Abstract
The prevalence of cutaneous malignant melanoma (CMM) has increased significantly in most Caucasian populations in recent decades. Both genetic and environment are significant risk factors involved in the development of CMM. A germline mutation in the Syntaxin 17 (STX17) gene was recently identified in horses causing premature hair gray and associated with susceptibility to melanoma. We hypothesized that common germline variants in the STX17 gene might be associated with predisposition to human CMM or might interact with other melanoma risk genes. We conducted a case-control study by genotyping 26 tagging single nucleotide polymorphisms (SNPs) across the STX17 gene region in an Australian sample and performed logistic regression analysis for predicting the possible SNP interactions in a combined dataset. Our results do not support an association between CMM and any of the STX17 SNPs and provide no evidence for interactions between the melanoma risk SNP rs910873 on chromosome 20 and any of the STX17 SNPs. We conclude that common variants in the STX17 gene region do not play a key role in the pathogenesis of human melanoma.
Keywords: Syntaxin 17, melanoma, polymorphisms
INTRODUCTION
Cutaneous malignant melanoma (CMM) is a form of skin cancer that arises from melanocytes. The prevalence and incidence of melanoma have increased significantly faster over the past few decades than any other cancer worldwide, particularly in Caucasian populations (Armstrong and Kricker, 1994; Berwick and Wiggins, 2006). The relative recurrence risk to siblings was estimated at 2.24 in a combined study of familial melanomas (Ford et al., 1995), suggesting that the familial cases carry an inherited susceptibility to CMM. Although the precise etiology of melanoma remains unknown, much data from molecular and epidemiologic studies clearly indicate that both genetic and environmental risk factors are involved (Hayward, 2003; Hussussian et al., 1994; Koh et al., 1990; Lejeune, 1986; Lynch and Fusaro, 1986).
Several different moderate to high risk melanoma genes have been identified (de Snoo and Hayward, 2005; Hussussian et al., 1994; Landi et al., 2006; Pollock et al., 2002; Zuo et al., 1996). Multiple somatic and/or germline mutations in these genes increase the risk of developing melanoma through altering normal programmes of cell proliferation, differentiation and apoptosis (Davies et al., 2002; Ranade et al., 1995). A genome-wide association study has recently identified and replicated a new melanoma risk locus on chromosome 20, close to the agouti signaling protein gene (ASIP) encoding an antagonist of the human melanocortin-1 receptor (MC1R) which regulates synthesis of melanin (Brown et al., 2008). In the study, SNP rs910873 was determined as a most highly associated SNP within the locus with human melanoma and the rs910873 risk allele was related to early onset of CMM (Brown et al., 2008).
Study of a horse model identified a germline mutation in the Syntaxin 17 (STX17) gene that causes premature hair graying is also associated with susceptibility to melanoma (Rosengren Pielberg et al., 2008). Horses homozygous for the mutation showed more rapid graying and have a higher incidence of melanomas in glabrous skin. Strong associations between the STX17 germline mutation, ASIP genotype and melanoma development were observed in gray horses (Rosengren Pielberg et al., 2008). One question that arises from the finding is whether variation in STX17 either on its own or through interaction with other melanoma risk SNPs such as rs910873 is associated with human CMM.
In the STX17 genomic region, four genes are located in close proximity to each other including NR4A3 (nuclear receptor subfamily 4, group A, member 3; OMIM 600542), STX17 (OMIM 604204), TXNDC4 (thioredoxin domain containing 4; OMIM 609170) and INVS (inversin; OMIM 243305). They map to a region of horse chromosome 25 which is syntenic with human chromosome band 9q31.1 (Locke et al., 2002; Pielberg et al., 2005). The long arm of chromosome 9 has been documented as a region to which a combined ocular-cutaneous melanoma risk gene has been located (Cannon-Albright et al., 1992; Zhu et al., 2007). Additionally, loss of heterozygosity (LOH) on 9q21-33 has been documented in 49% of 76 melanoma cell lines and homozygous deletion of markers in this region were observed in 5 samples (Stark and Hayward, 2007). These data, together with the cis-acting regulatory mutation that causes premature hair graying and susceptibility to melanoma in the horse, provide strong evidence for a locus on 9q in human CMM. The interaction between the variations of these genes may represent a common pathway linked to both human melanoma and horse melanoma susceptibility. We thus hypothesized that common germline variants in the region of the STX17 gene might be associated with predisposition to human melanoma and conducted a case-control study to test for association between SNPs in the STX17 genomic region and melanoma risk.
RESULTS
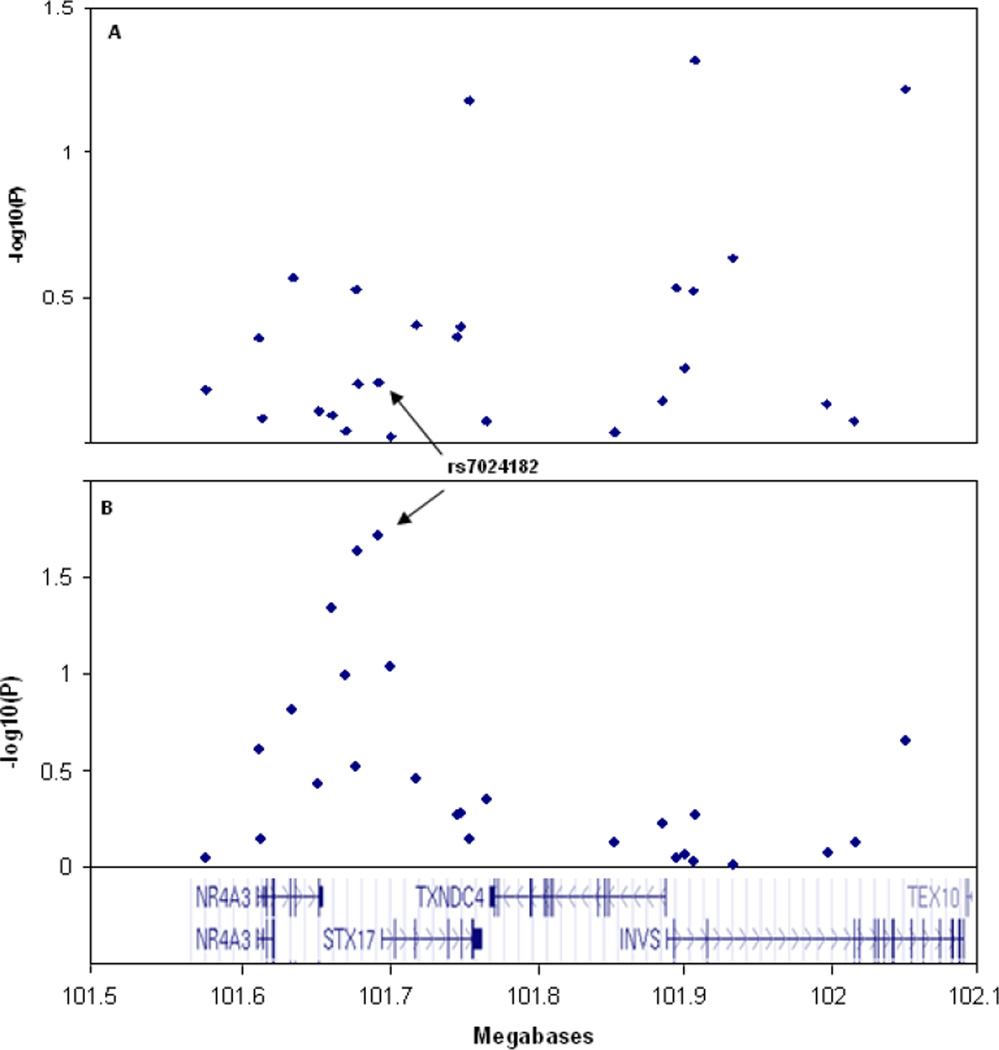
We genotyped 26 SNPs across the STX17 gene in 1560 melanoma cases and 1650 controls after selecting correlated tagging SNPs from the HapMap database (γ2 <0.9) aiming to completely cover this genomic region. All control genotype frequencies were in Hardy–Weinberg equilibrium. The overall genotype completion rate was 98.2%. Strong linkage disequilibrium (LD) between SNPs were detected in STX17 (rs7024182 with rs10988912, γ2=0.95; with rs4742776, γ2=0.82; rs10760704 with rs7038506 γ2=0.999), NR4A3 (rs7023690 with rs2416878 γ2=0.84), TXNDC4 (rs12552646 with rs1361668 γ2=0.94; with rs1535667 γ2=0.89) and INVS (rs7020636 with rs16918878 γ2=0.95). Allele frequencies did not differ significantly between cases and controls for any of the SNPs (Table 1) and none were significantly associated with melanoma (Table 1, Figure 1A).
Table 1.
Association analyses of the polymorphisms genotyped in 1560 familial melanoma cases and 1650 controls.
| Minor Allele Frequency | Allelic Association | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| dbSNP ID | Position | Gene | Role | Alleles | Case | Control | X2 | P | OR (95% CI) |
| rs10819693 | chr9:101577981 | NR4A3 | Upstream | C<A | 0.209 | 0.214 | 0.223 | 0.637 | 0.97 (0.86–1.10) |
| rs4743365 | chr9:101613594 | NR4A3 | Upstream | C<T | 0.123 | 0.129 | 0.537 | 0.464 | 0.95 (0.82–1.10) |
| rs13295822 | chr9:101615501 | NR4A3 | Promoter | A<G | 0.170 | 0.168 | 0.080 | 0.778 | 1.02 (0.89–1.16) |
| rs2900223 | chr9:101636142 | NR4A3 | 3' UTR | C<T | 0.235 | 0.247 | 1.060 | 0.303 | 0.94 (0.84–1.06) |
| rs2416879 | chr9:101653493 | NR4A3 | Intron | G<A | 0.107 | 0.109 | 0.101 | 0.751 | 0.97 (0.83–1.14) |
| rs7023690 | chr9:101663149 | NR4A3 | Intron | C<A | 0.440 | 0.443 | 0.025 | 0.874 | 0.99 (0.90–1.10) |
| rs2416878 | chr9:101672056 | NR4A3 | Downstream | A<G | 0.398 | 0.399 | 0.000 | 0.993 | 1.10 (0.90–1.11) |
| rs12554558 | chr9:101679119 | NR4A3 | Downstream | C<T | 0.083 | 0.090 | 1.139 | 0.286 | 0.91 (0.76–1.08) |
| rs10988912 | chr9:101680853 | STX17 | Upstream | G<C | 0.332 | 0.338 | 0.176 | 0.675 | 0.98 (0.88–1.09) |
| rs7024182 | chr9:101694268 | STX17 | Upstream | T<C | 0.343 | 0.349 | 0.187 | 0.665 | 0.98 (0.88–1.08) |
| rs4742776 | chr9:101702452 | STX17 | Promoter | T<C | 0.358 | 0.358 | 0.000 | 0.987 | 1.00 (0.90–1.11) |
| rs2416938 | chr9:101720516 | STX17 | Intron | A<G | 0.310 | 0.320 | 0.637 | 0.425 | 0.96 (0.86–1.07) |
| rs10760704 | chr9:101748385 | STX17 | Intron | G<A | 0.325 | 0.335 | 0.751 | 0.386 | 0.95 (0.86–1.06) |
| rs7038506 | chr9:101749953 | STX17 | Intron | T<C | 0.324 | 0.334 | 0.852 | 0.356 | 0.95 (0.86–1.06) |
| rs7865257 | chr9:101755787 | STX17 | Intron | G<A | 0.365 | 0.343 | 3.430 | 0.064 | 1.10 (0.99–1.22) |
| rs10116142 | chr9:101767497 | STX17 | Intron | G<C | 0.374 | 0.377 | 0.064 | 0.800 | 0.99 (0.89–1.09) |
| rs3824510 | chr9:101854271 | TXNDC4 | Intron | T<A | 0.084 | 0.083 | 0.019 | 0.890 | 1.01 (0.85–1.21) |
| rs16918878 | chr9:101886451 | TXNDC4 | Intron | T<C | 0.296 | 0.300 | 0.128 | 0.720 | 0.98 (0.88–1.09) |
| rs1361668 | chr9:101896127 | TXNDC4 | Intron | G<C | 0.461 | 0.474 | 1.231 | 0.267 | 0.95 (0.86–1.04) |
| rs7024375 | chr9:101901434 | TXNDC4 | Promoter | G<T | 0.166 | 0.171 | 0.424 | 0.515 | 0.96 (0.84–1.09) |
| rs12552646 | chr9:101908296 | TXNDC4 | Promoter | T<C | 0.445 | 0.458 | 1.219 | 0.270 | 0.95 (0.86–1.04) |
| rs12346672 | chr9:101908590 | TXNDC4 | Promoter | G<A | 0.310 | 0.287 | 3.911 | 0.048 | 1.12 (1.00–1.24) |
| rs1535667 | chr9:101934076 | INVS | Intron | T<C | 0.471 | 0.486 | 1.585 | 0.208 | 0.94 (0.85–1.04) |
| rs7020636 | chr9:101998420 | INVS | Intron | G<A | 0.302 | 0.306 | 0.115 | 0.734 | 0.98 (0.88–1.09) |
| rs4273907 | chr9:102017116 | INVS | Intron | T<C | 0.085 | 0.083 | 0.022 | 0.882 | 1.01 (0.85–1.21) |
| rs16919019 | chr9:102051408 | INVS | Intron | G<C | 0.092 | 0.107 | 3.365 | 0.067 | 0.86 (0.73–1.01) |
Figure 1.
Association analyses of the 26 polymorphisms genotyped in 1650 controls compared with (A) 1560 familial melanoma cases, (B) 117 site-restrict melanomas on external ear and face.
Because the horse melanomas occur primarily as jet black firm nodules well circumscribed in the dermis of glabrous skin (non-hair bearing), we hypothesized that variation in the STX17 gene region may contribute to risk of CMM in a site-specific manner. The allele frequency differences between melanoma body site (glabrous versus non-glabrous) and controls were analysed. A weak allelic association with melanoma for the STX17 promoter SNP rs7024182 was detected in the 117 melanomas located on the external ear and face (Figure 1B). Allele frequency difference of SNP rs7024182 between the subset of melanomas and controls gave a significant P = 0.019 (Case frequency = 0.274; Control frequency = 0.349). However, the difference was not significant after correcting for multiple testing. We expected if rs7024182 is associated with glabrous melanomas, other SNPs correlated with rs7024182 should also show evidence of association. We therefore conducted a HapMap database searching for statistically similar SNPs (ssSNP) to rs7024182 using the web-based program ssSNPer (Nyholt, 2006). Four statistically similar SNPs typed in our samples were identified: rs10988912, r2 = 0.921; rs4742776, r2 = 0.702; rs7023690, r2 = 0.654; rs2416878, r2 = 0.512. Analysis of LD between SNP rs7024182 and the four ssSNPs confirmed they are correlated in our sample with a decreased evidence of association with glabrous melanomas compared to rs7024182 (rs10988912 P = 0.023; rs4742776 P = 0.091; rs7023690 P = 0.045; rs2416878 P = 0.102). However, the allele frequency differences were not significant for these ssSNPs after correcting for multiple testing. Haplotype analyses on the 26 SNPs identified 5 haplotype blocks in both case and control samples (Figure 2). Tests of association with the haplotypes either in 1560 melanoma cases or in 117 glabrous melanomas indicated none significantly contributed to disease susceptibility after adjusting for multiple testing.
Figure 2.
Twenty-six polymorphisms genotyped in the human STX17 gene region. The linkage disequilibrium plot of single nucleotide polymorphisms estimated as r2 using Haploview (above) and common haplotypes and association analysis with melanomas (below). Shading key: white γ2=0; shades of grey 0>γ2<1; black γ2=1
To further evaluate the association signal observed from SNP rs7024182, we performed analyses based on primary clinical phenotypic data available (Table 2). Stratification of cases according to site of melanoma produced the smallest P-values of 0.008 and 0.023 on the face for allelic association and genotypic association tests, respectively. Genotyping frequency differences between 96 facial tumours and 1650 controls gave a significant P = 0.033 (χ2 = 4.54) for a dominant model and a significant P = 0.022 (χ2 = 5.19) for a recessive model. However, the differences were not significant after correcting for multiple testing.
Table 2.
Allelic and genotypic association analyses of the STX17 promoter SNP rs7024182 on the anatomic site of melanomas compared with 1650 controls.
| Allelic association | Genotypic association | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Site of CMM | No cases | Minor allele frequency | P | X2 | TT | TC | CC | P | X2 |
| External ear | 21 | 0.357 | 0.910 | 0.200 | 2 | 11 | 8 | 0.906 | 0.200 |
| Skin of other and unspecified parts of face | 96 | 0.255 | 0.008 | 7.045 | 4 | 41 | 51 | 0.023 | 7.520 |
| Glabrous (non-hairy) skin | 117 | 0.273 | 0.019 | 5.490 | 6 | 52 | 59 | 0.049 | 6.010 |
| Skin of scalp and neck | 71 | 0.357 | 0.326 | 2.240 | 11 | 28 | 31 | 0.326 | 2.240 |
| Skin of trunk | 538 | 0.359 | 0.406 | 1.800 | 70 | 243 | 219 | 0.406 | 1.800 |
| Skin of upper limb and shoulder | 356 | 0.343 | 0.667 | 0.810 | 41 | 156 | 150 | 0.667 | 0.810 |
| Skin of lower limb and hip | 399 | 0.334 | 0.552 | 1.190 | 43 | 175 | 173 | 0.552 | 1.190 |
| Non-glabrous skin | 1364 | 0.348 | 0.294 | 2.450 | 165 | 602 | 573 | 0.294 | 2.450 |
A recent study from our group identified a new melanoma risk locus close to the ASIP gene on chromosome 20 (Brown et al., 2008). To identify possible SNP interactions between the STX17 common variation and the SNP rs910873 which was strongly associated with CMM on chromosome 20, we conducted logic regression analysis in an attempt to identify the CMM risk conferred by SNP interactions. In a combined data, we did not detect SNP-SNP interactions between SNP rs910873 and any of the STX17 SNPs. Moreover, we investigated the effects of common variation at STX17 on melanocytic naevus count and on pigmentation classifications of eye and hair colours. We found no evidence for association between any of the STX17 SNPs and any phenotypes.
DISCUSSION
A germline mutation in intron 6 of the STX17 gene has been reported to cause premature hair graying and susceptibility to melanoma in gray horses (Rosengren Pielberg et al., 2008). We screened STX17 for association with melanoma risk because of the recent report of strong association between the STX17 germline mutation, ASIP genotype and melanoma development in horses and the location of the STX17 gene corresponds to a human melanoma susceptibility region on chromosome 9q.
STX17, together with neighbouring NR4A3 (encoding a member of the NR4A orphan nuclear receptor family) which has been associated with cell cycle regulation and has an established link with carcinogenesis (Maxwell and Muscat, 2006), were highly expressed in horse melanomas. STX17 belongs to the syntaxin family, which encodes membrane proteins involved in synaptic vesicle fusion. To determine whether an association exists between common variants of STX17 and human CMM, we genotyped 26 tag-SNPs across the four genes in the region. The tagging SNPs we selected for this study could capture 100 percent of alleles with γ2 cut off at 0.9. If the genetic predisposition of melanoma is influenced by common variation in the region, we would expect to detect evidence of association in the SNPs and SNP haplotypes in our sample. Our results do not support an association between melanoma and common variation in the STX17 gene in human melanoma predisposition. We found no evidence for SNP-SNP interactions between SNP rs910873 (ASIP) and any of the STX17 SNPs. Haplotype analyses using sliding windows of 2–5 contiguous SNPs did not identify any evidence for association between the tested variants and melanoma. However, our results do not exclude the possibility that either unknown variants in weak LD with the genotyped SNPs or rare variants of large effect in this region influence melanoma risk.
Determination of the anatomic site distribution of CMM is not fully understood, although sun exposure is believed to be associated causally with the disease. A study of 844 patients with head and neck melanoma compared with 4858 patients with other anatomical region’s melanoma estimated a significant increased risk of 2.6 for the patients with melanoma on face (Hoersch et al., 2006). Similar association was observed in an Australian population (Green, 1992; Green et al., 1988), whereas others did not support site specific theory (Chen et al., 1996; Randi et al., 2006; Rieger et al., 1995; Rodenas et al., 1997). It has been postulated that the STX17 duplication leads to proliferation of dermal melanocytes in glabrous (non-hairy) skin, thus predisposing to melanoma development (Rosengren Pielberg et al., 2008). We hypothesized that variation in the STX17 gene may contribute to risk of CMM in a site-specific manner. Data analysis using site-restricted cases provided weak evidence for association between the STX17 promoter SNP rs7024182 and CMM on the face, but no association was observed for this SNP with CMM on the external ear. This result may be due to a site-restricted sample size which is too small to have sufficient power to detect a true association. Since the overall results did not support an association between common variation in the STX17 gene region and CMM, we conclude that if the risk of melanoma on glabrous skin is influenced by the STX17 promoter SNP rs7024182, the effect size would not be large. Replication studies are required to confirm or refute this result.
In this study, we examined the association between CMM and common SNPs or haplotypes in human in the region syntenic to the horse gray-causing mutation containing the STX17, NR4A3, TXNDC4 and INVS genes. Our data do not support an association between common variation in these genes and melanoma risk. We found no evidence for SNP-SNP interactions between SNP rs910873 (ASIP) and any of the STX17 SNPs. However, our results can not exclude genomic deletions or insertions in the region which are not in LD with the genotyped SNPs. We conclude that common variants in the STX17 gene region do not play a key role in the pathogenesis of human cutaneous malignant melanoma.
MATERIALS AND METHODS
Participants
An Australian case–control panel was made up of 1560 familial melanoma cases drawn from Queensland, unselected for age at onset (Queensland study of Melanoma: Environment and Genetic Associations, (Baxter et al., 2008), and 1650 controls drawn from parents of twins enrolled in the Brisbane Twin Nevus Study (Zhu et al., 1999). All cases had incident primary melanomas. Tumour location and thickness were recorded. None of the controls have been diagnosed with melanoma. All samples are of European descent. The project was approved by the Human Research Ethics Committee of the Queensland Institute of Medical Research and the Australian Twin Registry.
Genotyping
To cover the region of Gray-causing mutation, we selected 26 functional and tagging SNPs (γ2 cut off 0.9) based on data from Pielberg’s paper (Rosengren Pielberg et al., 2008) and public databases including the International HapMap Project (http://www.hapmap.org/), and NCBI (http://www.ncbi.nlm.nih.gov/). There were 8 SNPs selected from the NR4A3 gene, 8 SNPs selected from the STX17 gene, 6 SNPs selected from the TXNDC4 gene and 4 SNPs selected from INVS gene. A region of 473.4 kb on chromosome 9 was covered by these SNPs. SNP sequences were downloaded from the Chip Bioinformatics database (http://snpper.chip.org/) and the sequences were cross-checked with NCBI before assay design. Multiplex assays were designed for the 26 SNPs using the Sequenom MassARRAY Assay Design software (version 3.1). SNPs were typed using iPLEX™ Gold chemistry and analyzed using a Sequenom MassARRAY Compact Mass Spectrometer (Sequenom Inc, San Diego, CA, USA). Briefly, The 2.5 µl PCR reactions were performed in 384-well plates using 12.5 ng genomic DNA, 0.9 unit of Taq polymerase (HotStarTaq, Qiagen, Valencia, CA), 500 µmol of each dNTP, 1.625 mM of MgCl2, and 100 nmol of each PCR primers (Bioneer, Korea). PCR thermal cycling was 15 min at 94°C, followed by 45 cycles of 20 sec at 94°C, 30 sec at 56°C, 60 sec at 72°C. The post-PCR reactions were performed in a final 5 µl of extension reaction containing 1× of termination mix, 1× of DNA polymerase, and 570 nM to 1240 nM extension primers. A two-step 200 short cycles program was used for the iPLEX Gold reaction as described in our previously study (Zhao et al., 2006). The products were spotted on a SpectroChip (Sequenom Inc, San Diego, CA, USA), and data were processed and analysed by MassARRAY TYPER 3.4 software (Sequenom Inc, San Diego, CA, USA).
Statistical analysis
SNP genotypes were tested for departures from Hardy–Weinberg equilibrium (HWE) for 1650 controls using Haploview version 4.1 (Barrett et al., 2005). Allelic association between melanoma and the SNPs were tested using the PLINK program (http://pngu.mgh.harvard.edu/purcell/plink/). Associations between categorical groups were tested by use of χ2 statistics. The global significance level was derived from multiple tests and values < 0.05 were considered to be statistically significant. Linkage disequilibrium (LD), haplotype frequencies and blocks were determined by Haploview using the default method of Gabriel et al (Gabriel et al., 2002). We conducted logic regression mythology for predicting the possible SNP interactions between the common variation in the STX17 gene region and the SNP rs910873 (Kooperberg and Ruczinski, 2005; Schwender and Ickstadt, 2008).
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute (NCI) of the US National Institutes of Health (CA88363), and the National Health and Medical Research Council of Australia (NHMRC) (380385, 389892, 496675, 402761). N.K.H. and G.W.M. are supported by the NHMRC Fellowships scheme.
Abbreviations
- CMM
cutaneous malignant melanoma
- STX17
Syntaxin 17
- SNPs
single nucleotide polymorphisms
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- Armstrong BK, Kricker A. Cutaneous melanoma. Cancer Surv. 1994;19–20:219–240. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baxter AJ, Hughes MC, Kvaskoff M, Siskind V, Shekar S, Aitken JF, et al. The Queensland Study of Melanoma: environmental and genetic associations (Q-MEGA); study design, baseline characteristics, and repeatability of phenotype and sun exposure measures. Twin Res Hum Genet. 2008;11:183–196. doi: 10.1375/twin.11.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick M, Wiggins C. The current epidemiology of cutaneous malignant melanoma. Front Biosci. 2006;11:1244–1254. doi: 10.2741/1877. [DOI] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon-Albright LA, Goldgar DE, Meyer LJ, Lewis CM, Anderson DE, Fountain JW, et al. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13-p22. Science. 1992;258:1148–1152. doi: 10.1126/science.1439824. [DOI] [PubMed] [Google Scholar]
- Chen YT, Dubrow R, Holford TR, Zheng T, Barnhill RL, Fine J, et al. Malignant melanoma risk factors by anatomic site: a case-control study and polychotomous logistic regression analysis. Int J Cancer. 1996;67:636–643. doi: 10.1002/(SICI)1097-0215(19960904)67:5<636::AID-IJC8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- de Snoo FA, Hayward NK. Cutaneous melanoma susceptibility and progression genes. Cancer Lett. 2005;230:153–186. doi: 10.1016/j.canlet.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Ford D, Bliss JM, Swerdlow AJ, Armstrong BK, Franceschi S, Green A, et al. Risk of cutaneous melanoma associated with a family history of the disease. The International Melanoma Analysis Group (IMAGE) Int J Cancer. 1995;62:377–381. doi: 10.1002/ijc.2910620403. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Green A. A theory of site distribution of melanomas: Queensland, Australia. Cancer Causes Control. 1992;3:513–516. doi: 10.1007/BF00052747. [DOI] [PubMed] [Google Scholar]
- Green A, Beardmore G, Hart V, Leslie D, Marks R, Staines D. Skin cancer in a Queensland population. J Am Acad Dermatol. 1988;19:1045–1052. doi: 10.1016/s0190-9622(88)70270-4. [DOI] [PubMed] [Google Scholar]
- Hayward NK. Genetics of melanoma predisposition. Oncogene. 2003;22:3053–3062. doi: 10.1038/sj.onc.1206445. [DOI] [PubMed] [Google Scholar]
- Hoersch B, Leiter U, Garbe C. Is head and neck melanoma a distinct entity? A clinical registry-based comparative study in 5702 patients with melanoma. Br J Dermatol. 2006;155:771–777. doi: 10.1111/j.1365-2133.2006.07455.x. [DOI] [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- Koh HK, Kligler BE, Lew RA. Sunlight and cutaneous malignant melanoma: evidence for and against causation. Photochem Photobiol. 1990;51:765–779. [PubMed] [Google Scholar]
- Kooperberg C, Ruczinski I. Identifying interacting SNPs using Monte Carlo logic regression. Genet Epidemiol. 2005;28:157–170. doi: 10.1002/gepi.20042. [DOI] [PubMed] [Google Scholar]
- Landi MT, Bauer J, Pfeiffer RM, Elder DE, Hulley B, Minghetti P, et al. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 2006;313:521–522. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- Lejeune FJ. Epidemiology and etiology of malignant melanoma. Biomed Pharmacother. 1986;40:91–99. [PubMed] [Google Scholar]
- Locke MM, Penedo MC, Bricker SJ, Millon LV, Murray JD. Linkage of the grey coat colour locus to microsatellites on horse chromosome 25. Anim Genet. 2002;33:329–337. doi: 10.1046/j.1365-2052.2002.00885.x. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Fusaro RM. Hereditary malignant melanoma: a unifying etiologic hypothesis. Cancer Genet Cytogenet. 1986;20:301–304. doi: 10.1016/0165-4608(86)90087-7. [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. ssSNPer: identifying statistically similar SNPs to aid interpretation of genetic association studies. Bioinformatics. 2006;22:2960–2961. doi: 10.1093/bioinformatics/btl518. [DOI] [PubMed] [Google Scholar]
- Pielberg G, Mikko S, Sandberg K, Andersson L. Comparative linkage mapping of the Grey coat colour gene in horses. Anim Genet. 2005;36:390–395. doi: 10.1111/j.1365-2052.2005.01334.x. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Walker GJ, Glendening JM, Que Noy T, Bloch NC, Fountain JW, et al. PTEN inactivation is rare in melanoma tumours but occurs frequently in melanoma cell lines. Melanoma Res. 2002;12:565–575. doi: 10.1097/00008390-200212000-00006. [DOI] [PubMed] [Google Scholar]
- Ranade K, Hussussian CJ, Sikorski RS, Varmus HE, Goldstein AM, Tucker MA, et al. Mutations associated with familial melanoma impair p16INK4 function. Nat Genet. 1995;10:114–116. doi: 10.1038/ng0595-114. [DOI] [PubMed] [Google Scholar]
- Randi G, Naldi L, Gallus S, Di Landro A, La Vecchia C. Number of nevi at a specific anatomical site and its relation to cutaneous malignant melanoma. J Invest Dermatol. 2006;126:2106–2110. doi: 10.1038/sj.jid.5700334. [DOI] [PubMed] [Google Scholar]
- Rieger E, Soyer HP, Garbe C, Buttner P, Kofler R, Weiss J, et al. Overall and site-specific risk of malignant melanoma associated with nevus counts at different body sites: a multicenter case-control study of the German Central Malignant-Melanoma Registry. Int J Cancer. 1995;62:393–397. doi: 10.1002/ijc.2910620406. [DOI] [PubMed] [Google Scholar]
- Rodenas JM, Delgado-Rodriguez M, Farinas-Alvarez C, Herranz MT, Serrano S. Melanocytic nevi and risk of cutaneous malignant melanoma in southern Spain. Am J Epidemiol. 1997;145:1020–1029. doi: 10.1093/oxfordjournals.aje.a009058. [DOI] [PubMed] [Google Scholar]
- Rosengren Pielberg G, Golovko A, Sundstrom E, Curik I, Lennartsson J, Seltenhammer MH, et al. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nat Genet. 2008;40:1004–1009. doi: 10.1038/ng.185. [DOI] [PubMed] [Google Scholar]
- Schwender H, Ickstadt K. Identification of SNP interactions using logic regression. Biostatistics. 2008;9:187–198. doi: 10.1093/biostatistics/kxm024. [DOI] [PubMed] [Google Scholar]
- Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- Zhao ZZ, Nyholt DR, Le L, Martin NG, James MR, Treloar SA, et al. KRAS variation and risk of endometriosis. Mol Hum Reprod. 2006;12:671–676. doi: 10.1093/molehr/gal078. [DOI] [PubMed] [Google Scholar]
- Zhu G, Duffy DL, Eldridge A, Grace M, Mayne C, O'Gorman L, et al. A major quantitative-trait locus for mole density is linked to the familial melanoma gene CDKN2A: a maximum-likelihood combined linkage and association analysis in twins and their sibs. Am J Hum Genet. 1999;65:483–492. doi: 10.1086/302494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Montgomery GW, James MR, Trent JM, Hayward NK, Martin NG, et al. A genome-wide scan for naevus count: linkage to CDKN2A and to other chromosome regions. Eur J Hum Genet. 2007;15:94–102. doi: 10.1038/sj.ejhg.5201729. [DOI] [PubMed] [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]