Abstract
We have recently proposed a mathematical framework for crowd-sourcing of biomedical image analysis and diagnosis through digital gaming. Here we review our recent progress on this gaming platform and demonstrate its viability for telediagnosis of malaria, achieving an accuracy that is within less than 2 percent of that of a trained expert.
Introduction
Medical imaging has gone through a co-evolution along with the computer industry over the past three decades, with each medical imaging modality benefiting in major ways from the ever-increasing abilities of modern computers. The possibility to capture, store, and manipulate images digitally has brought upon a new age of medical imaging with a significant shift in focus toward more complex analysis software. Through shear computation and clever mathematical algorithms, modern medical imaging devices are capable of producing higher-quality images faster while exposing patients to much less harmful radiation. As part of this trend, an emerging field where we have focused most of our own efforts over the past few years is that of computational microscopy.1–7
Another dimension of medical imaging's evolution has been a consequence of rapid advances in telecommunications and the coming of age of the Internet. These days an X-ray or a microscope slide image can be viewed almost instantaneously thousands of miles away from the point of capture by an expert who had no involvement in the imaging procedure. This unprecedented level of access to medical images and data is now opening up new approaches to medical diagnosis, heralding the age of telemedicine, where one can outsource medical diagnosis to doctors in faraway locations, while making it significantly easier to get a second opinion on a particular diagnosis.
This brings an interesting question to mind: What if instead of getting a second opinion, one could quickly get tens or hundreds of opinions all at once? Would it be possible to combine responses from many individuals to arrive at an accurate diagnosis decision? Put differently: Could we crowd-source medical diagnosis?
One of the first records on the use of the “wisdom of the crowd” goes back to the 19th century statistician Sir Francis Galton, who in 1907 reported on a peculiar contest that he encountered at a livestock fair.8 For a small cost, the participants entered a contest to guess the weight of an ox on display, with those coming closest to the true weight winning prizes. Having about 800 contestants, no one guessed the exact weight. However, Galton observed that the median of the weights guessed by all the participants was only 9 lbs more than the true weight of 1,198 lbs—or just off by 0.8 percent!
Over the past decade, there have been several projects that have crowd-sourced difficult pattern analysis and recognition tasks to individuals around the world. Perhaps one of the most successful of these is reCAPTCHA9—a crowd-sourcing project for digitizing books and other nondigital prints. FoldIt10,11 and EteRNA12 are two other projects that have crowd-sourced the task of scientific discovery to ordinary individuals through entertaining games. They all make use of the superior pattern-recognition capabilities of humans to solve tasks that would be difficult and time-consuming to solve by computers.
We have recently taken a similar approach to test the idea of crowd-sourcing medical diagnosis and initially tackled the problem of identifying malaria-infected blood cells, a task that normally demands professional training.13,14 Malaria is a major health problem in many tropical and subtropical climates, including much of sub-Saharan Africa. It is a disease that affects a rather large number of people every year. According to the World Health Organization's estimate, there were 174 million cases of malaria in 2010 that resulted in 655,000 deaths, where >90% of these deaths occurred in Africa.15
For diagnosis of malaria, conventional light microscopy remains as one of the gold standard methods, with 165 million cases having been diagnosed through this method in 2010.15 A pathologist must typically check on the order of 1,000 individual red blood cells under a high-magnification light microscope before being able to reliably call a sample healthy or negative. This, unfortunately, is a time-consuming and challenging task given the large number of cases observed, resulting in a false-positive rate of, for example, approximately 60 percent in some developing countries.16 Such a high false-positive rate can lead to unnecessary treatments and hospitalizations.
Materials and Methods
To test our idea, we started by creating entertaining digital games (termed “BioGames”) (Fig. 1) where the players were presented with a set of red blood cell images taken from potentially infected samples.13,14 They were allowed to choose to digitally “kill” or “bank” the infected and healthy cells, respectively. To be able to later combine the information generated by multiple gamers, we had to know how they were doing in terms of diagnosis accuracy as they went along playing the game. Toward this end, by carefully embedding some known images (i.e., control images) in our games, we were able both to assign scores to gamer performances and to quantify how well they could diagnose individual cells. Once the responses from all the gamers were collected, we could then combine them using techniques borrowed from telecommunications theory (by mathematically treating each gamer as a noisy telecommunication channel13) to yield much more reliable diagnoses for the unknown blood cell images. To quantify the performance of our gamers and our fusion algorithms, we also asked trained medical experts to individually check and label all of the images in our database, creating a gold standard label for each cell.
FIG. 1.
“BioGames” can be played on multiple platforms. The gamer can digitally “kill” or “bank” infected and healthy cells, respectively.14 Color images available online at www.liebertonline.com/g4h
Results and Discussion
Our initial experiments were conducted at the University of California, Los Angeles with a set of 31 students (from the School of Engineering [i.e., nonexperts]) as gamers.13 In these experiments, the combined performance of the gamers diagnosing 7,045 individual red blood cell images reached an accuracy of 98.78 percent when compared against a trained professional's responses, resulting in a sensitivity and specificity of 97.81% and 99.05%, respectively.13
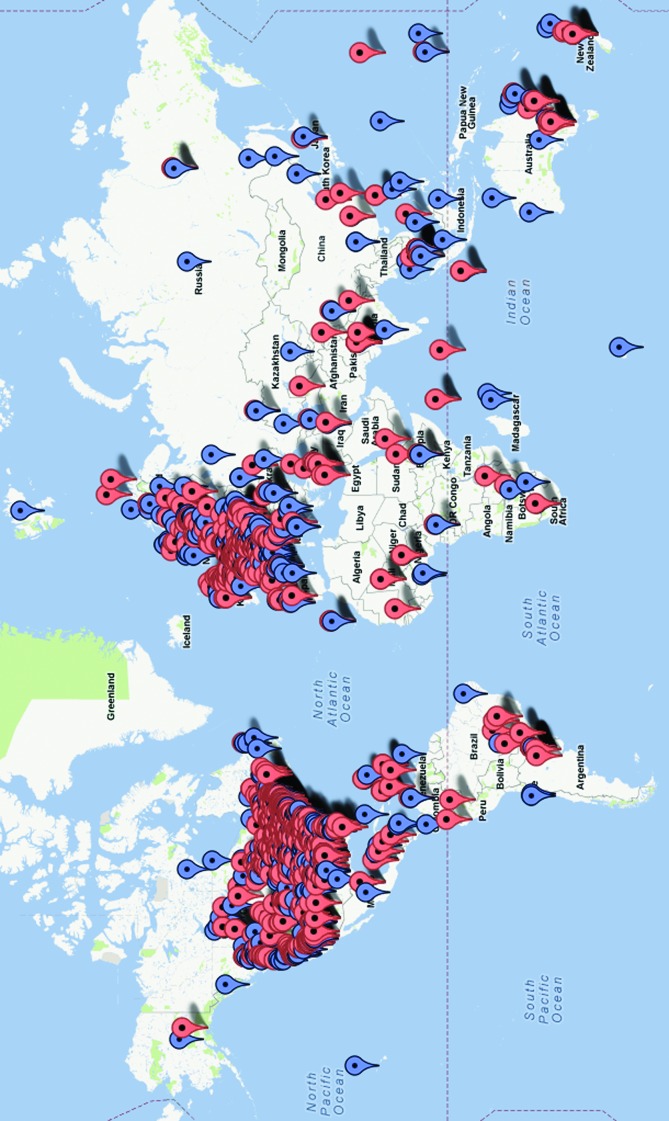
Following the success of our initial internal trials, in the spring of 2012 we made the “BioGames” platform public through a Web interface (Fig. 1).14 This time we used a database of red blood cell images (comprising approximately 8,500 individual cells) provided by the U.S. Centers for Disease Control and Prevention with each cell image labeled by nine independent experts, creating our gold standard diagnoses. Within a span of less than 4 months, we were able to collect approximately 1.5 million individual cell diagnoses, made by gamers from more than 70 countries around the world as illustrated in Figure 2.14
FIG. 2.
Each balloon in this map indicates an individual gamer, where a blue balloon symbolizes a gamer with ≥100 cell diagnoses and a red balloon symbolizes a gamer with <100 cell diagnoses. Since May 2012, we have had more than 2,150 gamers from 77 countries, who collectively generated >1.5 million individual cell diagnoses.14 Color images available online at www.liebertonline.com/g4h
The nature of this public “BioGames” experiment was different from the initial one, mainly in that only a few individuals had fully completed the game, labeling all 8,500 cell images. The large majority of the gamers had only labeled a relatively small portion of the entire image set, and therefore we only used the diagnosis data collected from those “dedicated” individuals who had labeled at least 100 cells. This yielded more than 1 million cell labels collected from approximately 1,000 untrained individuals scattered across 60 different countries (see the blue balloons in Fig. 2). Once the responses of these gamers were combined using a maximum a posteriori probability approach,13 the collective diagnosis accuracy of our crowd was once again remarkable: We achieved an overall accuracy of 98.13 percent, where 98.78 percent of cells labeled as healthy and 76.85 percent of cells labeled as infected were in fact correctly labeled.14
Looking forward, we believe that the “BioGames” platform can be extended to other biomedical image analysis and diagnosis problems, such as microscopic analysis of Pap smears for diagnosis of, for example, cervical cancer. Furthermore, in addition to binary diagnostic decisions, it can also accommodate a wider range of nonbinary diagnostic possibilities. A component of this platform that requires further work, and is crucial for its success and wide-scale deployment, is the game itself. In order to attract and keep the interest of gamers, highly entertaining games that can be played on a multitude of platforms such as PCs, mobile phones, tablets, and other gaming devices need to be designed/created. As such, a possible avenue that is worth exploring toward continuous development of entertaining games is to open up the “BioGames” platform to developers around the world and allow for a game-developer community to form.
We should also note that there are important legal issues that need to be addressed before such a platform can become readily available for mainstream use in clinical settings.13,14 As a matter of fact, in our next work we will be addressing this important question by shedding more light on implementation of the “BioGames” platform for a crowd of medical experts, where the gold standard diagnoses are inferred by combining individual responses of these experts, without any prior diagnostic information. Such a professional crowd (put together using, for example, monetary incentives such as a pay-per-image scheme), which remotely diagnoses, for instance, pathology samples based on microscopic images of specimens, could open up new business opportunities for telemedicine and could even be useful for training and/or monitoring of other medical experts.
In summary, we believe that such innovative uses of digital gaming technologies for telemedicine applications could in the very near future open up new avenues for delivering faster, more accurate, and cost-effective diagnosis to the masses globally, as well as for wide-scale and efficient training of medical professionals and diagnosticians.
Acknowledgments
A.O. gratefully acknowledges the support of the Presidential Early Career Award for Scientists and Engineers, an ARO Young Investigator Award, an NSF CAREER Award, the ONR Young Investigator Award 2009, and the NIH Director's New Innovator Award DP2OD006427 from the Office of the Director, National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Tseng D. Mudanyali O. Oztoprak C, et al. Lensfree microscopy on a cellphone. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishara W. Su TW. Coskun AF. Ozcan A. Lens- free on-chip microscopy over a wide field-of-view using pixel super-resolution. Optics Express. 2010;18:11181–11191. doi: 10.1364/OE.18.011181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isikman SO. Bishara W. Sikora U, et al. Field-portable lensfree tomographic microscope. Lab Chip. 2011;11:2222–2230. doi: 10.1039/c1lc20127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu H. Mavandadi S. Coskun AF, et al. Optofluidic fluorescent imaging cytometry on a cell phone. Anal Chem. 2011;83:6641–6647. doi: 10.1021/ac201587a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isikman SO. Bishara W. Mavandadi S, et al. Lensfree optical tomographic microscope with a large imaging volume on a chip. Proc Natl Acad Sci U S A. 2011;108:7296–7301. doi: 10.1073/pnas.1015638108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H. Sikora U. Ozcan A. Quantum dot enabled detection of Escherichia coli using a cell-phone. Analyst. 2012;137:2541–2544. doi: 10.1039/c2an35071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenbaum A. Sikora U. Ozcan A. Field-portable wide-field microscopy of dense samples using multi-height pixel super-resolution based lensfree imaging. Lab Chip. 2012;12:1242–1245. doi: 10.1039/c2lc21072j. [DOI] [PubMed] [Google Scholar]
- 8.Galton F. Vox Populi [reprint from 1907] Nature. 1949;75:450–451. [Google Scholar]
- 9.von Ahn L. Maurer B. McMillen C, et al. reCAPTCHA: Human-based character recognition via Web security measures. Science. 2008;321:1465–1468. doi: 10.1126/science.1160379. [DOI] [PubMed] [Google Scholar]
- 10.Cooper S. Khatib F. Treuille A, et al. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatib F. Cooper S. Tyka MD, et al. Algorithm discovery by protein folding game players. Proc Natl Acad Sci U S A. 2011;108:18949–18953. doi: 10.1073/pnas.1115898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.EteRNA. Eterna—played by humans, scored by nature. 2011. http://eterna.cs.cmu.edu/eterna_page.php?page=me_tab. [Sep 15;2012 ]. http://eterna.cs.cmu.edu/eterna_page.php?page=me_tab
- 13.Mavandadi S. Dimitrov S. Feng S, et al. A. Distributed medical image analysis and diagnosis through crowd-sourced games: A malaria case study. PLoS One. 2012;7:e37245. doi: 10.1371/journal.pone.0037245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavandadi S. Dimitrov S. Feng S, et al. Crowd-sourced BioGames: Managing the big data problem for next-generation lab-on-a-chip platforms. Lab Chip. 2012 Aug 23; doi: 10.1039/C2LC40614D. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. Geneva: World Health Organization; 2011. World Malaria Report 2011. [Google Scholar]
- 16.Amexo M. Tolhurst R. Barnish G. Bates I. Malaria misdiagnosis: Effects on the poor and vulnerable. Lancet. 2004;364:1896–1898. doi: 10.1016/S0140-6736(04)17446-1. [DOI] [PubMed] [Google Scholar]