
| Age (yr) | 53±13 | 52±16 |
| Men/women, n/n (% men) | 29/22 (56.8) | 10/20 (33.3) |
| Recipients | ||
| Age (yr) | 52±11 | 48±16 |
| Men/women, n/n (% men) | 34/17 (66.6) | 17/13 (56.6) |
| Kidney graft/pancreas and kidney grafts, n/n (% kidney) | 47/4 (92.1) | 20/10 (66.6) |
| Postgraft follow-up time (mo) | 43.1±23.8 | 49.8±30.3 |
| Total HLA-A-B-DR mismatches (n) | 3.6±1.2 | 4.0±1.0 |
| Immunization anti-MICA (%) | None | 30 (100) |
| Time to post-transplant MICA antibody appearance (mo) | NA | 16.4±23.1 |
| Immunization anti-class I (%) | None | 5 (16.6) |
| Immunization anti-class II (%) | None | 4 (13.3) |
| Original disease (n) | ||
| Polycystic kidney disease | 16 | 4 |
| Diabetes mellitus | 7 | 12 |
| Hypertensive nephritis | 6 | 0 |
| Chronic renal insufficiency | 6 | 2 |
| Chronic interstitial nephritis | 4 | 2 |
| IgA nephritis | 2 | 2 |
| Glomerular disease | 2 | 2 |
| Other | 8 | 6 |
Values expressed with a plus/minus sign are the mean ± SD. NA, not applicable.
Figure 6.
MICA A5.1 mismatch is a major determinant for anti-MICA immunization and graft function. (A) A total of 81 first kidney transplant recipients were separated into two groups according to Luminex analysis data. The first group contains recipients with no immunization against MICA or HLA class I or II antigens after transplantation (n=51). The second group contains recipients who have developed antibodies against MICA antigens (n=30). Genomic DNA from both donors and recipients was sequenced for MICA A5.1 mutation, and D/R pairs were classified in four groups as follows: D WT/R A5.1 (white panel); D WT/R WT (light gray panel); D A5.1/R A5.1 (dark gray panel); D A5.1/R WT (black panel). The respective percentages of D/R pairs are depicted. Statistical analysis of association between D/R pair MICA A5.1 mutation mismatch and anti-MICA alloimmunization was done using a Pearson chi-squared test (*P<0.05). (B) Comparison of serum anti-MICA antibody relative titer (Luminex MFI ratio) in MICA-immunized recipients according to the four D/R genotype combinations. (C–E) Post-transplant serum creatinine and proteinuria were analyzed in a subset of 14 of 31 MICA-immunized patients (recipients with anti-HLA antibodies [n=8] were excluded). A comparative analysis of serum creatinine and proteinuria values before (C, left panel) and at time (D, left panel) of anti-MICA immunization appearance is shown. Serum creatinine and proteinuria levels at time to anti-MICA antibody peak value were compared according to MICA genotype combinations in D/R pairs (C and D, right panels, respectively). (E) Correlation analysis between post-transplant anti-MICA antibody relative titers and proteinuria values. Statistical analysis was performed using linear regression analysis and Pearson correlation (R2=0.40; *P<0.05). Servier Medical Art (http://www.servier.fr/servier-medical-art) was used in the creation of this figure.
Clinical and biologic data have been examined and compared according to MICA A5.1 mismatch between donor and recipient, with a particular focus on mismatched D/R pairs in which donors bear the A5.1 mutation. For all patients included in the study, post-transplantation time course of proteinuria, serum creatinine, relative titer of anti-MICA antibodies (expressed as a ratio of MFI), rejection episodes, and C4d deposition were collected and analyzed. Recipients with anti-HLA antibodies (n=8) were excluded from the analysis. Of note, titers of anti-MICA antibodies at peak of immunization seem globally higher in the group of mismatched D/R pairs with donors bearing the A5.1 mutation; there was a 2-to 9-fold increase compared with other groups (Figure 6B).
Although not significant because of the small size of our cohort, these data further support the idea that kidney transplants bearing the A5.1 mutation both qualitatively and quantitatively promote anti-MICA immunization. Functionally, we found no significant association with serum creatinine (Figure 6C). However, a significant increase in proteinuria concomitant with the peak in anti-MICA antibodies was observed (P=0.03) (Figure 6D). Finally, our data suggest a significant correlation between anti-MICA antibody titer (expressed in MFI ratio) and proteinuria (R2=0.40; P=0.02) in recipients (Figure 6E). This analysis further suggests, in our conditions, an MFI ratio of 50 as a threshold value to reach a clinical effect on proteinuria. Rejection episodes and biopsies have been examined in our cohorts (data not shown). Thirty-five biopsies (24 protocol biopsies and 11 biopsies for diagnosis) have been performed in the nonimmunized group (n=51 recipients). No C4d-positive biopsy was reported in this group. In the MICA-immunized group (n=22 recipients after exclusion of patients with anti-HLA antibodies), 12 biopsies have been done (6 protocol biopsies and 6 biopsies for diagnosis); 3 C4d-positive biopsies have been found. Two of three C4d-positive biopsies correspond to MICA A5.1 transplants in WT recipients, including one associated with rejection (Banff score, 2b).
Discussion
MICA*008 is the predominant allele in several populations27,28 and is consequently highly represented in transplant donor and recipient populations, as demonstrated here. MICA*008 is characterized by a mutation (A5.1) leading to a premature stop codon resulting in a truncated transmembrane and absent cytoplasmic tail.11 A previous study by Suemizu and colleagues reported on cellular changes associated with MICA A5.1 mutation in epithelial cells.13 However, except in recent studies,29,30 the functional effect of MICA genetic variant and the endothelial expression and regulation of MICA have yet not been explored.
By comparing MICA transcripts and proteins on cultured ECs from WT or MICA A5.1 heterozygous and homozygous donors, we showed that A5.1 mutation causes elevated MICA protein levels in the cell membrane (7- to 10-fold increase versus WT MICA). MICA A5.1 overexpression was not due to an increase in protein stability but rather reflects specific regulatory control at the transcription or post-transcription level. Multiple checkpoints operate to control MICA expression, including post-transcriptional mechanisms that allow faster regulation than could be achieved via transcriptional regulation. Several miRNAs play a key regulatory role in MICA post-transcriptional control.24 In our study, no significant quantitative change was found for miR-20a, -105, -106b, -373, -520b, -520f, or -636. Nevertheless, we cannot exclude the possibility that cell-specific (i.e., endothelial-specific) miRNAs contribute to MICA regulation in ECs. However, these miRNAs still remain to be identified. We investigated whether a specific polymorphism in the 5′ and 3′UTR regions of the MICA A5.1–associated alleles could, independently of miRNA level, impair miRNA binding and regulatory action. Genotyping the 5′ and 3′UTRs from various MICA alleles reveals several SNPs in these regions. However, among the SNPs that we found none was specifically associated with the A5.1 mutation. We speculate that changes in ubiquitination and proteasome degradation processes, previously reported as important events regulating NKG2DL expression31 altered in cell lines homozygous for MICA*008 allele,32 could account, at least partially, for accumulated MICA in A5.1 ECs.
Functionally, and consistent with a high level of MICA expression, A5.1 ECs increase NKG2D interaction and activity in NK cells compared with ECs expressing full-length MICA. As a consequence, activation of NK cells leads to an elevated NKG2D-dependent lysis of allogeneic A5.1 EC targets. In the transplantation setting, our data suggest that donors bearing the A5.1 allele could be more susceptible to cell lysis mediated by NKG2D-positive effector cells (NK and T CD8+). The role of NK cells in transplant immunology has probably been underestimated, and the contribution of NK cells to graft rejection is an emerging concept supported by recent studies.33–35 Our findings support the importance of endothelial expression of polymorphic MICA molecules and of possible mismatch between NK receptors and ligands expressed on recipients and donors, respectively.
Shedding of MICA is a mechanism by which human tumors evade NKG2D-mediated immune destruction.7,36 Soluble MICA induces host immune suppression by downregulation of surface NKG2D expression on NK cells and cytotoxic T lymphocytes influencing immune response and patient outcome in cancer7 and in cardiac transplantation.37 Exosomes derived from cancer cells also express ligands for NKG2D.38 Consistent with previous data in a tumor cell line,23 MICA*008(A5.1) accumulates in the EC supernatant as exosomes and mutated A5.1 ECs fail to release soluble cleaved MICA. We also show, for the first time, that primary ECs secrete large quantities of exosomes (even more than immature dendritic cells39). Palmitoylation of two cysteine residues in the intracellular domain of MICA is necessary for proteolytic shedding of MICA.40 The absence of these two cysteines in A5.1 mutated proteins could explain the lack of MICA A5.1 protein shedding from ECs. Together, these data suggest that the common MICA*008(A5.1) allele provides specific immunoregulatory properties to ECs with exacerbated NKG2D interactions mediated via cellular contact but also via exosomes released by ECs. The respective functional effect of soluble versus exosomal MICA remains to be defined.41
An initial report showing that MICA molecules, expressed at the endothelial cell surface, are recognized by specific antibodies in recipients14 has been sustained by clinical studies showing the detrimental effect of anti-MICA sensitization in kidney, heart, and lung transplantation.15,17,42,43 Nevertheless, the molecular basis for MICA alloimmunization remains to be established.44 Here, we show that MICA A5.1 molecules are primary targets for post-transplant antibodies in kidney allograft recipients because of higher MICA expression associated with this mutation. Our results also indicate that anti-MICA antibodies are alloreactive antibodies that bind to graft ECs in an allele-specific manner. These findings further support a role for MICA*008(A5.1) molecules as major antigenic determinants and targets for recipient sensitization. Clinically, our retrospective cohort study, which aimed to determine the incidence of an A5.1 mismatch, provides the first evidence of a statistically significant association between MICA A5.1 mutation in transplant donors and anti-MICA sensitization of kidney recipients. Further, we observed a statistical correlation between anti-MICA immunization and increased proteinuria in A5.1 mismatched combinations.
To conclude, the A5.1 mutation related to the common MICA*008 allele elicits an endothelial phenotype characterized by an exacerbated expression of MICA at cell surface, the exclusive production of exosomes expressing a high level of MICA as circulating MICA molecules. We propose that MICA mismatch with MICA A5.1 phenotype on the graft ECs in MICA WT recipients promotes NKG2D-dependent effector cellular and anti-MICA humoral responses, suggesting that particular MICA mismatching between donor and recipient may be a risk factor to consider in long-term transplant outcomes.
Concise Methods
Patients and Samples
Since 1999, primary EC cultures at ITUN (Nantes, France) have been prospectively isolated from transplant donors at the time of kidney transplantation and stored for research purposes26 in the DIVAT Sample Biocollection (French Health Minister project number 02G55).
To investigate the effect of MICA A5.1 mutation on recipient alloimmunization, 81 patients who underwent kidney transplantation between 2001 and 2010 at ITUN (CHU de Nantes, France) were included in the study. This cohort includes 30 D/R pairs with post-transplant anti-MICA antibodies. A control group (n=51) consisted of transplant D/R couples with no antibodies against HLA class I or class II or MICA antigens after transplantation. To allow matching analyses, only D/R pairs with genomic DNA available for MICA typing were selected. Serologic testing for anti-HLA class I and class II and anti-MICA antibodies before and after transplantation was performed by Luminex assays (Labscreen; One Lambda, Canoga Park, CA) at the Laboratoire HLA, EFS Pays de la Loire, Nantes, France.
To evaluate the effect of anti-MICA antibodies on graft function, clinical data, including time to anti-MICA appearance, relative titer of anti-MICA antibodies, serum creatinine, proteinuria, C4d deposition, and rejection episodes, were collected and were available for 14 of the 22 MICA-immunized patients (recipients with anti-HLA antibodies [n=8] were excluded from the analysis). Serum creatinine and proteinuria values were analyzed and compared before and at the time of anti-MICA appearance.
The study was performed according to the guidelines of the local ethics committee (CCPRB, CHU de Nantes, France). Before inclusion of the study, patients consented to the collection and storage of cells and DNA (BioCollection INSERM, French Health Minister project no. 02G55).
EC Isolation and Cell Culture
Isolation and establishment of primary cultures of vascular ECs were performed and characterized as we previously described.45 ECs were cultured in EC basal medium supplemented with 10% FCS, 0.004 ml/ml EC growth supplement/heparin, 0.1 ng/ml human epidermal growth factor, 1 ng/ml human basic fibroblast growth factor, 1 µg/ml hydrocortisone, 50 µg/ml gentamicin, and 50 ng/ml amphotericin B (C-22010, PromoCell, Heidelberg, Germany). ECs were used between passages 2 and 5. The human NK cell line, NKL, was grown in RPMI 1640 media supplemented with 10% FCS, 4 mM glutamine, 1 mM sodium pyruvate, and 200 U/ml recombinant IL-2 (R&D Systems). The NKL cell line was kindly provided by Dr. Eric Vivier (Marseille, France). For NKG2D analysis, NKLs were incubated with confluent EC monolayers, and NKG2D expression by NKL was then measured by FACS.
Reagents and Antibodies
The following mAbs were used: anti-pan HLA class I (anti-HLA-A, -B, and -C; clone W6/32) (American Type Cell Culture), anti-MICA (AMO1) and MICA/B (BAM01, BAMO3) were for BamOmab (Tubingen, Germany), anti-ULBP1, -2, and -3 (R&D Systems, Lille, France), anti–α5 integrin and anti–glyceraldehyde 3-phosphate dehydrogenase (both from Chemicon, Val de Fontenay, France). Anti-NKG2D mAbs as well as FITC and phycoerythrin-conjugated antimouse F(ab′)2 and antihuman IgG were from Jackson Immunoresearch Laboratories (West Grove, PA). For protein stability analysis, confluent EC monolayers were incubated with cycloheximide (50 µM, Sigma-Aldrich, St. Louis, MO) for the indicated period. For inhibition of soluble MICA release, ECs were treated with galardin (GM6001, 50 µg/ml, Sigma-Aldrich) for the indicated period.
MICA Genotyping, 5′ and 3′UTR Analysis
MICA typing of transplant donors and recipients was performed as we previously described.21 MICA proximal promoter, exons, and 3′UTR region were amplified with the following primers: MICApromo-F5′-ACGCGTTGTCTGTCCTGGAA-3′, MICApromo-R5′-GAGGTGCAAAAGGGAAGATG-3′ for the proximal promoter, MICA2-F 5′-ATTTCCTGCCCCAGGAAGGTTGG-3′ and MICA2-R 5′-AGACAGGTCCCTGCTCTCTG-3′ for exon2, MICA3-F 5′-TTCGGGAATGGAGAAGTCACTGC-3′, MICA3-R 5′-AAATGCCTTCATCCATAGCACAG-3′ for exon3; MICA4-F 5′-GACTTGCAGGTCAGGGGTCCC-3′, MICA4-R 5′-TGTCCCTACCCTGGCCTGACC-3′ for exon 4, MICA5-F 5′-CCTTTTTTTCAGGGAAAGTGC-3′, MICA5-R 5′-CCTTACCATCTCCAGAAACTGC-3′ for exon5, and MICA6-F; 5′-GATGTTGATGGAGTGATGGGA-3′, MICA6-R; 5-’ATGTTGATCAGGATGGTCTCGATC-3′for exon 6 and 5′UTR region.
PCR for MICA promoter, exons 5 and 6, and 5′UTR was performed using 100 ng DNA, 12.5 mM deoxyribonucleotides, 1× Taq buffer, 2 mM MgCl2, 0.1U Taq DNA polymerase (Invitrogen, Carlsbad, CA), and 10 pM of each oligonucleotide. For MICA exons 2, 3, and 4, we first performed PCR using 100 ng DNA, 15 pM of each primer,46 12.5 mM deoxyribonucleotides, 1 U of Herculase Taq (Stratagene, La Jolla, CA). Then, nested PCR was performed using 1 µl of PCR product and conditions reported above for exons 1, 5, and 6. PCR amplifications were carried out on PTC200 (Bio-Rad Laboratories, Hercules, CA) thermocycler. PCR products were run on 1% agarose gels for control. DNA sequencing was performed (Sequencing Core Facility INSERM/IFR26, Nantes, France) using a 48-capillary AB 3730 automatic system (Applied Biosystems, Foster City, CA) and analyzed using ChromasPro 1.5 software (Digital River GmbH, Shannon, Ireland).
RNA and Quantitative Real-Time PCR
Total RNA was isolated using the Trizol reagent (Invitrogen). After phenol-chloroform extraction and ethanol precipitation, total RNA (2 µg) was treated with ribonuclease-free Turbo-DNase (Ambion) before reverse transcription. Treated RNA was then reverse-transcripted with Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time quantitative PCR was performed in an ABI PRISM 7900 sequence detection application program using labeled TaqMan probes (Applied Biosystems). The following commercial ready-to-use primers/probe mixes were used (Applied Biosystems): MICA (Hs00792_m1) and hypoxanthine guanine phosphoribosyl transferase (Hs99999909_m1); the latter was used as an endogenous control to normalize RNA amount. Relative expression between a given sample and a reference sample was calculated according to the 2−ΔΔCt method, in which the reference represents 1-fold expression, as previously described.47
siRNAs and Silencing
siRNAs were designed, synthesized, and purchased from Ambion (Applied Biosystems). Cells were transiently transfected with 25 nM of nontargeting (siCONTROL) or MICA-specific siRNAs (siRNA#s8771, Ambion, Applied Biosystems) using LipofectAMINE RNAiMAX reagent according to the manufacturer's instructions (Invitrogen). The efficiency of silencing, determined by flow cytometry analyses in each experiment, ranged from 70% to 90%.
Immunoblotting
Cells or exosomes were lysed on ice in 20 mmol/L Tris-HCl (pH, 7.4), 137 mmol/L NaCl, 0.05% Triton X-100, 1 mmol/L supplemented with protease inhibitors (PIC, Sigma-Aldrich). Deglycosylation with endoglycosidase H and peptide:N-glycosidase F (Sigma-Aldrich) was performed as we described previously.48 Lysates (20 µg) were resolved by SDS-PAGE (12%) and subjected to Western immunoblot analysis using specific antibodies for MICA/MICB (BAMO1), α5 integrin, or glyceraldehyde 3-phosphate dehydrogenase, as well as secondary horseradish peroxidase–labeled antimouse antibodies (Cell Signaling Technology, St. Quentin-en-Yvelines, France). Antibody-bound proteins were detected using an ECL kit (Amersham) and luminescent image analyzer LAS-4000 (Fujifilm, Tokyo, Japan). Image analysis was performed with Multi Gauge software (Fujifilm).
Flow Cytometry, Luminex, and ECXM
For phenotype analysis, cells (1–2×105 cells/sample) were harvested, washed twice with PBS containing 1% BSA and 0.1% NaN3, and then incubated on ice for 30 minutes with a saturating concentration of primary antibodies. After three washes, cells were incubated with phycoerythrin-labeled goat antimouse F(ab′)2 IgG (Jackson Immunoresearch Laboratories) at 4°C for 30 minutes. Cells were fixed in 1% paraformaldehyde. Negative controls were performed using an isotype-matched IgG control.
For experimental endothelial cell cross-match, target ECs (1–2×105 cells/sample) isolated from MICA*002(WT), *004(WT), *008(A5.1), *009(WT), or *010(null) homozygous donors were suspended with Trypsin-EDTA (Gibco BRL), washed twice with PBS containing 1% BSA and 0.1% NaN3, and then incubated on ice for 30 minutes with 25 µl of the patient’s sera (dilution 1:4 in PBS/BSA/NaN3). After three washes, cells were incubated with FITC-labeled goat antihuman F(ab′)2 IgG (Jackson Immunoresearch Laboratories) at 4°C for 30 minutes. Cells were fixed in 1% paraformaldehyde. A positive ECXM result was obtained when MFI was at least twice the MFI obtained with EC control expressing MICA*010 (null), a MICA allele associated with no membrane-bound MICA due to unstable proteins.49 Negative controls were performed using a pool of normal human AB sera from 250 healthy male donors (EFS, Nantes, France) or isotype-matched IgG control (Jackson Immunoresearch Laboratories). Experiments were repeated at least three times. Sera from kidney recipients (n=28) tested in ECXM were provided by Laboratoire HLA (EFS Pays de la Loire, Nantes, France). These sera contain defined anti-MICA as determined by Luminex Single Antigen (LABScreen, One Lambda) but no anti-HLA class I or II antibodies. MICA single antigen kit allows detection of antibodies against MICA*001, *002, *004, *007, *009, *012, *017, *018, *019, and *027. MICA*027 and MICA*008 share the same extracellular region and have been considered equivalent in this study. Fluorescence was measured on 10,000 cells/sample using an FACS (FACScantoII; Becton Dickinson, Mountain View, CA) and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR) and Cytobank.50 Data are depicted in histograms plotting median ou geomean fluorescence intensity (MFI) on a four-decade logarithmic scale (x-axis) versus cell number (y-axis).
Cell-Mediated Cytotoxicity Assays
ECs labeled with 51Cr were incubated with NKL cells for 4 hours at various effector cell–to–target cell ratios. The supernatants were obtained after incubation and being subjected to gamma counting. The maximum or spontaneous release was defined as counts from samples incubated with 5% Triton X-100 or medium alone, respectively. Cytolytic activity was calculated with the following formula:
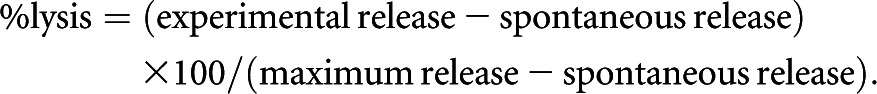 |
The spontaneous release in all assays was <20% of the maximum release.
Soluble MICA and Exosome Analysis
Detection of sMICA was done using a sandwich ELISA from BamoMab (Tubingen, Germany). For exosome purification, confluent EC monolayers were cultured in EC basal medium without serum or growth factor supplementation for 48 hours. Cell culture media were centrifuged twice for 10 minutes at 300 g and then centrifuged for 30 minutes at 10,000 g. Supernatants were centrifuged for 70 minutes at 100,000 g. Pellets containing exosomes were washed in PBS and centrifuged for 70 minutes at 100,000 g. Exosomes were stored at −80°C before analysis.
Statistical Analyses
The data are expressed as mean ± SEM and compared using nonparametric Mann-Whitney test or Kruskal-Wallis test (with Dunn multiple comparison post-test) if there were more than two conditions. Statistical analysis of the association between D/R MICA WT or A5.1 genotype mismatch and MICA alloimmunization was performed using Pearson chi-squared test. A correlation analysis between serum anti-MICA relative titer and proteinuria was performed using linear regression analysis and Pearson correlation. Statistical analysis was performed using GraphPad Prism software (GraphPad, San Diego, CA). A P value <0.05 was considered to represent a statistically significant difference. In all figures, one asterisk (*) denotes P<0.05, two asterisks (**) denotes P<0.01, three asterisks denotes P<0.001.
Disclosures
None.
Acknowledgments
The authors would like to thank Marie-Luce Chesneau (EFS, Nantes) for excellent technical assistance and Dr. Katia Gagne (EFS, Nantes) for helpful discussions.
This work was supported by grants from “L’Agence de Biomédecine” (Recherche et Greffes 2009) and “La Société de Néphrologie.” This work was also in part supported by a European Union–funded Integrated Project in Life Sciences, Genomics and Biotechnology for Health LSHB-CT-2006-037377. P.T. was supported by grants from “La région Pays de la Loire” (PROVASC project) and “La Société Francophone de Transplantation” (SFT).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2012080814/-/DCSupplemental.
References
- 1.Bahram S, Mizuki N, Inoko H, Spies T: Nucleotide sequence of the human MHC class I MICA gene. Immunogenetics 44: 80–81, 1996 [DOI] [PubMed] [Google Scholar]
- 2.Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T: Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A 96: 6879–6884, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T: Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285: 727–729, 1999 [DOI] [PubMed] [Google Scholar]
- 4.Ehrlich LIR, Ogasawara K, Hamerman JA, Takaki R, Zingoni A, Allison JP, Lanier LL: Engagement of NKG2D by cognate ligand or antibody alone is insufficient to mediate costimulation of human and mouse CD8+ T cells. J Immunol 174: 1922–1931, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Zwirner NW, Fernández-Viña MA, Stastny P: MICA, a new polymorphic HLA-related antigen, is expressed mainly by keratinocytes, endothelial cells, and monocytes. Immunogenetics 47: 139–148, 1998 [DOI] [PubMed] [Google Scholar]
- 6.Gasser S, Orsulic S, Brown EJ, Raulet DH: The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436: 1186–1190, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groh V, Wu J, Yee C, Spies T: Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 419: 734–738, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Groh V, Bruhl A, El-Gabalawy H, Nelson JL, Spies T: Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc Natl Acad Sci U S A 100: 9452–9457, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodil N, Laloux L, Wanner V, Pellet P, Hauptmann G, Mizuki N, Inoko H, Spies T, Theodorou I, Bahram S: Allelic repertoire of the human MHC class I MICA gene. Immunogenetics 44: 351–357, 1996 [DOI] [PubMed] [Google Scholar]
- 10.Stephens HA: MICA and MICB genes: can the enigma of their polymorphism be resolved? Trends Immunol 22: 378–385, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Mizuki N, Ota M, Kimura M, Ohno S, Ando H, Katsuyama Y, Yamazaki M, Watanabe K, Goto K, Nakamura S, Bahram S, Inoko H: Triplet repeat polymorphism in the transmembrane region of the MICA gene: A strong association of six GCT repetitions with Behçet disease. Proc Natl Acad Sci U S A 94: 1298–1303, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto K, Ota M, Ohno S, Mizuki N, Ando H, Katsuyama Y, Maksymowych WP, Kimura M, Bahram S, Inoko H: MICA gene and ankylosing spondylitis: Linkage analysis via a transmembrane-encoded triplet repeat polymorphism. Tissue Antigens 49: 503–507, 1997 [DOI] [PubMed] [Google Scholar]
- 13.Suemizu H, Radosavljevic M, Kimura M, Sadahiro S, Yoshimura S, Bahram S, Inoko H: A basolateral sorting motif in the MICA cytoplasmic tail. Proc Natl Acad Sci U S A 99: 2971–2976, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zwirner NW, Marcos CY, Mirbaha F, Zou Y, Stastny P: Identification of MICA as a new polymorphic alloantigen recognized by antibodies in sera of organ transplant recipients. Hum Immunol 61: 917–924, 2000 [DOI] [PubMed] [Google Scholar]
- 15.Sumitran-Holgersson S, Wilczek HE, Holgersson J, Söderström K: Identification of the nonclassical HLA molecules, mica, as targets for humoral immunity associated with irreversible rejection of kidney allografts. Transplantation 74: 268–277, 2002 [DOI] [PubMed] [Google Scholar]
- 16.Mizutani K, Terasaki P, Bignon JD, Hourmant M, Cesbron-Gautier A, Shih RNJ, Pei R, Lee J, Ozawa M: Association of kidney transplant failure and antibodies against MICA. Hum Immunol 67: 683–691, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Zou Y, Stastny P, Süsal C, Döhler B, Opelz G: Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med 357: 1293–1300, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Hankey KG, Drachenberg CB, Papadimitriou JC, Klassen DK, Philosophe B, Bartlett ST, Groh V, Spies T, Mann DL: MIC expression in renal and pancreatic allografts. Transplantation 73: 304–306, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Suárez-Alvarez B, López-Vázquez A, Gonzalez MZ, Fdez-Morera JL, Díaz-Molina B, Blanco-Gelaz MA, Pascual D, Martínez-Borra J, Muro M, Alvarez-López MR, López-Larrea C: The relationship of anti-MICA antibodies and MICA expression with heart allograft rejection. Am J Transplant 7: 1842–1848, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Terasaki PI, Ozawa M, Castro R: Four-year follow-up of a prospective trial of HLA and MICA antibodies on kidney graft survival. Am J Transplant 7: 408–415, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Tonnerre P, Gérard N, Chatelais M, Charreau B: MICA gene polymorphism in kidney allografts and possible impact of functionally relevant variants. Transplant Proc 42: 4318–4321, 2010 [DOI] [PubMed] [Google Scholar]
- 22.Bahram S, Inoko H, Shiina T, Radosavljevic M: MIC and other NKG2D ligands: From none to too many. Curr Opin Immunol 17: 505–509, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Ashiru O, Boutet P, Fernández-Messina L, Agüera-González S, Skepper JN, Valés-Gómez M, Reyburn HT: Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res 70: 481–489, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stern-Ginossar N, Mandelboim O: An integrated view of the regulation of NKG2D ligands. Immunology 128: 1–6, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee S, Pal JK: Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell 101: 251–262, 2009 [DOI] [PubMed] [Google Scholar]
- 26.Canet E, Devallière J, Gérard N, Karam G, Giral M, Charreau B, Coupel S: Profiling posttransplant circulating antibodies in kidney transplantation using donor endothelial cells. Transplantation 93: 257–264, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Petersdorf EW, Shuler KB, Longton GM, Spies T, Hansen JA: Population study of allelic diversity in the human MHC class I-related MIC-A gene. Immunogenetics 49: 605–612, 1999 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Lazaro AM, Lavingia B, Stastny P: Typing for all known MICA alleles by group-specific PCR and SSOP. Hum Immunol 62: 620–631, 2001 [DOI] [PubMed] [Google Scholar]
- 29.Shafi S, Vantourout P, Wallace G, Antoun A, Vaughan R, Stanford M, Hayday A: An NKG2D-mediated human lymphoid stress surveillance response with high interindividual variation. Sci Transl Med 3: 113ra124, 2011 [DOI] [PMC free article] [PubMed]
- 30.Lin D, Lavender H, Soilleux EJ, O’Callaghan CA: NF-κB regulates MICA gene transcription in endothelial cell through a genetically inhibitable control site. J Biol Chem 287: 4299–4310, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coscoy L, Sanchez DJ, Ganem D: A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol 155: 1265–1273, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, Wills M, Lehner PJ: Down-regulation of NKG2D and NKp80 ligands by Kaposi’s sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci U S A 105: 1656–1661, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hidalgo LG, Campbell PM, Sis B, Einecke G, Mengel M, Chang J, Sellares J, Reeve J, Halloran PF: De novo donor-specific antibody at the time of kidney transplant biopsy associates with microvascular pathology and late graft failure. Am J Transplant 9: 2532–2541, 2009 [DOI] [PubMed] [Google Scholar]
- 34.Hidalgo LG, Sis B, Sellares J, Campbell PM, Mengel M, Einecke G, Chang J, Halloran PF: NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: Evidence for NK cell involvement in antibody-mediated rejection. Am J Transplant 10: 1812–1822, 2010 [DOI] [PubMed] [Google Scholar]
- 35.Todorova D, Sabatier F, Doria E, Lyonnet L, Vacher Coponat H, Robert S, Despoix N, Legris T, Moal V, Loundou A, Morange S, Berland Y, George FD, Burtey S, Paul P: Fractalkine expression induces endothelial progenitor cell lysis by natural killer cells. PLoS ONE 6: e26663, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salih HR, Rammensee H-G, Steinle A: Cutting edge: Down-regulation of MICA on human tumors by proteolytic shedding. J Immunol 169: 4098–4102, 2002 [DOI] [PubMed] [Google Scholar]
- 37.Suárez-Alvarez B, López-Vázquez A, Díaz-Molina B, Bernardo-Rodríguez MJ, Alvarez-López R, Pascual D, Astudillo A, Martínez-Borra J, Lambert JL, González S, López-Larrea C: The predictive value of soluble major histocompatibility complex class I chain-related molecule A (MICA) levels on heart allograft rejection. Transplantation 82: 354–361, 2006 [DOI] [PubMed] [Google Scholar]
- 38.Clayton A, Tabi Z: Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol Dis 34: 206–213, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA: Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29: 341–345, 2011 [DOI] [PubMed] [Google Scholar]
- 40.Agüera-González S, Gross CC, Fernández-Messina L, Ashiru O, Esteso G, Hang HC, Reyburn HT, Long EO, Valés-Gómez M: Palmitoylation of MICA, a ligand for NKG2D, mediates its recruitment to membrane microdomains and promotes its shedding. Eur J Immunol 41: 3667–3676, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Théry C, Ostrowski M, Segura E: Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9: 581–593, 2009 [DOI] [PubMed] [Google Scholar]
- 42.Mizutani K, Terasaki P, Rosen A, Esquenazi V, Miller J, Shih RNJ, Pei R, Ozawa M, Lee J: Serial ten-year follow-up of HLA and MICA antibody production prior to kidney graft failure. Am J Transplant 5: 2265–2272, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q, Cecka JM, Gjertson DW, Ge P, Rose ML, Patel JK, Ardehali A, Kobashigawa JA, Fishbein MC, Reed EF: HLA and MICA: Targets of antibody-mediated rejection in heart transplantation. Transplantation 91: 1153–1158, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou Y, Qin Z, Silveus A, Fan Y, Stastny P: Polymorphisms of MICA recognized by human alloantibodies. Immunogenetics 61: 91–100, 2009 [DOI] [PubMed] [Google Scholar]
- 45.Coupel S, Leboeuf F, Boulday G, Soulillou J-P, Charreau B: RhoA activation mediates phosphatidylinositol 3-kinase-dependent proliferation of human vascular endothelial cells: An alloimmune mechanism of chronic allograft nephropathy. J Am Soc Nephrol 15: 2429–2439, 2004 [DOI] [PubMed] [Google Scholar]
- 46.Katsuyama Y, Ota M, Ando H, Saito S, Mizuki N, Kera J, Bahram S, Nose Y, Inoko H: Sequencing based typing for genetic polymorphisms in exons, 2, 3 and 4 of the MICA gene. Tissue Antigens 54: 178–184, 1999 [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 48.Coupel S, Moreau A, Hamidou M, Horejsi V, Soulillou J-P, Charreau B: Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood 109: 2806–2814, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Li Z, Groh V, Strong RK, Spies T: A single amino acid substitution causes loss of expression of a MICA allele. Immunogenetics 51: 246–248, 2000 [DOI] [PubMed] [Google Scholar]
- 50.Kotecha N, Krutzik PO, Irish JM: Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom Chapter 10: Unit10.17, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]