Abstract
Polymeric chains of a small protein ubiquitin are involved in regulation of nearly all vital processes in eukaryotic cells. Elucidating the signaling properties of polyubiquitin requires the ability to make these chains in vitro. In recent years, chemical and chemical-biology tools have been developed that produce fully natural isopeptide-linked polyUb chains with no need for linkage-specific ubiquitin-conjugating enzymes. These methods produced unbranched chains (in which no more than one lysine per ubiquitin is conjugated to another ubiquitin). Here we report a nonenzymatic method for the assembly of fully natural isopeptide-linked branched polyubiquitin chains. This method is based on the use of mutually orthogonal removable protecting groups (e.g., Boc- and Alloc-) on lysines combined with an Ag-catalyzed condensation reaction between a C-terminal thioester on one ubiquitin and a specific ε-amine on another ubiquitin, and involves genetic incorporation of more than one Lys(Boc) at the desired linkage positions in the ubiquitin sequence. We demonstrate our method by making a fully natural branched tri-ubiquitin containing isopeptide linkages via Lys11 and Lys33, and a 15N-enriched proximal ubiquitin, which enabled monomer-specific structural and dynamical studies by NMR. Furthermore, we assayed disassembly of branched and unbranched tri-ubiquitins as well as control di-ubiquitins by the yeast proteasome-associated deubiquitinase Ubp6. Our results show that Ubp6 can recognize and disassemble a branched polyubiquitin, wherein cleavage preferences for individual linkages are retained. Our spectroscopic and functional data suggest that, at least for the chains studied here, the isopeptide linkages are effectively independent of each other. Together with our method for nonenzymatic assembly of unbranched polyubiquitin, these developments now provide tools for making fully natural polyubiquitin chains of essentially any type of linkage and length.
Keywords: ubiquitin, polyubiquitin, branched chain, isopeptide bond, unnatural amino acids, deubiquitination
1. Introduction
Among various post-translational modifications of proteins in the cell perhaps the most fascinating is ubiquitination, i.e. a covalent attachment of a small, 76-a.a. protein ubiquitin (Ub), whose role is to act as a molecular signaling tag. The ubiquitination process involves formation of an isopeptide bond between G76 of Ub and a ε-NH2 of a lysine of the target protein (could be another Ub) and generally involves a series of enzymatic reactions catalyzed by Ub-activating (E1) and Ub-conjugating (E2) enzymes, and Ub-ligases (E3)1, 2. Ub-mediated signaling pathways are involved in regulation of essentially all aspects of eukaryotic cell life3–6. The versatility of Ub as a molecular signal stems from its ability to form polymers (polyUb) of various lengths and linkages, in which the protomers – ubiquitins -- are linked to each other through one of the seven lysines in Ub or head to tail (via α-NH2 of M1)6, 7. For example, K48-linked polyUb chains act as a universal signal targeting proteins for degradation by the 26S proteasome, whereas chains linked via K63 act as regulatory, non-proteolytic signals in various processes, including trafficking, DNA repair, and inflammatory response, and K11-linked chains were reported to act as both degradative and non-degradative signals5, 6. The biological roles of polyUb chains linked through other lysines are yet unclear. To add to the complexity of Ub-signaling, recent in vivo and in vitro studies indicate that more than one linkage type can be present in the same polyUb chain8–13. Furthermore, these so-called mixed-linkage chains can be unbranched (i.e., no more than one lysine- or methionine-linkage per Ub) or branched (i.e. more than one such linkage per Ub)8, 12, 13. This raises an exciting possibility that linkage mixing and branching could enhance the signaling capabilities of the chain or perhaps even bestow polyUb with novel signaling properties not available to homogeneously-linked or unbranched chains.
Despite the significant advances in elucidating the biological/cellular roles of various Ub-signaling pathways in the last decades, understanding of the mechanisms that allow differently linked Ub chains to serve as versatile and yet distinct molecular signals is missing. The current, ‘linkage-conformation-signal’ hypothesis is that the Ub-Ub linkage defines the conformational ensemble for a given polyUb chain, which in turn determines the ability of the chain (through conformational selection or induced fit) to adopt the proper conformation required for binding to a specific receptor. Studies of the structural properties and receptor recognition of polyUb require the ability to form these chains in vitro. This however has been a major challenge, primarily because lysine-specific E2 enzymes for linkages other than through K11, K63, or K48 are currently not known.
The established biochemical procedures for controlled assembly of polyUb chains from recombinant Ub protomers mostly follow the scheme, designed by Cecile Pickart and co-authors1, 14. This method relies on the availability of E2 enzymes to form a desired Lys-linkage and uses chain terminating mutations (i) to control the chain length and (ii) to avoid formation of unwanted linkages due to promiscuity of some E2s. Using genetic incorporation of unnatural amino acids15 (UAA) with removable protecting groups (e.g., Lys(Boc)) we have recently designed a new strategy that circumvents the need for permanent mutations and allows formation of fully natural polyUb chains using linkage-specific E2 enzymes16. However, performing ubiquitination enzymatically is inevitably limited by the availability of linkage-specific E2s and/or substrate-specific E3s.
Enormous progress has been made in recent years in developing chemical or chemical-biology approaches that circumvent the need for Ub-conjugating E2 enzymes to form isopeptide-linked Ub chains17–25 (reviewed in26, 27). In addition, several groups reported assembly of Ub chains using various non-native linkages, including isopeptide-bond replacements or lysine modifications28–30.
In particular, we focus on developing tools for nonenzymatic assembly of polyUb chains from recombinant Ub monomers that could be implemented in every biochemical laboratory. Using recombinant monomers is particularly critical for NMR and other structural or biophysical studies that require unit-specific or other types of isotopic labeling of Ub chains, which is essentially impractical for chemically synthesized chains because of the high cost of incorporating isotopic labeling into chemical synthesis. Previously, we have demonstrated that by combining UAA incorporation with the use of mutually orthogonal removable protecting groups (Boc- and Alloc-) and Ag-catalyzed condensation reaction between a C-terminal thioester on one Ub and a specific ε-amine on another Ub, it is possible to assemble polyUb chains of essentially any desired length, linkage composition, and isotope-labeling schemes, and in sufficient quantities even for structural studies25. Moreover, the resulting chains are fully natural, i.e. they are linked via isopeptide bonds and contain no mutations. Specifically, we successfully made di-Ub chains linked via K11, K27, or K33, as well as K11-linked tri-, and tetra-Ubs, and a mixed-linkage K33 and K11-linked unbranched tri-Ub. By enriching a specific Ub unit in the chain with 15N, we were able to characterize by NMR individual Ub units in some of these chains in order to reveal intra-chain Ub-Ub interactions.
That method relies on incorporation of a single UAA (e.g. Lys(Boc)) at the desired position in the protein sequence, and is inevitably limited to making unbranched chains of either homogeneous or mixed linkages. Making a branched Ub chain requires addressing yet another challenge, i.e. incorporation of UAA at more than one lysine site within the same Ub molecule, which has not been achieved before. In this paper, we describe and successfully demonstrate a method for assembly of fully natural branched polyUb chains that builds upon and extends our previous method for non-enzymatic assembly of unbranched Ub chains. Together, these two methods now allow assembly of essentially any polyUb chain, homogeneously-linked or with mixed linkages, branched or unbranched.
2. Materials and Methods
2.1. Assembly of [Ub]2–11,33Ub
2.1.1. Preparation and 15N enrichment of Ub K11Boc,K33Boc
Ub with the (unnatural) amino acid Lys(Boc) at both positions 11 and 33 was expressed in BL21(DE3) E. coli cells. The plasmid pTXB1, carrying ampicillin resistance, contained the E. coli codon-optimized Ub gene with TAG codon at positions 11 and 33, fused to an intein tag with a chitin binding domain (CBD). Incorporation of the Lys(Boc) at the TAG sites was accomplished with the plasmid pSUP-PylT-PylS containing the cellular machinery for recognition of the TAG codon and incorporation of Lys(Boc). The latter plasmid carried chloramphenicol resistance, allowing for double transformation into the BL21(DE3) chemically competent cells16. 15N-enriched Ub K11Boc,K33Boc was produced in autoinducing minimal media31 supplied with Lys(Boc) (final concentration 4 mM) in powder form16 and containing 15NH4Cl as the sole source of nitrogen. Ub protein was cleaved from the intein tag with 50 mM dithiothreitol at 25 °C. Two elutions at 48 hours and 120 hours were collected. The protein was purified via size exclusion chromatography, buffer exchanged into dH2O, and lyophilized. The presence of two Lys(Boc) groups was confirmed by ESI-MS.
2.1.2. Thioesterification of Ub
E1 Ub-activating enzyme was used to add a sodium 2-mercaptoethane-sulfonate (MESNA)-derived thioester group to the C-terminus of unlabeled wild-type ubiquitin as described previously25. Two 5 mg Ub reactions were performed in a 1 mL solution of 20 mM sodium phosphate buffer (pH 8.0) containing 10 mM ATP, 10 mM MgCl2, 100 mM MESNA, and 250 nM E1 enzyme. The reaction was incubated at 37 °C for 16 hours. Acid-sensitive E1 enzyme was precipitated from the reaction with two drops of glacial acetic acid. Thioesterification was confirmed by ESI-MS.
2.1.3. Alloc protection of Ub monomers
Two 5 mg portions of lyophilized thioesterified Ub (Ub-COSR) and 4.2 mg Ub K11Boc,K33Boc were each dissolved in 450 µL dimethyl sulfoxide (DMSO) and allowed to react with 17 µL N,N-diisopropylethylamine (DIEA) and 75 µL freshly prepared 40 mg/mL N-(Allyloxycarbonyloxy) succinimide (Alloc-OSu). Solutions were allowed to react at room temperature on an orbital shaker for two hours, followed by cold ether precipitation of the protein. Complete Alloc protection was confirmed by ESI-MS.
2.1.4. Removal of Boc groups
The Alloc-protected Ub K11Boc,K33Boc was dissolved on ice in 500 µL 60% TFA and placed on a shaker for four hours at 4 °C. Protein was precipitated with cold ether. Removal of both Boc protection groups was verified by ESI-MS.
2.1.5. Ligation
10 mg of Alloc-protected Ub-COSR and 5 mg TFA-treated Ub K11Boc,K33Boc were combined in a volume of 300 µL DMSO. To this, 13.2 µL DIEA, 7.5 µL each of freshly prepared 390 mg/mL N-hydroxysuccinimide (H-OSu) and 57 mg/mL silver nitrate were added. The reaction was incubated in the dark on an orbital shaker for 30 hours. The now gel-like solution was split into three tubes and reduced to a rosy orange powder by four rounds of cold ether precipitation. Formation of tri-Ub was confirmed by SDS-PAGE.
2.1.6. Global Alloc deprotection
All Alloc groups were removed from the Ub ligation mixture with 50 mol % ruthenium catalyst, chloro-pentamethylcyclopentadienyl-cyclooctadiene-ruthenium(II) ([Cp*Ru(cod)Cl]) and 50 equivalents of thiophenol. The reaction assumed a slight excess of over nine amines per Ub monomer, and 16 mg of overall protein. The protein was dissolved in 1680 µL of DMSO, to which were added 960 µL dH2O, 465 µL freshly prepared 20 mM [Cp*Ru(cod)Cl] in DMSO, and 95 µL thiophenol. The reaction was incubated at 50 °C for two hours in a thermal cycler, before being allowed to sit at 25 °C overnight. The solution was creamy orange with a black crust precipitate at the bottom. The supernatant was aliquoted into over 20 tubes and then reduced to a flaky brown precipitate after being subjected to over 20 rounds of cold ether precipitation.
2.1.7. Renaturation and purification
Each pellet was dissolved in 100 µL 6 M guanidine hydrochloride (GdnHCl), 20 mM sodium phosphate (pH 6.8) with periodic gentle agitation for 30 minutes, and centrifuged to remove precipitate. Supernatant was combined in a 50 mL conical tube and slowly diluted with 25 mL 20 mM sodium phosphate, 130 mM NaCl (pH 6.8). This solution was dialyzed against 2 L of the same buffer at room temperature overnight in 3K MWCO dialysis tubing. The dialysate was concentrated to a volume of 1.2 mL in a 3K MWCO Amicon concentrator and purified through size exclusion. The fractions corresponding to trimer were verified in purity via SDS-PAGE, concentrated and buffer exchanged into 20 mM sodium phosphate buffer (pH 6.8). Molecular weight of the purified branched tri-Ub was verified via ESI-MS.
2.2. Chain disassembly assays with Ubp6
Ubp6-catalyzed disassembly of various polyUb chains was assessed via SDS-PAGE analysis. Assays were performed in 50 µL phosphate-buffered saline (pH 7.4) using 25 µM of the polyUb chain and a sub-stoichiometric amount (8 µM) of Ubp6. The reactions were incubated at 30 °C for 21 hours. Time points were taken by quenching 4 µL reaction in 7 µL reducing SDS-PAGE loading buffer, then storing at −20 °C.
2.3. Enzymatic assembly of [Ub]2–11,63Ub
Tri-Ub branched at K11 and K63 was prepared using controlled-length chain assembly with chain-terminating K-to-R mutations and linkage-specific E2 enzymes14. K63-linked Ub2 was first constructed by reacting Ub K11R,K63R (distal Ub) and 15N Ub D77 (proximal Ub) in the presence of K63-specific E2 enzyme complex of Ubc13 and Mms2. After cation purification, the Ub2 was reacted with Ub K11R,K63R (distal Ub) in the presence of K11-specific E2 enzyme Ube2S to form a K11 isopeptide linkage on the proximal Ub.
2.4. NMR experiments
All NMR measurements were performed on an Avance III 600 MHz spectrometer (Bruker Biospin) equipped with TCI cryoprobe using standard or in-house pulse sequences32; sample temperature was set to 23 °C. All protein NMR samples were in pH 6.8, 20 mM sodium phosphate buffer containing 5% D2O and 0.02% (w/v) NaN3. Combined amide chemical shift perturbations (CSPs) were determined from differences in peak positions in 1H-15N NMR spectra using the equation Δδ = [(ΔδH)2 + (ΔδN/5)2]1/2, where ΔδH and ΔδN are differences in the 1H and 15N chemical shifts, respectively, for a given residue. For 15N T1 experiment, errors in intensity measurements were estimated using median noise calculations within Sparky33. Errors in 15N T1 relaxation times were determined using 500 Monte Carlo trials in the in-house program, RELAXFIT.
2.5. Mass spectrometry
For all proteins, high-resolution mass spectra of m/z 250–2500 were acquired with a JEOL AccuTOF-CS mass spectrometer in electrospray (ESI-MS) positive mode using flow injection. To determine the molecular weight, spectra were deconvoluted using MagTran software with the maximum charge set to 35.
2.5.1 Tryptic digestion of [Ub]2–11,33Ub, 15N-labeled on the proximal Ub
The purified [Ub]2–11,33Ub (40 µM in PBS pH 7.4 buffer) was digested with 0.3 µg trypsin (Promega) at 37 °C overnight. The digest was then acidified with 1 µL formic acid and stored at −20 °C until analysis. For LC-MS/MS analysis, 2 µL digest was injected and trapped into an Agilent Zorbax C18 trapping column (0.3 × 5 mm), desalted for 10 min before eluted onto an Agilent Zorbax C18 nano column (0.075 × 150 mm) with a gradient of 5–35% B in 40 min (Solvent A: 97.5% water, 2.5% ACN, 0.1% formic acid, solvent B: 2.5% water, 97.5% ACN, 0.1% formic acid). The LTQ Orbitrap was set to analyze 1 full scan MS with R=30,000 at m/z 400 followed by 5 data dependent MS/MS of the most abundant ions using the ion trap. The resulting data were searched against a human protein database using in-house mascot server to locate tryptic peptides of 14N Ub. Existence of 15N-enriched Ub was verified by pairs of unmodified tryptic peptides corresponding to the molecular ion of 14N and 15N peptides; ratios of 14N/15N peptide are appropriately 2/1 (data not shown). This verifies that for each molecule of 15N Ub, there are 2 molecules of 14N Ub.
3. Results
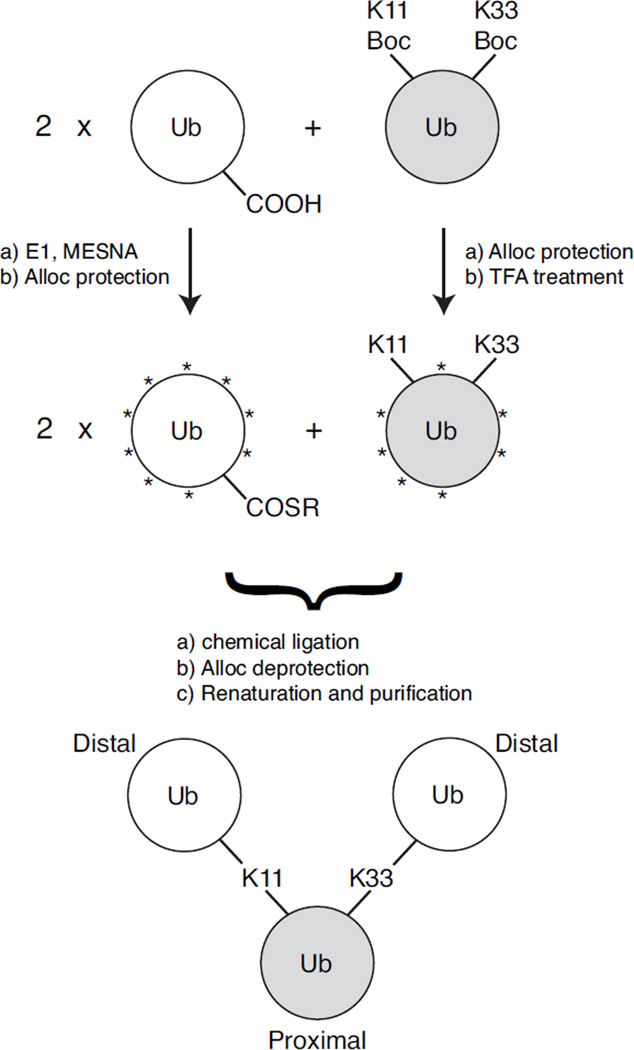
Here we describe a method to assemble all-natural branched ubiquitin chains via Ag-mediated condensation reactions that allow isopeptide linkages to form between available ε-amines of specific lysines on one Ub and the activated C-termini of other Ub monomers. This procedure is an extension of our work published recently25 that uses UAA incorporation and orthogonal removable lysine-protecting groups. Specifically, we will illustrate this method for the construction of a branched tri-Ub chain comprising both K11 and K33 isopeptide linkages on one Ub (Scheme 1). To distinguish the architecture of branched chains from unbranched mixed-linkage chains comprising the same linkages, we will use recently proposed nomenclature34 designating the branched chain as [Ub]2–11,33Ub and the unbranched chain as Ub–33Ub–11Ub, where the proximal Ub is shown on the right, the distal Ub is on the left, and the superscripts indicate the lysines involved in the linkages and, in the case of unbranched chain, also the linkage order. Note that in the branched chain, the proximal Ub is the one with the free C-terminus and the ε-amines of K11 and K33 covalently attached via isopeptide bonds to the C-termini of two other (distal) Ubs. Critical to the characterization of these polyUb chains with high-resolution structural methods, our scheme permits the isotopic labeling of either the distal Ubs collectively or the proximal Ub. Selective isotopic labeling of specific Ub units in polyUb chains is essential to avoid signal overlap in NMR spectra due to the homo-polymeric nature of polyUb25.
Scheme 1.
Non-enzymatic assembly of K11,K33 branched tri-Ub ([Ub]2–11,33Ub). Gray shading indicates Ub unit that is 15N-enriched. Asterisks (*) represent the Alloc protecting group.
3.1 Preparation and characterization of K11Boc,K33Boc Ub monomer
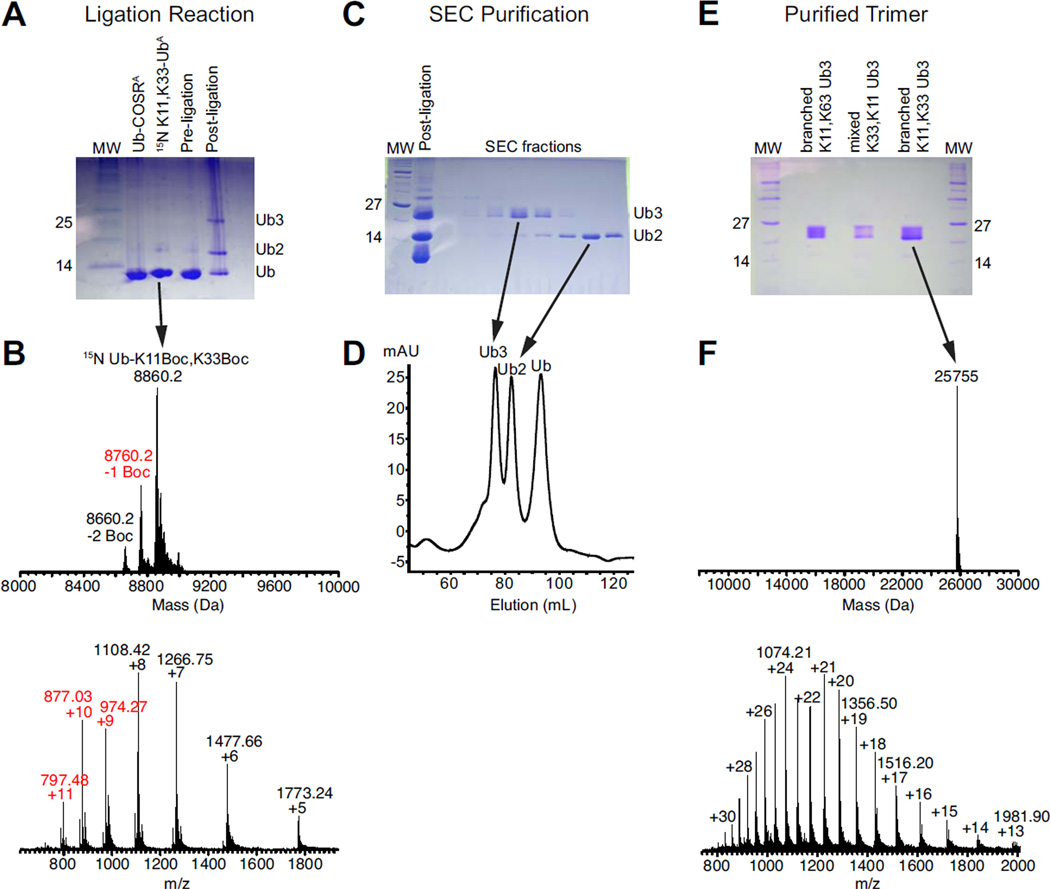
The most critical aspect of our method lies in the successful preparation of the proximal Ub with the genetic incorporation of Lys(Boc) amino acid at the specific lysine positions targeted for the isopeptide linkages. Previously, we demonstrated successful incorporation of a single Lys(Boc) at a specific lysine through encoding with an (amber) TAG stop codon. The genetic code of E. coli strain is expanded by adding an orthogonal pyrrolysyl-tRNA-synthetase/tRNAPyl pair that specifically recognizes the TAG codon and incorporates Lys(Boc) at that position in the emerging peptide16, 35. Here we introduced two TAG stop codons, at residues 11 and 33 in Ub via site-directed mutagenesis. Using procedures previously described16, 25, we produced ~5 mg of pure Ub containing both K11Boc and K33Boc mutations (Ub K11Boc,K33Boc). To enable NMR characterization of this protein, we enriched it with 15N by overexpressing it in cells growing in minimal media containing 15NH4Cl as the sole source of nitrogen. By SDS-PAGE, expression of Ub K11Boc,K33Boc appeared comparable to that of Ub containing a single Lys(Boc) (Supplementary Figure S1), but the overall yield of the purified Ub was 70–75% lower. Mass spectrometric analysis of the purified Ub K11Boc,K33Boc confirmed that the protein indeed contained two Boc protecting groups (Figure 1B).
Figure 1.
Assembly of [Ub]2–11,33Ub. (A) SDS-PAGE of the ligation reaction. Ub-COSRA is a fully Alloc-protected Ub containing a C-terminal thioester. 15N K11,K33 UbA is Ub that is Allocprotected except for K11 and K33, where the genetically-incorporated Boc protecting groups have been removed via TFA treatment. After 16 hours, ligation was complete and shows a tri-Ub band. (B) ESI-MS analysis of the 15N Ub K11Boc,K33Boc protein prior to Alloc protection or TFA treatment. The observed molecular weight is 8,860 Da, consistent with a 15N-enriched Ub containing two 14N-labeled Lys(Boc) amino acids. While the MS data also indicate the presence of a Ub species containing a single Boc group, this protein would terminate at the dimer stage during a ligation reaction. (C,D) Size-exclusion chromatography of fully Alloc-deprotected and renatured ligation product, indicating separation of tri-Ub from incompletely reacted Ub2 and Ub. (E) [Ub]2–11,33Ub is >95% by SDS-PAGE. (F) ESI-MS analysis of purified [Ub]2–11,33Ub, indicating a molecular weight of 25,755 Da, consistent with a tri-Ub species containing one 15N Ub, two 14N Ub units, and the loss of two H2O molecules upon isopeptide bonds formation.
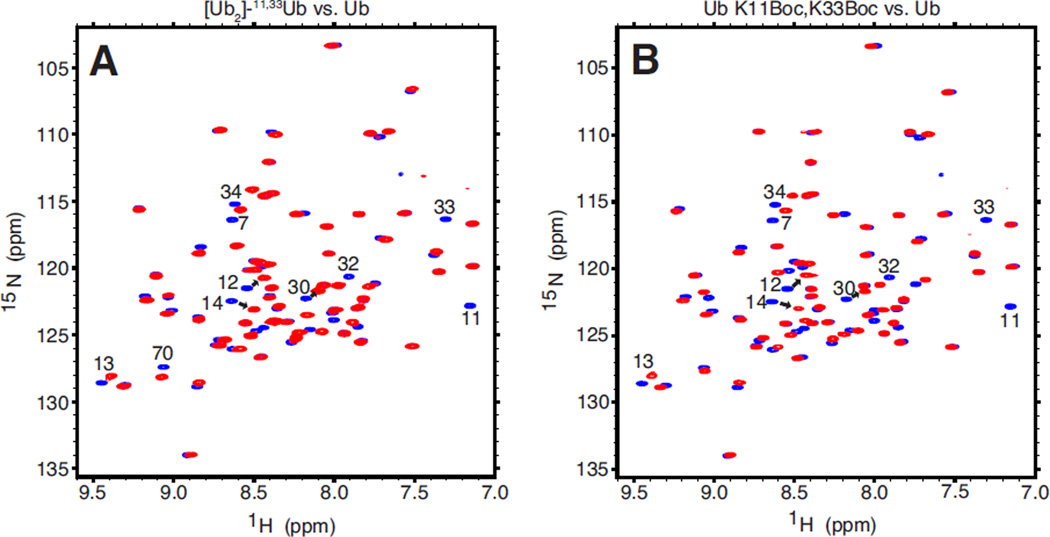
To further characterize the Ub K11Boc,K33Boc monomer, we collected a 1H-15N NMR spectrum of this protein and compared it with the spectrum of Ub (Figure 2B). The similarity of the two spectra indicates that Ub K11Boc,K33Boc is indeed folded and retains a spectral “fingerprint” pattern characteristic of monomeric Ub (monoUb). Importantly, the backbone amide resonances of K11 and K33 are absent in the spectrum of Ub K11Boc,K33Boc, indicating the successful incorporation of unlabeled Lys(Boc) amino acid at those positions. Furthermore, we quantified the shifts in the position of the NMR signals (otherwise known as chemical shift perturbations, or CSPs) on a per residue basis (Figure 3B). Most spectral perturbations are localized to those residues surrounding K11 and K33 in the 3D structure of Ub (not shown).
Figure 2.
NMR characterization of [Ub]2–11,33Ub and Ub K11Boc,K33Boc. (A) Overlay of 1H-15N TROSY spectra of the proximal Ub in [Ub]2–11,33Ub (red) and of monomeric wild type Ub (blue). (B) Overlay of 1H-15N TROSY spectra of Ub K11Boc,K33Boc monomer (red) and of wild type Ub (blue). Note the absence of (red) NMR signals for residues 11 and 33 in A and B due to the incorporation of unlabeled Lys(Boc) at these positions.
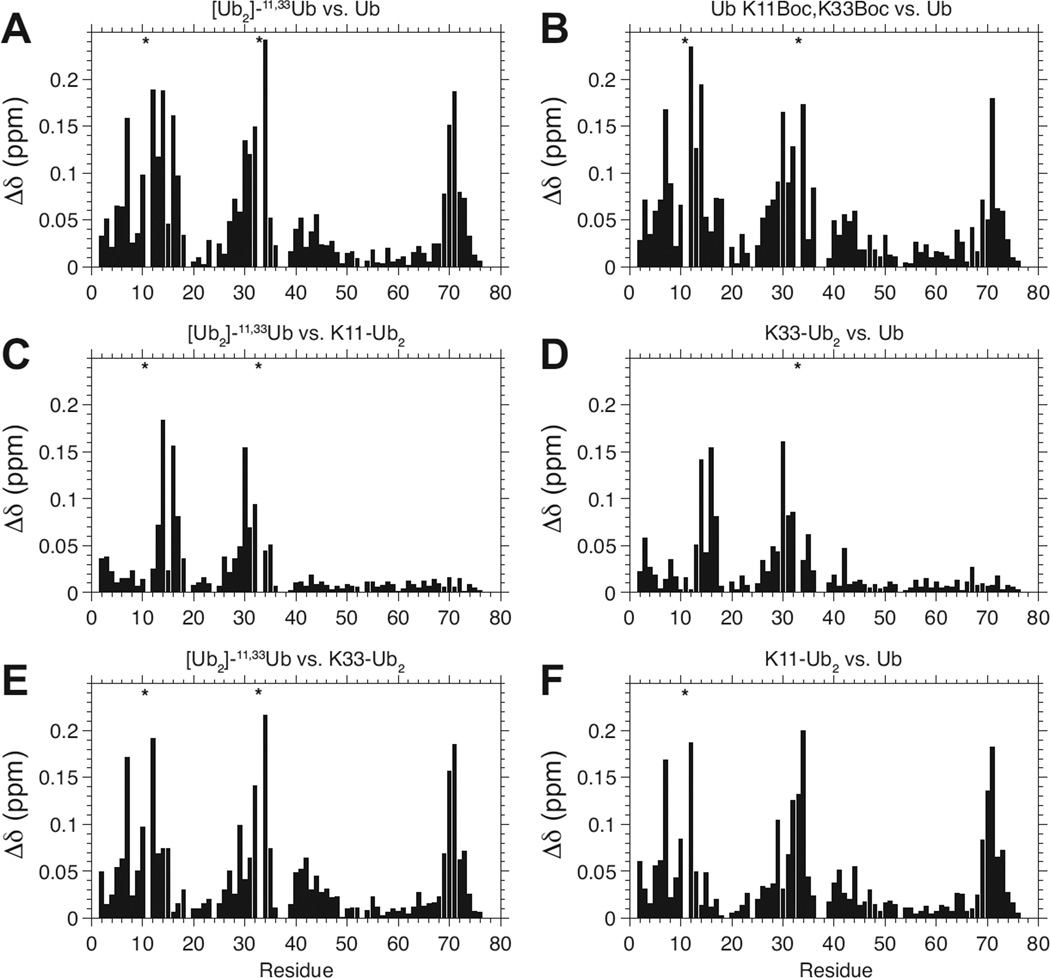
Figure 3.
Spectral differences, quantified as amide chemical shift perturbations (CSPs, Δδ), of the proximal Ub in [Ub]2–11,33Ub versus (A) monoUb, (C) the proximal Ub in K11-linked Ub2, and (E) the proximal Ub in K33-linked Ub2. (B) CSPs of Ub K11Boc,K33Boc monomer versus Ub. (D,F) CSPs of the proximal Ub in K33-linked Ub2 (D) and proximal Ub in K11-linked Ub2 (F) versus Ub. Absent NMR signals due to the incorporation of unlabeled Lys(Boc) are marked with asterisks.
3.2 Assembly of branched K11,K33-linked tri-Ub chain
Preparation of the distal Ub component(s) was achieved via overexpression and purification of monoUb from E. coli cells. To ready the monoUb for chemical ligation, its C-terminus was modified with a thioester moiety by reacting with Ub-activating E1 enzyme and the MESNA compound20, 25. To prevent all other available amines on both monoUb and Ub K11Boc,K33Boc from reacting in the chemical ligation process, these two proteins were reacted with Alloc-OSu, which specifically modifies the amines with the orthogonal protecting group Alloc25. Subsequent treatment of Ub K11Boc,K33Boc with TFA then selectively removed the Boc protecting group from the K11 and K33 side chains, rendering these particular ε-amines available for the ligation reaction, while keeping all other amines protected with the Alloc group (Scheme 1).
To generate the branched tri-Ub chain, a mixture of both monoUb and TFA-treated Ub K11Boc,K33Boc in a 2:1 stoichiometric ratio was reacted in the presence of H-OSu and AgNO3. The Ag-mediated condensation reaction resulted in formation of isopeptide bonds between the activated C-termini of the distal Ubs and the exposed ε-amine groups of K11 and K33. Successful formation of a tri-Ub product was monitored via SDS-PAGE (Figure 1A). The Alloc protection groups were then chemically removed, and the protein was renatured as previously described25. Branched tri-Ub was separated from incompletely reacted dimer and unreacted monomer by using size exclusion chromatography (Figure 1C,D). A final yield of 0.5 – 1 mg of purified tri-Ub was achieved from a total input of 14 mg of Ub units (4 mg of Ub K11Boc,K33Boc and ~10 mg of monoUb). The purity of the branched tri-Ub product ([Ub]2–11,33Ub) was assessed via SDS-PAGE analysis of this and other tri-Ub chains, including a mixed-linkage unbranched tri-Ub chain containing K11 and K33 linkages (Ub–33Ub–11Ub) (made as detailed in25) and another branched tri-Ub (enzymatically-assembled [Ub]2–11,63Ub) linked via K11 and K63 (Figure 1E). The observed molecular weight of [Ub]2–11,33Ub from mass-spectrometric analysis was 25,755 Da (expected 25,756 Da) (Figure 1F). This number is consistent with a tri-Ub species comprising two 14N-Ubs (each 8,564 Da) and a near-uniformly 15N-enriched Ub (~8,660 Da) with two water molecules (2×18 Da) released upon isopeptide formation at K11 and K33 side chains. The existence of both K11 and K33 linkages on the 15N-enriched proximal Ub of [Ub]2–11,33Ub was confirmed via tryptic digestion and subsequent LC-MS/MS analysis (Supplementary Figure S2).
3.3 NMR characterization of the branched K11,K33-linked tri-Ub
To examine [Ub]2–11,33Ub by NMR, a 1H-15N TROSY spectrum was collected of the 15N-enriched proximal Ub in this chain and overlaid with the corresponding spectrum of monoUb (Figure 2A). The overall similarity between the spectra indicates that the proximal Ub in the branched tri-Ub exhibits spectral properties similar to those of monoUb. The differences between the two spectra (quantified as CSPs) are illustrated in Figure 3A. Strikingly, the CSP pattern is nearly identical to the CSP pattern of monomeric Ub K11Boc,K33Boc versus wild type Ub, shown in Figure 3B. In other words, the effects of the K11- and K33-isopeptide linkages on the backbone amide resonances of the proximal Ub in the branched tri-Ub are quite similar to the effects of the Boc protection groups on K11 and K33 in the Ub K11Boc,K33Boc monomer. Thus, we conclude that the Boc moieties on K11 and K33 serve as isopeptide mimics. The data then suggest that the breadth of CSPs observed in the proximal Ub of [Ub]2–11,33Ub is primarily a consequence of the chemical modifications to K11 and K33 resulting from the isopeptide linkage formation, and the additional effect of non-covalent interactions (if they exist) between the proximal Ub and the two distal Ubs is minimal.
To investigate the effects of the individual isopeptide linkages on the spectral properties of the proximal Ub in [Ub]2–11,33Ub, we compared its spectrum with the spectra of proximal Ubs in K11-linked Ub2 (Figure 3C) and in K33-linked Ub2 (Figure 3E). Strikingly, the CSPs shown in Figure 3C are nearly identical to the CSPs of the proximal Ub in K33-linked Ub2 versus monoUb (Figure 3D). This is because the comparison with K11-linked Ub2 (Figure 3C) essentially has “subtracted” the contributions of the K11 isopeptide linkage and revealed the effects of the K33 isopeptide linkage on the spectrum of the branched tri-Ub. Similarly, the CSPs between the spectra of the proximal Ub in [Ub]2–11,33Ub and the proximal Ub of K33-linked Ub2 (Figure 3E), are nearly identical to those of the proximal Ub in K11-linked Ub2 versus monoUb (Figure 3F). This means that when we have accounted for the K33 isopeptide linkage effects, we observe CSPs resulting from the K11 isopeptide linkage, and vice versa. Importantly, this suggests that the effects of the individual K11 and K33 isopeptide linkages can be considered independent of each other. Thus, the CSPs in Figure 3A appear to be a sum of the perturbations arising from the K33 isopeptide linkage (Figure 3D) and the K11 isopeptide linkage (Figure 3F). This suggest that if any interactions exist between the two distal Ubs in [Ub]2–11,33Ub, they have little effect on the proximal Ub. Revealing contacts between the distal units by NMR would require the ability to isotopically label them individually, which is beyond the current methodology.
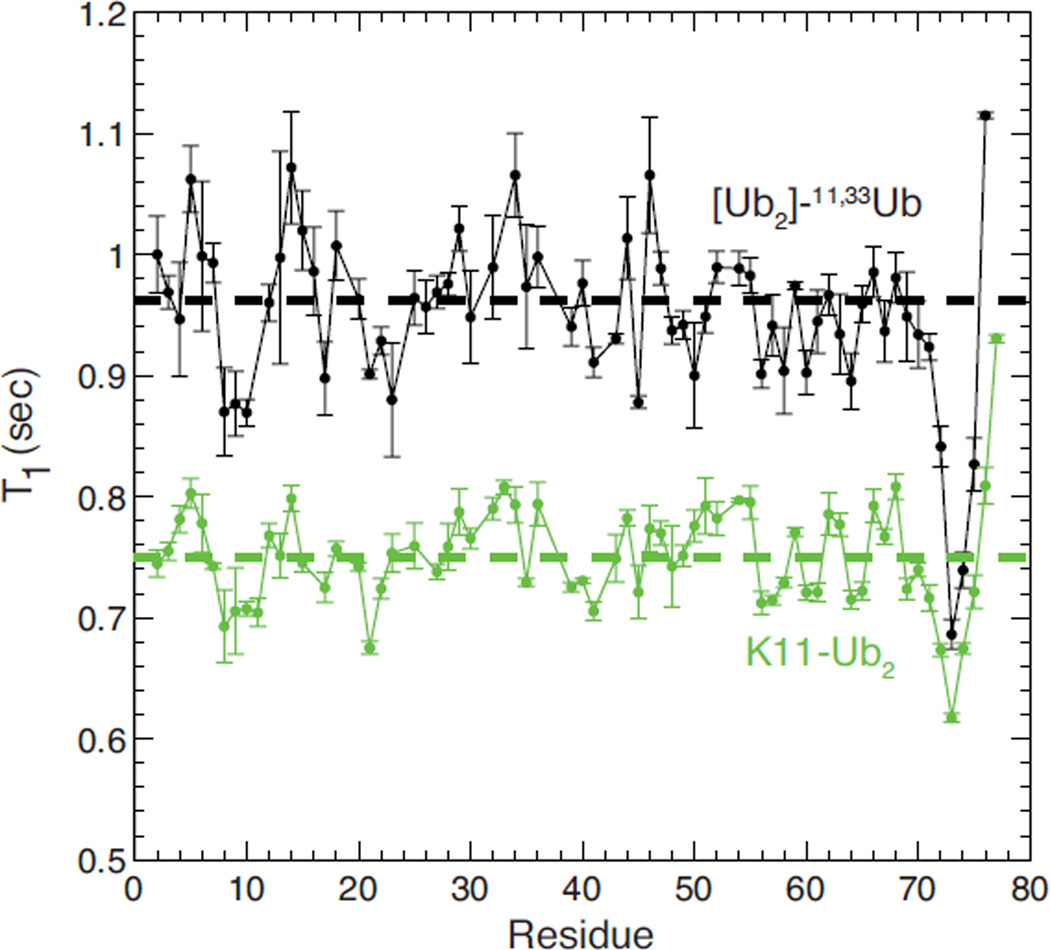
As a first attempt to characterize the structure and dynamics of the [Ub]2–11,33Ub at atomic level resolution, we measured 15N longitudinal relaxation time (T1) for each backbone amide of the proximal Ub (Figure 4). The 15N T1 relaxation time reflects the overall tumbling rate of a protein, and is correlated with its molecular weight36. As its size increases, protein tumbles more slowly in solution, and this increases the T1. For the proximal Ub in [Ub]2–11,33Ub, the average 15N T1 for residues in secondary structure elements is 0.962 ± 0.044 s. This number is consistent with a species of a molecular weight between 24 and 28 kDa36. This range is in excellent agreement with the reported molecular weight (25.5 kDa) of the branched tri-Ub from mass spectrometric analysis. For comparison, this 15N T1 for the branched tri-Ub is significantly higher than that of a (smaller in size) Ub2 species (Figure 4). On a residue-specific level, the overall pattern of 15N T1 relaxation times for the proximal Ub in [Ub]2–11,33Ub is very similar to the 15N T1 pattern observed in the proximal Ub of K11-linked Ub2. This indicates that the local backbone dynamics within the proximal Ub units of both Ub2 and the branched tri-Ub are largely identical.
Figure 4.
15N T1 relaxation time measured for backbone amides in the proximal Ub of [Ub]2–11,33Ub (black) and in the proximal Ub in K11-linked Ub2 (green). The dashed horizontal lines represent the average levels of 15N T1 for residues in secondary structure elements.
3.4 Disassembly of branched tri-Ub chains
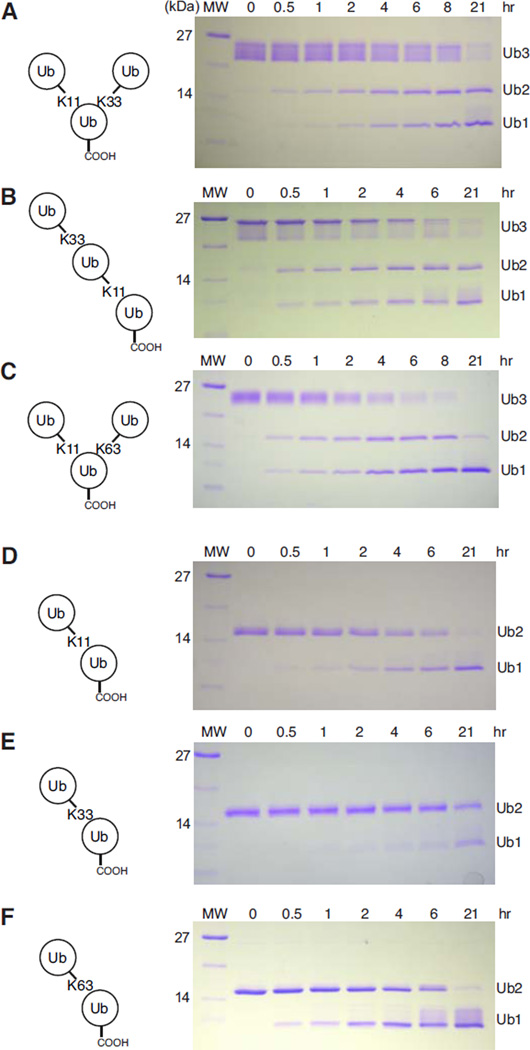
To assess the recognition properties of the branched tri-Ub, we performed a disassembly assay using yeast Ubp6 (USP14 in humans), a proteasome-associated deubiquitinating enzyme (DUB). Ubp6, together with other DUBs, regulates and maintains the amount of free Ub in the cell. We monitored the time course of the deubiquitination reaction by SDS-PAGE analysis (Figure 5). As controls, we monitored disassembly of the Ub2 chains comprising different linkages, particularly K11, K33, and K63 (Figure 5D–F). K11-linked and K63-linked Ub2 are mostly reduced to Ub monomer after 21 hours, whereas a significant amount of K33-linked Ub2 relative to Ub monomer is still present at this endpoint. The varying disassembly rates indicate that these chains are differentially processed by Ubp6.
Figure 5.
Disassembly of polyubiquitin chains by a proteasome-associated deubiquitinase Ubp6. A cartoon representation of each polyUb species is depicted on the left side of the corresponding gel.
The Ubp6-catalyzed disassembly of [Ub]2–11,33Ub was compared with that of the unbranched mixed-linkage chain, Ub–33Ub–11Ub (Figure 5A, 5B). It is clear that the apparent rate of cleavage for [Ub]2–11,33Ub nearly matches that of the mixed-linkage chain comprising the same, K11 and K33 linkages. This strongly suggests that Ubp6 is able to independently identify and cleave the K11 and K33 linkages in the branched [Ub]2–11,33Ub chain, and that branching of K11 and K33 linkages does not present any new recognition motifs for Ubp6. We observed that even at the final time point (21 hours), a significant amount of Ub2 was still present. From a comparison with the Ub2 disassembly results, we speculate that most of the dimer at this time point is K33-linked Ub2, since nearly all K11-linked Ub2 is reduced to monomer by this time (Figure 5E).
To compare the rate of disassembly of [Ub]2–11,33Ub with another branched tri-Ub, we performed a Ubp6 disassembly assay on a branched tri-Ub containing K11 and K63 linkages ([Ub]2–11,63Ub) (Figure 5C). Interestingly, only Ub monomer was present at the final time point for that chain. This contrasts with the slower rate of cleavage for branched [Ub]2–11,33Ub. It is interesting to note that the apparent rate of cleavage for [Ub]2–11,63Ub is consistent with the rates of cleavage for K11-linked Ub2 and K63-linked Ub2. Both of these Ub2s are effectively reduced to monomer after 21 hours, just as [Ub]2–11,63Ub. In summary, our disassembly data suggest that Ubp6 can distinguish the different isopeptide linkages in the two particular branched tri-Ubs assayed here. Moreover, from the gels in Figure 5 it appears that the two linkages in a branched tri-Ub are cleaved independently from each other.
4. Discussion
In this work, we have demonstrated the successful assembly of an all-natural branched tri-ubiquitin chain ([Ub]2–11,33Ub), verified via both NMR and tryptic digestion combined with LC-MS/MS analyses. Importantly, because K33-linkage Ub-conjugating E2 enzymes are not available, our method provides a unique opportunity to make this chain, and the ability to specifically isotopically label the proximal Ub allowed us to study it by NMR, which would be impractical when using total chemical synthesis. Our method is an extension of the previous work, in which we demonstrated the ability to build all-natural homogeneous- or mixed-linkage polyUb chains of any desired length and isotopic labeling scheme25. That work relied upon successful incorporation of a single unnatural amino acid (Lys(Boc)) at a specific site targeted for the isopeptide linkage. For branched Ub chains, our method necessitates successful incorporation of multiple Lys(Boc) amino acids at the same time into a single Ub. Here, for the first time, we have demonstrated that at least two UAAs can be incorporated into Ub. This is a notable achievement because of the very nature of how UAAs are incorporated in proteins expressed in E. coli cells. In most cases (here as well), UAAs are introduced into proteins by recoding a stop codon sequence (e.g. the amber codon, TAG) to be recognized by an orthogonal tRNA-synthetase/tRNA pair harboring the desired UAA. Thus, the orthogonal tRNA machinery competes with the natural release mechanism that recognizes the stop codon and terminates translation. As a result, the efficiency of incorporation of multiple UAAs into a single protein is dramatically reduced. In fact, we observed a significant drop in yield for Ub containing two Lys(Boc) amino acids (1 mg/L culture) versus Ub containing one Lys(Boc) amino acid (5 mg/L culture). However, recent advances in this field37, 38 may circumvent the competition between UAA incorporation and the release machinery in cells, allowing effective incorporation of multiple UAA in a single Ub without a significant decrease in protein yield. These advances could facilitate the assembly and structural and biochemical studies of polyUb chains branched at more than two lysines.
One limitation of the branched chains assembled using our method with regard to monomer-specific analysis (e.g., by NMR) is that it allows specific isotopic labeling of the proximal Ub, but not selective labeling of only one of the distal Ubs. The reason is that our method does not distinguish the Lys(Boc) side chains from each other nor the distal Ubs attached to them (Scheme 1). For example, in the case of [Ub]2–11,33Ub assembly, the TFA treatment of Ub K11Boc,K33Boc deprotects the ε-amines of both K11 and K33 indiscriminately. To achieve specific labeling of the distal Ubs, one of the lysines will have to be selectively deprotected at a time and then reacted with a selectively labeled distal Ub. This would require incorporation of different lysine variants (bearing different protecting groups) at specific positions in the sequence, which would necessitate further development of tools for expansion of E.coli genetic code.39
From our NMR studies of the proximal Ub in [Ub]2–11,33Ub, we can make several important observations regarding branched polyUb chains. First, the CSPs in the proximal Ub of [Ub]2–11,33Ub nearly match the CSPs of the Ub K11Boc,K33Boc variant vs. monoUb. This observation suggests that the majority of the CSPs seen in the proximal Ub of this branched tri-Ub stem not from Ub/Ub interactions but rather from the chemical modification of the K11 and K33 side chains with isopeptide linkages. We demonstrated previously that the Boc moiety on Lys side chains acts as an isopeptide mimic25. Second, the CSPs in the proximal Ub of [Ub]2–11,33Ub appear to be essentially a sum of the CSPs observed previously in the proximal Ubs of K11-linked Ub2 and K33-linked Ub2. This is an important observation as it suggests that the K11 and K33 linkages in the branched tri-Ub act almost independently of each other (at least in their effect on the proximal Ub). Interestingly, we observed similar behavior for the CSPs in the proximal Ub of another branched tri-Ub comprising K11 and K63 linkages (Figure S3). The CSPs of the proximal Ub in [Ub]2–11,63Ub versus monoUb, nearly match those of the proximal Ub in K11-linked Ub2 versus monoUb, with the exception of the CSPs present for residues 62–65. Because the K63-linkage only affects residues adjacent to (or in the vicinity of) K63 in the proximal Ub40, we can consider the CSPs in the proximal Ub of [Ub]2–11,63Ub to be a linear superposition of CSPs arising from the K11 and K63 isopeptide linkages.
The functional studies of both branched tri-Ubs via Ubp6-catalyzed disassembly certainly suggest that each branched tri-Ub exhibits properties of their respective isopeptide linkages, rather than a new set of properties that could result from linkage branching. In other words, at least for the two branched tri-Ubs assayed here, their recognition by Ubp6 and the rate of cleavage is dependent only on the lysine-specific properties of the individual isopeptide linkages present in each chain. Taken together, our functional and NMR studies suggest, that at least for both [Ub]2–11,33Ub and [Ub]2–11,63Ub, the isopeptide linkages are effectively independent of each other, i.e. the linkages appear to be recognized and cleaved independently. It is important to note that the observed linkage independence in these branched tri-Ubs is not necessarily present in all other branched polyUb chains, especially ones whose lysines are spatially closer to each other than the K11–K33 or K11–K63 pairs. However, we can infer that in [Ub]2–11,33Ub and [Ub]2–11,63Ub, the locations of the lysine residues permit the attachment of Ub without introducing any structural perturbations on the proximal Ub or introducing any new interactions between the proximal Ub and the distal Ub units.
That Ubp6 can recognize and disassemble branched tri-Ubs is an important finding because it suggests that the proteasomal machinery has a mechanism for recognizing and treating such chains. It remains to be seen if this holds for other branched chains, including the so-called “forked” chains, branched on two closely positioned lysines (K6 & K11, K27 & K29, K29 & K33), which have been reported to resist disassembly by purified proteasomes12, 41. The ability to assemble fully natural branched polyUb chains without the need for linkage-specific E2 enzymes provides tools for addressing these and other questions in order to elucidate and understand the mechanisms involved in Ub-mediated signaling pathways.
5. Conclusions
In conclusion, we developed a method for nonenzymatic assembly of all-natural branched polyUb chains. We demonstrated this method by making, for the first time, a branched tri-Ub chain linked via K11 and K33 and isotopically enriched on a specific Ub unit. Based on our spectroscopic and functional data, the two linkages in the branched trimer appear to act independently of each other. Because our method does not rely on the existence of linkage-specific E2 enzymes, branched chains of virtually any linkage composition can be constructed. This includes chains containing linkages such as K33, for which a specific E2 is unavailable. Together with our method for nonenzymatic assembly of unbranched polyUb chains, these developments now provide tools for making fully natural polyUb chains of essentially any type of linkage and length. As recent advances in the detection of more complex Ub chains have indicated the existence and involvement of mixed-linkage or branched polyUb chains in cellular signaling pathways, we anticipate that the method presented here will facilitate structural and biochemical studies of these branched Ub chains.
Supplementary Material
Acknowledgements
This work was supported by NIH grant GM065334 to D.F. We thank Michael Glickman for Ubp6 plasmid, Mark Nakasone for providing Ubp6 and advice regarding deubiquitination assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data include three supplementary figures.
References
- 1.Piotrowski J, Beal R, Hoffmann L, Wilkinson KD, Cohen RE, Pickart CM. J. Biol. Chem. 1997;272:23712. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Cell. 2004;116:181. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda F, Dikic I. EMBO Rep. 2008;9:536. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komander D, Rape M. Annu Rev Biochem. 2012;81:203. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 6.Fushman D, Wilkinson KD. F1000 Biol Rep. 2011;3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickart CM, Fushman D. Curr Opin Chem Biol. 2004;8:610. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. Mol Cell. 2006;24:701. doi: 10.1016/j.molcel.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Cell. 2006;127:1401. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 10.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Nat Cell Biol. 2006;8:700. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 11.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Cell. 2008;134:668. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 12.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. J Biol Chem. 2007;282:17375. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 13.Kravtsova-Ivantsiv Y, Ciechanover A. J Cell Sci. 2012;125:539. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 14.Pickart CM, Raasi S. Methods Enzymol. 2005;399:21. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 15.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. Science. 2003;301:964. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 16.Castaneda CA, Liu J, Kashyap TR, Singh RK, Fushman D, Cropp TA. Chem Commun (Camb) 2011;47:2026. doi: 10.1039/c0cc04868b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar KS, Spasser L, Erlich LA, Bavikar SN, Brik A. Angew Chem Int Ed Engl. 2010;49:9126. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]
- 18.Kumar KS, Bavikar SN, Spasser L, Moyal T, Ohayon S, Brik A. Angew Chem. 2011;50:6137. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]
- 19.Kumar KS, Spasser L, Ohayon S, Erlich LA, Brik A. Bioconjug Chem. 2011;22:137. doi: 10.1021/bc1004735. [DOI] [PubMed] [Google Scholar]
- 20.El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Angew Chem Int Ed Engl. 2010;49:10149. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang R, Pasunooti KK, Li F, Liu XW, Liu CF. Chem Commun (Camb) 2010;46:7199. doi: 10.1039/c0cc01382j. [DOI] [PubMed] [Google Scholar]
- 22.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Nat Chem Biol. 2010;6:750. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 23.Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. J Am Chem Soc. 2011;133:10708. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castaneda CA, Spasser L, Bavikar SN, Brik A, Fushman D. Angew Chem Int Ed Engl. 2011;50:11210. doi: 10.1002/anie.201104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castaneda CA, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. J. Am. Chem. Soc. 2011;133:17855. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spasser L, Brik A. Angew Chem Int Ed Engl. 2012;51:6840. doi: 10.1002/anie.201200020. [DOI] [PubMed] [Google Scholar]
- 27.Strieter ER, Korasick DA. ACS Chem Biol. 2012;7:52. doi: 10.1021/cb2004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eger S, Scheffner M, Marx A, Rubini M. J Am Chem Soc. 2010;132:16337. doi: 10.1021/ja1072838. [DOI] [PubMed] [Google Scholar]
- 29.Weikart ND, Sommer S, Mootz HD. Chem Commun (Camb) 2012;48:296. doi: 10.1039/c1cc15834a. [DOI] [PubMed] [Google Scholar]
- 30.Valkevich EM, Guenette RG, Sanchez NA, Chen YC, Ge Y, Strieter ER. J Am Chem Soc. 2012;134:6916. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studier FW. Protein Expr Purif. 2005;41:207. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Hall JB, Fushman D. J Biomol NMR. 2003;27:261. doi: 10.1023/a:1025467918856. [DOI] [PubMed] [Google Scholar]
- 33.Goddard TD, Kneller DG. SPARKY3. San Francisco: University of California; [Google Scholar]
- 34.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Structure. 2013 doi: 10.1016/j.str.2013.02.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Chem Biol. 2008;15:1187. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Mol Cell. 2005;18:687. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L. Nat Chem Biol. 2011;7:779. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson DB, Wang C, Xu J, Schultz MD, Schmitz RJ, Ecker JR, Wang L. ACS Chem Biol. 2012;7:1337. doi: 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Nature. 2010;464:441. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 40.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. J Biol Chem. 2004;279:7055. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 41.Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. EMBO J. 2009;28:1867. doi: 10.1038/emboj.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.