Summary
Next generation sequencing has dramatically changed the landscape of microbial ecology, large-scale and in-depth diversity studies being now widely accessible. However, determining the accuracy of taxonomic and quantitative inferences and comparing results obtained with different approaches are complicated by incongruence of experimental and computational data types and also by lack of knowledge of the true ecological diversity. Here we used highly diverse bacterial and archaeal synthetic communities assembled from pure genomic DNAs to compare inferences from metagenomic and SSU rRNA amplicon sequencing. Both Illumina and 454 metagenomic data outperformed amplicon sequencing in quantifying the community composition, but the outcome was dependent on analysis parameters and platform. New approaches in processing and classifying amplicons can reconstruct the taxonomic composition of the community with high reproducibility within primer sets, but all tested primers sets lead to significant taxon-specific biases. Controlled synthetic communities assembled to broadly mimic the phylogenetic richness in target environments can provide important validation for fine-tuning experimental and computational parameters used to characterize natural communities.
Introduction
For over two decades, amplification and sequencing of the small subunit ribosomal RNA (SSU rRNA or 16S rRNA) gene has been the primary approach to assess the abundance and taxonomic identity of microbes in the environment. Based on its universal presence and relatively uniform rate of evolution, SSU rRNA enabled the discovery and classification of a vast diversity of uncultivated microorganisms spanning all phylogenetic levels (DeLong and Pace, 2001; Tringe and Hugenholtz, 2008; Pace, 2009). With increasing sequencing depth and throughput, statistically robust quantitative comparisons between communities have become feasible. Direct, metagenomic sequencing of the community DNA pool complements rRNA gene-based characterization by providing insights into physiological potentials and expanding phylogenetic diversity characterization into protein sequence space (Tringe et al., 2005) while metatranscriptomics and metaproteomics offers a direct access to community physiology (Verberkmoes et al., 2009; McCarren et al., 2010). A variety of experimental and data analysis alternatives have been developed to allow in-depth characterization and large-scale comparative studies of complex microbial communities (Schloss and Westcott, 2011a; Sun et al., 2011). Each experimental and computational step in diversity characterization is prone, however, to errors (Polz and Cavanaugh, 1998; Hong et al., 2009; Engelbrektson et al., 2010; Huse et al., 2010; Haas et al., 2011b). Amplification of different hypervariable rRNA gene regions can lead to inconsistent taxonomic coverage and incongruence between datasets. In addition, short read amplicon sequencing requires specific considerations for the methodology, data analysis and interpretation of microbial diversity (Caporaso et al., 2010; Quince et al., 2011; Schloss et al., 2011; Gihring et al., 2012; Werner et al., 2012).
Metagenomic sequencing avoids some of the limitations of rRNA amplicon sequencing by directly accessing the community genomic information. Diversity interpretation is however, complicated by uncertainties in assigning genes to specific organisms (especially for taxa with no cultured representatives) and by bias introduced during sequencing (Gomez-Alvarez et al., 2009). Direct, quantitative comparisons between known diversity and that inferred by rRNA gene and metagenomic data are scarce (Morgan et al., 2010; Caporaso et al., 2011b; Caporaso et al., 2011a; Haas et al., 2011b; Schloss and Westcott, 2011b) and have been limited in taxonomic coverage. However, as the depth of sequence coverage continues to increase and methods for binning and assembling are improving, some recent studies have reported near complete genomes from complex environmental samples or laboratory enrichments (e.g. (Iverson et al., 2012; van de Vossenberg et al., 2012; Wrighton et al., 2012).
Here we quantitatively compared the accuracy of SSU rRNA gene based microbial diversity analyses with metagenomic sequencing using a controlled “synthetic community” approach. The communities consisted of 64 laboratory-mixed microbial genomic DNAs (gDNA) of known sequence, representing a broad diversity of bacteria and archaea. Species from nearly all phyla with cultured representatives were included, isolated from many different environments, covering a wide range of genetic variation at different taxonomic levels (strains to phyla) and spanning the full spectrum of genome sizes, (G+C)% (GC) content, genomic divergence, and rRNA operon copy numbers. The effects of varying experimental procedures and data analysis strategies on rRNA based diversity and composition were compared with those determined using two metagenomic sequencing platforms, 454 and Illumina.
Results and Discussion
Synthetic archaeal and bacterial community characteristics
By combining known amounts of purified gDNAs we constructed two diverse synthetic communities representing the domains Archaea and Bacteria, respectively. These communities included most phyla with cultured representatives, as well as contained closely related species and strain pairs. All included organisms have complete or high quality draft genomic sequences. Sixteen members of Crenarchaeota, Euryarchaeota and Nanoarchaeota represented Archaea, while Bacteria included 48 organisms from 18 phyla (Table S1). The organisms covered a wide variety of metabolic strategies and adaptations to the human body, marine and terrestrial aquatic environments, soils and the subsurface, and extreme physical or chemical conditions. Unlike environmental communities, each gDNA was individually purified and quantified prior to being mixed with others, thus true community composition was known, and extraction-based biases were eliminated. The genomes span a broad range of GC content (27–70%), sizes (0.5–10 Mbp) and rRNA operon number (1–10). Based on these known parameters and quantification by Q-PCR, we validated the representation (cell equivalents) of each species. The Archaea community contained one dominant species (Nanoarchaeum equitans, 30% of genome copies), with the others present at abundances between 1–10%. Due to differences in genome size and rRNA gene copy number, the actual contribution of individual organisms to the metagenome complexity spanned a 20-fold range (e.g. N. equitans, with a genome of 0.5 Mbp represented 10% of the metagenome). The Bacteria community contained no single dominant organism however there was a 25-fold variation in the gDNA abundance among the individual taxa. For several tests, we also combined them into a 64-member Archaea-Bacteria (AB) community in which the genomic abundance of taxa spanned a 200-fold range, from 0.05–1% (36 taxa), 1–5% (23 taxa) to 5–8% (6 taxa). These communities were not aimed at reproducing any specific type of natural diversity, but to broadly represent the phylogenetic and genomic heterogeneity within Bacteria and Archaea that is often encountered in complex community assessments. Many communities contain a vastly greater number of taxa at all levels (e.g. soil communities) or are scarcer in number of high taxa but much more diverse at genus level and below (e.g. human gut microbiota). Other communities (e.g. marine sponge-associated bacteria) are rich in uncultured taxa at all taxonomic levels and contain ecologically important low abundance members (Webster and Taylor, 2012), difficult to mimic in the laboratory. However, the synthetic community used here, by combining taxa adapted to many different types of environments, should provide a good initial assessment of the power of taxonomic and quantitative determinations to be expected when using a broad range of natural samples.
Metagenomic characterization of the synthetic Archaea-Bacteria community
For metagenomic 454 and Illumina sequencing we used the AB community as it contained the broadest variation in phylogenetic diversity and genomic characteristics. These two platforms differ in output and data characteristics but both have been widely used for environmental sequencing (McCarren et al., 2010; Hess et al., 2011).
The 454 sequencing library was generated using Nextera™ in vitro transposition (Caruccio, 2011), which has low DNA, input requirement compared to physical shearing methods (50 ng vs. >1 μg). Analyses using single organism libraries have shown relatively similar coverage in such libraries compared to ones obtained by shearing (Adey et al., 2010). Because in many environmental studies the availability of DNA is limited, determining the accuracy of community composition inferences using metagenomic sequencing of such samples is important. A recent study demonstrated that extensive, phi29 polymerase amplification significantly alters the composition of metagenomic libraries (Yilmaz et al., 2010). While the transposition-generated library requires only mild amplification, a bias risk remains and was therefore investigated. The Nextera™ AB library was sequenced on one quarter of a 454 FLX Titanium plate and generated 2.9×105 reads (85.5 Mbp of sequence). For the Illumina platform, a standard sheared library was constructed and bidirectional sequencing on one lane resulted in 107 million reads (>10 Gbp of sequence).
Using local alignment, >97% of the 454 data (83.5 Mbp of sequence) was mapped to the 64 reference genomes (205.6 Mbp total length) and 2% to plasmids of those organisms (32 plasmids, totaling 4.2 Mbp), reaching average metagenome coverage of 0.39 fold. Similarly, over 92% of the Illumina reads were mapped to the reference genomes and plasmids, achieving 46–50 fold metagenome coverage. The combination of a 200-fold range variation in individual genome abundance with the 20-fold variation in genome size generated distinct sequence frequency distributions for the different members of the community. Consequently, the observed average fold coverage of individual genomes by mapped reads ranged from 0.01–1.3 for the 454 data to 6–300 for Illumina (Table S1). With Illumina, 53 of the 64 organisms had coverage overlaying >95% of their determined chromosomal sequences.
To determine how accurately the metagenomic data described the actual community composition, we compared the expected representation of each species based on qPCR quantification with the observed coverage. Overall, both metagenomic sequence sets described the community genomic composition remarkably well, with 70% (454) and 78% (Illumina) of the individual species/strains estimated within a factor of two-fold or less from their actual abundance (Figure 1). Importantly, both sequencing platforms appear relatively unaffected by the abundance of individual genomes spanning two orders of magnitude variation. However, the 454 metagenome showed a measurable bias towards oversampling of genomes with low GC (<40%) and under sampling for those with high GC values (>60%), including in-depth of coverage across the individual genomes (Figures S1 and S2). Potentially, such bias could be attributed to enzymatic steps in library construction. In comparison, the Illumina metagenome was less influenced by GC content, with abundances of most of the individual genomes <2-fold of their expected levels and with GC-based coverage better tracking the actual metagenome composition than 454. Pairwise t-tests confirmed a higher differential GC-linked bias in low-GC organisms (27–40% GC; p=0.027) and lower in high-GC organism (40–70% GC; p=2.7e-05) for 454 data, while representation of mid-range GC organisms (40–60%) was not significantly different between 454 and Illumina (p=0.170). The abundance of some thermophilic taxa (Pyrococcus, Dictyoglomus, Sulfurihydrogenibium) was overestimated by both platforms (2–5 fold). This may be linked to extensive regions of very low GC in their genomes, which displayed an inflated coverage with both platforms, although other sources of bias could be involved. However, when we analyzed each individual genome in the community in terms of sequence coverage, the only bias detected was linked to local GC content. A comparison of the intra-genome coverage with both 454 and Illumina for representative genomes with different GC content is presented in Figure S3 (similar plots were obtained for the other genomes of the community and also for genomes we sequenced independently).
Figure 1.
Characterization of the Archaea-Bacteria community by 454-FLX-T (A) and Illumina-HighSeq (B) metagenomic sequencing. The accuracy of retrieving the known composition of the metagenome is indicated for each organism as a ratio of the observed genomic coverage to the known genome abundance in the community and is plotted against its known abundance in the community. Shading zones indicate a low level of bias (dark: <1.5 fold; light: 1.5–2 fold) from the perfect value of 1.
The sequences were uploaded into two widely used metagenomic analysis systems, IMG/M (Markowitz et al., 2012) (454 data) and MG-RAST (Glass et al., 2010) (454 and Illumina data). Because all individual genomes of the synthetic community are integrated in these systems, we evaluated IMG/M and MG-RAST for accuracy in predicting and quantifying the genomic diversity of the synthetic metagenome. Although corrections for individual genome size and coverage are difficult to apply on metagenomic datasets, both systems recovered the bacterial phyla representation quite well, with most taxa estimated to within a factor of two of their actual abundance (Figure S4). Archaea were less accurately quantified, some being either under (N. equitans), or over estimated (Crenarchaeota and Euryarchaeota). Inferences based on the Illumina data were generally consistent with those based on 454, with some discrepancies potentially due to GC coverage differences between platforms. However, both IMG/M and MG-RAST predicted a higher diversity than actually present, at most taxonomic levels (MG-RAST alpha diversity was overestimated by >6 fold for both datasets)(Table S2). Most of the spurious groups of organisms at high taxonomic levels (some bacterial and archaeal phyla as well as fungal, plant and metazoan lineages) were based however on few sequences and had relatively low confidence values but numerous sequences were also incorrectly assigned to taxa closely related to those present in the community. This appears to be due to a combined effect of the short read data and the variable analysis stringency, limiting phylogenetic resolution and leading to incorrect assignments between closely related organisms and, for conserved genes, even across high-rank taxa. Such, overestimations are an important factor to consider, especially in studies that aim to identify rare organisms. Default predictions by the current versions of these two widely used analysis platforms reported organisms and taxa (e.g. Eukarya, several bacterial and archaeal phyla) that cannot be linked to the community we used here. Some issues may be addressable by sequence assembly, which should improve gene prediction and functional annotation in addition to taxonomic assignments, especially at high sequence coverage (Pignatelli and Moya, 2011). At present, neither IMG-M nor MG-RAST provides Illumina sequence assembly options although MG-RAST allows upload and assignment of raw sequence reads. It is important to note, however, that the Illumina short reads provided a very good estimates of taxonomic distribution above the species level, with only a 2–3 fold overestimation of the actual number of genera and orders. For the 454 data, however, the use of the default parameters severely overestimated higher level diversity (~20 fold for bacterial genera and identified >100 spurious eukaryotes). Increasing the stringency of the analysis produced much more accurate results, inline with the Illumina output (Table S2). In analyzing environmental metagenomic datasets selection of the various cutoff parameters is therefore an important consideration and, the results presented here may serve as initial guidance in developing such procedures.
We also evaluated the accuracy of taxonomic assignments under the hypothesis that none of the exact genomes of the community are represented in the database. While in natural communities one often times identifies closely related organisms to those that have a genome sequence determined, a large fraction of the Bacteria and Archaea still have poor genomic coverage, even often at phylum level. Therefore, determining where the metagenomic sequence data maps and how accurate the assignments are to higher taxon level (e.g. family, order, phylum) is of interest to expand results obtained with synthetic communities to natural ones. Because the genomes of all organisms we included in the synthetic community are part of the public databases used by IMG-M and MG-RAST, with no option to be excluded from the analysis, we performed a local metagenomic analysis with MEGAN (Huson et al., 2007). We compared taxonomic assignments to a Bacteria-Archaea database containing all available sequenced genomes, and to a version of that from which we removed the genomes represented in the community. In the first case, the accuracy was verified to species, genus and family levels. When the reference organism was excluded from the community, the accuracy was analyzed to genus and family levels. When taxa were poorly represented in the genomic database (e.g. the reference genome in the community was the single sequenced representative at genus, order or even phylum level, such as Ignicoccus, Nanoarchaeum, Gemmatimonas), eliminating the reference genome from the database affected the assignment of those sequences, most having no match, especially using the stringent megablast algorithm. As a result, the abundances of those taxa were underestimated in the community. The more permissive Blastn-based analysis produced a more accurate representation, especially at family level for both 454 and Illumina data. Figure S5 summarizes the result of analyses using the two different blast-mapping criteria in comparison to the known taxonomic composition of the community. In addition, we analyzed the use of a single marker gene (SSU rRNA) for explaining the taxonomic and quantitative composition of the community using the 454 and Illumina metagenomes. The reads corresponding to that gene were identified and analyzed using the RDP Classifier (Cole et al., 2009). While many of the taxa were identified to genus level, the quantitative recovery of the relative community composition was very poor, especially with the Illumina data, and there was a severe overestimation of the Archaea (Figure S5). We explain this by a combination of low taxonomic resolution of the short reads that randomly cover the rRNA sequence (and therefore carry different phylogenetic signal depending on the degree of variability within the gene) and by the GC bias present in the rRNA operons relative to the average genome content, especially in the many hyperthermophilic archaea present in the community. In addition, because single gene coverage by 454 data in complex metagenome is generally low, taxon identification and quantification is statistically weak. The result of this analysis indicates that a single gene marker such as rRNA is a poor determinant of the community structure in metagenomic sequence data from complex communities, especially when one desires quantitative estimates. A deeper metagenomic coverage combined with the concerted use of single copy genes that provide sufficient taxonomic resolution (e.g. RNA polymerase subunits, translation factors) may overcome such limitations. Synthetic metagenomes can provide important controls in selecting algorithms and parameters used for interpretation of actual environmental data on various platforms and software and should be explored in conjunction with in situ benchmark studies (Mavromatis et al., 2007; Pignatelli and Moya, 2011).
SSU rRNA gene amplification, pyrosequencing and data processing
For rRNA gene-based taxonomic characterization of the three synthetic communities, multiple, variable-length fragments of the SSU rRNA genes spanning most hypervariable regions were amplified and sequenced using the 454 platform. The selection of primers was based on their use in prior Sanger and 454 sequencing studies and included five pairs for Bacteria, three pairs for Archaea and a pair that we developed to simultaneously capture both domains (Frank et al., 2008; Engelbrektson et al., 2010; Porat et al., 2010; Wu et al., 2010; Bates et al., 2011; Haas et al., 2011a; Kan et al., 2011). Because some were limited in taxonomic coverage, we introduced modifications or supplemental variants employed in primer mixtures, to expand their breadth (Figure S6 and Table S3). While degenerate positions in primers were predicted to enable annealing to almost all taxa included in the simulated communities, mismatches to some of the target sequences existed. Such mismatches allowed us to identify their effects in detecting those taxa, and reflect the important reality that there are no truly universal primer sets. Effects of polymerase fidelity, amplification cycle number and amplicon length on inferred taxonomic diversity as well as the variability between replicate amplifications were also tested. Amplicons were sequenced using either FLX or Titanium chemistry and the resulting data processed for barcode-based de-multiplexing, removal of amplification or sequencing artifacts, and diversity analyses using a combination of software packages (mothur, ChimeraSlayer, AmpliconNoise, RDP, ESPRIT, QIIME). AmpliconNoise analysis involved PyroNoise removal of 454 errors and SeqNoise removal of PCR single base misincorporations(Quince et al., 2011). We estimated the proportion of errors attributable to these two sources by calculating the reduction in error rate after applying each algorithm (Table S4). Raw per-base error rates varied from 0.1% to 0.25% for FLX and 0.15% to 0.9% for Titanium, with some differences noted between the various amplicons. The error rate following noise and chimera removal was remarkably low, at less than 0.1% for most regions.
A commonly reported artifact in SSU rRNA amplicon analyses is the formation of chimeras during PCR (Suzuki and Giovannoni, 1996; Haas et al., 2011b; Quince et al., 2011), which inflates the inferred richness. Overall, the frequency of chimeras detected by ChimeraSlayer or Perseus in AmpliconNoise was very low (<1% of reads) with the exception of the bacterial V13 dataset for which it ranged between 7–10%. The rate decreased (2–3 fold) with fewer PCR cycles (<3% at 24 cycles for V13) and also when using highly accurate enzymes with additives for increasing PCR specificity (High-GC mix, Accuprime-Pfx). Whilst the proportion of chimeric reads was generally low, they could form a large proportion of the unique sequences present following noise removal, implying that their contribution to the over estimation of diversity is significant.
Community diversity analysis using sequence similarity
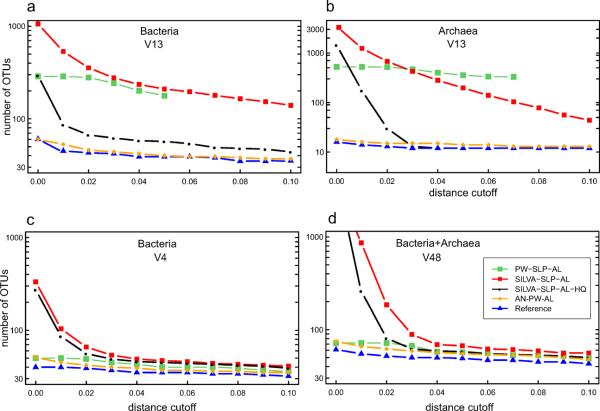
Because SSU rRNA gene sequence similarity decreases with increasing phylogenetic distance between organisms, quantifying the differences between individual sequences in microbial community datasets provides a metric of phylogenetic diversity that can be standardized and applied in an ecological and statistical framework. Though approximate and not without pitfalls (Stackebrandt and Ebers, 2006), pairwise similarity values have been adopted in comparing distance-based classifications to phylogenetically defined taxonomic ranks (e.g. 97% similarity corresponding to “species” level). For the synthetic communities analyzed here, we determined the actual sequence similarity level for each sequenced region of the SSU rRNA gene and each pair of species and strains from the same genus of Archaea and Bacteria (Figure S7). These values were used to determine the maximum resolution of the sequence analysis step and, in conjunction with the pairwise distances between all the members of the community, to calculate the actual number of taxonomic units at various levels of sequence similarity. For some genera the 97% value holds relatively well and is uniform between the various regions. For other genera however (e.g. Thermotoga, Sulfurihydrogenibium, Salinispora) inter-species similarity values were significantly higher (>99%), which limited the taxonomic resolution and underestimated diversity. However, as OTU similarity cutoffs approach 100%, effective resolution of species and strains in natural communities is confounded by rare sequence errors. Parameters and methods used for sequence processing and clustering into OTUs can additionally impact the inferred diversity OTUs (Huse et al., 2010; Kunin et al., 2010; Schloss et al., 2011; Sun et al., 2011). To exemplify these effects the number of OTUs at 97% similarity (3% distance) is shown in Figure 2 for bacterial and archaeal V1–V3 region. The frequency of OTUs with only one or two sequences is compared with those consisting of multiple sequences as well as with the actual OTUs determined by clustering of reference sequences. Less stringent sequence processing leads to severe diversity overestimation, primarily by singletons. Sequence trimming to common coordinates and quality filtering eliminated a large proportion of the singletons and reduced the number of spurious OTUs. However, even after OTU calculation using the mothur implementation of SLP-AL(Huse et al., 2010; Schloss and Westcott, 2011b), some 30% (19–21 out of 61–63) of the bacterial OTUs were still attributable to noise although only one contained more than two sequences. This could be reduced to just 6% (2–3 out of 44–47) if AmpliconNoise is used instead of single linkage pre-clustering for noise removal. This effect is even more dramatic at lower similarity cut-offs. A summary of the number of OTUs at progressive distances for each SSU rRNA gene amplicon is shown in Figure 3 and Figure S8. However, combined removal of sequencing/PCR noise and chimeras by AmpliconNoise and Perseus followed by pairwise alignments and average linking clustering, eliminated most spurious OTUs at virtually all distance settings and best represented the community diversity.
Figure 2.

Effect of sequence processing parameters on OTUs. Sequenced amplicons from V13 region of SSU rRNA of both Archaea (A) and Bacteria (B) were filtered, trimmed, and clustered using the parameters specified in a table form (A–H). Sequences were trimmed to the same coordinates after alignment against the SILVA database and clustered using either complete linkage clustering (CLC) or average linkage clustering (AL) with distances based on Infernal alignment or SILVA based alignment in mothur, respectively. The numbers of OTUs at 97% sequence similarity (distance 0.03) are shown with distinguished contribution from OTUs consisting of one, two or more sequences.
Figure 3.
OTU-based diversity estimation as a function of genetic distance and analytical approach relative to the reference genomic SSU rRNA sequences. Bacterial V13, V4, archaeal V13 and the combined archaeal-bacterial V48 amplicon datasets are shown. The results for the other amplicons are shown online in the Supplementary Figure 8. Silva–SLP–AL (red) and Silva–SLP–AL-HQ (black): single-linkage pre-clustering 2% and average linkage clustering of SILVA alignment of sequences not purged of errors and on sequences with the chimeras removed (parameters B and G in Figure 2, respectively). PW–SLP–AL (green): single-linkage pre-clustering 2% and average linkage clustering of Needleman-Wunsch (NW) pairwise alignment of sequences not purged of errors. PW-AN-AL (orange): average linkage clustering of pairwise alignment of sequences after denoising and chimera removal using AmpliconNoise/Perseus. For comparison, OTUs obtained by clustering the reference sequences using Silva–SLP–AL (blue) are shown. Note that the y-axis in (A) is scaled logarithmically.
Community diversity analysis using taxonomic mapping
In addition to diversity estimation using similarity clustering, relating SSU rRNA gene sequences to taxonomically classified organisms provides important information about the composition and, to some extent, potential physiological and ecological characteristics of a community. Because the actual composition of natural communities is not known a priori, the accuracy and resolution of sequence based taxonomic inferences remains undetermined, and most often, is not verified by independent measurements/techniques in ecological studies. Using the synthetic Archaea and Bacteria communities we analyzed how the different SSU rRNA regions reflect the known quantitative taxonomic composition and in comparison to the frequencies obtained by metagenomics. For each sequence dataset and each organism, an accuracy ratio (observed versus predicted sequence frequency) was determined and the average of three replicates is represented as a heat map in Figure 4, with a value of one corresponding to perfect agreement. The technical reproducibility between replicates for each primer set ranged from an average of 2.5 fold variation for the bacterial V4 amplicon to 1.5 fold for V13 amplicon. A higher variation in inferred abundance between the replicates was correlated with decreasing actual organism abundance in the synthetic community, especially at levels below 1%. This closely followed the expected patterns associated with Poisson distribution noise and as such may be mostly be attributable to undersampling. However, stochastic variation in PCR amplification efficiencies may play a role as well (Figure S9). In general, over or underestimating the abundance of the different taxa in a community by up to two fold may be considered a resolution limit of these approaches, however these would likely be greater in the absence of averaging independent sequencing data or pooling of PCR products before sequencing. Such noise can be expected to be even more pronounced in natural communities with higher diversity and many low abundance organisms(Zhou et al., 2011). Our analysis indicates that although for many organisms the inferred abundance is within a factor of two or less from the actual value, no primer set was ideal for quantitatively representing the entire diversity of even our relatively simple community and biases did occur. Some taxon underestimation could be explained by mismatches between primers and the target sequence, as no primers are universal, especially at species level. Primer alignments for all tested organisms and identification of mismatches likely associated with underestimation or lack of detection are shown in Figure S6. Surprisingly, some phylum-level detection problems in several amplicon regions could not be directly attributed to primer mismatches. The most apparent discrepancies were underestimation of Bacteroidetes and Actinobacteria by the V4 amplicons and the lack of detection of most thermophilic Aquificales and Thermotogales by V69 amplicons. Because these group are important members of specific communities (e.g. mammalian gut, soils, hydrothermal environments), the choice of primers can significantly impact diversity estimation in ways not always predictable by primer sequence analysis(Morales and Holben, 2009). Therefore, caution should be applied when analyzing diversity with novel sets of primers and, if feasible, testing using a synthetic community of gDNA or SSU rRNA plasmid clones from that environment should be considered.
Figure 4.

Taxonomic diversity and abundance inferences based on shotgun metagenomic and amplicon sequencing. The accuracy ratio (observed abundance/expected abundance) is represented as a heat map diagram with values for each organism and data set. Bias values of >1.5-fold are represented as a heat map of increasing color intensity (red for underestimated and green for overestimated abundance). A value of 0.0 indicates >10 fold underestimated abundance, but detection at low levels. ND indicates that no sequence for that organism was identified in that amplicon dataset. Values are averages of three independent amplification and sequencing replicates.
Similarly for the Archaea community, significant differences occurred between primer sets and no combination quantitatively reproduced the composition of the synthetic community. The V13 region amplicon performed best for most of the Euryarchaeota lineages but failed to detect two of the Pyrobaculum species that have an intron in that region, and underestimated other Crenarchaeota as well as N. equitans. Conversely, combinations of primers that amplify the V4 or V48 region tend to overestimate Crenarchaeota and underestimate Euryarchaeota, including the methanogens. The explanation for these biases is unclear as, with the exception of Methanopyrus kandleri, no clear mismatches occur for any primer combinations with their target species. One potential reason for these fluctuations could be the high GC content of the SSU rRNA sequence of these mostly thermophilic and hyperthermophilic organisms, that in some cases contrasts sharply with that of the overall gDNA. Differences in local melting kinetics in such genomes combined with PCR competition between primers may be one explanation for such bias. Species with extreme genome GC composition (N. equitans and H. volcanii) were indeed most affected, both in amplicon and metagenomic sequencing. We did not observe any correlation between the degree of bias in either Bacteria or Archaea communities that can be traced back to the number of rRNA operons in individual genomes. One has to consider nevertheless that accurate quantification of an organism's presence is dependent on rRNA copy number estimation. Because in natural communities the actual number of rRNA associated with each organism is unknown, inferences have to rely on using genome sequence of related species.
To evaluate the reproducibility of replicates and accuracy of community representation between each rDNA amplicon region, and each metagenomic sequencing approach, relative to the expected community structure we calculated Bray-Curtis similarity matrices using the species detection ratios for each dataset. Principal coordinate analysis and hierarchical clustering derived from these matrices indicated that both Illumina and 454 metagenomic data closely recovered the known taxonomic and quantitative composition of both Bacteria and Archaea communities (Figure 5). Among the rRNA amplicons, V13 and V35 for Bacteria and V13 and V4a for Archaea best represented the overall composition of the two communities and displayed lowest variability among replicates. The amplicon that captures Bacteria and Archaea (V48) also appears to be a viable option for diversity surveys that target both domains simultaneously.
Figure 5.
(A) Principal Coordinate Analysis (Bray-Curtis similarity) of Bacteria and Archaea community composition inferred using metagenomics (454-M and ILM-M) and SSU rDNA amplicon sequencing relative to the known composition based on community assembly (REF). Replicates for each amplicon are presented, with closer grouping indicating less variability. The V48 data is presented separately for Archaea and Bacteria in those respective panels but was obtained using the combined AB community. (B) Hierarchical clustering (Bray-Curtis similarity) of community composition accuracy indexes for each amplicon region and sequencing strategy.
Conclusions
With the dramatic decline in cost and increase in output, NGS technologies have changed the scale of microbial ecological studies and have made deep metagenomic sequencing much more feasible, affordable and enabled statistically replicated designs that can be quantitatively robust. As 454 and Illumina sequencing probe deeper into the structure of complex communities, determining the real diversity and distinguishing novel or rare organisms from experimental and computational artifacts continues to be a challenge, even though methods and algorithms are continuously improving. Results presented here allow direct and quantitative comparisons within a defined taxonomic space of two complementary and widely used approaches in microbial ecology, shotgun metagenomics and SSU rRNA gene-based diversity characterization. Both metagenomic strategies recovered the quantitative distribution of the various archaeal and bacterial taxa remarkably well even though organisms spanned two orders of magnitude in abundance. A certain degree of bias was observed for genomes with extreme genomic GC content in transposon based library sequencing but because that method enables analysis of samples with reduced biomass, such potential bias may be acceptable and could be accounted for when required by sample/environmental constraints. Additional challenges in analyses of actual environmental metagenomic datasets remain, such as taxonomic assignments for sequences that belong to uncultured taxa, distinguishing closely related organisms, and genome scale assemblies for low abundance species. Advances in taxonomic binning and assembly algorithms(Koren et al., 2011; Liu et al., 2011; Patil et al., 2011), expanding the repertoire of genome sequences to understudied taxa and uncultured single cells(Wu et al., 2009) and very deep sequencing using the Illumina platform(Hess et al., 2011) indicate that even more complex communities are becoming amenable to comprehensive metagenomic characterization.
Although metagenomic sequencing outperformed most SSU rRNA gene primer sets used in this study, diversity characterization using this traditional phylogenetic marker is an important approach to compare complex communities in ecological studies where large numbers of samples are required. With the decline in cost and the development of Illumina amplicon sequencing (Caporaso et al., 2011a), deeper coverage and more extensive technical replication has become feasible even for highly diverse communities (Prosser, 2010; Zhou et al., 2011). Among the bacterial primer sets for rRNA gene regions, V13 recovered most accurately the composition of the synthetic community. None of the archaeal sets tested performed comparably to the bacterial ones and presented biases that generally occurred at high taxonomic levels but a modified set of V4 primers (V4a) produced good results. The universal Archaea-Bacteria primer sets (V48) that we tested here, although suboptimal for several taxa, allowed simultaneous comparisons of the representation of the two of the three domains of cellular life in environmental samples. In addition, this was the longest amplicon tested and could provide increased taxonomic resolution with future improvements in read lengths. Each of the primer sets presented phylum-specific biases, not all of which were easily predictable computationally even within the known genomic context of this synthetic community. In particular, the suboptimal detection of Bacteroidetes and Actinobacteria by the V4 primer set can impact analysis of human microbiota and some soil samples, while V12, V13 and V35 each has difficulties in recovering phyla that are often times highly abundant in some free living communities (e.g. Aquificae, Thermotogae, Planctomycetes) or in some human microbiota samples (e.g. Verruccomicrobia). Concerted use of two distinct primer pairs for different rDNA regions is therefore important for revealing such biases or even missed detection that may occur for certain taxa (Griffen et al., 2011; Campbell et al., 2012). Since many natural communities contain a much higher taxonomic richness and include uncultured taxa not represented here separate primer sets can provide an independent measure of the accuracy of diversity inferences.
An important aspect in microbial ecology studies is richness and evenness estimation and its comparison between communities (alpha and beta diversity). Severe alpha diversity over estimation, especially at low divergence levels (<0.03), can result from sequence errors and from clustering artifacts that are unaccounted for in QA/QC procedures (Quince et al., 2009; Huse et al., 2010; Kunin et al., 2010; Reeder and Knight, 2010; Quince et al., 2011; Schloss and Westcott, 2011a). At the same time, the high sequence similarity of SSU rRNA genes between clearly distinct organisms indicates that a component of the diversity may be lost if sequence data is analyzed at distances above the generally applied 0.03 threshold. The results presented here demonstrate that the use of quality-filtered data can nearly eliminate diversity artifacts in SSU rRNA amplicon data. Addressing diversity overestimation in metagenomic analyses is more computationally difficult and may require simultaneous advances in sequence assembly combined with sequence composition analysis and classification improvements. In environmental datasets, distinguishing rare but real OTUs or metagenomic signatures of uncultured taxa from experimental and computational artifacts remains a challenge. As more and more microbial groups are being sequenced based on pure cultures or single cell genomic DNA, the uncertainty in recognizing and quantifying currently uncultured organisms in metagenomic data is diminishing. In addition, metagenomic sequence binning and assembly is becoming an effective method to identify uncultured taxa and reconstitute their metabolic capabilities (Iverson et al., 2012; Wrighton et al., 2012). Diverse synthetic communities and validation datasets such as the ones presented here enable direct comparison of sequencing, data processing accuracy and effectiveness in sequence binning and assembly for representing the environmental microbial composition and genomic information. Tailored to more closely resemble the expected taxonomic diversity from a specific environment, additional synthetic communities could provide important analytical controls, weather for single gene-type or, increasingly, for metagenomic studies.
Experimental Procedures
Collection of gDNA for the synthetic communities
Three distinct synthetic communities with gDNAs from representatives of 17 bacterial and 5 archaeal phyla were assembled. Except for four bacteria that have genomes in high quality draft stage (Sulfurihydrogenibium yellowstonense SS-5, Sulfitobacter sp. EE-36, Sulfitobacter sp.NAS-14.1 and Desulfovibrio piger), all other species and strains included in the study have their genome sequences closed. Pure cultures of 27 archaea and bacteria were grown as part of this study in liquid using stocks from ATCC (American Type Culture Collection), DSMZ (Deutsche Sammlung von Mikroorganismen) or from collaborators, using the published media and conditions for each organism. High molecular weight DNA was extracted using a mechanical and organic cell lysis method as described in Ley et al. (Ley et al., 2008), dissolved in TE buffer (pH 8) and measured spectrophotometrically for quality and concentration. For 37 archaea and bacteria we received either purified gDNA or cell cultures from collaborators (Table S1), from which we extracted the gDNA. All gDNA solutions were stored in nuclease-free sylanized tubes (Ambion, Austin TX), to minimize loss by adsorption to tube walls.
DNA quantification and assembly of synthetic communities
Three different methods were used to determine the quality and concentration of each gDNA. The initial concentration of each gDNA preparation was measured by fluorescence assay against a set of standards using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad CA). For an estimation of the molecular weight, approximately 50 ng DNA was separated and visualized on 1.2% agarose E-gels (Invitrogen) with a set of lambda phage DNA mass standards (10–100 ng). All DNAs used in the assembly of the synthetic communities had average molecular weight exceeding that of the lambda phage and no RNA or small, degraded nucleic acids were detected.
Because the DNAs were isolated from very diverse organisms, grown in different media and potentially still contained molecules that could interfere with accurate fluorescence and gel quantification, we used generalized quantitative PCR (qPCR) assays for Bacteria (Fierer et al., 2005) and Archaea (Reysenbach et al., 2006) to guide assembly of the synthetic communities. For each organism to be represented in the community, qPCR was performed on its purified DNA with either the archaeal or the bacterial primer pair. Sequences of SSU rRNA genes from each organism were screened against the published primer sequences (Eub338- Eub518 and Arc915f-Arc1059r) in silico prior to performing qPCRs. To broaden the specificity of the primers so that the SSU rRNA genes of all the species targeted for inclusion in the synthetic community could be amplified, we modified both forward primers Eub338 and Arc915f (see below).
DNA SYBR Green qPCR assays (20 μL) were performed in a Bio-Rad CFX96TM (Hercules, CA) thermal cycler using primers synthesized by IDT (Coralville, IA) and Eurofins MWG Operon (Huntsville, AL) and Bio-Rad iQ Supermix.
Archaeal assays used primers arc915fmc (5'-AGGAATTGGCGGGRGRGCAC -3') and arc1059r (5'-GCCATGCACCWCCTCT-3') at a final concentration of 350 nM each. Cycling parameters included an initial denaturation at 95C for 5 min followed by 45 amplification cycles of 95C for 30 sec, 61C for 30 sec, 72C for 1 min and a fluorescence reading. Following amplification cycles, products were denatured at 95C for 10 sec, and a melt curve was determined over a range of 60–95C. Standard curves were constructed using Methanococcus maripaludis S900 genomic DNA diluted from 1 ×107–1×102 SSU rRNA gene copies per reaction.
Bacterial assays used primers Eub338mc (5'-ACTCCTACGGGDGGCWGCAG-3') and Eub518 (5'-ATTACCGCGGCTGCTGG-3') at a final concentration of 500 nM each. Cycling parameters included an initial denaturation of 95C for 5 min followed by 45 amplification cycles of 95C for 30 sec, 53C for 30 sec, 72C for 1 min and a fluorescence reading. Following amplification cycles, products were denatured at 95C for 10 sec, and a melt curve was determined over a range of 50–95C. Standard curves were constructed using Escherichia coli K12 genomic DNA diluted from 1×108–1×102 SSU rRNA gene copies per reaction
After individual organism DNA quantification, in order to achieve a diverse community composition in both taxonomic distribution and abundance we mixed individual gDNAs, obtaining two primary synthetic communities (a bacterial and an archaeal one). The organisms for which we had low amounts of gDNA were represented at lower abundances in the final mix. The genomic abundance for each organism in the two communities was calculated based on the qPCR-determined concentration and the known number of rRNA operons present in each genome (1–10 copies; Table S1). To obtain the Archaea-Bacteria community, aliquots of the two were mixed and the individual genomic abundances were calculated based on those in the primary communities.
Metagenomic sequencing and analysis
Two metagenomic libraries were constructed for sequencing using the 454 and Illumina platforms. For 454 sequencing, 50 ng of the Archaea-Bacteria synthetic community gDNA was used to prepare an FLX Titanium compatible library using a Nextera™ DNA sample prep kit (Epicentre Biotechnologies, Madison WI) and following manufacturer's instructions. Briefly, the DNA was fragmented (“tagmented”) using the transposase enzyme mix and purified. 454 sequencing primers, a bar-coded Titanium Adaptor 1 (MID3: AGACGCACTC), were incorporated using 15 cycles of PCR followed by purification and size distribution analysis on a an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Insert sizes varied between 500 and 1500 nt. The library was unidirectionally sequenced in-house on one fourth of an FLX Titanium sequencing plate using standard 454/Roche reagents and protocols.
The 454.sff sequence file was loaded into the CLC Genomics Workbench 4.8 (CLCBio, Cambridge MA). Low quality reads (limit =0.05), ambiguous nucleotides, 454 and Nextera adaptors were removed and any further remaining reads shorter than 20 nt were discarded. The resulting dataset contained 291,146 reads with an average length of 320 nt, totaling 85.5Mbb. The sequences were mapped to a database containing the 64 reference genomes (combined total length of 205.6 Mbp) using the CLC local aligner algorithm, with a similarity threshold of 0.9, length fraction of 0.5 and default mismatch/indel cost values. The average coverage of the metagenome was 0.39 fold, with 261,385 reads mapped the genomes. A breakdown of the number of reads mapped to each genome and their coverage is shown in Table S1. The number of reads mapped to each genome was used to calculate the coverage distribution relative to expected values, taking also in account the variable genome size among the represented organisms. Un-mapped reads were further analyzed for mapping either to known plasmids of the included organisms (32 plasmids totaling 4.2 Mbp, ranging in size from 3.6 kbp for a Caldicellulosiruptor bescii plasmid to >635 kbp for a Haloferax volcanii megaplasmid) or back to the reference genomes by decreasing the similarity threshold in order to accommodate unfiltered sequence artifacts. A total of 5,455 reads mapped to plasmids, reaching the same average coverage obtained for the genomic component (0.39 fold). The identity of reads that did not map to either genomes or plasmids was not further explored but likely include both reads with a higher mutations or sequencing errors frequency and reads that belong to a Clostridium sp. contaminant identified in the Desulfovibrio vulgaris culture, for which a genome sequence is not available.
For Illumina sequencing, 1 μg of the Archaea-Bacteria synthetic community gDNA was physically sheared by Covaris Inc. (Woburn, MA), to an average fragment size of 250bp. The fragmented DNA was sequenced bi-directionally (100 bp each direction) on a lane of Illumina HighSeq 2000 using V3 sequencing reagents at the Genome Sciences Resource Center of Vanderbilt University (Nashville, TN). Read quality was analyzed using FastQC (Brabahan Bioinformatics). Filtering out sequences shorter than 50 nt, removal of low-quality reads and of those with ambiguous nucleotides in CLC Genomics Workbench 4.8 resulted in two datasets (forward-reverse reads) of >53.5 and >53.7 million reads, respectively, with an average length of 100 nt and totaling over 10.7 Gbp.
Mapping reads to reference genomes with CLC Genomics Workbench 4.8 followed the same approach except that a higher sequence fraction match (0.8) was used as threshold. Over 96 million reads were mapped, achieving an average 46-fold coverage across the metagenome (1,500-fold maximum region coverage), with many genomes being covered over >95% of their length (Table S1). Two million reads mapped to the 32 known plasmids, with some regions reaching >1,000-fold coverage (average 50-fold).
An “accuracy of detection” ratio for each species within each sample was calculated by dividing the fraction of its sequences in the metagenomic dataset by its known abundance (Q-PCR-based), normalized to genome size, within the corresponding synthetic community. A matrix containing species accuracy detection within each sample sequenced was constructed relative to the standard Q-PCR-based estimates (always = 1), with separate analyses performed for Archaeal and Bacterial data. PRIMER-E v6 (Clarke and Warwick, 2001) was used to calculate Bray-Curtis resemblance matrices for each dataset. These matrices were used to generate Principal Coordinate Analysis (PCoA) plots and hierarchical clustering dendrograms to visualize reproducibility of replicates and accuracy of community representation (based upon Q-PCR) for each amplicon region and sequencing strategy.
Comparison of bias due to GC content
The bias in metagenomic coverage on 454 and Illumina platforms was calculated across the range of genomic GC content of synthetic community constituents (R v2.14; stats package). For each genome, the Illumina accuracy factor was subtracted from the corresponding 454 values, and the resulting difference was regressed against the genomic GC content. Matched, pairwise t-tests were used to compare these accuracy differences between the sequencing platforms across the GC spectrum in three window intervals (27–40%, 40–60% and 60–70%). To determine the coverage bias across the metagenome, we analyzed the 454 and Illumina reads coverage for each genome in the community separately. We did not observe coverage fluctuations linked to genome size. However, the intra-genome sequence coverage matches what we observed at the level of the community with local GC-content having a strong influence on the number of reads depending on the sequencing platform.
MEGAN Analysis
The available genomes of all Archaea and Bacteria were downloaded from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes/). We created three different blast-able datasets with those genomes. First, a database that exclusively contained genomes of the synthetic community organisms (REF); second, a database that contains the genome of all organisms, including the genomes of synthetic community organisms (ALL); third, a database that excludes the members of the synthetic community, but includes all other organisms (X Reference). We used megablast (-v 1 -b 1 -a 10 -m 7) with either 454 or Illumina metagenomic sequences against these three databases. Additionally, we also used the less stringent blastn (-v 1 -b 1 -a 10 -m 7) against the `X Reference' database. We analyzed and quantified the taxonomic abundance of the community of each blast results using MEGAN (MEtaGenome ANalyzer) (Huson et al., 2007). For combined visualization of the result, ratios between the known composition and those determined for each database type and blast approach were displayed as a heatmap and included species, genus and family taxonomic levels (Figure S5). The sequences known to belong to individual genomes (based on CLC-Bio genome mapping) were identified in terms of their predicted taxonomic affiliation by blast-MEGAN and the distribution was projected as histograms for each individual genome (Figure S5 B,C) or globally (Figure S5 D).
IMG-M and MG-RAST analysis
To submit the 454 data into IMG/M we used a fasta format file containing the CLC-Bio quality filtered reads. Data processing used default parameters including gene prediction and functional annotation. For MG-RAST v3 analysis we uploaded both the 454 sff file and the Illumina fastq data files. Quality filtering and sequence analysis followed the default MG-RAST pipeline flow. To analyze the taxonomic composition of the community based on IMG-M and MG-RAST we extracted the inferred abundance at phylum level for reach dataset and also the number of taxonomic units predicted by both systems. MG-RAST enables changing cutoff parameters for the taxonomic mapping and we explored the effect those changes have on the types and numbers of predicted taxa (Table S2). To evaluate the community composition based on SSU rRNA sequences present in the metagenomes we extracted the sequences assigned to that gene from both IMG-M and MG-RAST and analyzed their affiliation using the RDP Classifier. The metagenomes are publicly available in those systems for further analyses. The raw data files have been deposited in the NCBI Sequence Read Archive under accession number SRA059004. Sequences from various filtering stages are also available from the authors upon request.
PCR amplification and 454 sequencing of SSU rRNA amplicons
Sets of amplification primers were chosen to cover most of the hypervariable regions of SSU rRNA (Table S2)(Lane, 1991; Weisburg et al., 1991; Muyzer et al., 1996; Nubel et al., 1996; Suzuki and Giovannoni, 1996; Ovreas et al., 1997; Takai and Horikoshi, 2000; Watanabe et al., 2001; Baker et al., 2003; McCutcheon and Moran, 2007; Frank et al., 2008; Bates et al., 2011). Some of the primers were modified from their original published sequence or additional variants were added to broaden their taxonomic coverage. Still, some primer-rRNA gene mismatches to some of the species represented in the synthetic communities remained, allowing estimation of their effects on amplification efficiency (Figure S6). Amplification primers targeting bacterial V4, V12 and archaeal V4 regions were designed with FLX adapters and rest of the primers were designed with FLX Titanium adapters. To allow multiplexing, the sequencing primers contained 6–8 nt long barcodes. The primers were synthesized by IDT (Coralville, IA) and Eurofins MWG Operon (Huntsville, AL) and were HPLC or HPSF purified.
Polymerase chain reaction (PCR) was performed in 50-μl reactions with 1× High Fidelity PCR buffer (Invitrogen, Carlsbad CA), 2 mM MgSO4, 300 nM of each primer, 200 mM dNTPs, and 1 unit of Platinum Taq DNA Polymerase High Fidelity (Invitrogen; Carlsbad, CA). Between 2.5–10 ng template gDNA was used for the different synthetic communities. All reactions were performed in duplicate or triplicate, and separate reactions with different number of cycles, annealing temperature, and different polymerases were also conducted. The range of amplicon lengths obtained with each primer pair, based on the genomic sequences, is shown in Table S3 and a summary of amplification parameters is shown in Table S5. For the Bacteria-Archaea community, three different polymerases, TaqHiFi (Invitrogen), High GC (Roche Diagnostics, Indianapolis, IN) and Accuprime Pfx (Invitrogen) were used to compare effects of polymerase fidelity and annealing specificity on resulting sequences. Amplicons were purified using AMPure paramagnetic beads (Agencourt Biosciences Corporation, Beverly, MA) followed by concentration and size analysis using DNA 1000 chips on an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Amplicon libraries were then prepared for unidirectional sequencing using the emPCR Kit II (Roche) followed by sequencing on a 454 FLX Life Sciences Genome Sequencer (Roche Diagnostics, Indianapolis, IN). Pyrosequencing using the FLX chemistry and Titanium chemistry was done according to manufacturer's instructions.
Amplicon Sequence Processing
For SSU rRNA amplicon sequence data processing we used primarily the software packages mothur (v1.1639)(Schloss et al., 2009), QIIME (v1.3.040)(Caporaso et al., 2010), RDP(Cole et al., 2009), and AmpliconNoise v1.25(Quince et al., 2011). Sequences were processed using different filtering parameters for respective analyses.
For Low Quality filtering (LQ), sequences were removed from the analysis if they were <200 nt, had ambiguous bases, had a non-exact barcode match, or showed more than two mismatches for the amplification primer. Quality score was not used for this filtering. Remaining sequences were assigned to samples based on the barcode matches, trimmed and reads that were sequenced from the reverse end were reverse complemented so that all sequences begin with the 5' end of the amplicon. Potential chimeras were identified using mothur implementation of ChimeraSlayer. Reference sequences were aligned against a bacterial or archaeal SILVA database using the Needleman-Wunsch algorithm in mothur. The aligned reference sequence was then used as the template for flagging chimeric sequences.
For High Quality filtering (HQ), sequences were removed from the analysis if they were < 200 nt, had ambiguous bases, had a non-exact barcode and primer match, and had a homopolymer >9 nt. If a sequence quality score fell below 20 for a 50-nt window, then it was trimmed at previous position where the average quality score >20. Similarly, sequences were binned to corresponding sample based on corresponding barcodes and reverse complemented. Potential chimeras were identified using Chimeraslayer. Reference sequences were aligned against the Greengenes database (greengenes.lbl.gov) using Pynast. The aligned reference sequences were then used as template for flagging chimerical sequences.
For sequences that were filtered using AmpliconNoise (AN), all samples were run through the AmpliconNoise pipeline, which consists of removal of both sequencing and PCR errors and removal of chimeras using its in built Perseus algorithm(Quince et al., 2011). AmpliconNoise analysis consists of two stages, PyroNoise removal of 454 errors, and SeqNoise removal of PCR single base misincorporations. We have, therefore, estimated the proportion of errors attributable to these two sources by calculating the reduction in error rate after applying each algorithm (Table S4). Raw, per-base error rates varied from 0.1–0.25% for FLX and 0.15–0.9% for Titanium chemistry. For FLX, the V12 region (~0.25%) was associated with a higher raw rate than V4 (~0.1%). For Titanium, higher error rates are associated with V13 (~0.8%) region, mostly due to PCR chimeras. Both 454 sequencing errors and PCR errors are responsible for around 0.05% of the overall error rate each, but there is some variation between regions. The V6 region appears particularly prone to PCR noise with a 0.1% error rate attributable to this source, and the V13 region has a higher rate of 454 errors (0.1%). Frequencies of chimeras were also calculated as a percentage of unique types of sequences following noise removal by AmpliconNoise. This method(Quince et al., 2011) was used to classify sequences as `good', `chimeric', or `trimeric' by direct comparison with databases composed of the corresponding region extracted from genome sequences. These results confirmed those from ChimeraSlayer, and all the shorter FLX amplicons chimera frequencies were low among erroneous reads (<7%). However, Titanium sequences showed higher frequencies for V13 (>60%) than V35 (~10%) or V69 (<5%), and a twofold reduction in frequency for V13 was observed when the cycle number was reduced to 24.
For downstream OTU analysis, except for the sequences that were denoised using AmpliconNoise, sequences were then trimmed so that all sequences began and ended at the same coordinates. Sequences were aligned in mothur against the SILVA database and trimmed at the same alignment position. The position for trimming was manually selected to conserve number of sequences per sample and also have an approximate average length of 200 nt for FLX and 400 nt for Titanium amplicons (Table S3). An example of mothur batch file for each amplicon that was used to trim sequences is included. A summary of number of raw reads and processed reads is shown in Table S4.
OTU Diversity Analysis
On both the LQ and HQ datasets we applied three different clustering algorithms as implemented by RDP, mothur, and ESPRIT/SLP. For the RDP-based analysis, sequences were aligned using the secondary-structure-aware Infernal aligner and clustered using complete-linkage clustering. In mothur-based OTU analysis, trimmed sequences were aligned against the SILVA database using Needleman-Wunsch alignment, pre-clustered using the mothur implementation of single-linkage preclustering algorithm from Huse et al. 2010(Huse et al., 2010) and clustered using average linkage clustering. Batch files that list the commands that were used to cluster the sequences are provided. For the SLP-PW/AL analysis, trimmed sequences were aligned using the pairwise alignment algorithm in ESPRIT, pre-clustered using the single linkage script from Huse et al. 2010(Huse et al., 2010) and clustered based on pairwise distances using average-linkage clustering in mothur. A shell script was used to generate pairwise distance and cluster sequences (http://alrlab.research.pdx.edu/aquificales/pyrosequencing.html). To identify which of the clusters at different distance levels corresponded to which taxa (strain, species, genus etc.) in the synthetic community, the trimmed reference sequences were also clustered with pyrosequence data. Sequences that did not co-cluster with reference sequences were analyzed for potential mutations, chimeras and by taxonomic affiliation to identify potential unexpected contaminants.
Sequences that were denoised using the AmpliconNoise pipeline were clustered based on the distance matrices generated as a result of pairwise alignment similar to ESPRIT package. Average linkage clustering was implemented in this case, essentially as described (Quince et al., 2011).
Taxonomic diversity analysis
Because each sequence in the dataset should correspond to a SSU rRNA gene sequence from the represented genomes, accuracy of SSU rDNA-based diversity estimation was also investigated directly by matching pyrosequence data to references and comparing observed diversity and abundance with those known based on the assembly of each synthetic community. Each processed amplicon dataset was top hit matched to a corresponding reference database by Megablast. As few as single nucleotide differences were sufficient for accurate matching to the corresponding reference sequence, as determined empirically. For some closely related strains or species, however some of the SSU rRNA region amplicons were 100% identical (Fig S7) and assignment to a specific organism in the community was not possible. In those cases, the numbers of hits to the group were assigned to the organisms based on their Q-PCR-based representation. Two pairs of organisms could not be discriminated with any amplicon (Pyrococcus furiosus - P. horikoshii and Sulfitobacter sp. EE-36 - Sulfitobacter sp. NAS-14.1), pointing to limitations of the SSU rRNA gene for comprehensive diversity estimation. For each amplicon dataset, we calculated a ratio between the observed reads-based abundance of each organism and that known based on qPCR-guided community assembly.
Supplementary Material
Acknowledgements
This research was sponsored by NIH National Human Genome Research Institute grant 1R01HG004857-01A1 and by the U.S. Department of Energy, Office of Science - Biological and Environmental Research as part of the Plant Microbe Interfaces Scientific Focus Area (http://pmi.ornl.gov). Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725. CQ is funded by an EPSRC Career Acceleration Fellowship EP/H003851/1. We thank our colleagues that provided us archaeal, bacterial cultures or purified gDNA for this study (Table S1).
References
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6:2033–2044. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J, Kuczynski J, Stombaugh J, Bittinger K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:336–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011a;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011b;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- Clarke K, Warwick R. Change in marine communities: an approach to statistical analysis and interpretation. 2nd edition PRIMER-E; Plymouth, UK: 2001. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Pace NR. Environmental diversity of bacteria and archaea. Syst Biol. 2001;50:470–478. [PubMed] [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of Soil Microbial Community Structure by Use of Taxon-Specific Quantitative PCR Assays. Appl Environ Microbiol. 2005;21:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gihring TM, Green SJ, Schadt CW. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol. 2012;14:285–290. doi: 10.1111/j.1462-2920.2011.02550.x. [DOI] [PubMed] [Google Scholar]
- Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5368. pdb prot5368. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2011;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011a;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011b;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- Hong S, Bunge J, Leslin C, Jeon S, Epstein SS. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009;3:1365–1373. doi: 10.1038/ismej.2009.89. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- Kan J, Clingenpeel S, Macur RE, Inskeep WP, Lovalvo D, Varley J, et al. Archaea in yellowstone lake. ISME J. 2011;5:1784–1795. doi: 10.1038/ismej.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Treangen TJ, Pop M. Bambus 2: scaffolding metagenomes. Bioinformatics. 2011;27:2964–2971. doi: 10.1093/bioinformatics/btr520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. John Wiley and Sons; New York: 1991. pp. 115–175. [Google Scholar]
- Ley RE, Hamady M, Lozupone CA, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Gibbons T, Ghodsi M, Treangen T, Pop M. Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genomics. 2011;12(Suppl 2):S4. doi: 10.1186/1471-2164-12-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Research. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci U S A. 2010;107:16420–16427. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales SE, Holben WE. Empirical testing of 16S rRNA gene PCR primer pairs reveals variance in target specificity and efficacy not suggested by in silico analysis. Appl Environ Microbiol. 2009;75:2677–2683. doi: 10.1128/AEM.02166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Darling AE, Eisen JA. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One. 2010;5:e10209. doi: 10.1371/journal.pone.0010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Hottentrager S, Teske A, Wawer C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA. A new molecular approach to analyze the genetic diversity of mixed microbial communities. In: Akkermans ADL, van Elsas JD, de Bruijn FJ, editors. Molecular Microbial Ecology Manual. Kluwer Academic Publishing; Dordrecht: 1996. pp. 3.4.4.1–3.4.4.22. [Google Scholar]
- Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, et al. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovreas L, Forney L, Daae FL, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. Mapping the tree of life: progress and prospects. Microbiol Mol Biol Rev. 2009;73:565–576. doi: 10.1128/MMBR.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil KR, Haider P, Pope PB, Turnbaugh PJ, Morrison M, Scheffer T, McHardy AC. Taxonomic metagenome sequence assignment with structured output models. Nat Methods. 2011;8:191–192. doi: 10.1038/nmeth0311-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli M, Moya A. Evaluating the fidelity of de novo short read metagenomic assembly using simulated data. PLoS One. 2011;6:e19984. doi: 10.1371/journal.pone.0019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat I, Vishnivetskaya TA, Mosher JJ, Brandt CC, Yang ZK, Brooks SC, et al. Characterization of archaeal community in contaminated and uncontaminated surface stream sediments. Microb Ecol. 2010;60:784–795. doi: 10.1007/s00248-010-9734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI. Replicate or lie. Environ Microbiol. 2010;12:1806–1810. doi: 10.1111/j.1462-2920.2010.02201.x. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing Noise From Pyrosequenced Amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A-L, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, et al. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:1–4. doi: 10.1038/nature04921. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL. Assessing and Improving Methods Used in Operational Taxonomic Unit-Based Approaches for 16S rRNA Gene Sequence Analysis. Appl Environ Microbiol. 2011a;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL. Assessing and improving methods used in OTU-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol. 2011b;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiology Today. 2006:152–155. [Google Scholar]
- Sun Y, Cai Y, Huse SM, Knight R, Farmerie WG, Wang X, Mai V. A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Brief Bioinform. 2011;13:107–121. doi: 10.1093/bib/bbr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- van de Vossenberg J, Woebken D, Maalcke WJ, Wessels HJ, Dutilh BE, Kartal B, et al. The metagenome of the marine anammox bacterium 'Candidatus Scalindua profunda' illustrates the versatility of this globally important nitrogen cycle bacterium. Environ Microbiol. 2012 May 9; doi: 10.1111/j.1462-2920.2012.02774.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Kodama Y, Harayama S. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods. 2001;44:253–262. doi: 10.1016/s0167-7012(01)00220-2. [DOI] [PubMed] [Google Scholar]
- Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol. 2012;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, et al. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz S, Allgaier M, Hugenholtz P. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat Methods. 2010;7:943–944. doi: 10.1038/nmeth1210-943. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, et al. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 2011;5:1303–1313. doi: 10.1038/ismej.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.