Abstract
Catechol estrogens are carcinogenic, probably because of their estrogenicity and potential for further oxidative metabolism to reactive quinones. Estrogenic quinones cause oxidative DNA damage as well as form mutagenic depurinating adenine and guanine adducts. O-Methylation by catechol-O-methyltransferase (COMT) blocks their estrogenicity and prevents their oxidation to quinones. A single gene encodes both membrane bound (MB) and soluble (S) forms of COMT. The COMT gene contains 34 single nucleotide polymorphisms (SNPs). The valine108 (S-COMT)/158 (MB-COMT) SNP encodes a low activity form of COMT and has been widely studied as a putative risk factor for breast cancer, with inconsistent results. Investigations of two other SNPs in the promoter of MB-COMT that may affect its expression have also provided mixed results. Future studies on the role of COMT in breast cancer should incorporate measurement of biomarkers that reflect COMT activity and its protective effects.
Introduction
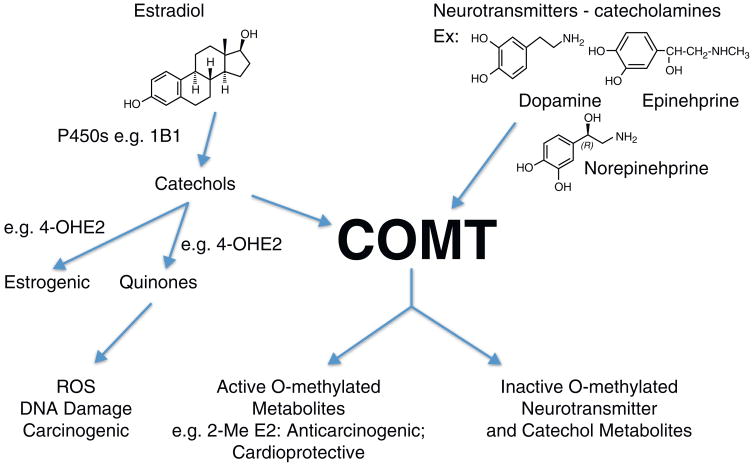
Axelrod and Tomchick first described and characterized catechol-O-methyltransferase (COMT) in the cytosolic fraction of liver and other organs as the enzyme responsible for catalyzing the O-methylation of catecholamine neurotransmitters [1]. COMT also methylates catechol xenobiotics and catechol metabolites of estradiol and estrone (E2, E1) [2] (Fig. 1).
Figure 1.
Endogenous COMT substrates. O-Methylation of endogenous catechols, catecholamine neurotransmitters and catechol estrogens by COMT results in their inactivation. However, 2-MeE2 has been shown to be anticarcinogenic and cardioprotective.
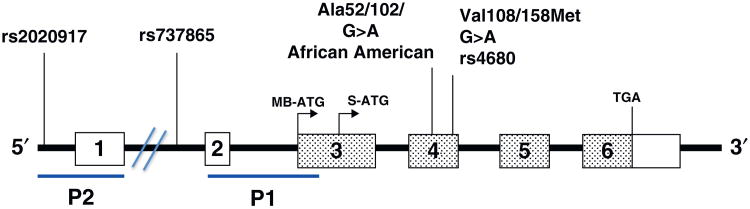
A single gene encodes COMT, Fig. 2, with 80% protein sequence similarity between rat and human [3]. COMT is expressed in two forms from two promoters, a 24 kDa cytosolic protein (S-COMT) and a 30 kDa membrane-bound protein (MB-COMT) [2,3]. The additional N-terminal region in MB-COMT contains membrane anchoring hydrophobic amino acids [4]. In humans, with the exception of brain, S-COMT is expressed at greater levels than MB-COMT. MB-COMT has been localized to the endoplasmic reticulum and variably on the plasma membrane and nuclear envelope in a variety of cell types [5,6] and in breast epithelial normal and tumor tissue [7]. COMT expression is greatest in liver and kidney.
Figure 2.
Human COMT gene structure and selected polymorphisms. The COMT gene comprises six exons indicated by the boxes. Open boxes represent untranslated exons and stippled boxes translated exons. Common SNPs discussed in the text are indicated along with the translation initiation and termination sites for MB- and S-COMT.
O-Methylation of catechol estrogens
COMT catalyzes O-methylation of 2-OHE2/E1 at the 2-OH and 3-OH positions and of 4-OHE2/E1 at the 4-OH position with the methyl group derived from S-adenosylmethionine. The catechol metabolites of E2 and E1, particularly 4-OH E2/E1, have been shown to be carcinogenic [8,9]. They are estrogenic with the relative estrogenicity of 4-OHE2 > 2-OHE2 = E2[10–12]. O-Methylation by COMT eliminates their estrogenicity. The E2/E1 catechols also undergo oxidative metabolism to quinones that can redox cycle to form reactive oxygen species (ROS). ROS cause oxidative DNA damage and the estrogen quinones, especially the 4-OH E2/E1-derived quinone, form depurinating adducts with adenine and guanine [13].
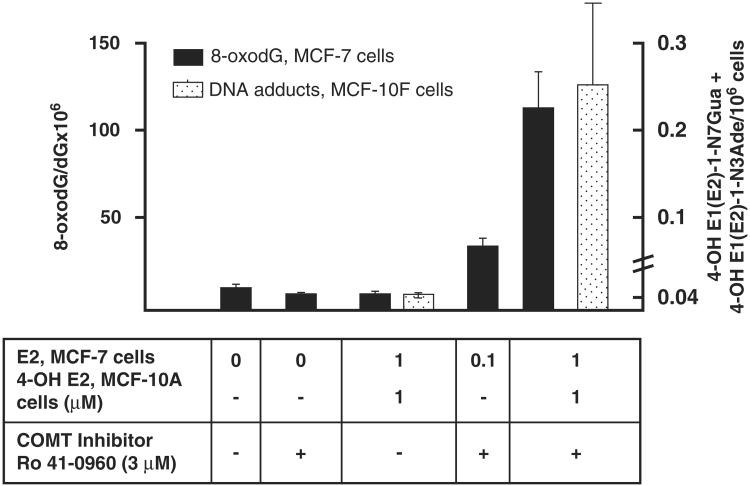
COMT plays a central role in protection from formation of DNA damage as shown in Fig. 3. In MCF-7 cells, inhibition of COMT increased the amount of oxidative DNA damage (8-oxo-dG) [14]. In MCF-10F cells, COMT inhibition dramatically increased the amount of estrogen 4-OH-quinone depurinating adducts [15]. These results are consistent with the notion that COMT is a ‘gate keeper’ phase II enzyme responsible for blocking the estrogenicity of catechol estrogens and preventing their oxidative metabolism to genotoxic quinone metabolites. Furthermore, as illustrated in Fig. 1, 2-MeE2 has anticarcinogenic and cardioprotective effects mediated through anti-angiogenic and growth inhibitory effects [11,16–19]. Thus, COMT-mediated O-methylation of the 2-OHE2 catechol estrogen not only blocks its potential estrogenic and genotoxic effects but generates a potentially protective metabolite.
Figure 3.
Effect of COMT inhibition on E2 or 4-OH E2-induced DNA damage. The amount of oxidative DNA damage represented by 8-Oxo-dG is indicated on the left ordinate and solid bars; the amount of 4-OHE1/E2 quinone-derived DNA adducts is indicated on the right ordinate and stippled bars. Data in this figure were taken from [14,15].
COMT gene structure and polymorphisms
The COMT gene is located on chromosome 22 and contains six exons, the first two of which are noncoding [2–4], Fig. 2. Promoter P1 regulates expression of a 1.3 kb S-COMT mRNA. Promoter P2 regulates the expression of a 1.5 kb mRNA that includes noncoding exons 1 and 2 [3,4].
The COMT gene is polymorphic. While COMT activity was known to vary among individuals, a seminal familial study of human erythrocytes revealed that a low activity phenotype was inherited as an autosomal recessive trait [20,21]. The low activity form of COMT had one-third to one-quarter of the activity of the wild type (WT) enzyme and exhibited thermolability [20]. The single nucleotide polymorphism (SNP) responsible [22] is a G>A nonsynonymous mutation in exon 4 causing a change of valine108 (S-COMT)/158 (MB-COMT) to methionine, Fig. 2. The mutation is not in the catalytic domain and does not affect its kinetic capacity [23–25]. The Val108/158Met SNP is common in the Caucasian population with an allele frequency of approximately 50%. Thus 25% of Caucasians are homozygous for the low activity form of COMT. This mutation has been widely studied with regard to its association with neurological diseases and cancer (see below).
Numerous additional polymorphisms have been identified. The COMT gene was resequenced from exon 2 through 6 [26]. Of the 24 SNPs detected, one was a novel second nonsynonymous polymorphism, Ala52/102Thr only observed in African American women (Fig. 2). Saito et al. analyzed DNA from 96 Japanese subjects and detected 34 SNPs [27]. Ji et al. resequenced DNA from Caucasian subjects with inclusion of 1 kb on each side of exon 1 and detected 33 SNPs [28]. In particular, two SNPs designated 5′FR(−628; C>T; rs2020917) and I1(701; A>G; rs737865) located in the P1 promoter (Fig. 2) were associated with reduced breast cancer risk ([28] and see below).
Determinants of COMT expression and activity levels
Expression vectors for the WT and the two nonsynonymous alleles encoding the variant forms of S-COMT, Thr102 and Met108 (Fig. 2) were transfected into COS-1 and HEK293 cells for functional analysis [26]. Compared to WT, COMT activity was reduced in cells expressing Met108 but not in cells expressing the Thr52 variant. The Met108 variant demonstrated thermo instability, as expected, whereas the Thr52 variant did not. Kinetic analysis revealed small differences in Km that were not sufficient to account for differences in activity. However, Western blot analysis demonstrated a reduced amount of Met108 S-COMT protein and a somewhat increased amount of Thr52 protein.
These studies were extended to 33 human liver biopsy samples genotyped for the Met108 S-COMT polymorphism. The amount of COMT protein present in homogenates was significantly reduced in subjects homozygous for the Met108/158 allele [26]. In a similar study of human hepatocytes from 31 female Caucasian subjects we observed significantly reduced S-COMT activity and protein in samples from individuals homozygous for the Met108/158 allele compared to heterozygous and WT individuals [29]. In a subsequent study using human breast tumor-derived cell lines we found the half-life of WT S-COMT to be unexpectedly long, 4.7 days, and to be significantly shortened to 3.0 days in Met108/158 variant allele homozygous cells, consistent with their reduced level of S-COMT protein and activity [30]. No significant difference in the rate of S-COMT synthesis was detected. These studies [26,29,30] confirm that the Val108/158Met polymorphism, while not affecting catalytic activity, affects protein stability, with the increased turnover perhaps due to altered conformation making it more susceptible to recognition by cellular protein degradation processes.
A study by Ji et al. [28] demonstrated that the SNPs mentioned above in the promoter region can alter nuclear protein binding and affect COMT expression. Similarly, COMT expression can also be affected by synonymous SNPs that alter mRNA secondary structure potentially affecting its translation and/or degradation [31]. While it is accepted that the val108/158 SNP is responsible for the most of the interindividual differences in COMT activity levels, these results clearly demonstrate that haplotypes comprising other nonsynonymous and synonymous SNPs can alter protein levels and demonstrate the importance of functional haplotype analysis in attempts to associate polymorphisms with diseases.
COMT levels exhibit a gender difference, with expression in liver greater in males than females, a reduction in hepatic levels following estrogen exposure as well as variations in level during the estrus cycle [2,32]. Treatment of estrogen receptor-expressing MCF-7 cells with low physiologically relevant concentrations of E2 for 48 hours caused a concentration-dependent reduction in COMT mRNA [32]. Sequencing of promoter P1 and P2 regions revealed the presence of putative transcription factor binding elements including five half palindromic estrogen response elements (ERE) in P1 and three in P2. Functional analysis demonstrated that ERE 1 and 2 in P1 and ERE 8 in P2 are crucial for E2 responsiveness [32]. These results indicate that endogenous estrogen levels modulate COMT gene expression.
COMT polymorphisms in breast cancer
Given the potentially important protective role of COMT, we hypothesized that the Val108/158Met SNP encoding the low activity form would be a risk factor for breast cancer and conducted one of the first nested case control studies to test this hypothesis [33]. Postmenopausal women homozygous for the low activity allele had increased breast cancer risk with an Odds Ratio (OR) of 2.2 that approached significance (95% Confidence Interval [CI], 0.93–5.11). The risk was increased in postmenopausal women with a Body Mass Index >24.47 kg/m2 (OR, 3.58; 95% CI, 1.07–11.98) and in post-menopausal women who were either glutathione S-transferase (GST) M1 null (OR, 4.10; 95% CI, 1.17–14.27) or GSTP1 intermediate/low activity (OR, 3.40; 95% CI, 1.17–12.33). Shortly thereafter several additional studies examining the effect of this low activity polymorphism on breast cancer risk were published, with three showing an increased risk [34–36] and one not finding an association [37].
There have been several recent reports describing results from meta-analyses of the many subsequent studies on the association of the COMT low activity SNP with breast cancer risk. Ding et al. analyzed data from 26 studies, 16,693 breast cancer subjects and 18,261 controls [38]. The authors concluded that the ‘COMT val158Met polymorphism may be a low penetrant risk factor for breast cancer development in the European population’ [38]. Actually, this conclusion is somewhat misstated in that their results suggest that the COMT low activity SNP is a protective factor, not a risk factor. Xi et al.[39] criticized several aspects of this study including that it presented data that differed from those presented in the original studies. Furthermore, the result indicating a possible protective role for the low activity genotype runs counter to the protective role of COMT as important in the inactivation of estrogenically active and genotoxic catechol metabolites of E2/E1. In another study, Mao et al. [40] analyzed data from 41 studies, 25,627 breast cancer cases and 34,222 controls and found no association between the low activity val108/158met SNP and breast cancer. Finally, He et al. [41] conducted a meta-analysis of 44 studies, 30,199 breast cancer cases and 38,922 controls and found no association of the low activity allele with breast cancer risk. By contrast, when conducting a stratified analysis by ethnicity, menopausal status and family history they observed a small protective effect of borderline significance among Caucasian subjects, OR, 0.96; 95% CI, 093–1.00 [41]. However, because this SNP lowers COMT levels, as mentioned previously, from a biological perspective, these results are inconsistent with what we currently know about the protective role of COMT. Together, these results demonstrate that without some measure of COMT expression or activity it is difficult to interpret results from these types of SNP association studies.
What are potential confounding factors that could account for these inconsistent results? First and foremost, these meta-analyses have only analyzed individual studies with this single SNP. As mentioned previously, the effect of the mutation is relatively small as indicated by the low ORs in most studies. There are multiple genetic and other determinants of the levels of estrogen catechols and reactive quinone metabolites that may be formed. Even in our first study, we found other factors that influenced breast cancer risk associated with the low activity SNP, for example BMI [33]. Other genetic determinants include the activity of the cytochrome P450s involved in catechol and quinone formation (e.g. CYP1B1), the activities of Phase II enzymes involved in quinone inactivation (e.g. GSTs, quinone reductase), DNA repair capacities, among others.
Some studies have combined the COMT low activity SNP with putative high risk SNPs of other genes involved in the metabolism of E2 and the quinone metabolites and protection from ROS. For example, Cerne et al. [42] analyzed the combined effect of the putative high-risk genotypes for CYP1B1, COMT, GSTP1 and MnSOD on breast cancer risk in postmenopausal women, 530 cases and 270 controls. There was no association of risk with any of the four high risk variant SNPs alone. However, the presence of both the CYP1B1 high activity val432 and COMT low activity met108/158 SNP was associated with an increased risk, OR 2.0, 95% CI 1.1–3.5. A similar increased risk was observed in the presence of high risk SNP for MnSOD and COMT, OR 2.0; 95% CI, 1.0–3.8 [42]. A limitation of the studies on associations of polymorphisms in genes involved in estrogen metabolism is that given limits on the number of subjects that can be included, only combinations of the SNPs of a few genes at most can be analyzed.
Another factor that could contribute to differences between individual studies is the presence of other polymorphisms within the COMT gene that may alter the conformation of COMT mRNA and affect its ability to be translated as mentioned above [28, 31]. Ji et al. [28] reported the results from a study in Caucasians of the association of 15 COMT SNPs that were genotyped in 750 breast cancer cases and 732 controls at the Mayo Clinic. They found no association of increased risk with the low activity Met108/158 SNP, but as mentioned previously, they did observe a reduction in risk associated with the two common SNPs present in promoter P2 for MB-COMT mentioned above that functional studies indicated could effect COMT gene expression. The allele specific ORs were: SNP rs2020917, OR, 0.70, 95% CI, 0.51–0.92; SNP rs737865, OR, 0.68, 95% CI, 0.51–0.92. They repeated this analysis in two other independent case control studies and found similar results in one but not in another, again suggesting that other factors can affect the expression of the phenotype, and again pointing to the need for determination of some direct measure of COMT expression or activity.
It is also possible that exposures to various drugs or environmental chemicals that are catechols or are metabolized to catechols could affect COMT activity and SNP penetrance. Chemicals such as metabolites of zeranol [43], flavonoids, quercetin and selected polychlorinated biphenol catechols are COMT substrates (see [4,25] for references) and could affect COMT protein levels or inhibit its activity toward the endogenous estrogen catechols and thus modify risk associated with COMT SNPs.
Conclusions
Estrogen catechol metabolites are carcinogenic and it is probable that the mechanism involves both their estrogenicity and oxidative metabolism to genotoxic quinones. COMT is a gatekeeper phase II enzyme that O-methylates the catechols blocking their estrogenicity and further metabolism. Common SNPs in COMT that affect its activity and expression have been extensively studied as risk factors for breast cancer. However, the penetrance of the phenotype encoded by these SNPs is low and their impact on breast cancer risk in different populations is not consistent, perhaps because SNPs in other genes and environmental chemicals that are COMT substrates may also vary and impact the effects of COMT. Future investigations into the role of estrogen catechols, COMT and other genes involved in estrogen metabolism and their SNPs must collect samples for the measurement of specific biomarkers of metabolites hypothesized to be on the causative pathway. For estradiol, this would involve determination of the levels of the E2/E1 catechol metabolites, in particular 4-OHE2/E2 or even more appropriately, the specific adenine and guanine DNA adducts and markers of oxidative DNA damage that can be detected in urine and plasma. It is only through this approach that the role of catechol estrogens and SNPs in the enzymes involved in their formation and inactivation, for example COMT, GSTs and others, can be determined.
Acknowledgments
Research done by the author's laboratory was supported by grants CA77550, NIEHS Center Grant ES03819 and NIEHS TG T32 ES07141.
Footnotes
Conflict of interest: The author has no conflict of interest to declare.
References
- 1.Axelrod J, Tomchick R. Enzymatic O-methylation of epinephrine and other catechols. J Biol Chem. 1958;233:702–705. [PubMed] [Google Scholar]
- 2.Mannisto PT, Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev. 1999;51:593–628. [PubMed] [Google Scholar]
- 3.Lundstrom K, et al. Cloning, expression and structure of catechol-O-methyltransferase. Biochim Biophys Acta. 1995;1251:1–10. doi: 10.1016/0167-4838(95)00071-2. [DOI] [PubMed] [Google Scholar]
- 4.Zhu BT. Catechol-O-methyltransferase (COMT)-mediated methylation metabolism of endogenous bioactive catechols and modulation by endobiotics and xenobiotics: importance in pathophysiology and pathogenesis. Curr Drug Metab. 2002;3:321–349. doi: 10.2174/1389200023337586. [DOI] [PubMed] [Google Scholar]
- 5.Ulmanen I, et al. Expression and intracellular localization of catechol O-methyltransferase in transfected mammalian cells. Eur J Biochem. 1997;243:452–459. doi: 10.1111/j.1432-1033.1997.0452a.x. [DOI] [PubMed] [Google Scholar]
- 6.Tilgmann C, et al. Expression of recombinant soluble and membrane-bound catechol O-methyltransferase in eukaryotic cells and identification of the respective enzymes in rat brain. Eur J Biochem. 1992;207:813–821. doi: 10.1111/j.1432-1033.1992.tb17112.x. [DOI] [PubMed] [Google Scholar]
- 7.Weisz J, et al. Nuclear localization of catechol-O-methyltransferase in neoplastic and nonneoplastic mammary epithelial cells. Am J Pathol. 2000;156:1841–1848. doi: 10.1016/S0002-9440(10)65057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liehr JG, et al. Carcinogenicity of catechol estrogens in Syrian hamsters. J Steroid Biochem. 1986;24:353–356. doi: 10.1016/0022-4731(86)90080-4. [DOI] [PubMed] [Google Scholar]
- 9.Newbold RR, Liehr JG. Induction of uterine adenocarcinoma in CD-1 mice by catechol estrogens. Cancer Res. 2000;60:235–237. [PubMed] [Google Scholar]
- 10.Schutze N, et al. Catecholestrogens are MCF-7 cell estrogen receptor agonists. J Steroid Biochem Mol Biol. 1993;46:781–789. doi: 10.1016/0960-0760(93)90319-r. [DOI] [PubMed] [Google Scholar]
- 11.Lottering ML, et al. Effects of 17 beta-estradiol metabolites on cell cycle events in MCF-7 cells. Cancer Res. 1992;52:5926–5932. [PubMed] [Google Scholar]
- 12.Van Aswegen CH, et al. Binding of 2-hydroxyestradiol and 4-hydroxyestradiol to estrogen receptors from human breast cancers. J Steroid Biochem. 1989;32:485–492. doi: 10.1016/0022-4731(89)90380-4. [DOI] [PubMed] [Google Scholar]
- 13.Cavalieri EL, Rogan EG. Unbalanced metabolism of endogenus estrogens in the etiology and prevention of human cancer. J Steroid Biochem Mol Biol. 2011;123:169–180. doi: 10.1016/j.jsbmb.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavigne JA, et al. The effects of catechol-O-methyltransferase inhibition on estrogen metabolite and oxidative DNA damage levels in estradiol-treated MCF-7 cells. Cancer Res. 2001;61:7488–7494. [PubMed] [Google Scholar]
- 15.Zahid M, et al. Inhibition of catechol-O-methyltransferase increases estrogen-DNA adduct formation. Free Radic Biol Med. 2007;43:1534–1540. doi: 10.1016/j.freeradbiomed.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barchiesi F, et al. Candidate genes and mechanisms for 2-methoxyestradiol-mediated vasoprotection. Hypertension. 2010;56:964–972. doi: 10.1161/HYPERTENSIONAHA.110.152298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubey RK, et al. 2-Methoxyestradiol: a potential treatment for multiple proliferative disorders. Endocrinology. 2007;148:4125–4127. doi: 10.1210/en.2007-0514. [DOI] [PubMed] [Google Scholar]
- 18.Klauber N, et al. Inhibition of angiogenesis and breast cancer in mice by the microtubule inhibitors 2-methoxyestradiol and taxol. Cancer Res. 1997;57:81–86. [PubMed] [Google Scholar]
- 19.Fotsis T, et al. The endogenous oestrogen metabolite 2-methoxyoestradiol inhibits angiogenesis and suppresses tumour growth. Nature. 1994;368:237–239. doi: 10.1038/368237a0. [DOI] [PubMed] [Google Scholar]
- 20.Scanlon PD, et al. Catechol-O-methyltransferase: thermolabile enzyme in erythrocytes of subjects homozygous for allele for low activity. Science. 1979;203:63–65. doi: 10.1126/science.758679. [DOI] [PubMed] [Google Scholar]
- 21.Weinshilboum RM, Raymond FA. Inheritance of low erythrocyte catechol-O-methyltransferase activity in man. Am J Hum Genet. 1977;29:125–135. [PMC free article] [PubMed] [Google Scholar]
- 22.Lachman HM, et al. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6:243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Cotton NJ, et al. Oxidative inhibition of human soluble catechol-O-methyltransferase. J Biol Chem. 2004;279:23710–23718. doi: 10.1074/jbc.M401086200. [DOI] [PubMed] [Google Scholar]
- 24.Vidgren J, et al. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 25.Goodman JE, et al. Characterization of human soluble high and low activity catechol-O-methyltransferase catalyzed catechol estrogen methylation. Pharmacogenetics. 2002;12:517–528. doi: 10.1097/00008571-200210000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Shield AJ, et al. Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Mol Psychiatry. 2004;9:151–160. doi: 10.1038/sj.mp.4001386. [DOI] [PubMed] [Google Scholar]
- 27.Saito S, et al. Identification of 197 genetic variations in six human methyltranferase genes in the Japanese population. J Hum Genet. 2001;46:529–537. doi: 10.1007/s100380170035. [DOI] [PubMed] [Google Scholar]
- 28.Ji Y, et al. Breast cancer risk reduction and membrane-bound catechol O-methyltransferase genetic polymorphisms. Cancer Res. 2008;68:5997–6005. doi: 10.1158/0008-5472.CAN-08-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyle AE, et al. Cancer Lett. 2004;214:189–195. doi: 10.1016/j.canlet.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 30.Doyle AE, Yager JD. Catechol-O-methyltransferase: effects of the val108met polymorphism on protein turnover in human cells. Biochim Biophys Acta. 2008;1780:27–33. doi: 10.1016/j.bbagen.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nackley AG, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 32.Xie T, et al. Characterization and implications of estrogenic down-regulation of human catechol-O-methyltransferase gene transcription. Mol Pharmacol. 1999;56:31–38. doi: 10.1124/mol.56.1.31. [DOI] [PubMed] [Google Scholar]
- 33.Lavigne JA, et al. An association between the allele coding for a low activity variant of catechol-O-methyltransferase and the risk for breast cancer. Cancer Res. 1997;57:5493–5497. [PubMed] [Google Scholar]
- 34.Yim DS, et al. Relationship between the Val158Met polymorphism of catechol O-methyl transferase and breast cancer. Pharmacogenetics. 2001;11:279–286. doi: 10.1097/00008571-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Huang CS, et al. Breast cancer risk associated with genotype polymorphism of the estrogen-metabolizing genes CYP17, CYP1A1, and COMT: a multigenic study on cancer susceptibility. Cancer Res. 1999;59:4870–4875. [PubMed] [Google Scholar]
- 36.Thompson PA, et al. Genetic polymorphisms in catechol-O-methyltransferase, menopausal status, and breast cancer risk. Cancer Res. 1998;58:2107–2110. [PubMed] [Google Scholar]
- 37.Millikan RC, et al. Catechol-O-methyltransferase and breast cancer risk. Carcinogenesis. 1998;19:1943–1947. doi: 10.1093/carcin/19.11.1943. [DOI] [PubMed] [Google Scholar]
- 38.Ding H, et al. COMT Val158Met polymorphism and breast cancer risk: evidence from 26 case-control studies. Breast Cancer Res Treat. 2010;123:265–270. doi: 10.1007/s10549-010-0759-5. [DOI] [PubMed] [Google Scholar]
- 39.Xi B, et al. Catechol-O-methyltransferase Val158Met polymorphism in breast cancer risk. Breast Cancer Res Treat. 2011;126:839–840. doi: 10.1007/s10549-010-1337-6. author reply 841. [DOI] [PubMed] [Google Scholar]
- 40.Mao C, et al. Lack of association between catechol-O-methyltransferase Val108/158Met polymorphism and breast cancer risk: a meta-analysis of 25,627 cases and 34,222 controls. Breast Cancer Res Treat. 2010;121:719–725. doi: 10.1007/s10549-009-0650-4. [DOI] [PubMed] [Google Scholar]
- 41.He XF, et al. Association between the COMT Val158Met polymorphism and breast cancer risk: a meta-analysis of 30,199 cases and 38,922 controls. Mol Biol Rep. 2012;39:6811–6823. doi: 10.1007/s11033-012-1506-2. [DOI] [PubMed] [Google Scholar]
- 42.Cerne JZ, et al. Combined effect of CYP1B1, COMT, GSTP1, and MnSOD genotypes and risk of postmenopausal breast cancer. J Gynecol Oncol. 2011;22:110–119. doi: 10.3802/jgo.2011.22.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleck SC, et al. Catechol metabolites of zeranol and 17beta-estradiol: a comparative in vitro study on the induction of oxidative DNA damage and methylation by catechol-O-methyltransferase. Toxicol Lett. 2012;210:9–14. doi: 10.1016/j.toxlet.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Diatchenko L, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet. 2005;14:135–143. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]