Abstract
The CD8 memory T cell repertoire to the influenza A derived M158–66 epitope shows a restricted V genes and CDR3 sequences usage. The repertoire is highly polyclonal and the clonotype distribution has been described as consisting of two components, one showing a power law-like distribution and the other composed of a few clonotypes with a very high relative frequency. The question is whether the complex repertoire defined by its ability to flourish in a short term recall culture corresponded to functional cells. Here we show that there is a relation between expression of the degranulation marker CD107 and cytotoxicity or IFN-γ production in CD8 T cell lines and clones. We then examine recently degranulated CD8 cells from recall cultures from four middle aged HLA-A2 subjects and show that these functional cells are polyclonal. The clonotype distributions of the CD8 + CD107+ repertoires are complex in the same manner as previously reported. The clonotype composition of CD8 + CD107+ repertoires is also very similar to CD8 only repertoires, and to CD8 + HLA-A2-M158–66 pentamer positive repertoires. We postulate that multiple exposures during childhood to this conserved influenza A epitope has generated a complex functional repertoire in HLA-A2 individuals.
INTRODUCTION
CD8 T cells can play an important role in controlling and resolving viral infections. Because they focus on intracellular proteins CD8 cells can recognize conserved epitopes associated with viral replication or assembly. The response of HLA-A2 individuals to the conserved M158–66 epitope is a case in point. We have studied over 30 healthy middle aged HLA-A2 individuals and they all mount a strong in vitro recall response to this peptide.
The response to M158–66 is characterized by cells that predominantly use the BV19 TCR gene with a CDR3 length of 11 amino acids which show a conserved RS motif (1–4). Early studies used CTL lines and clones for assessment of functional activity such as cytotoxicity or cytokine release (1, 2). Availability of HLA-A2-M158–66 tetramers and pentamers have provided a useful tool to isolate T cells whose TCR have a high avidity for this complex (5). Short term (two to three week) recall cultures have been used with some success to analyze this response. These studies have shown that the recall repertoire to M158–66 consists of multiple clonotypes (3, 6, 7). Using clonotype specific probe hybridization we have shown that for the higher frequency clonotypes, the relative frequency of a clonotype in short term recall cultures is similar to that in the starting PBMC (3). In many individuals, the RS sequence in the CDR3 can be encoded in multiple ways. The frequency distribution of the recall repertoire is interesting in that there can be a few clonotypes present at high frequency, more clonotypes at intermediate frequencies, and a large number, often over 50%) at very low frequencies. The intermediate and low frequency component can be described as power law-like. There is a self-similarity to these distributions that has led to our describing the repertoire as fractal (6). Such complex repertoires have also been observed in the mouse (7, 8) and cow (9).
The significance of such a polyclonal repertoire is not completely understood. However, different clonotypes appear to respond at different epitope concentrations (10), indicating that some clonotypes may be more useful at particular stage of infection than others. A major difficulty in appreciating the significance of such complex repertoires is that it is unclear to what extent these clonotypes actually define functional T cells as opposed to T cells that merely grow in culture. While complex repertoires are associated with multimer binding (10), this just reposes the question as to what portion of cells that grow in culture and have sufficient avidity to bind multimers are functional.
The two most understood functions of CD8 cells are their cytotoxicity and their secretion of IFN-γ. However, neither cytotoxicity assays nor intracellular staining for Ifn-γ are easily amenable for molecular clonotype analysis. Degranulation is associated with cytotoxicity and can be indicative of cytokine secretion. A surface marker for degranulation associated with cytotoxicity exists in the form of CD107 (12, 13). Expression of the marker and isolation of recently degranulated cells can facilitate clonotype analysis of these cells.
In this study we used CD107 mobilization as an indication of the cytotoxic potential of the responding CD8+ T cells. We first compared CD107 expression with the lytic potential measured by traditional chromium (51Cr) release assay for flu specific lines and clones and found strong correlation between the two techniques. We then used this technique to isolate functional flu-specific CD8 T cells in from a short term recall response. Sequencing the BV19 TCR from CD1-7 positive cells showed that the repertoire of the functional flu specific memory CD8 T cells is indeed polyclonal. In addition we show that the repertoire of cells with avidity for HLAA2- M158–66 pentamers is similar to those that have undergone degranulation.
Materials and Methods
Cells
PBMC were obtained from HLA-A2 blood donors under BloodCenter of Wisconsin IRB-approved protocols. Cells were stored frozen under liquid N2 until used.
T2 cells that is deficient in the transporter-associated protein (TAP), was obtained from Dr. Malarkannan.
Peptide
Influenza matrix peptide M158–66 peptide GILGFVFTL was synthesized by The Blood Research Institute Peptide Core.
Monoclonal antibodies
CD8-Pacific Blue, BV17 (19)-FITC or –PE from (Invitrogen, Carlsbad, CA), (CD107a, and CD107b)-FITC or -APC, GolgiStop, and IFNγ intracellular staining Kit from (B.D. Pharmingen, San Diego, CA), Fc Block Miltenyi Biotech (Germany
Reagents
HLA-A2- M158–66 pentamer-PE (Proimmune, Bradenton, FL)
Phytohaemagglutanin PHA (Sigma , Saint Louis, MO)
51Cr (sodium Chromate) was purchased from Perkin Elmer (Boston, MA)
CD8 positive isolation kit (Invitrogen)
Primers
βV19primer; 5’CCAAAAGAACCCGACAGCTTTC , Fam labeled βC primer; 5’Fam-GCTTCTGATGGCTCAAACACAG, and Poly T primer 5’TTTTTTTTTTTTTTTTTT were made by Integrated DNA Technologies (Coralville, Iowa)
In vitro stimulation of PBMC in a short term recall culture
PBMC were cultured at 1×106 cell/ml with1 µM of influenza A matrix peptide M158–66 and 10 U/ml of recombinant IL-2 for 7 days. Primed cells were re-stimulated in the presence of recombinant IL-2 (10U/ml) and equal number of irradiated (3000 R) autologous PBMC that had been preloaded with M158–66 peptide. At the end of the second week, the cells were used for generation of CTL lines, single cell clones or for repertoire analysis as described below. Longer culture times result in a decrease in viable cells most likely due to the HLA-A2 expressing T cells becoming targets after the addition of peptide.
Generation of Influenza virus M158–66-specific, HLA-A2-restricted CTL lines
At the end of the second week, the cultured cells were stained for CD8 and βV19. The CD8+βV19+ cells were sorted using FACSAria (BD Biosciences). The sorted cells were put into culture with 40 U/ml IL-2 and re-stimulated every other week for total of 4–6 weeks.
Generation of Influenza virus M158–66-specific, HLA-A2-restricted single cell clones
At the end of the second week, the cultured cells were stained for CD8 and βV19. The CD8+βV19+ single cells were sorted into each well of five 96 well plate, and maintained with PHA and autogenic stimulator/allogenic feeder cells. The cultures were continued until each clone number reached about 0.5 million cells.
Repertoire analysis
For repertoire analysis the cultures were split. One portion was used for CD8 isolation using Dynal CD8 positive isolation kit (Invitrogen) according to manufacture instructions. The other portion was used for CD107 mobilization as described below. In two cases, a third portion was stained with CD8 and M158–66-loaded HLA-A2 Pentamer. Clonotyping was performed as described below.
RNA and cDNA preparation
mRNA was isolated from recall cultures using Dynal Oligo (dT) Beads according to manufacture instruction (Invitrogen, Carlsbad, CA), and cDNA was prepared using Poly T primer and MMLV reverse transcriptase ( Invitrogen).
Clonotyping
Clonotyping was done using the colony counting technique (3). In summary, cDNA from cultured cells was amplified using βV19 – βC primers, the PCR product was cloned into E. coli using TOPO Cloning Kit (Invitrogen). Bacterial colonies (192) were grown overnight and sent to Agencourt Bioscience (Beverly, MA) for sequencing. Clonotype sequences were analyzed using CDR3Reader software, which counts identical sequences and assigns clonotype names according to the convention described by Yassai et al (14).
Although the colony counting procedure involves ligation and bacterial transformation steps, the results are reproducible as can be seen from a test experiment in which a two week culture was divided in three and each portion subject to CD8 selection and clonotyping. Supplementary Figure 1 shows that there was excellent clonotype overlap between the three measurement assays. As might be expected, the best overlap was observed in the high frequency clonotypes. The non overlapping clonotypes were predominantly clonotypes only observed once (38/47 ≈ 81%) and at most observed three times (2/47). This experiment is representative of three such tests.
Intercellular IFNγ staining
Clones and cell lines were stained with CD8- Pacific blue, the cells fixed, permeabilized, and stained with IFN γ-APC using a BD Pharmingen IC staining kit according to the manufacturer’s instructions.
Pentamer staining
Cells were washed and re-suspended in PBS with 0.1% BSA (10 million /ml). Fc block was added (2ul/million) and incubated for 15 min on ice. Pentamer reagent was added (10 ul/million), and incubated for 20 minutes on ice. Cells were washed once and stained with anti CD8 pacific blue and CD3 FITC antibodies.
51Cr release assay
Standard 51Cr-release assays (15) were performed using 51Cr-labelled T2 target cells (2000/well). Target cells were pulsed with influenza A Matrix M158–66 peptide and incubated overnight. After washing, the target cells were labeled with 51Cr for 1 hour. PBMC cell lines or clones and T2 target cells were then incubated for 4 hours at titrated E:T ratios. Control wells containing only T2 target cells without peptide were also set up with each assay. Percent specific lysis was calculated as (100*experimental release - spontaneous release)/(maximum release – spontaneous release).
CD107 mobilization assay
CD107 mobilization assay was performed as previously described (12). Briefly, CD8+ T cells from two week culture and T cell clones or lines were incubated with influenza A Matrix M158–66 peptide preloaded T2 target cells at E:T ratio of 1:2. FITC conjugated anti-CD107a (4ul/ml) and anti-CD107b (4ul/ml) monoclonal antibodies were added. After 1 hour incubation GolgiStop was added to each sample. Control wells containing only T2 target cells were also set up with each assay. Cells were harvested 4–5 hours later, Fc block was added to inhibit non-specific binding of antibodies, and cells were stained with fluorescently conjugated monoclonal antibodies specific for CD8 pacific blue, BV19 PE. The fluorescence intensities were determined using a LSRII (BD Biosciences). Data were analyzed using Flowjo software.
Repertoire measures and characteristics
There are a number of measures that constitute a clonotypic repertoire analysis.
M = Total number of sequences
N = Total number of different CDR3 sequences (Clonotypes)
Rmax = Maximum number of observations for any clonotype(s) in the repertoire
Ns = number of clonotypes that appeared only once (singletons)
These measures are used to calculate certain repertoire characteristics as described by Naumova et al (16).
M/N = observations per clonotype
Ns/N = fraction of singletons
Rmax/M = proportion of measurements due to the clonotype observed most frequently
There are a number of measures specific for the M158–66 response:
NRS = number of clonotypes with R (Arginine ) and S ( Serine) at position 5 and 6 in the CDR3
NRSs = number of singleton RS clonotypes
NJ2.7 = number of clonotypes that used J2.7
The fraction of each of these clonotypic measures is given by dividing by N. e.g. NJ2.7/N is the fraction of clonotypes using J2.7.
Selection comparisons.
Nov = Clonotypes that are observed in two different selected repertoires e.g. CD8+ or CD8+CD107+ are considered to be overlapping
Mov = the number of observations attributable to overlapping clonotypes
The above designations can be combined; for example:
NRSov = number of overlapping RS clonotypes
Clonotype distributions
Clonotype frequency was changed into a rank frequency measure by counting all the clonotypes that were observed the same number of times. As described above, clonotypes observed once (singletons) represent a boundary condition in the description of the repertoire, as does the clonotype(s) that has the highest rank (observed most frequently). As described previously (6) the repertoire can be analyzed by plotting the natural logarithm of the rank vs rank-frequency. Starting with the singletons the repertoire distribution decreases linearly over a number of ranks, this is followed a component consisting of a few high ranking clonotypes. A line was fitted to the first component and the Pearson Correlation calculated using MS Excel. The p value was obtained from the formula; t = r/((1—r2)/(N—2))1/2, where n is the number of observations and r the correlation.
RESULTS
Specificity and functional potential of CD8 T cell lines
To analyze the correlation between cytotoxicity and CD107 expression we generated influenza virus M158–66-specific BV19 positive long term cell lines from three HLA-A2 individuals. From one individual (RM1) CD8 clones were also obtained. The specificity of the clones and cell lines was checked by staining with HLA-A2-M158–66 pentamer, and their ability for IFNγ production was measured by IFNγ intracellular staining. The lytic activity of the cell lines was measured by two techniques; CD107 mobilization which transiently is expressed on the surface of CD8 T cells upon de-granulation of the cytotoxic granules, and by traditional chromium release assay.
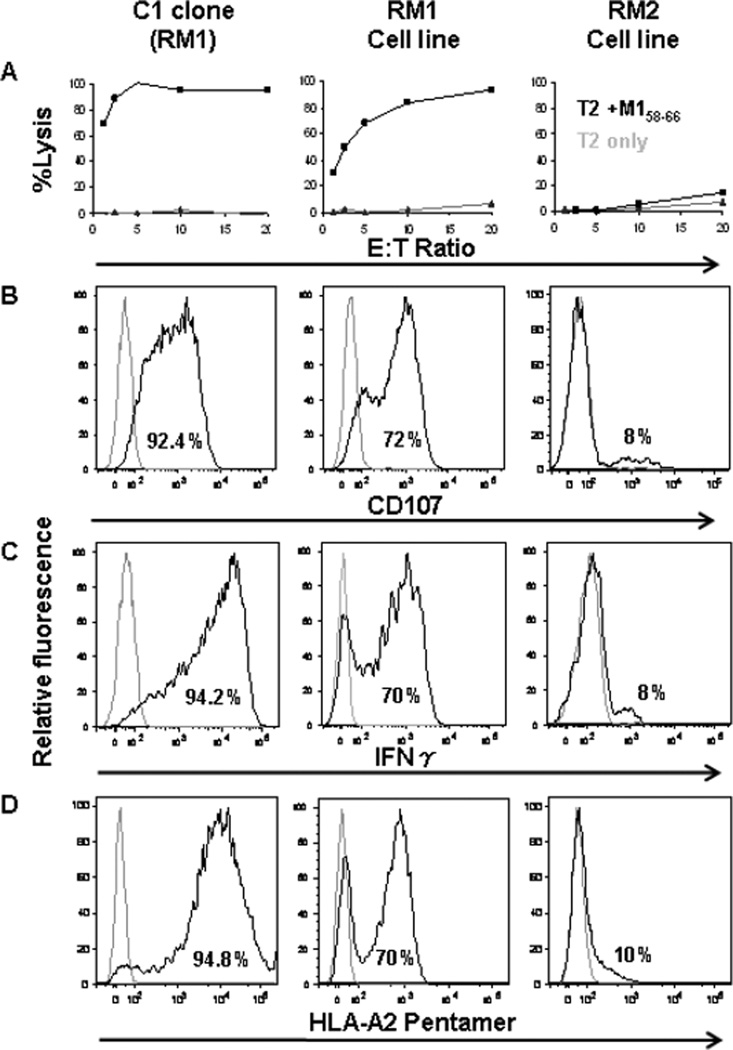
An example of the analysis of one clone and two cell lines with different functions is shown in Figure 1. The C1 (RM1) clone can lyse 100% of the target cells at E:T ratio of 5, the RM1 cell line, from which C1 clone was generated, could lyse 90% of the target cells at E:T ratio of 20. It has a high level of CD107 expression, can produce IFNγ, and binds to pentamer. The data suggests that RM1 cell line is a flu specific cytotoxic cell line with protective function against influenza. On the other hand, the RM2 cell line while expanding in culture, shows very low levels of cytotoxicity at the highest E:T ratio, has very low levels of degranulation, does not produce high levels of interferon, and does not bind to pentamer well.
Figure 1. Phenotypic and functional analysis of a clone and two cell lines.
The first column from the left shows the analysis of C1 clone, and the 2nd and the 3dr columns show the analysis of the two cell lines (RM1 and RM2 respectively). A. Lysis of T2 target cells; X-axis shows E:T ratio, and Y-axis shows the % of lysis. Black line shows the % of lysis at different E:T ratio, and the gray line shows the background lysis. B. Relative fluorescence of CD107 expression presented as histograms. C. Relative fluorescence of IFNγ intercellular staining presented as histogram, D. Relative fluorescence of HLA-A2-M158–66- Pentamer staining is presented as histogram. In all histograms, the percentage of relative fluorescence for each staining is shown inside the histogram, and the gray histogram is the control.
The phenotypic characteristics of lines and clones from three recall cultures are summarized in Table 1. The RM1 cell line showed 50% cytoxicity at an E:T ratio of 2. About 72% of the cells expressed CD107, with similar levels of cells showing intracellular IFNγ production, and specificity for flu peptide assessed by HLA-A2-M158–66 pentamer binding. The line is heterogeneous as can be directly evidenced from the four clones derived from it. Two clones C1, and C2 had strong lytic potential, a high percentage of CD107 expression, IFNγ production, and pentamer binding (all >90%), reflecting the purity of the cell population. Two other RM1- derived clones (C3 and C4) had no lytic potential. The absence of lytic potential was accompanied by an absence of the other measured functions. The cell line RM2 had very low lytic ability, and very low degranulation, Ifn-production and pentamer binding. The F2 cell line showed no lysis, or other phenotypic markers. The data indicate the expected correlation between cytotoxicity measured by chromium release and degranulation as measured by CD107 expression. Ifn-γ production and pentamer binding were also associated with cytotoxicity.
Table 1.
| ID | ED50 E:T |
% CD107 |
% IFNγ |
% Pentamer |
|---|---|---|---|---|
| RM1 | 2:1 | 72 | 73 | 70 |
| C1 (RM1) |
1:1 | 92.4 | 94.2 | 94.8 |
| C2 (RM1) |
1:1 | 90.4 | 90.9 | 93.9 |
| C3 (RM1) |
No Lysis |
0 | 0 | 0 |
| C4 (RM1) |
No Lysis |
0 | 0 | 0 |
| RM2 | >20:1 | 8 | 8 | 10 |
| F2 | No Lysis | 0 | 0 | 2 |
The repertoire of functional flu specific CD8 T cells generated in a short term culture is polyclonal and complex
The above data indicate that the CD107 mobilization assay is useful for identifying functional M158–66 specific CD8 T cells. To analyze functional repertoires as defined by degranulation we chose PBMC from four HLA-A2 individuals who we had studied previously using our standard approach of analyzing CD8 cells from recall cultures. The PBMC were cultured with M158–66 peptide in the presence of IL2 for 2 weeks. A portion of each culture was remobilized with antigen followed by isolation of CD8+ CD107+ cells (referred to as the CD107 repertoire). Clonotypes were identified on the basis of the CDR3 nucleotide sequence of the BV19 TCR using our previously described colony counting approach (3) and named using our clonotype naming convention (14).
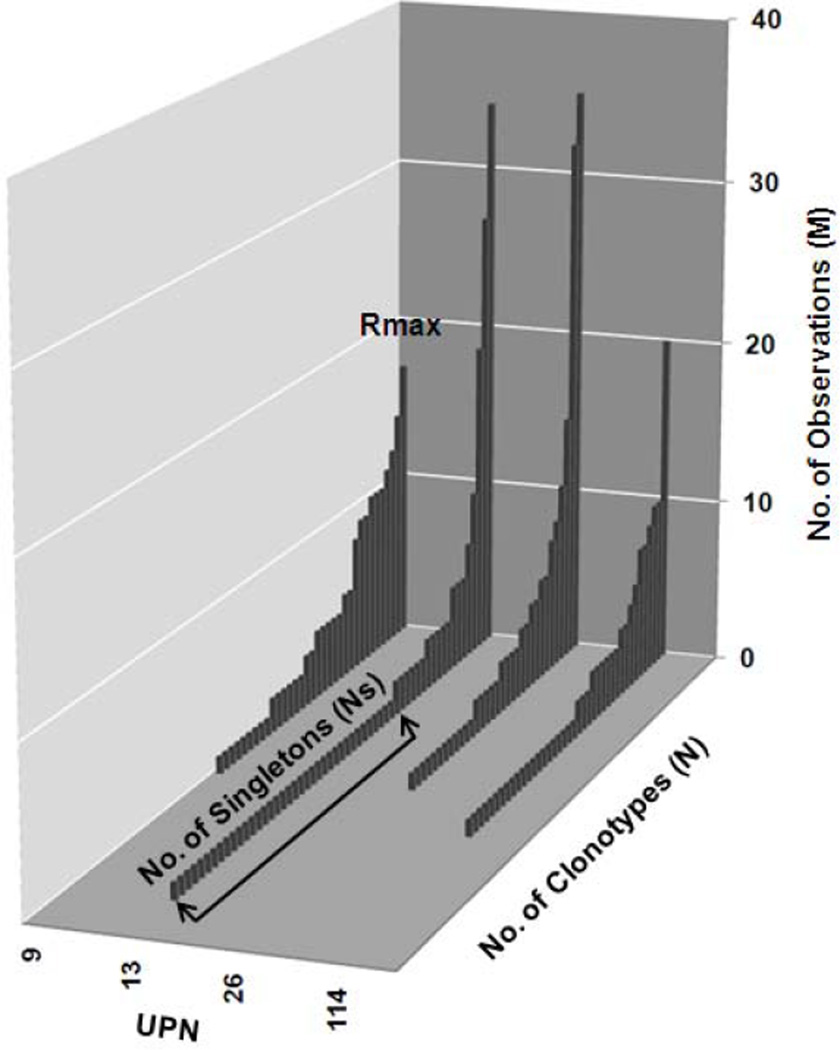
The CD107 repertoire of the four subjects showed a high degree of polyclonality (Figure 2). For each subject, the clonotypes are displayed in descending order of frequency of observation. For each subject the repertoires were comprised of a number of different clonotypes (N). The clonotype distribution was variable but in each individual there was a clonotype with a maximum number of observations (Rmax), and a number of clonotypes that were observed only once (singletons, Ns).
Figure 2. Clonotype distribution of CD8+CD107+ cells for four subjects.
The X-axis shows the subject unique identification numbers. The Y-axis shows the number of observations of each clonotype. Z-axis shows the clonotypes identified for each donor in decreasing order.
We have previously introduced measures and characteristics for repertoires analyzed at the clonotype level by CDR3 sequencing (16) and these are defined in Materials and Methods. The functional M158–66-specific repertoire defined as CD8+CD107+ was analyzed and the measurements and characteristics for the four subjects are shown in Table 2. The TCRβ CDR3 of each clonotype was given a unique ID based on the CDR3 nucleotide sequences (14), and the sum of the number of times each clonotype was observed generated (M). The number of clonotypes (N) for the CD107 repertoire varied between 33 and 56, indicating a high level polyclonality. Since M, can vary between each analysis, we normalized the clonotype count by expressing it in terms of observations per clonotype (M/N). This gives us an estimation of clonotype abundance. The M/N values varied between 3.1 and 5.3 (avg 4.1) depending on the subject analyzed. The higher the value the fewer clonotypes observed in the repertoire. Our previous examinations of the M158–66 response showed a high fraction of singleton clonotypes, often approaching 60% (6). The fraction of singletons (Ns/N) in the CD107 repertoires, varied between 26% and 65%. The fraction of clonotypes that used the RS motif (NRS/N ) varied between 60% and 88% (avg 74%), indicating that the use of this motif is highly correlated with effector function.
Table 2.
| Repertoire Measurements & Characteristics |
UPN9 | UPN13 | UPN26 | UPN114 |
|---|---|---|---|---|
| M | 167 | 176 | 174 | 129 |
| N | 34 | 55 | 33 | 40 |
| Rmax | 17 | 34 | 35 | 20 |
| Ns | 9 | 36 | 12 | 21 |
| NRS | 25 | 33 | 25 | 35 |
| M/N | 4.9 | 3.2 | 5.3 | 3.2 |
| Ns/N | 0.26 | 0.65 | 0.36 | 0.53 |
| Rmax/M | 0.1 | 0.19 | 0.2 | 0.16 |
| NRS/N | 0.74 | 0.60 | 0.76 | 0.88 |
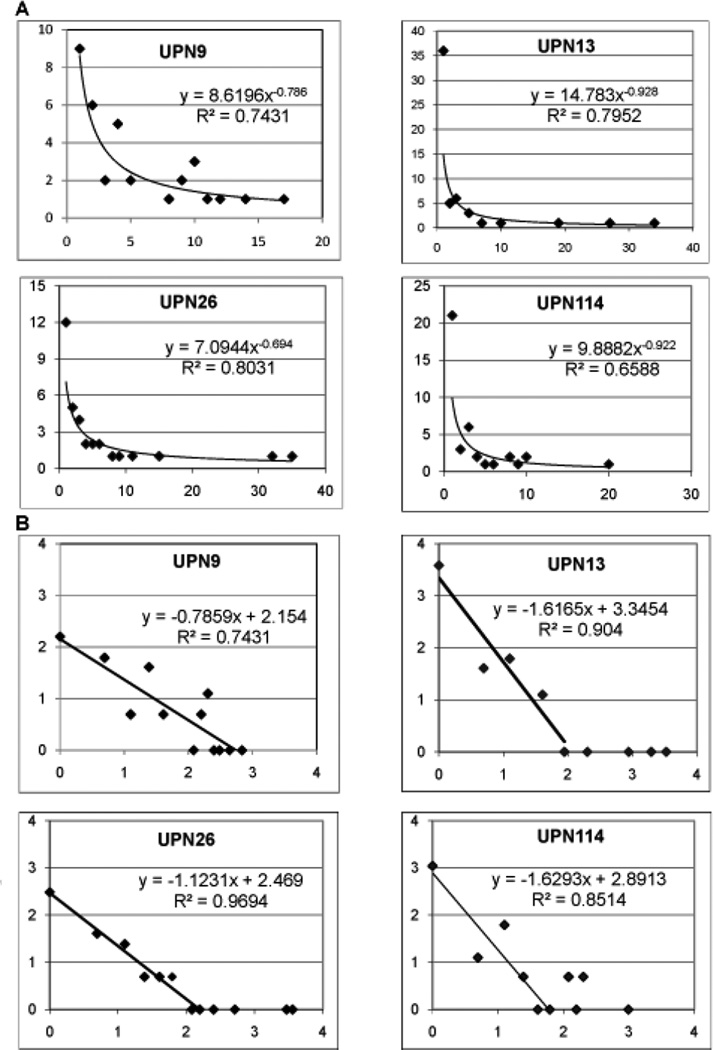
We analyzed the CD107 repertoires to determine if they showed the complex clonotype distribution pattern that we had reported earlier for CD8 T cell repertoires. First, the clonotype distribution is converted into a rank vs rank frequency format by counting all the clonotypes present once, twice, three-times, etc. The rank-frequency plots for the data for the four subjects are shown in Figure 3A and a power-law trend line is fit to the complete data set. A better appreciation for the data can be obtained by plotting the natural logarithm of the rank and rank frequency. We have observed that the clonotype distribution is better fit to a two component description and this is more readily observed with the ln-ln plots (Figure 3B). The CD107 repertoires of UPN13, UPN26, and UPN114 are examples of two component descriptions. The fit to the power law-like portions becomes higher when two components are used. This is especially evident for UPN26 and UPN13. The clonotype distribution of UPN9 fits best to a single power law-like component.
Figure 3. Rank – frequency analysis of clonotype distributions in CD107 and CD8 recall repertoires from UPN13.
A. Rank – frequency plot of CD107 positive cells isolated after reactivation of a two week recall culture for the four subjects analyzed. Line is fit to a power law for the entire dataset. B. Natural log transformation of the rank-frequency data. The natural log of the rank and the rank frequency are plotted on the X- and Y-axes respectively. For three subjects, the data can be divided into two components, a portion described by a linear decrease in the frequency and a portion consisting of a few high ranking clonotypes. The slope of the first component is shown.
Clonotype analysis and overlap between the functional CD107 CD8 T cell repertoire and the CD8 T cell repertoire from recall cultures
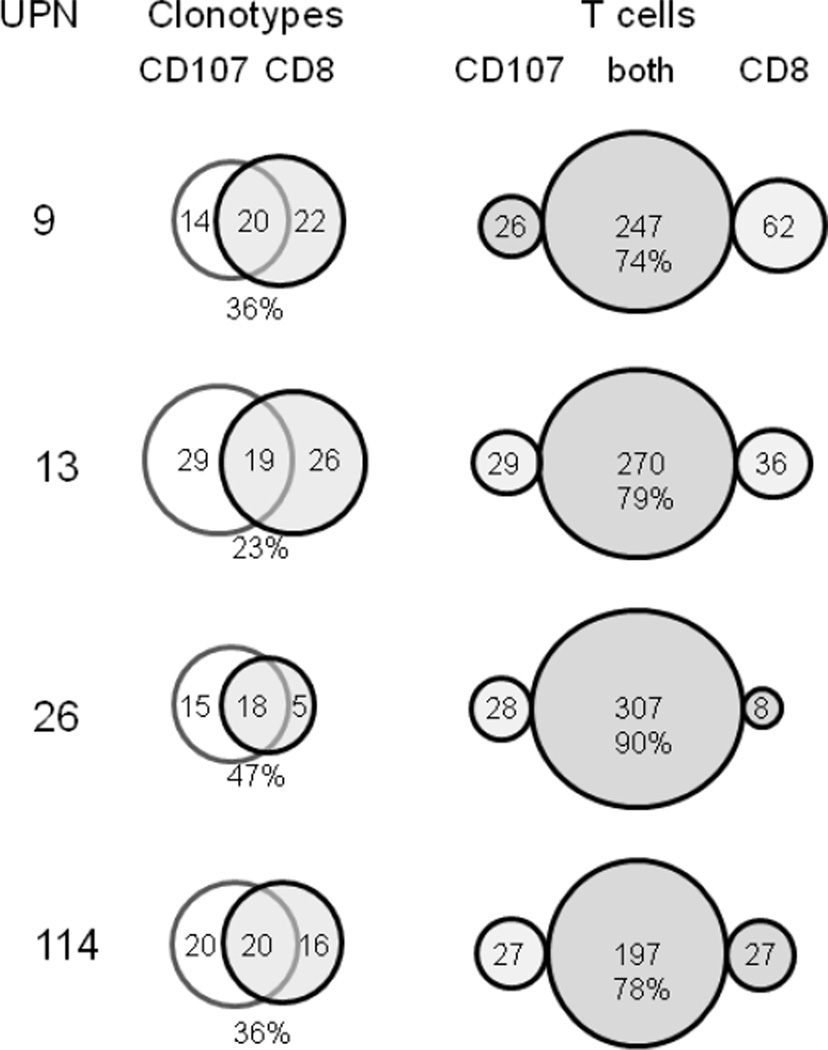
We continued our examination of the functional CD107 repertoire at the clonotype level by comparing it with the CD8 repertoire from the same M158–66 stimulation (Table 3). Repertoire overlap at the clonotype level is defined by observing the same clonotype in both repertoires. The clonotype overlap for the four subjects is presented in the form of Venn diagrams in Figure 4. The clonotype overlap range is between 23% and 47% with an average of 35%.
Table 3.
Repertoire overlap between the CD107 and CD8 repertoires for four subjects.
| Characteristic of Overlapping Repertoire |
UPN 9 |
UPN 13 |
UPN 26 |
UPN 114 |
|---|---|---|---|---|
| M | 335 | 340 | 343 | 251 |
| N | 56 | 82 | 38 | 56 |
| Mov | 247 | 270 | 307 | 197 |
| Nov | 20 | 27 | 18 | 20 |
| Nov/N | 0.36 | 0.33 | 0.47 | 0.36 |
| Mov/M | 0.74 | 0.79 | 0.90 | 0.78 |
| NRS | 42 | 45 | 27 | 49 |
| NRSov | 14 | 19 | 14 | 18 |
| NRSov/NRS | 0.33 | 0.42 | 0.52 | 0.37 |
Figure 4. Clonotype and T cell overlap between CD8 CD107 repertoires and CD8 repertoires in recall analyses.
The UPN are identified at the left. The Venn diagrams show the clonotype overlap. The percent of overlapping clonotypes is given below the overlap zone. On the right the distribution of T cells as measured by the number of sequences is shown in the form of bubble graphs. The percent of T cells observed in both repertoires is shown below the actual measured number.
The T cell overlap between the two repertoires, as measured by the number of observations (M), is presented in the last column of Figure 4 in the form of bubble plots representing the number of observations associated with clonotypes that were present only in the CD107 repertoire, in both repertoires, and only in the CD8 repertoire. The three bubbles correspond to the three segments of the Venn diagram. The proportion of the T cell repertoire that overlaps ranges from 74% to 90%, with an average of 80%. This shows that a majority of CD8 cells are in fact CD107 positive. It also indicates that in general the clonotype overlap predominantly corresponds to high frequency clonotypes. With distributions that contain low frequency components, the probability of observing an overlap decreases as the frequency of observation decreases.
Repertoire overlap of functional CD107+CD8+ T cells with HLA-A2 M158–66 pentamer+ CD8+ T cells
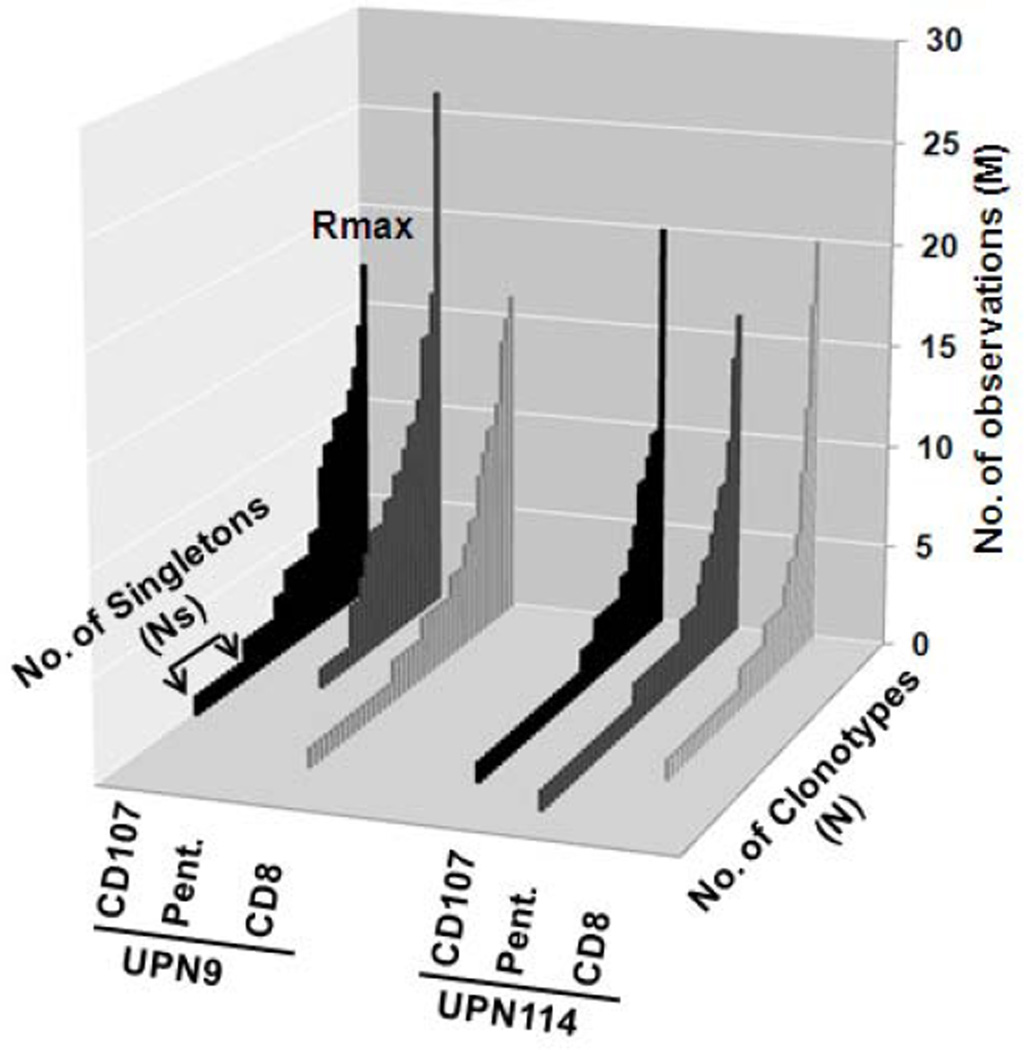
For two of the subjects, UPN9 and UPN114, the recall cultures were divided into three, with a million cells used for CD8 isolation, and the rest equally divided for CD107 mobilization and for pentamer staining. The clonotype distribution of functional CD107+ cells, pentamer positive cells, and CD8 cells is shown in Figure 5. The clonotype distribution of the CD107 and pentamer repertoires is very similar in both subjects.
Figure 5. Clonotype distribution of CD107+CD8+, HLA-A2 M158–66 pentamer+ CD8+, and CD8+ T cells for two subjects.
The legend is as described for Figure 2.
The repertoire measures and characteristics for the three analyses are summarized in Table 4. Overall the data for the three modes of repertoire analysis are very similar. There is an increased M/N value for the pentamer selected repertoire in UPN9 which could indicate that we had started to oversample this repertoire. The fraction of RS clonotypes (NRS/N) is high for both subjects, UPN9 (avg 0.73) and UPN114 (avg 0.87).
Table 4.
The repertoire measures and characteristics for the three parallel analyses of recall cultures.
| UPN 9 |
UPN 114 |
|||||
|---|---|---|---|---|---|---|
| CD8 CD107 |
CD8 | CD8 Pent |
CD8 CD107 |
CD8 | CD8 Pent |
|
| M | 167 | 168 | 179 | 129 | 122 | 142 |
| N | 34 | 42 | 25 | 40 | 36 | 46 |
| Rmax | 17 | 16 | 26 | 20 | 20 | 16 |
| Ns | 9 | 16 | 6 | 21 | 17 | 21 |
| NRS | 25 | 32 | 18 | 35 | 32 | 39 |
| M/N | 4.91 | 4.00 | 7.16 | 3.23 | 3.39 | 3.09 |
| Rmax/M | 0.10 | 0.10 | 0.15 | 0.16 | 0.16 | 0.11 |
| Ns/N | 0.26 | 0.38 | 0.24 | 0.53 | 0.47 | 0.46 |
| NRS/N | 0.74 | 0.76 | 0.72 | 0.88 | 0.89 | 0.85 |
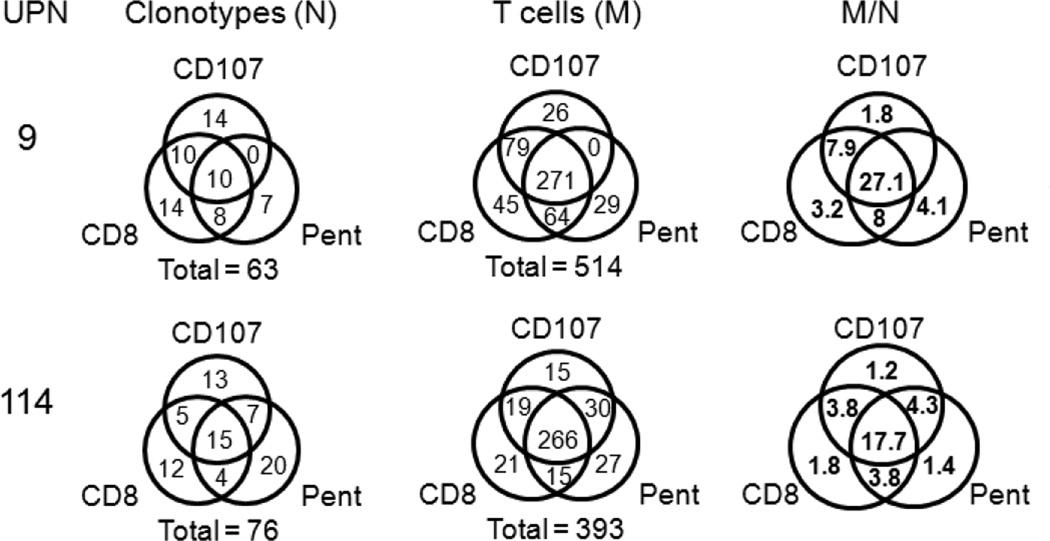
The clonotype overlap and the number of observations (T cells) in the repertoires selected using the three different approaches is shown in Figure 6 in the form of Venn diagrams. The level of triple overlaps in the two subjects is similar (UPN9: 10/63 = 0,16 and UPN114: 15/76=0.2). The level of double overlaps is also similar between the two subjects (18 vs 16). While the double overlaps are equally distributed between the three possibilities in UPN114, there are no examples of clonotypes observed only in the CD107 and pentamer repertoires in UPN9. The high M/N values for the triple overlaps indicates that these are predominantly high frequency clonotypes.
Figure 6. Clonotype and T cell overlap between CD107+CD8+-selected, HLA-A2 M158–66 pentamer+CD8+-selected, and CD8+-selected repertoires from recall analyses.
Venn diagrams are used to show the data for each selection-based repertoire subset. The selection methods are identified next to each circle. All the selection methods included CD8+ but this is dropped in the labeling of the CD107-selected and M158–66 HLA-A2 pentamer (Pent)-selected subsets. The UPN is identified on the left of each series. For the diagrams showing the clonotype in each subset (N) and those for the actual T cell observations (M) the total number of clonotypes or measurements is shown below each diagram. The leftmost set of diagrams shows the observations per clonotype for each selection-based subset.
Other repertoire considerations
The clonotype data for all the repertoires examined here are available as Supplementary Figure 2. The relation between overlapping clonotypes and high frequencies can be readily observed, even though some of the overlap does extend down to lower frequencies. Public clonotypes are highlighted with various colors. Clonotype names with an asterisk represent RS clonotypes that may have arisen by long P nucleotide addition (17). The number of public clonotypes can vary by individual. For example UPN13 only has two. Clonotypes that could be generated by long P nucleotide addition are highly represented among public clonotypes.
Also of interest are clonotypes that do not encode RS or RA. If such clonotypes are present at low frequencies in CD8 repertoires, they may represent high-frequency non-specific clonotypes that have not been overgrown by specific cells. However there are examples of non-RS/RA clonotypes in the overlap repertoire. For example, in UPN9 we observe three overlapping clonotypes that do not use these sequences in the CDR3 (sTYs.21, sMVSLd.1126, and sSHGTLy.62434). For UPN13 there are a number of examples of possible alternative motifs VSF and FSV (underlined).
Taken together, these data indicate that the recall repertoire to the M158–66 epitope in HLA-A2 individuals is polyclonal whether the analysis is only based on the growth/survival of CD8 cells in the culture, the presence of TCR with sufficient avidity to bind peptide loaded HLA-A2 tetramers, or on the expression of a marker of degranulation.
DISCUSSION
Recall responses have long been used as a measure of memory T cells in man. The underlying assumption is that in a short term culture only memory cells are being mobilized. The precursor frequency of T cells against nominal antigens is very low in naïve cells and these need special conditions to expand in vitro (18). We use two week cultures to optimize response and to minimize in vitro deviations. Longer culture times show decreased survival of CD8 T cells. For HLA-A2 responses to the conserved M158–66 epitope, the BV19 RS clonotypes are not observed in recall cultures from newborn PBMC (4), indicating a requirement for pathogen exposure to recruit these clonotypes into the memory pool.
We have previously shown that the recall repertoire is polyclonal and that the distribution of the clonotypes is complex, with a portion that can be described as power law-like, and often a second component of higher frequency (6). The clonotype distribution pattern can remain similar but the actual clonotypes change in recall cultures where the concentration of the stimulating peptide change (7) or with the passage of time (16). We examined the relation between the relative frequency of clonotypes in recall cultures and in the PBMC used for the cultures by clonotype specific probe hybridization. We observed that in general the clonotypes maintain the same relative frequency (3). This indicates that the short term cultures do not overly bias the distribution measures. We are currently working on defining in more detail the extent of bias introduced in recall cultures.
The relation between the frequency of a clonotype in a recall culture and it avidity and function is unclear. For example one could hypothesize that there is a direct relationship between a clonotype’s frequency in recall culture and its avidity for the MHC:peptide and this is related to its ability to perform effector functions. Under such a hypothesis only the high frequency clonotypes of the power law component or of the second component may actually constitute functional clonotypes. The low frequency component would consist of clonotypes that can survive in culture or replicate at low levels and which do not represent “genuine” effectors.
Rapid degranulation has been found to be observed with either effector or central memory even though the levels of immediate cytotoxicity may differ between these two populations (19). Therefore, degranulation in response to antigen exposure can be considered a proxy for effector function. Using the degranulation marker, CD107, we show that CD107 selected repertoires are polyclonal and have many of the characteristics associated with our previous analysis of these repertoires. This includes their complex clonotype distribution that is best described as power law-like.
In analyzing T cell lines, others (1–4) and we (6, 7) have noted that not all clonotypes responding to M158–66 encode whose CDR3 is 11 amino acids in length the RS sequence or less frequently the RA at positions 5 and 6. Until now however, all the T cell clones that have been generated and shown to be cytotoxic have used this CDR3 (1, 2). It is of note that the two M158–66 specific cytotoxic clones generated in our study did not encode the RS sequence in the CDR3 (sMVs.11B19B23L11 for C1 and iAITGe.2341B19B27L11 for R2). Furthermore, there were examples of high frequency clonotypes in the CD107 repertoires that did not encode RS. Thus, while the use of RS is a general rule, exceptions do exist.
Our results have some practical implication for the study of human memory repertoires. Such studies will often include longitudinal analyses, using an individual as their own control. An example would be repertoire analysis prior to and after vaccination or pathogen exposure. To compensate for individual differences such studies will also require examination of populations. Both these approaches will often require a large number of analyses. Our data would indicate that for strong recall responses, such as the one studied here, examining the CD8 cells that are expected to be responding is sufficient to generate a clear description of the functional repertoire. It has also been shown that tetramer selected repertoires and IFN-γ repertories are very similar (20). Thus, not having to include MHC-multimer selection or activation and selection for CD107 can increase throughput and decrease costs. These implications may not extend to other repertoires where the number of clonotypes is smaller or for which the standard recall culture conditions do not provide the correct growth conditions for the cells.
The response under study here may be a special case in which multiple exposures to a conserved viral epitope have greatly expanded the repertoire during childhood, the time of extensive thymic output. This would result in an unusually large repertoire that allows a reasonable sampling under recall culture conditions that are far from perfect. Nevertheless, with respect to the HLAA2 restricted M158–66 response, our data clearly show that the observations made previously; a high degree of polyclonality, a high contribution of RS clonotypes, a long tail of clonotypes observed once, and a predominant use of Jβ 2.7, are true for cells that have degranulated in response to activation by the peptide. We propose that this complexity is the result of multiple rounds of exposures that accumulate over the individual’s childhood. With each exposure memory cells are reactivated but newer memory may also be generated. This results in a functional memory repertoire that is polyclonal and complex. During middle age such complex memory repertoires are still available for responses to influenza exposure. Aging changes these clonotype distributions in these memory repertoires (21). However, to determine if aging changes the functional properties of the M158–66- specific repertoire will require that samples from older subjects to be analyzed in a manner similar to that performed here.
Supplementary Material
Supplemental Figure 1. Three independent clonotype analyses from aliquots of the same recall culture. Clonotype names and the number of observations (colonies) corresponding to each overlapping clonotypes are shown. The triplicate analyses are identified. The header at the top shows the number of observations, M, and the number of clonotype, N, for each of the analyses. The clonotypes that were observed only in one of the analyses are counted at the bottom.
Supplemental Figure 2. Clonotype information for the analyses of the four subjects. Clonotype names are provided in the left column. The number of observations of each clonotype in each of the selection protocols is shown below the column identifying the selection protocol used. For UPN13 and UPN26 cultures were split and clonotypes selected on the basis of 1) CD8 & CD107 expression, or 2) CD8 only CD8 expression. For UPN9 and UPN114, the cultures were split into three and an additional selection based on CD8 expression and binding of HLA-A2 pentamers loaded with the M158–66 peptide. For each UPN the fourth column identifies whether the clonotype used the RS (1) or RA (2) CDR3 amino acid motifs. The fifth column labeled “Total” is the sum for each clonotype. Clonotype measures for each mode of selection are presented as header material. This can be used to calculate repertoire characteristics such as; M/N, Nov/N, etc. Clonotype names identified by colored boxes represent public clonotypes (clonotypes observed in more than one subject). A clonotype name with an asterisk indicates that the RS sequence was generated by the proposed long P-nucleotide addition mechanism. The number of public, P-nucleotide, and public p-nucleotide clonotypes is shown below the clonotype count data.
Acknowledgements
This work was funded by the National Institutes of Health Grant U19 AI062627. The contents of this publication are solely the responsibility of the authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Moss PA, Moots RJ, Rosenberg WM, Rowland-Jones SJ, Bodmer HC, McMichael AJ, Bell JI. Extensive conservation of and β-chains of the human T cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc. Natl. Acad. Sci. USA. 1991;88:8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the Vβ17 gene segment. J. Exp. Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection: implications for establishing memory T cell repertoires in"real-world" conditions. J. Immunol. 1998;160:2842–2852. [PubMed] [Google Scholar]
- 4.Lawson TM, Man S, Williams S, Boon AC, Zambon M, Borysiewicz LK. Influenza A antigen exposure selects dominant Vβ17+ TCR in human CD8+ cytotoxic T cell responses. Int. Immunol. 2001;13:1373–1381. doi: 10.1093/intimm/13.11.1373. [DOI] [PubMed] [Google Scholar]
- 5.Dunbar PR, Ogg GS, Chen J, Rust N, van der Bruggen P, Cerundolo V. Direct isolation, phenotyping and cloning of low-frequency antigen-specific cytotoxic T lymphocytes from peripheral blood. Current Biology. 1998;8:413–416. doi: 10.1016/s0960-9822(98)70161-7. [DOI] [PubMed] [Google Scholar]
- 6.Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A fractal clonotype distribution in the CD8+memory T cell repertoire could optimize potential for immune responses. J. Immunol. 2003;170:3994–4001. doi: 10.4049/jimmunol.170.8.3994. [DOI] [PubMed] [Google Scholar]
- 7.Pewe LL, Netland JM, Heard SB, Perlman S. Very diverse CD8 T cell clonotypic responses after virus infections. J Immunol. 2004;172:3151–3156. doi: 10.4049/jimmunol.172.5.3151. [DOI] [PubMed] [Google Scholar]
- 8.Kedzierska K, Day EB, Pi J, Heard SB, Doherty PC, Turner SJ, Perlman S. Quantification of repertoire diversity of influenza-specific epitopes with predominant public or private TCR usage. J Immunol. 2006;177:6705–6712. doi: 10.4049/jimmunol.177.10.6705. [DOI] [PubMed] [Google Scholar]
- 9.Connelley TK, MacHugh ND, Pelle R, Weir W, Morrison WI. Escape from CD8+ T cell response by natural variants of an immunodominant epitope from Theileria parva is predominantly due to loss of TCR recognition. J Immunol. 2011;187:5910–5920. doi: 10.4049/jimmunol.1102009. [DOI] [PubMed] [Google Scholar]
- 10.Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J. Immunol. 2006;177:2006–2014. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- 11.Williams MA, Bevan MJ. Effector and memory T cell differentiation. Ann Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 12.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. JIM. J. Immunol Methods. 2003;28:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 13.Betts MR, Koup RA. Detection of T-cell degranulation: CD107a and b Methods. Cell Biol. 2004;75:497–512. doi: 10.1016/s0091-679x(04)75020-7. [DOI] [PubMed] [Google Scholar]
- 14.Yassai MB, Naumov YN, Naumova EN, Gorski J. A clonotype nomenclature for T cell receptors. Immunogenetics. 2009;61:493–502. doi: 10.1007/s00251-009-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunner KT, Mauel J, Cerottini JC, Chapuis B. Quantitative assay of the lytic action of immune lymphoid cells on 51Cr labelled allogenic target cells in vitro: Inhibition of isoantibody by drugs. Immunology. 1968;14:181–186. [PMC free article] [PubMed] [Google Scholar]
- 16.Naumova EN, Gorski J, Naumov YN. Two Compensatory Pathways Maintain Long-Term Stability and Diversity in CD8 T Cell Memory Repertoires. J. Immunol. 2009;183:2851–2858. doi: 10.4049/jimmunol.0900162. [DOI] [PubMed] [Google Scholar]
- 17.Yassai M, Bosenko D, Unruh M, Zacharias G, Reed E, Demos W, Ferrante A, Gorski J. Naive T Cell Repertoire Skewing in HLA-A2 Individuals by a Specialized Rearrangement Mechanism Results in Public Memory Clonotypes. J. Immunol. 2011;186:2970–2977. doi: 10.4049/jimmunol.1002764. [DOI] [PubMed] [Google Scholar]
- 18.Geiger R, Duhen T, Lanzavecchia A, Sallusto F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J Expt. Med. 2009;206:71525–71534. doi: 10.1084/jem.20090504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy M, Eirikis E, Davis C, Davis HM, Prabhakar U. Comparative analysis of lymphocyte activation marker expression and cytokine secretion profile in stimulated human peripheral blood mononuclear cell cultures: an in vitro model to monitor cellular immune function. J. Immunol. Methods. 2004;293:127–142. doi: 10.1016/j.jim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Cohen GB, Islam SA, Noble MS, Lau C, Brander C, Altfeld MA, Rosenberg ES, Schmitz JE, Cameron TO, Kalams SA. Clonotype tracking of TCR repertoires during chronic virus infections. Virology. 2002;304:474–484. doi: 10.1006/viro.2002.1743. [DOI] [PubMed] [Google Scholar]
- 21.Naumov YN, Naumova EN, Yassai MB, Gorski J. Selective T cell expansion during aging of CD8 memory repertoires to influenza revealed by modeling. J Immunol. 2011;186:6617–6624. doi: 10.4049/jimmunol.1100091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Three independent clonotype analyses from aliquots of the same recall culture. Clonotype names and the number of observations (colonies) corresponding to each overlapping clonotypes are shown. The triplicate analyses are identified. The header at the top shows the number of observations, M, and the number of clonotype, N, for each of the analyses. The clonotypes that were observed only in one of the analyses are counted at the bottom.
Supplemental Figure 2. Clonotype information for the analyses of the four subjects. Clonotype names are provided in the left column. The number of observations of each clonotype in each of the selection protocols is shown below the column identifying the selection protocol used. For UPN13 and UPN26 cultures were split and clonotypes selected on the basis of 1) CD8 & CD107 expression, or 2) CD8 only CD8 expression. For UPN9 and UPN114, the cultures were split into three and an additional selection based on CD8 expression and binding of HLA-A2 pentamers loaded with the M158–66 peptide. For each UPN the fourth column identifies whether the clonotype used the RS (1) or RA (2) CDR3 amino acid motifs. The fifth column labeled “Total” is the sum for each clonotype. Clonotype measures for each mode of selection are presented as header material. This can be used to calculate repertoire characteristics such as; M/N, Nov/N, etc. Clonotype names identified by colored boxes represent public clonotypes (clonotypes observed in more than one subject). A clonotype name with an asterisk indicates that the RS sequence was generated by the proposed long P-nucleotide addition mechanism. The number of public, P-nucleotide, and public p-nucleotide clonotypes is shown below the clonotype count data.