Abstract
The Golgi apparatus is a membranous organelle in the cell that plays essential roles in protein and lipid trafficking, sorting, processing and modification. Its basic structure is a stack of closely aligned flattened cisternae. In mammalian cells, dozens of Golgi stacks are often laterally linked into a ribbon-like structure. Biogenesis of the Golgi during cell division occurs through a sophisticated disassembly and reassembly process that can be divided into three distinct but cooperative steps, including the deformation and reformation of the Golgi cisternae, stacks and ribbon. Here, we review our current understanding of the protein machineries that control these three steps in the cycle of mammalian cell division: GRASP65 and GRASP55 in Golgi stack and ribbon formation; ubiquitin and AAA ATPases in post-mitotic Golgi membrane fusion; and golgins and cytoskeleton in Golgi ribbon formation.
Keywords: Golgi, biogenesis, cell cycle, ribbon linking, stacking, membrane budding and fusion
Overview of Golgi membrane dynamics during the mammalian cell cycle
The Golgi apparatus is a membranous organelle essential for intracellular protein and lipid trafficking and modification. In mammalian cells, the Golgi consists of dozens of stacks of 5–7 parallel-aligned flattened cisternae; Golgi stacks are highly concentrated in the pericentriolar region of the cell. In most cells, the enrichment of the Golgi stacks near the centrosome -- mediated by the microtubule minus end-directed motor protein dynein -- is likely required for lateral linking of the Golgi stacks into a ribbon, as treatment of cells with microtubule depolymerizing drugs disperses the Golgi ribbon into individual stacks. The Golgi structure is well maintained in a dynamic equilibrium between input and output of membranes from and to other organelles, including the endoplasmic reticulum (ER), the endosome-lysosome system, and the plasma membrane 1.
The mitotic division of cells requires the duplication and partitioning of all cellular components. It is generally believed that the Golgi apparatus exists as an autonomous organelle and its division does not rely on other cellular organelles such as the ER 2, 3. In mammalian cells, biogenesis of the Golgi during each cycle of cell division occurs through a unique disassembly and reassembly process. The Golgi is fragmented at the onset of mitosis, first with the stacks unlinked, which then undergo further unstacking and vesiculation, yielding thousands of vesicles that are distributed throughout the cytoplasm and equally into the two daughter cells. In telophase, Golgi vesicles fuse to form new cisternae that are aligned as stacks. The Golgi stacks then accumulate in the pericentriolar region and form a ribbon in each daughter cell during cytokinesis. Thus mitotic Golgi disassembly could be divided into three steps: ribbon unlinking, cisternal unstacking and vesiculation. Conversely, post-mitotic Golgi reassembly includes membrane fusion/cisternae regrowth, cisternae stacking, and ribbon linking 4, 5. Many factors are involved in the regulation of Golgi membrane dynamics during the cell cycle; these include Golgi matrix proteins, kinases and phosphatases, ubiquitin ligases and deubiquitinating enzymes, vesicle budding and fusion machineries, as well as actin and microtubule cytoskeleton. In this review we summarize recent findings on the mechanisms that regulate these three processes in mitotic Golgi disassembly and post-mitotic Golgi reassembly during the mammalian cell cycle. We focus particularly on Golgi stacking proteins in Golgi stack formation, ubiquitin in post-mitotic Golgi membrane fusion, and golgins and cytoskeleton in Golgi ribbon linking.
Golgi cisternal stacking and its regulation in the cell cycle
Golgi stacks are the basic structural and functional units of the Golgi in mammalian cells. Our current understanding of the mechanism of Golgi stacking largely benefits from an in vitro assay that reconstitutes the cell cycle-regulated Golgi disassembly and reassembly process. Briefly, highly purified rat liver Golgi stacks 6 are incubated with mitotic cytosol to generate mitotic Golgi fragments (MGFs). After re-isolation, these MGFs reassemble into Golgi stacks upon incubation with interphase cytosol or purified cytosolic components. This provides a readily manipulatable biochemical system within which the sequence of morphological events can be precisely followed by quantitative electron microscopy (EM) or biochemical analysis 7, 8. This approach has contributed to the discovery and characterization of many proteins that mediate Golgi membrane tethering, fusion, cisternal stacking, and regulation 4, 9–11.
The role of GRASP65 and GRASP55 in Golgi cisternal stacking
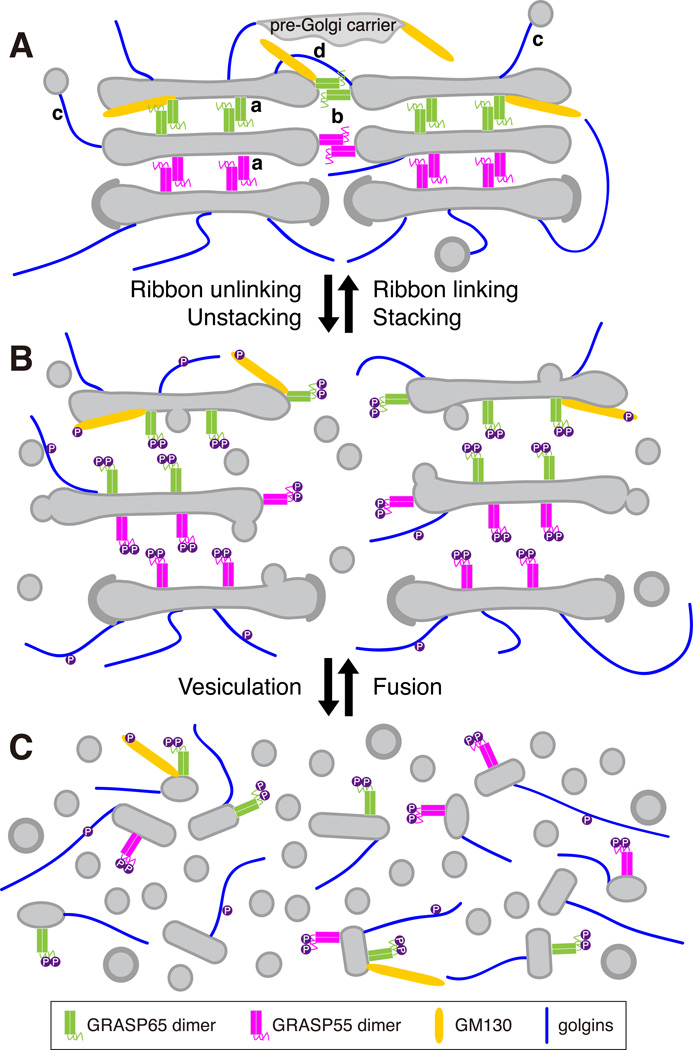
The mechanism and biological significance of Golgi stacking in protein trafficking and processing remain as interesting and significant questions to be answered in the field. So far the only proteins that are shown to directly participate in Golgi stacking are the Golgi reassembly stacking proteins (GRASPs), which include GRASP65 and GRASP55 in mammalian cells (Fig. 1A). GRASP65 was first identified as a peripheral Golgi protein that is accessible to N-ethylmaleimide (NEM) only when the Golgi stacks are disassembled, and NEM treatment of the Golgi membranes abolished Golgi cisternae re-stacking in the in vitro reassembly assay described above. GRASP65 localizes to the cis- Golgi; its homologue GRASP55,s subsequently discovered based on sequence similarity, stacks the medial-to-trans cisternae 12. Several lines of evidence support the idea that GRASPs are involved in Golgi stacking. Antibodies against GRASP65 or GRASP55 inhibited the stacking of newly formed cisternae in the in vitro assay 12, 13, whereas microinjection of anti-GRASP65 antibodies into mitotic cells inhibited subsequent Golgi stack formation in the daughter cells 14. Furthermore, depletion of GRASP65 or GRASP55 in mammalian cells by RNA interference (RNAi) reduced the number of cisternae per stack 15–17. Since GRASP65 and GRASP55 localize to the cis- and medial-to-trans Golgi, respectively 12, the reduced cisternae number caused by knocking down each individual GRASP protein was possibly due to losing either the cis or the medial-to-trans cisternae from the Golgi stacks. Simultaneous depletion of both GRASPs resulted in complete disassembly of the Golgi stacks 16, suggesting that they play complementary roles in the formation of the polarized stacked structure of the Golgi 4, 16. GRASPs are peripheral membrane proteins that form stable homodimers, and dimers from adjacent cisternae oligomerize in trans through their N-terminal GRASP domains to zip the cisternae into stacks. The oligomerization state of GRASPs is regulated by mitotic phosphorylation at the C-terminal Serine/Proline-Rich (SPR) domain discussed below (Fig. 1) 4, 16, 18.
Fig. 1. GRASPs and golgins in Golgi stacking and ribbon linking during the cell cycle.
A. GRASP65 (locates to cis-Golgi) and GRASP55 (locates to medial-to-trans-Golgi) homodimers on adjacent membranes form trans-oligomerizers, bring the membrane closer to each other to (a) form stacks when GRASPs localize between the cisternae, and to (b) form a ribbon when GRASPs are at the rims of the Golgi stacks. In addition to GRASPs, golgins are also required for the maintenance of Golgi ribbon integrity by either (c) tethering vesicles/transport carriers to Golgi membranes, such as GM130, or by (d) tethering membranes from neighboring stacks, as GMAP210.
B. During mitosis, GRASPs and GM130 are phosphorylated by mitotic kinases, which disrupts their oligomerization and allows ribbon unlinking and cisternae unstacking.
C. The resulting single cisternae by unstacking undergo continuous vesicle budding to further disassemble Golgi membranes into vesicles and tubular structures. In cells, Golgi ribbon unlinking occurs in G2 phase, while Golgi unstacking and vesiculation take place in prophase and prometaphase. The reverse processes, Golgi membrane fusion, stacking and ribbon linking, occur in telophase and cytokinesis.
Cell cycle regulation of GRASP65 and GRASP55 in Golgi stacking
The Golgi apparatus undergoes cisternae unstacking and restacking during the cell cycle 14, 16, 17. This process depends on the mitotic phosphorylation and post-mitotic dephosphorylation of the GRASP proteins. The phosphorylation of GRASP65 on multiple sites in the SPR domain by cdc2 (cdk1) and polo-like kinase 1 (Plk1) during mitosis changes its conformation, leads to GRASP65 de-oligomerization and thereby allows the Golgi cisternae to separate 18. After mitosis, GRASP65 is dephosphorylated by the phosphatase PP2A, reforms trans-oligomers and stacks Golgi cisternae 7. GRASP55 is regulated in a similar way 16; it is phosphorylated by MEK1/ERK during mitosis and is dephosphorylated by a phosphatase yet to be identified 16. When phospho-deficient mutants of GRASP65 or GRASP55 are expressed in cells, mitotic Golgi fragmentation is at least partially inhibited 17, 18. Conversely, expression of their phosphomimetic mutants, or inhibition of their dephosphorylation, accelerates mitotic Golgi disassembly and reduces Golgi reassembly 16.
GRASP65 has multiple phosphorylation sites localized to the C-terminal SPR domain that are phosphorylated by cdc2 during mitosis 19, 20. It was recently shown that these sites are sequentially phosphorylated and dephosphorylated during the cell cycle; and the phosphorylation of a Threonine 220/Threonine 224 site, which is physically close to the GRASP domain at the amino acid level, is likely the most important in regulating Golgi unstacking 20–23. Besides the many phosphorylation sites in the SPR domain, Serine 189 in the N-terminal GRASP domain has been identified as a plk1 site in both GRASP65 and GRASP55. Phosphorylation of this site also interrupts GRASP oligomerization and is required for mitotic Golgi disassembly 24, 25. Since expressing the GRASP domain including S189 inhibits Golgi fragmentation 16, 17, it is possible that S189 phosphorylation alone may not be sufficient to regulate GRASP oligomerization. Alternatively, phosphorylation of the SPR domain by cdc2 may be a prerequisite of S189 phosphorylation by plk1, as it has been shown that phosphorylation by cdc2 is required for docking plk1 onto GRASP65 19.
It is worth noting that GRASPs have multiple functions. Their roles in Golgi ribbon formation will be discussed in later sections in this review. Other functions, including unconventional secretion and cell cycle regulation have been discussed elsewhere 22, 23. GRASPs are also involved in vesicle-mediated Golgi transport. Inhibition of GRASP65 function by microinjecting GRASP65 antibodies into the cell accelerated protein trafficking, possibly because unstacked Golgi cisternae have more surface area to generate vesicles 26. During mitosis, Golgi unstacking by GRASP phosphorylation facilitates the vesiculation of Golgi membranes, as discussed below.
Golgi cisternae vesiculation and regeneration during the cell cycle
During interphase, COPI vesicles are generated from one Golgi subcompartment and fuse with another for intra-Golgi and Golgi-to-ER trafficking 27. During mitosis, the Golgi membranes disassemble via continuous COPI vesicle budding while fusion is inhibited. In post-mitotic Golgi reassembly, those COPI vesicles fuse to generate new cisternae (Fig. 1). This vesiculation and cisternae regeneration process is mediated by vesicle budding and fusion machineries, which are modulated by the vesicle tethers.
Golgi vesiculation during mitosis
During prophase and prometaphase, Golgi cisternae undergo extensive COPI-dependent vesiculation 28, 29. Treatment of unstacked Golgi membranes (induced by kinase treatment) with purified Arf1 and coatomer is sufficient to transform the Golgi cisternal membrane into vesicles in vitro 7, suggesting that Arf1 and coatomer are sufficient to trigger Golgi membrane vesiculation. Not all of the Golgi membranes are turned into vesicles during mitosis; some remain as tubulovesicular structures. These membranes contain Golgi matrix proteins including GRASPs and golgins and may serve as templates in late mitosis for the formation of new Golgi apparatus in the daughter cells 30, 31, but the underlying mechanism remains elusive.
Post-mitotic Golgi membrane fusion
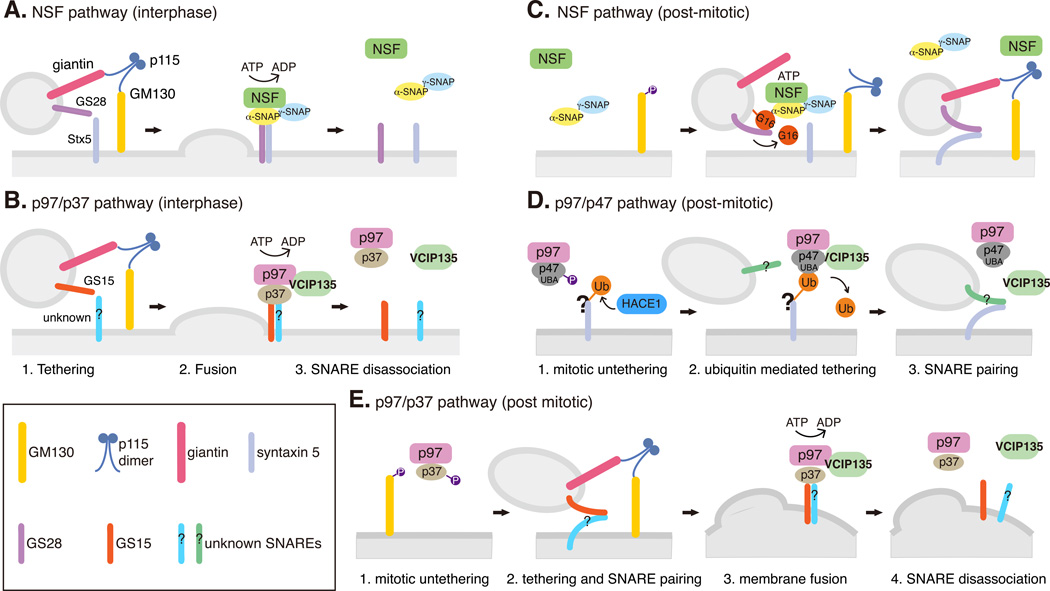
Post-mitotic Golgi membrane fusion requires tethering proteins like p115, giantin and GM130, soluble NSF attachment protein (SNAP) receptors (SNAREs), and two AAA ATPases, N-ethylmaleimide sensitive fusion protein (NSF) and valosin-containing protein p97 [VCP] with their adaptors (Fig. 2) 32. NSF and its adaptors α/γ-SNAPs catalyze the disassembly of cis-SNARE complex after membrane fusion therefore allowing a new round of fusion to take place. In interphase, this process is required for both ER-to-Golgi and intra-Golgi transport (Fig. 2A). In post-mitotic Golgi reassembly, two steps of NSF function are required for post-mitotic Golgi reassembly. At the onset of mitosis when the Golgi is vesiculated, NSF drives SNARE dissociation by hydrolyzing ATP, similar to its function in interphase. At the second step in late mitosis, NSF catalyzes the complex formation between GATE-16 and the v-SNARE GS28. The binding with GATE-16 precludes GS28 from binding its cognate t-SNARE syntaxin-5. This process appears to be important for post-mitotic Golgi membrane fusion and is independent of ATP hydrolysis (Fig. 2C) 32.
Fig. 2. NSF and p97 pathways in interphase Golgi membrane fusion and post-mitotic Golgi reassembly.
A-B. Interphase Golgi membrane fusion mediated by NSF (A) and p97/p37 (B). Vesicles or Golgi membranes are tethered by the p115/GM130 complex, which facilitates SNARE-mediated fusion. The SNARE complexes are disassociated by NSF or p97 by ATP hydrolysis to allow next round vesicle fusion.
C-D. Ubiquitin-mediated post-mitotic Golgi fusion by NSF (C) and p97/p47 (D). During mitosis, GM130 phosphorylation interrupts its interaction with p115 and facilitates Golgi vesiculation. After mitosis, ubiquitination operates as a general mechanism for Golgi cisternae regrowth. In the NSF pathway (C), GATE-16, a ubiquitin-like protein, recruits NSF onto the membranes. NSF then catalyzes GATE-16 to form a complex with GS28 and inhibits GS28-syntaxin 5 interaction in an ATP-hydrolysis independent manner. GATE-16 is released from the membrane by an unknown mechanism to allow SNARE complex assembly and membrane fusion. In the p97/p47 pathway (D), HACE1 attaches ubiquitin onto unknown substrates on the Golgi membranes during mitosis; ubiquitination of these Golgi proteins may inhibit vesicle fusion. In late mitosis, the ubiquitin bound to a Golgi protein interacts with the UBA domain of p47 and recruits the p97/p47/VCIP135 complex onto Golgi vesicles. VCIP135 removes ubiquitin from Golgi proteins and allows p97/p47 to fuse the Golgi fragments into new cisternae. How p97/p47 regulates SNARE pairing and membrane fusion in this process is still unclear.
E. Ubiquitin is not involved in post-mitotic Golgi fusion by p97/p37. During mitosis, p37 is phosphorylated and thus membrane fusion is inhibited. After mitosis, dephosphorylation of p37 allows membrane fusion similar to that in interphase (B). In this pathway, SNARE complex is disassembled by p97 ATP hydrolysis, while the p97/p37 complex is disassembled by VCIP135.
The other AAA ATPase, p97, with its adaptor p47, promotes post-mitotic cisternae regrowth but not interphase trafficking 33. Unlike NSF, p97/47-mediated post-mitotic Golgi reassembly requires neither p115 tethering nor GS28; however it uses syntaxin 5 as a shared receptor. Therefore, it is proposed that the NSF pathway mediates heterotypic fusion between vesicles and Golgi remnants, while the p97 pathway mediates homotypic fusion between Golgi remnants (Fig. 2D) 32. Another p97 adaptor, p37, is involved in both post-mitotic reassembly and interphase maintenance of the Golgi complex. The p97/p37 pathway for Golgi reassembly requires p115 and GS15 but not syntaxin 5, suggesting a mechanism different from either the NSF pathway or the p97/p47 pathway (Fig. 2B&E) 34.
The role of ubiquitin in post-mitotic Golgi membrane fusion
One exciting finding in cell cycle-regulated Golgi dynamics concerns the involvement of ubiquitin in post-mitotic membrane fusion 32. The first evidence that indicates a role of ubiquitin in post-mitotic Golgi fusion came from the domain analysis of p47. p47 contains a ubiquitin-associated (UBA) domain that binds mono-ubiquitin; inhibition of p47-ubiquitin interaction suppresses p97/p47- mediated Golgi membrane fusion 35. Mono-ubiquitination of Golgi proteins occurs during mitotic Golgi disassembly and is required subsequently at the end of mitosis for targeting p97/p47 complex onto the Golgi remnants for reassembly 36, 37. This process does not involve the proteasome 36. Both the ubiquitin ligase and the deubiquitinating enzyme (DUB) have been recently discovered. The ubiquitin E3 ligase was identified as HACE1 (the HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1) 37, a HECT (Homologous to the E6-AP Carboxyl Terminus) domain-containing ubiquitin ligase that is down regulated in a variety of tumors 38. VCIP135 (valosin-containing protein p97/p47 complex-interacting protein, p135) was identified as the deubiquitinating enzyme (DUB). VCIP135 forms a quaternary complex with syntaxin 5 and the p97/p47 complex via a ubiquitin fold domain and its enzymatic activity is required for post-mitotic Golgi membrane fusion 36. Furthermore, VCIP135 interacts with WAC, which enhances the DUB activity of VCIP135. Depletion of WAC by RNAi in cells resulted in extreme Golgi vesiculation and VCIP135 mis-localization from the Golgi 39. These results indicate a cycle of ubiquitination and deubiquitination in p97/p47-mediated Golgi dynamics.
The ubiquitin moieties on these substrates, added by HACE1 during mitosis, likely serve as receptors for the p97/p47/VCIP135 complex at the end of mitosis to carry on fusion reactions after the removal of the ubiquitin tags by VCIP135 (Fig. 2D) 36, 37. It has been proposed that during mitosis when the Golgi membranes are disassembled, HACE1 attaches monoubiquitin onto a Golgi membrane protein(s), which subsequently recruits the p97/p47 complex onto the Golgi fragments through the UBA domain of p47. The deubiquitinase VCIP135, which is associated with the p97/p47 complex, removes the ubiquitin moiety and thus allows p97/p47 to fuse the membranes after mitosis (Fig. 2D) 37. The underlying mechanism, however, remains largely unknown. For example, it has not been determined whether p97/p47-mediated membrane fusion requires cognate SNARE pairing and whether p97 drives SNARE disassociation by hydrolyzing ATP. In addition, since the substrates of HACE1 and VCIP135 on the Golgi membranes are still unidentified, it is not clear whether membrane association of the p97/p47 complex depends on the level of ubiquitin on the Golgi membranes and whether ubiquitination of Golgi membrane proteins is the cause of shutting down membrane trafficking during mammalian cell division. Unlike p47, p37 lacks a ubiquitin-binding domain that interacts with ubiquitin, thus the p97/p37 pathway does not need ubiquitin and WAC. However, the p97/p37 pathway requires VCIP135 for disassociating the p37-GS15 complex, although its deubiquitinating activity is not required (Fig. 2E) 34, 39, suggesting that VCIP135 has a ubiquitin-independent role in regulating SNARE pairing.
The other AAA ATPase, NSF, although it does not interact with ubiquitin as the p97/p47 complex, requires a ubiquitin-like protein, GATE-16, to drive the post-mitotic cisternal regrowth 32. GATE-16 recruits NSF/SNAPs complex onto vesicles through direct interaction with NSF. NSF then catalyzes GATE-16 binding to the Golgi v-SNARE GS28, which blocks the interaction between GS28 and syntaxin-5 and thus negatively regulates their pairing 40. Releasing GATE-16 allows SNARE pairing and subsequent membrane fusion. In this way, GATE-16 serves as a receptor for the NSF/SNAPs complex, like ubiquitin for the p97/p47 complex, with the membrane association and dissociation of GATE-16 possibly through a lipidation/de-lipidation cycle 32, which requires further confirmation. So far it has not been shown whether GATE-16 can be covalently linked to proteins in a similar way as ubiquitin. In addition, NSF does not hydrolyze ATP to disassemble SNARE complexes during post-mitotic membrane fusion. Instead, it catalyzes GATE-16 to interact with GS28 to negatively regulate SNARE pairing. How this results in membrane fusion remains a mystery. Nevertheless, these observations suggest that ubiquitination operates as a general mechanism for Golgi cisternae regrowth after mitosis (Fig. 2C&D).
Cell cycle regulation of golgins in vesicle tethering
Golgins are long coiled-coil proteins that play key roles in vesicle tethering, trafficking and Golgi ribbon formation (Box 1). In interphase, vesicles are tethered to the Golgi membranes by golgins before SNARE-mediated fusion 41. Some golgins are phosphorylated during mitosis, which disrupts their tethering function and facilitates mitotic Golgi disassembly. For example, GM130, p115 and giantin form a complex that captures COPI vesicles to the cis-Golgi in interphase cells 42. During mitosis, phosphorylation of GM130 on Serine 25 by cdc2 prevents p115 binding, while subsequent dephosphorylation of GM130 by PP2A after mitosis resumes its interaction with p115 and restores NSF and p97/p37 mediated fusion. Another golgin, golgin-84, which tethers vesicle for intra-Golgi trafficking, may be regulated similarly to GM130 in the cell cycle 11, 43, 44. Besides their function in tethering vesicles, many golgins are required for maintaining Golgi ribbon integrity, as discussed in the next section.
Box 1. Golgins: incorporating Golgi ribbon formation with trafficking.
Golgins are long coiled-coil proteins that are associated with the cytoplasmic face of Golgi membranes either by a C-terminal transmembrane domain or through interaction with small GTPases (Rabs, Arf and Arls). Many golgins have multiple Rab binding sites, which may facilitate capturing membranes bearing specific Rabs and excluding other cellular organelles like ribosomes from the Golgi region 65. Based on their locations, golgins can be simply divided into cis-golgins, golgins on the rim of the stacks and trans-golgins 1, 66, 67.
Well-characterized cis-golgins include GM130, GMAP210 and golgin-160. GM130 targets to the Golgi membranes through its interaction with GRASP65 and other adaptors such as Rab1, mediates ER-to-Golgi transport and the formation of cis-Golgi cisternae. When GM130 is depleted, Golgi ribbon breaks down 54. GMAP210 contains an N-terminal ALPS motif that forms an amphipathic α-helical structure and interacts with the curved membrane structures (e.g. the Golgi rims and vesicles), and a C-terminal GRAB domain that can be recruited to the Golgi cisternae flat surface through interaction with Arf1 GTPase. The C-terminus can also bind to γ-tubulin in the centrosome 55, 68–70. GMAP210 is required for maintaining Golgi ribbon integrity, possibly by pulling one Golgi stack to a neighboring stack, and by directing Golgi stacks to the centrosome 55, 68–70. The ALPS motif of GMAP210 may also be required to tether vesicles to the Golgi for ER-to-Golgi transportation 71. The third cis-golgin, golgin-160, is the Golgi receptor for dynein, which is required for the localization of the Golgi membranes in the pericentriolar region and ribbon formation 59.
Three golgins that localize to the rim through the whole stack, giantin, golgin-84 and CASP, have transmembrane domains at their C-terminus required for membrane anchoring. Giantin is involved in anterograde whereas golgin-84 and CASP play a role in retrograde trafficking through the Golgi stack 1.
Trans-golgins include the GRIP domain golgins GCC88, GCC185, golgin-97 and golgin-245 (also known as p230 or tGolgin-1). These four golgins all contain C-terminal GRIP domains that dimerize and simultaneously interact with two small GTPases 66. Localization of golgin-97 and golgin-245 to the trans-Golgi is mediated by Arl1-GTP, whereas the recruitment of GCC88 and GCC185 is still controversial 72. GCC88 and GCC185 localize to different domains of the TGN and mediate different retrograde transport pathways from endosome to the TGN 73. GCC185 was also shown to recruit microtubule +end binding proteins CLASPs to the TGN, which is required for Golgi-derived microtubule formation and Golgi ribbon integrity 74. ARL4A, an Arf/Arl family protein, is shown to interact with GCC185 and regulate GCC185-mediated recruitment of CLASPs 75.
Golgi ribbon formation and cell cycle regulation
Unlinking of the Golgi ribbon at the onset of mitosis is likely to be important for partitioning the Golgi membranes into the two daughters 4 and for mitotic progression 45. Golgi ribbon unlinking occurs in late G2 phase, prior to Golgi cisternal unstacking and vesiculation which start in prophase 46. So far the exact mechanism for Golgi ribbon linking is not well understood, but involves a number of proteins including the GRASP proteins, several golgins, CtBP/BARS, as well as the microtubule and actin cytoskeleton 4, 47, 48.
Cell cycle regulation of GRASP65 and GRASP55 in Golgi ribbon linking
In addition to Golgi cisternal stacking, GRASP65 and GRASP55 also link the Golgi stacks laterally into a ribbon (Fig. 1A). Depletion of either GRASP65 or GRASP55 causes Golgi ribbon breakdown in the cell 16, 49–51. It has been proposed that GRASP trans-oligomers also exist at the rims of Golgi stacks for stack tethering, which is also regulated by phosphorylation and dephosphorylation during the cell cycle 24, 46, 50, 51. In late G2 phase, MEK and ERK kinases are activated and phosphorylate GRASP55, which promotes Golgi unlinking 46. GRASP65, on the other hand, is phosphorylated at Serine 277 and Serine 376 by cdc2 and Serine 189 by plk in late G2 phase, correlating with Golgi ribbon disassembly 19, 20, 24. Expressing a phospho-deficient mutant of GRASP55 inhibited Golgi disassembly in G2 phase 46, whereas expression of a phospho-mimic mutant of GRASP65 or GRASP55 leads to Golgi fragmentation in interphase cells 24, 51. As in cisternae stacking, GRASPs must be dephosphorylated at the end of mitosis to allow Golgi ribbon to reform (Figure 1).
CtBP/BARS in Golgi ribbon unlinking in G2 phase
CtBP/BARS is a protein that plays a dual role in gene transcription and Golgi membrane fission. In addition to its function in helping vesicle fission in interphase Golgi transport, CtBP/BARS is also required for Golgi ribbon unlinking at late G2 phase, earlier than the mitotic vesiculation of Golgi membranes. In cells where CtBP/BARS is depleted or inhibited by microinjected antibodies, the Golgi remains connected during mitosis and cell mitotic progression is arrested 52, 53. However, how CtBP/BARS coordinates with GRASPs and golgins in regulating Golgi ribbon linking in the cell cycle remains elusive.
Golgins in Golgi ribbon formation
Golgins are also ideal candidates in Golgi ribbon formation due to their long rod shape. The best example is perhaps GM130, a golgin that interacts with GRASP65. Knockdown of GM130 causes Golgi ribbon unlinking, similar to GRASP65 depletion 54. Since the gaps between the stacks in the ribbon are relatively larger and more heterogeneous (10s to 100s nm) compared to those between the cisternae in the stack, it is reasonable to speculate that the long coiled-coil proteins like GM130 are better candidates for stack tethering. Another possible mechanism for GRASP65 and GM130 in Golgi ribbon formation depends on their vesicle tethering function, as continuous membrane input from the ER to the Golgi is required to maintain Golgi ribbon integrity. A similar hypothesis applies to GRASP55 and its interacting protein golgin-45 as well as several other golgins 1, 4.
In addition to GM130, depletion of many other golgins including p115, GMAP210, golgin-160, giantin, golgin-84, GCC185 and golgin-245 by RNAi, or impairing golgin-97 function by microinjection of inhibitory antibodies, all lead to Golgi fragmentation 1. Since most of these proteins function in membrane tethering, their function in maintaining Golgi integrity may rely on membrane trafficking. Alternatively, some golgins including GM130, p115, golgin-160, GMAP210 and GCC185 interact with microtubules, which regulate the centralization and lateral linking of the Golgi stacks (see below and in Box 2). A third mechanism, suggested by the cis-Golgi golgin GMAP210, is to directly link membranes from adjacent stacks. In addition to its interaction with γ-tubulin, GMAP210 binds the cis- Golgi by interacting with Arf1 via its C-terminal GRAB domain and with highly curved membranes (vesicles or the rim of a neighboring stack) by its N-terminal ALPS motif, thereby directing homotypic fusion of cis-cisternae between two stacks (Fig. 1A) 55. Despite the findings about golgins in maintaining Golgi integrity in interphase, how they are regulated during the cell cycle is largely unknown.
Box 2. Golgi-derived microtubules and microtubule-regulated Golgi reassembly.
Golgi reassembly depends on microtubules. Conversely, Golgi serves as an unconventional MTOC and regulates microtubule organization. Several recent studies have demonstrated the formation of Golgi-derived microtubules and their roles in Golgi assembly and maintenance 58, 74, 76, 77. Microtubules require γ-tubulin complexes to initiate growth. Two cis-golgins, GM130 (through AKAP450) 77 and GMAP210 78, 79, anchor γ-tubulin complexes to the cis-side of the Golgi complex. Golgi-derived microtubules need to be stabilized by CLASPs (CLASP1/2) that are tethered to trans-Golgi membranes through trans-golgin GCC185 74, 75. In addition, CAP350 and Hook3 may be involved in microtubule stabilization on the Golgi 80, 81.
Golgi-derived microtubules cooperate with centrosome-derived microtubules in Golgi reassembly at mitotic exit and in the assay after nocodazole washout. The reassembly of the Golgi ribbon from cell periphery includes two stages. First, Golgi ministacks generated by simple vesicle fusion and stacking undergo initial clustering at the cell periphery, which is referred to as the Golgi stage. At this stage, the Golgi clusters stay far away from centrosome and spindle, but are linked by peripheral microtubules derived from the Golgi. During this stage of Golgi reassembly, the size of each Golgi particle is doubled and the total number of Golgi particles reduced. Depleting CLASP from the cell abolishes Golgi-derived microtubule formation and the initial clustering of Golgi ministacks. In the second stage, Golgi clusters move towards the cell center along centrosome-derived radial microtubule arrays to complete Golgi ribbon assembly. This stage is called centrosome stage. Without the Golgi-derived microtubules by CLASP depletion, the Golgi ministacks can still be relocated to the cell center by centrosome-derived microtubules, but remain more fragmented 58. On the other hand, when centrosome-derived microtubules were disrupted by laser ablation of the centrosome, a large portion of the Golgi clusters can still move towards each other and finally to the cell center 82. However, some small fragments are blocked by the nucleus and remain uncaptured in the absence of centrosomal microtubules. Interestingly, once the Golgi is present in the cell center, Golgi-derived microtubules are sufficient to support Golgi integrity, and the centrosome becomes dispensable 82. Both stages of Golgi assembly require the function of dynein 58, 82. This is consistent with the fact that when the Golgi anchor of dynein, golgin-160, is depleted, Golgi fragments into ministacks throughout the cell 59.
The role of microtubule organization in Golgi positioning and ribbon linking during the cell cycle
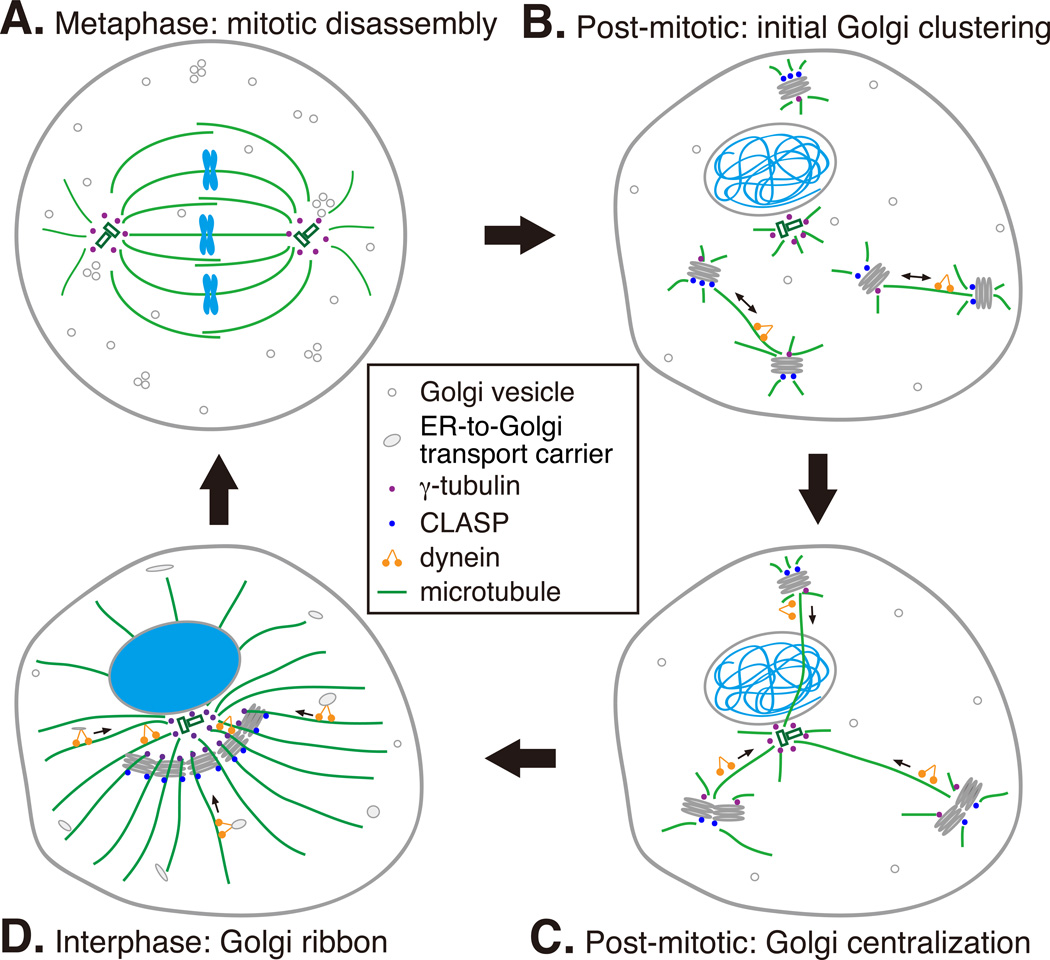
Assembly of Golgi ribbon in the cell center requires an intact microtubule network with microtubule arrays shooting from the microtubule organization centers (MTOCs) (including the centrosome and the Golgi membranes) to the cell periphery (Box 2). The microtubule minus end-directed motor, the dynein/dynactin complex, brings Golgi membranes and ER-derived vesicles to the cell center 56. During mitosis, microtubules are reorganized into a mitotic spindle and the Golgi disassembles. The derived vesicles disassociate from the spindle microtubules, while some Golgi remnants remain associated with the spindle, undergo ordered partitioning and may serve as Golgi templates in the reassembly 57. In telophase, Golgi ministacks form by membrane fusion and stacking in a microtubule-independent manner. Subsequently, microtubules generated from both the Golgi ministacks and the centrosome direct Golgi stacks moving towards each other and to the cell center for new ribbon assembly (Fig. 3) 58.
Fig. 3. Microtubule cytoskeleton in post-mitotic Golgi ribbon formation.
A. Golgi disassembly in mitosis. During mitosis, centrosome-derived microtubules are assembled into a mitotic spindle while Golgi-derived microtubules are depolymerized. Golgi membranes are disassembled into vesicles and mitotic Golgi clusters, the latter of which are partially associated with spindle microtubules.
B. Golgi clustering in telophase. At mitotic exit, Golgi mini-stacks start to form. In the initial stage, mini-stacks move by dynein towards each other along Golgi-derived microtubules that are stabilized by CLASP.
C. Golgi ribbon formation in later telophase and cytokinesis. After the initial Golgi clustering directed by Golgi-derived microtubules, Golgi clusters are captured by centrosome-derived microtubules and carried by dynein to the minus end of microtubules. The high concentration of Golgi membranes in the cell center allows further membrane fusion and ribbon formation.
D. Maintenance of the Golgi ribbon in interphase. Golgi is positioned surrounding the centrosome, with cis-golgins (GMAP210 or AKAP450/GM130) associate with microtubules derived from centrosomal γ-tubulin. Golgi-associated γ-tubulin also nucleates microtubules on the cis-Golgi. These microtubules are elongated through the trans-Golgi where they are stabilized by CLASP (through interaction with trans-golgin GCC185) towards the cell periphery. Dynein is required for maintaining Golgi ribbon in the pericentriolar region and for the directional movement of newly formed pre-Golgi transport carriers from cell periphery to the Golgi ribbon. Cis-golgin golgin-160 is the dynein receptor on Golgi apparatus.
The role of microtubule cytoskeleton in Golgi ribbon formation is highlighted by a recent finding that a cis-Golgi protein golgin-160 recruits dynein to the Golgi by direct interaction. Depletion of golgin-160 abolished ER-to-Golgi trafficking and caused Golgi ribbon unlinking, similar to the effect caused by nocodazole treatment. Golgin-160 is recruited to the cis-Golgi by interacting with Arf1 GTPase while its seventh coiled-coil domain, cc7, binds to dynein 59. During mitosis, golgin-160 disassociates from Golgi membranes through an unknown mechanism, which allows the Golgi to disperse.
Actin filaments in Golgi biogenesis
In addition to microtubules, actin filaments are also involved in Golgi trafficking and structure maintenance but the underlying mechanism remains largely unknown. Impairing actin dynamics by treating cells with either actin depolymerizing or stabilizing drugs both give a compact morphology of the Golgi 60. Actin polymerization facilitates membrane deformation to drive vesicle formation, fission and fusion, and short-range movement 61. Not much is known about the function of actin in Golgi dynamics in mitosis. The only indication has come from a study in Drosophila S2 cells, in which Golgi stacks are duplicated and exist as pairs before cell division. At late G2 phase, depolymerization of actin filaments induces scission of the paired-Golgi and the number of total Golgi stacks in the cell doubles. This process is mediated by inactivation of the actin nucleation promoting factors WAVE/Scar/Abi 62. These paired Golgi stacks have been observed in human cells, however whether Golgi pairing and unpairing is also regulated by the same mechanism in mammalian cells is yet to be determined 62.
Concluding remarks
Golgi inheritance during cell division is a dynamic process that requires the cooperation of many factors including Golgi matrix proteins, membrane tethers, vesicle budding and fusion machineries, cytoskeleton, as well as kinases and phosphatases that transmit signals onto the Golgi membranes. During the cell cycle, these factors coordinately regulate mitotic ribbon unlinking, cisternae unstacking and membrane vesiculation, as well as post-mitotic cisternae growth, cisternae stacking and ribbon formation. Although much progress has been made in elucidating the mechanisms of Golgi membrane dynamics during the cell cycle, a number of questions remain to be addressed. First, the GRASP proteins are certainly important for Golgi structure formation, but whether they are involved in Golgi stacking, ribbon linking or both is still under debate. Second, although ubiquitin is required for post-mitotic Golgi membrane fusion, several related questions are unclear, including: how do ubiquitin and GATE-16 promote p97/p47- and NSF-mediated membrane fusion; what are the ubiquitinated substrates in the p97/p47 pathway; and how is GATE-16 added onto or removed from the Golgi membranes. Third, whether the two functions of golgins, membrane tethering and Golgi ribbon linking, are coupled, and how they coordinate with the reorganization of the microtubule cytoskeleton during the cell cycle, requires further exploration. Fourth, actin filaments play important roles not only in regulating Golgi transport but also in Golgi morphology maintenance. However, the mechanism is largely unknown. Finally, Golgi defects have been observed in several diseases such as cancer, neurodegeneration and viral infection 63, 64. These defects may affect the trafficking, sorting and modification of a large number of proteins and cause global effects to the cell surface that compromise a variety of cellular functions. Defining the cause of the Golgi defects at the molecular level may provide insight into the pathogenesis of the related diseases.
Acknowledgments
We would like to thank Dr. Nobuhiro Nakamura (Kyoto Sangyo University), Dr. Robert Z. Qi (The Hong Kong University of Science and Technology) and Dr. Ayano Satoh (Okayama University) for critical reading the manuscript. This work was supported by the National Institute of Health (GM087364) and the American Cancer Society (RGS-09-278-01-CSM) to Y. Wang. D. Tang was supported by the Rackham Predoctoral Fellowship from the University Michigan and granted by Chinese Government Award for Outstanding Self-financed Students Abroad by China Scholarship Council (CSC). We apologize to those whose work we could not cite due to space constrains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Mironov AA, Beznoussenko GV. Molecular mechanisms responsible for formation of Golgi ribbon. Histol Histopathol. 2011;26:117–133. doi: 10.14670/HH-26.117. [DOI] [PubMed] [Google Scholar]
- 2.Barr FA. Golgi inheritance: shaken but not stirred. J Cell Biol. 2004;164:955–958. doi: 10.1083/jcb.200402011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altan-Bonnet N, et al. Golgi Inheritance in Mammalian Cells Is Mediated through Endoplasmic Reticulum Export Activities. Mol Biol Cell. 2006;17:990–1005. doi: 10.1091/mbc.E05-02-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330. doi: 10.1101/cshperspect.a005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang JS, et al. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. Embo J. 2005;24:4133–4143. doi: 10.1038/sj.emboj.7600873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Golgi: Methods for Preparation. Encyclopedia of Life Sciences. 2008 [Google Scholar]
- 7.Tang D, et al. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang D, et al. Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nat Protoc. 2010;5:758–772. doi: 10.1038/nprot.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y. Golgi apparatus inheritance. In: Mironov A, et al., editors. The Golgi apparatus. State of the art 110 years after Camillo Golgi’s discovery. Springer; 2008. pp. 580–607. [Google Scholar]
- 10.Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- 11.Satoh A, et al. Golgin-84 is a rab1 binding partner involved in Golgi structure. Traffic. 2003;4:153–161. doi: 10.1034/j.1600-0854.2003.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shorter J, et al. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. Embo J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barr FA, et al. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, et al. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. Embo J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutterlin C, et al. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang D, et al. The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression. Traffic. 2010;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preisinger C, et al. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. Embo J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang D, et al. Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol Open. 2012;1:1204–1214. doi: 10.1242/bio.20122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bisel B, et al. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinke FP, et al. The multiple facets of the Golgi reassembly stacking proteins. Biochem J. 2011;433:423–433. doi: 10.1042/BJ20101540. [DOI] [PubMed] [Google Scholar]
- 23.Giuliani F, et al. Unconventional secretion: a stress on GRASP. Curr Opin Cell Biol. 2011;23:498–504. doi: 10.1016/j.ceb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Sengupta D, Linstedt AD. Mitotic inhibition of GRASP65 organelle tethering involves Polo-like kinase 1 (PLK1) phosphorylation proximate to an internal PDZ ligand. J Biol Chem. 2010;285:39994–40003. doi: 10.1074/jbc.M110.189449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truschel ST, et al. Allosteric regulation of GRASP-dependent Golgi membrane tethering by mitotic phosphorylation. J Biol Chem. 2012 doi: 10.1074/jbc.M111.326256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Golgi Cisternal Unstacking Stimulates COPI Vesicle Budding and Protein Transport. PLoS ONE. 2008;3:e1647. doi: 10.1371/journal.pone.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popoff V, et al. COPI budding within the Golgi stack. Cold Spring Harb Perspect Biol. 2011;3:a005231. doi: 10.1101/cshperspect.a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang Y, et al. Active ADP-ribosylation factor-1 (ARF1) is required for mitotic Golgi fragmentation. J Biol Chem. 2007;282:21829–21837. doi: 10.1074/jbc.M611716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe M, Barr FA. Inheritance and biogenesis of organelles in the secretory pathway. Nat Rev Mol Cell Biol. 2007;8:429–439. doi: 10.1038/nrm2179. [DOI] [PubMed] [Google Scholar]
- 30.Seemann J, et al. Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science. 2002;295:848–851. doi: 10.1126/science.1068064. [DOI] [PubMed] [Google Scholar]
- 31.Puri S, et al. Dispersal of Golgi matrix proteins during mitotic Golgi disassembly. Journal of cell science. 2004;117:451–456. doi: 10.1242/jcs.00863. [DOI] [PubMed] [Google Scholar]
- 32.Meyer HH. Golgi reassembly after mitosis: the AAA family meets the ubiquitin family. Biochim Biophys Acta. 2005;1744:481–492. [PubMed] [Google Scholar]
- 33.Dalal S, et al. Distinct Roles for the AAA ATPases NSF and p97 in the Secretory Pathway. Mol Biol Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uchiyama K, et al. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Meyer HH, et al. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. Embo J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, et al. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang D, et al. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nature Communications. 2011;2:501. doi: 10.1038/ncomms1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat Med. 2007;13:1060–1069. doi: 10.1038/nm1621. [DOI] [PubMed] [Google Scholar]
- 39.Totsukawa G, et al. VCIP135 deubiquitinase and its binding protein, WAC, in p97ATPase-mediated membrane fusion. Embo J. 2011;30:3581–3593. doi: 10.1038/emboj.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller JM, et al. Sequential SNARE disassembly and GATE-16-GOS-28 complex assembly mediated by distinct NSF activities drives Golgi membrane fusion. J Cell Biol. 2002;157:1161–1173. doi: 10.1083/jcb.200202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diao A, et al. Coordination of golgin tethering and SNARE assembly: GM130 binds syntaxin 5 in a p115-regulated manner. J Biol Chem. 2008;283:6957–6967. doi: 10.1074/jbc.M708401200. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura N. Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J Pharmacol Sci. 2010;112:255–264. doi: 10.1254/jphs.09r03cr. [DOI] [PubMed] [Google Scholar]
- 43.Diao A, et al. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohda M, et al. Interaction of Golgin-84 with the COG complex mediates the intra-Golgi retrograde transport. Traffic. 2010;11:1552–1566. doi: 10.1111/j.1600-0854.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- 45.Colanzi A, Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr Opin Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Feinstein TN, Linstedt AD. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol Biol Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei JH, Seemann J. Unraveling the Golgi ribbon. Traffic. 2010;11:1391–1400. doi: 10.1111/j.1600-0854.2010.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sengupta D, Linstedt AD. Control of organelle size: the Golgi complex. Annu Rev Cell Dev Biol. 2012;27:57–77. doi: 10.1146/annurev-cellbio-100109-104003. [DOI] [PubMed] [Google Scholar]
- 49.Puthenveedu MA, et al. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 50.Duran JM, et al. The Role of GRASP55 in Golgi Fragmentation and Entry of Cells into Mitosis. Mol Biol Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feinstein TN, Linstedt AD. GRASP55 Regulates Golgi Ribbon Formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corda D, et al. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 2006 doi: 10.1016/j.tcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Colanzi A, et al. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. Embo J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marra P, et al. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardenas J, et al. Golgi localisation of GMAP210 requires two distinct cis-membrane binding mechanisms. BMC Biol. 2009;7:56. doi: 10.1186/1741-7007-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brownhill K, et al. Molecular motors and the Golgi complex: Staying put and moving through. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 57.Shima DT, et al. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller PM, et al. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yadav S, et al. Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev Cell. 2012;23:153–165. doi: 10.1016/j.devcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Egea G, et al. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006 doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Lanzetti L. Actin in membrane trafficking. Curr Opin Cell Biol. 2007;19:453–458. doi: 10.1016/j.ceb.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 62.Kondylis V, et al. The Golgi Comprises a Paired Stack that Is Separated at G2 by Modulation of the Actin Cytoskeleton through Abi and Scar/WAVE. Dev Cell. 2007;12:901–915. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Hu Z, et al. The study of Golgi apparatus in Alzheimer's disease. Neurochem Res. 2007;32:1265–1277. doi: 10.1007/s11064-007-9302-4. [DOI] [PubMed] [Google Scholar]
- 64.Ungar D. Golgi linked protein glycosylation and associated diseases. Semin Cell Dev Biol. 2009;20:762–769. doi: 10.1016/j.semcdb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Sinka R, et al. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol. 2008;183:607–615. doi: 10.1083/jcb.200808018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matteis M, et al. The Golgi ribbon and the function of the Golgins. In: Mironov A, Pavelka M, editors. The Golgi Apparatus. Springer Vienna; 2008. pp. 223–246. [Google Scholar]
- 68.Rios RM, et al. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 69.Drin G, et al. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 70.Drin G, et al. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320:670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- 71.Pernet-Gallay K, et al. The overexpression of GMAP-210 blocks anterograde and retrograde transport between the ER and the Golgi apparatus. Traffic. 2002;3:822–832. doi: 10.1034/j.1600-0854.2002.31107.x. [DOI] [PubMed] [Google Scholar]
- 72.Houghton FJ, et al. The localization of the Golgin GCC185 is independent of Rab6A/A' and Arl1. Cell. 2009;138:787–794. doi: 10.1016/j.cell.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 73.Luke MR, et al. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J Biol Chem. 2003;278:4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- 74.Efimov A, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin YC, et al. ARL4A acts with GCC185 to modulate Golgi complex organization. Journal of cell science. 2011;124:4014–4026. doi: 10.1242/jcs.086892. [DOI] [PubMed] [Google Scholar]
- 76.Chabin-Brion K, et al. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rivero S, et al. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang S, et al. Capacity of the Golgi Apparatus for Cargo Transport Prior to Complete Assembly. Mol Biol Cell. 2006 doi: 10.1091/mbc.E05-12-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kasap M, et al. Dynamic Nucleation of Golgi Apparatus Assembly from the Endoplasmic Reticulum in Interphase HeLa Cells. Traffic. 2004;5:595–605. doi: 10.1111/j.1398-9219.2004.00203.x. [DOI] [PubMed] [Google Scholar]
- 80.Hoppeler-Lebel A, et al. Centrosomal CAP350 protein stabilises microtubules associated with the Golgi complex. Journal of cell science. 2007;120:3299–3308. doi: 10.1242/jcs.013102. [DOI] [PubMed] [Google Scholar]
- 81.Walenta JH, et al. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol. 2001;152:923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vinogradova T, et al. Concerted effort of centrosomal and Golgi-derived microtubules is required for proper Golgi complex assembly but not for maintenance. Mol Biol Cell. 2012;23:820–833. doi: 10.1091/mbc.E11-06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]