Abstract
Alterations in cilia formation or function underlie a growing class of pleiotropic disorders termed ciliopathies. The genetic basis of ciliopathies is remarkably complex with an incomplete but expanding list of more than 89 loci implicated in various disorders. Current treatment of ciliopathies is limited to symptomatic therapy. However, our growing understanding of ciliopathy genetics coupled with recent advances in gene delivery and endogenous gene and transcript repair, demonstrated thus far in tissues of the eye, nose, and airway, offers hope for curative measures in the near future. This review highlights these advances as well as the challenges that remain with the development of personalized medicine for treating a very complex spectrum of disease, penetrant in a variety of organ systems.
Keywords: Cilia, viral vectors, genetic disorder, gene therapy, ciliopathies, treatment
Ciliopathies
Cilia are evolutionarily conserved organelles found on the surface of most vertebrate cell types. Motile cilia of the node, airway, and ventricles facilitate fluid movement while the sperm flagellum enables propulsion. In other cell types throughout the body immotile cilia are utilized for sensation of external stimuli such as odors, light, fluid flow, and/or a range of growth factors[1-3]. The emergence of a class of human diseases, termed ciliopathies, underscores the importance of cilia in mammalian development, sensory perception, and homeostasis (Fig 1). Ciliopathies are a diverse class of congenital diseases, with 23 recognized syndromes caused by mutations in at least 89 different genes [4-6]. These diseases are highly pleiotropic and the degree of penetrance within a given tissue can vary between patients. Clinical manifestations of ciliopathies can arise in nearly all tissue types during development and throughout life. Sensory impairments include the presence or onset of blindness, neurosensory hearing loss, altered nociception and anosmia. In addition, organ defects such as renal and liver cyst formation, airway distress, and hydrocephaly occur (for full review of ciliopathy genes and phenotypes, see [5]). Although current research is rapidly uncovering new causative mutations in genes encoding cilia related proteins and elucidating the molecular and functional basis of ciliopathies, there are currently limited therapeutic measures and no curative options for patients. With our expanding understanding of cilia biology and gene mutations, research attention must now begin to focus this basic knowledge on developing personalized therapeutic and curative solutions. For many patients health care options are restricted to symptom management and general lifestyle recommendations. As the disease progresses, effective treatment may require extreme measures such as whole organ transplant. Looking forward, viable therapeutic options that will reduce disease progression, provide palliative care, and/or offer a potential cure will likely require both pharmacology and biologics such as stem cells or gene therapy for maximum efficacy.
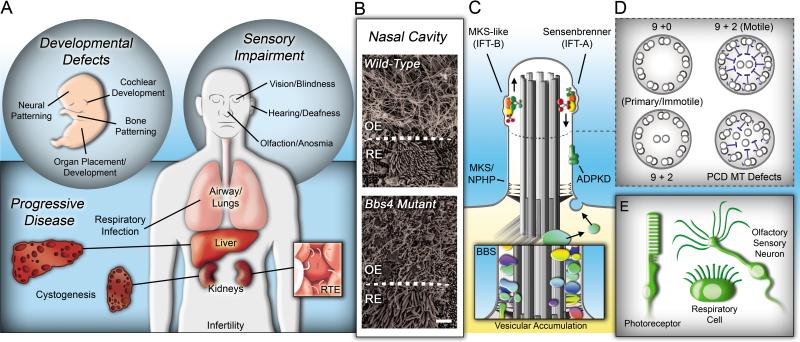
Figure 1. Gene therapy has been successful in several ciliated cell types.
(A) Ciliopathies encompass phenotypes in multiple organ systems that can alter development, impair sensory systems, and lead to tissue degeneration. RTE, renal tubule epithelium. (B) Ciliopathies show pleiotropic penetrance. In the nasal cavity, both olfactory sensory neurons and respiratory epithelium cells possess cilia. Scanning electron micrographs of the OE/RE border show a loss of olfactory cilia, but not respiratory cilia in Bbs4 null mice (Mice provided by Dr. Val Sheffield, University of Iowa). OE, olfactory epithelium; RE, respiratory epithelium; scale bar = 2.5μm. (C) Macrociliary complexes, including IFT, ADPKD, MKS, NPHP and BBS proteins regulate cilia development, maintenance and/or function. Mutations in genes encoding proteins involved in distinct complexes often share similarities in phenotypes. Inset depicts vesicular accumulation along the ciliary axoneme in Bbs4 mutant mice [79]. (D) Primary and motile cilia posses distinct axonemal microtubule structures. Disorganization of these structures can cause PCD, through defects in ciliary motility. MT, microtubule. (E) To date, cilium-focused gene therapy strategies have been successful in several cell types, including retinal photoreceptors (in both human and mouse models), respiratory epithelial cells (ex vivo), and olfactory sensory neurons (in mouse models).
Although to date there are very few pharmacological options that provide significant relief of ciliopathy phenotypes, aggressive drug research and clinical trials for symptomatic treatment of polycystic kidney disease (PKD) is ongoing [7, 8]. These trials mainly monitor kidney and liver volume changes expected from reduced cell proliferation and lowered cellular fluid secretion through inhibition of cyclic adenosine monophosphate (cAMP) and mammalian target of rapamycin (mTOR), two factors heavily implicated in pathogenesis downstream of cilia dysfunction. Several drugs have shown promise in animal testing [9-12]; however reports from early clinical trials have produced negative results or modestly beneficial outcomes accompanied by various side effects [8, 13]. While hope remains that ongoing trials will uncover highly effective treatments for PKD, overall utility of the drugs currently in testing are limited mainly to kidney and liver function as the they target specific pathways in cyst development, providing little to no efficacy in other organs affected in pleiotropic ciliopathies. Alternative drug treatments are also being explored, albeit to a lesser extent, for extra-renal/biliary problems such as the onset of blindness. To that end, recent research using a murine Bardet-Biedl syndrome (BBS) model has demonstrated that retinal degeneration as a result of cilia dysfunction can be delayed by pharmacological intervention [14]. In Bbs12 knockout mice, activation of an unfolded protein response (UPR) triggered by protein accumulation in the endoplasmic reticulum of photoreceptors was shown to promote apoptosis and retinal degeneration. Administration of a combination of drugs targeted against UPR-mediated effectors of apoptosis was able to slow photoreceptor death and increase electroretinography responses to light in mutant animals.
While pharmacology offers the hope of delaying disease progression in some tissues, this symptomatic treatment approach lacks the curative potential inherent in gene therapy or stem cell treatments. These latter options represent flexible, personalizable options that have the potential to be effectively targeted with high precision to specific organs for long term and perhaps even permanent remediation of genetic defects on a cellular level. The ongoing identification of ciliopathy genes and mutations provides a critical starting point for developing personalized genetic therapies for patients. In principle, pharmacological intervention could be used to help delay or prevent cellular loss and tissue degeneration in an individual while their personalized corrective gene therapy is developed. In this review we will discuss the current gene therapies that have been applied to ciliopathy disorders and address challenges that must be faced in order to make gene therapy approaches a viable therapeutic option for the complex pleiotropy of ciliopathies.
Recent advances in ciliopathy gene therapy
With the recent revival of gene therapy research, promising results from both ex vivo and in vivo studies using several different strategies have begun to emerge in the field of ciliopathy treatment [6, 15-17] (Fig 2). These reports build upon early successes demonstrated in clinical trials of gene therapy treatment of non-ciliary retinal diseases [18-20] and answer several fundamental questions concerning the primary challenges inherent in restoring function to a genetically and functionally diverse organelle. Together, the gene delivery methods utilized in these reports and described below have demonstrated the restoration of cilia formation or function in multiple cell types. These early studies provide evidence supporting the hypothesis that gene delivery is a viable means of correcting ciliopathy phenotypes in patients.
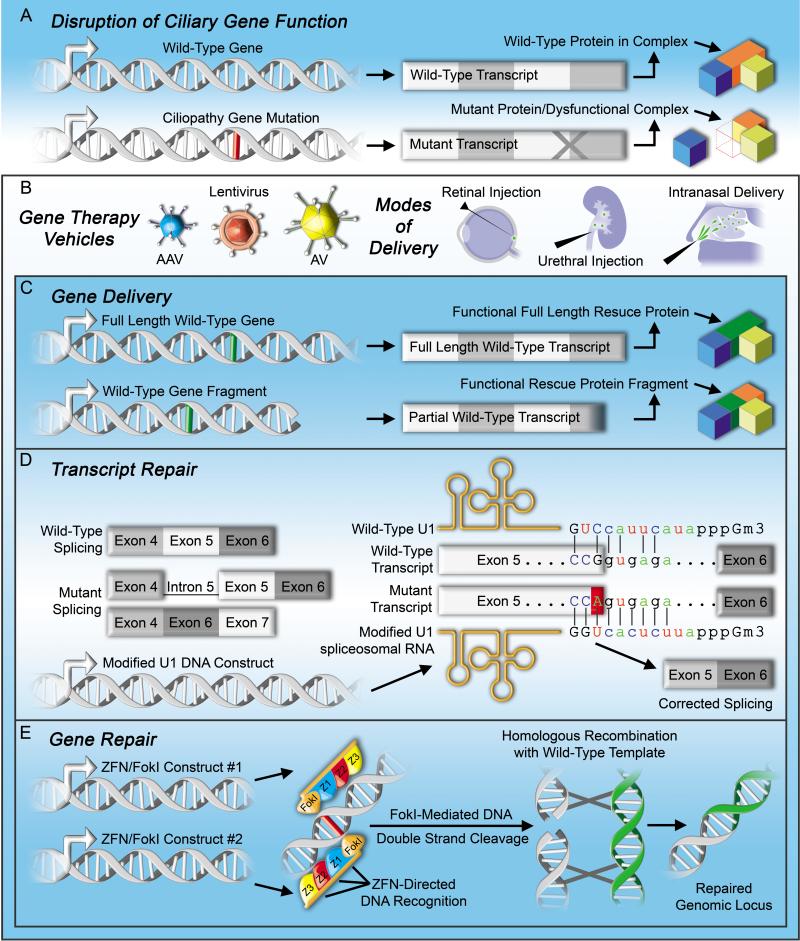
Figure 2. Strategies for rescuing mutant genes.
(A) Ciliopathy mutations can lead to altered gene expression or protein function through insertions/deletions or point mutations that disrupt mRNA splicing, cause premature termination, or change protein coding. (Bottom, right) Representation of a ciliopathy gene mutation causing protein truncation and disruption of a ciliary functional unit. (B) (Right) To date, delivery of genes in ciliopathy models has been mediated by adeno-associated virus (AAV), lentivirus, and adenovirus (AV). (Left) Delivery methods of gene therapies to the visual and olfactory systems (via retinal injection and intranasal delivery, respectively) are minimally invasive. Urethral injection of the kidney represents a feasible delivery method for treating ciliopathies affecting internal organs. (C) Viral vectors can be used to express (top) whole genes in targeted cells. (Bottom) In the event of a full gene surpassing the size limitations imposed by viral packaging, a partial gene fragment could potentially be used to specifically replace the missing portion of a truncated protein. (D) Mutations that occur in splice donor sites can prevent complex formation of the splice factor U1 snRNP with pre-mRNAs and result in (left) intronic read-through or exon skipping. Viral expression of a modified U1 snRNA, to increase its complementarity to the mutated splice site, can correct misspliced transcripts allowing expression of the full-length gene. Sequences shown correspond to the splice donor site of BBS1 exon/intron 5 affected by the c.479G-A mutation and the modified U1 snRNP virally expressed to restore splicing as reported in [41]. (E) Finally, targeted ciliopathy gene repair/genome editing machineries have been successfully delivered by viral mechanisms. Zinc-finger nucleases (ZFNs), encoded by virally delivered minigenes and fused to FokI nuclease, target genomic DNA surrounding a ciliopathy mutation and induce a double-stranded DNA break. Subsequent homologous recombination with a correct version of the sequence (presented via gene delivery) results in a repaired genomic locus.
Primary Ciliary Dyskinesia (PCD) arises as a consequence of compromised ciliary motility. PCD mutations can cause infertility in males due to diminished sperm flagellar propulsion and may result in situs inversus due to ciliary beating defects in the embryonic node. In addition, as a result of ciliary immotility in the respiratory tract, pulmonary infections arise that even with antibiotic treatment, eventually destroy the lungs. Many mutations causing PCD are found within genes encoding structural components of the dynein arm/radial spoke mechanism that is employed to generate ciliary beating [21]. Recently, in ex vivo cultures of respiratory epithelial cells harvested from PCD patients with DNAI1 mutations, a healthy copy of DNAI1 introduced via lentiviral transduction was sufficient to restore ciliary beating [15]. Importantly, this result demonstrates that structural/mechanical components can be introduced by gene therapy techniques to repair broken machinery in existing cilia to restore motility.
Differentiated cells that lack cilia due to a congenital mutation can be induced to build or restore cilia structures de novo. In the Oak Ridge Polycystic Kidney (ORPK) mouse in which cilia formation is compromised due to hypomorphic expression of the ciliogenic gene Ift88 [22], olfactory sensory neurons (OSNs) in the nasal cavity of mutant animals were unable to build or maintain cilia [6]. This resulted in anosmia as seen in several classes of ciliopathies [23-25]. Restoration of Ift88 expression via adenoviral (AV) gene delivery was sufficient to produce functional olfactory cilia; these new OSN cilia enabled electro-olfactogram responses to odorants and gene-treated animals showed a significant weight gain, indicating restoration of odor-guided suckling behavior. This study demonstrated for the first time a gene therapeutic approach to produce cilia on differentiated cells in vivo. Importantly, this study provides proof-of-concept that restoration of cilia, which are lost on many cell types in various human ciliopathies, is possible.
In some ciliopathies, cilia structures appear unaffected while the trafficking of specific proteins necessary for proper ciliary signaling is ablated [26]. Mutations in BBS genes can cause some signaling proteins to aberrantly localize or accumulate within cilia and flagella [27, 28]. Conversely, BBS mutations can also result in disrupted ciliary targeting of specific signaling proteins such as G protein-coupled receptors (somatostatin receptor 3 and opsins) [26, 29, 30]. As is the case when opsin function is disrupted [31], BBS gene mutations affecting opsin trafficking in the retinal photoreceptor outer-segment cause blindness and eventual photoreceptor death/retinal degeneration [16, 32, 33]. In a recent study using adeno-associated virus (AAV), BBS4 was introduced into Bbs4-/- mice via retinal injection prior to the onset of retinal degeneration [16]. This viral delivery was sufficient in restoring rhodopsin localization in photoreceptors, and more importantly, the treatment prevented photoreceptor death and improved both retinal electrophysiological responses and visually guided behaviors in treated animals. In addition, a recent report demonstrated successful gene therapy in a mouse model of Usher syndrome type 2d, a disease that features retinal degeneration and deafness [17]. Injection of AAV containing the Usher syndrome gene Dfnb31 into the retina of whirlin-/- mice restored a complex of Usher proteins (USH2A and VLGR1) to the pericillary membrane of photoreceptor cells, prevented cell loss, and enabled photoreceptor function [17]. Notably, these effects were still evident 6 months after injection.
Together, these studies all demonstrate that the introduction of wild-type genes through viral packaging and delivery can correct cilia dysfunction. They also show proof-of-concept for the treatment of multiple aspects of cilia disruption such as immotility, defective ciliogenesis/maintenance and altered protein trafficking.
Challenges and potential solutions for gene therapy in treatment of ciliopathies
Several barriers to developing effective gene therapies exist, any of which if not fully considered could derail an otherwise well-planned gene delivery strategy. The development of gene therapies will rely on successful integration of delivery vectors and repair strategies (Fig. 2). Some challenges apply to limitations of gene therapy in a broad sense while others pertain to concerns directly relating to gene therapy in ciliopathies. Common gene therapy difficulties include overcoming DNA insert length limitations, avoiding unwanted ectopic gene expression in off target cell types, and accounting for potential unintended consequences of gene overexpression. In addition the development of tissue specific delivery strategies, addressing obstacles posed by cell-type specific protein isoform expression, and combating tissue degeneration must also be addressed
While the aforementioned studies show success with delivering entire genes, expression of DNAs beyond a certain length is not possible with currently available viral delivery tools. Thus, insert size limitations present a significant challenge to the effective treatment of many ciliopathies. AAVs are an attractive vehicle due to their stable incorporation and low immunogenicity. However, AAVs are limited to DNA inserts of approximately 5.5 Kb. In contrast, while most AVs are capable of packaging up to 8 Kb of DNA, expression of genes delivered via AVs is transient in nature, limiting long-term efficacy. These size limitations preclude the packaging of at least 14% of the currently known ciliopathy genes. This list expands when space needed for cell type-specific promoter elements are factored into the size of the DNA requiring delivery. Of note, among the larger ciliopathy genes are PKD1 and DNAH5, which are the main contributors to ADPKD (~85%) and PCD (~50%), respectively [34-36]. This is particularly disconcerting in the case of ADPKD, which is by far the most common ciliopathy in the human population (affecting 1 in 400-1000 individuals) and thus represents a target for treating a significant number of patients. In addition, two other PCD genes and more than half the known Usher syndrome genes surpass the AV and AAV size limitations. This is unfortunate since the major pathologies of these two diseases are primarily restricted to the airways, eyes, and/or ears, all three of which are easily accessible and eminently treatable via gene therapy.
A promising means of bypassing size barriers was recently shown in zebrafish in which the function of the large CEP290 gene was inhibited by morpholino knockdown [37]. Similar to a relatively common human CEP290 partial deletion allele, this mutation resulted in the expression of truncated CEP290. Introduction of an N-terminal fragment of human CEP290 resulted in improved responses to visual stimulation in morphant animals. These experiments indicate that expression of a portion of a gene may be sufficient to overcome functional defects of the corresponding endogenous mutant protein. While this strategy may prove useful in some cases, a large body of research would be necessary to determine which gene fragments are capable of overcoming specific mutations in each given ciliopathy protein.
Lower organisms offer a means to rapidly assess consequences of specific gene deliveries prior to analysis in animal models or treatment in patients. In addition to examining whether gene fragments can restore function, important assessments in lower organisms could include analysis of human gene mutant pathogenicity and testing for potential deleterious effects of overexpressing ciliopathy genes. For example, an analysis of human gene point mutations in zebrafish BBS morphants surprisingly uncovered BBS gene variants that manifested dominant negative phenotypes_[38], suggesting that delivery of the wild-type gene in patients harboring these mutations may result in minimal or no therapeutic outcome, or worse, exacerbation of disease. Similar approaches using transgenic expression have been used in C. elegans to categorize the pathogenic potential of Nephronophthisis (NPHP) gene mutations [39]. Analysis of lower organisms has also provided information on how mutations in multiple genes can lead to cilia defects [40]. These experiments, along with genetic analysis of patients, raise the possibility that mutations in multiple ciliopathy genes may together influence the overall effectiveness of gene therapy strategies (Text box 1). Using model organisms however could provide quick insight into strategies that may be effective first in animals and then patients. For genes and mutations in which ectopic expression of wildtype copies is not a viable option, different treatment strategies will need to be devised and implemented.
Text Box #1: Epistatic interactions of ciliopathy gene mutations and implications on treatment.
Through the genetic analysis of ciliopathy disorders numerous genes regulating cilia function have been identified. This analysis has lead to the emergence of several distinct macromolecular ciliary complexes such as the ADPKD TRP channel complex [68], the IFT particles [69, 70], the BBSsome [29, 71], and the MKS and NPHP modules, [40, 72, 73] underling compartmentalized functions of certain ciliopathy proteins and highlighting the potential interdependence of the constituent proteins within the complex. It has also become apparent that in many cases, mutations in more than one gene underlie the presenting disease in an individual [74]. The identification of these epistatic interactions has promoted the idea of ‘mutational load’ in ciliopathy patients; mutations in multiple ciliary proteins often lead to more severe phenotypes. These interactions can impact the effectiveness of treatment, and thus should be a focus of future research. Interestingly, genetic suppression through mutations affecting two different ciliopathy gene products resulting in the reversal of some phenotypes has also been reported [62]. The rd16 mouse strain, harboring a mutation in the MKS/NPHP complex gene Cep290, was bred to the Mkks knockout mouse strain in which BBS6 (a component of the BBS6/10/12 secondary BBS complex) is disrupted to produce compound mutant animals. Remarkably, some combinations of these mutations were sufficient for remediation of ciliogenesis and sensory defects observed in either Cep290rd16/rd16 or Mkksko/ko single mutants. This analysis provides support to the notion that the ‘mutational load’ in an individual may not necessarily correlate with more pronounced disease penetrance. These findings also underscore the possibility that, opposed to targeting specific gene mutations, therapies may be developed that can generally modify the function of ciliary modules to produce a positive outcome. Further understanding the specific function and potential modular association of each ciliopathy protein will be necessary to verify the feasibility of this approach.
Approaches aimed at directly manipulating the endogenous gene mutation offer solutions to multiple problems such as gene size, cell type specificity, multiple isoforms and overexpression. To this end, gene therapies designed to target DNA splicing and gene repair are promising alternatives to full-length gene delivery. These methods attempt to repair the endogenous gene so that its expression is maintained under its own promoter. Given that many genes encode multiple isoforms these approaches would help conserve the processing of cell-type specific isoforms and maintain normal expression levels of the effected gene. While many ciliopathies are caused by mutations within coding regions of DNA, leading to point mutations, truncations, or frame shifts [41], mutations that occur outside of coding regions also contribute to disease phenotypes. For example, hypomorphic expression of Ift88 seen in ORPK mice is due to an insertion of a transgene in the intron region of Ift88 [22]. The expansion of genome sequencing outside of coding regions in patients will aid in determining the extent by which noncoding mutations contribute to ciliopathies and is necessary precursor to developing therapeutic strategies for these defects. Mutations in splice donor/acceptor sites can prevent the efficient binding of the splicing machinery and lead to read-through into introns or exon skipping, thus altering gene expression [42]. These types of mutations have been found to cause several ciliopathy disorders [42-46]. Within the last several years, it has been shown that specifically-engineered U1 small nuclear RNA (U1) isoforms can be generated to recognize and interact with mutant splice donor sites in order to enable proper splicing of the affected transcript [47]. This technology was applied to the ciliopathy gene BBS1 in which at least 7 different mutations are known to occur at splice donor sites causing splicing defects [42]. Via expression of specific U1 isoforms in patient derived fibroblasts, correction of aberrant splicing was observed with four of these mutations[42]. A combined treatment expressing both modified U1 and modified U6 small nuclear RNA (U6) improved endogenous BBS1 expression to varying degrees in the remaining 3 mutant cell lines [42, 48]. Thus, in ex vivo systems splice modulation is effective at correcting some ciliopathy mutations. Whether this strategy will show efficacy in vivo remains to be tested.
Gene therapy through repair of genomic DNA is a second potential avenue for correcting endogenous ciliopathy mutations while circumventing issues such as size limitations and overexpression concerns inherent in exogenous gene delivery systems. The ability to repair DNA mutations has been demonstrated using Zinc-finger nucleases (ZFN), enzymes that can be engineered to induce double strand breaks at specific sites in genomic DNA. Through the inclusion of a corrective DNA template, the induced break is then repaired by homologous recombination to correct the mutation [49]. The use of ZFNs has been successful in correcting defects found in the Usher syndrome gene, USH1C [50]. In cultured cells derived from patients with a nonsense truncating mutation in USH1C, expression of modified ZFNs was sufficient to repair the mutation and restore endogenous full length USH1c protein. Again, efficacy of this approach treating ciliopathies has not yet been verified in vivo.
A general challenge of gene therapy is the delivery of viral particles to only an organ of interest to achieve tissue specific expression. Some tissues are easily accessible for non-systemic virus delivery, such as the respiratory tract and sensory systems such as the retina and olfactory epithelium (OE). To date, all studies of gene therapy for ciliopathies in vivo have been performed in either the retina or olfactory epithelium; a much greater challenge is effective gene delivery to an internal organ such as the kidney, which is one of the tissues most broadly affected across the ciliopathy spectrum. To this end, AAVs can be delivered to renal tubules in mice using retrograde uretal virus injection [51]. As this technique minimizes infection of other tissues, its implementation or adaptation in studies examining corrective therapies would be a potential starting point for treating kidneys or other internal organs in ciliopathy models.
The existence of heterogeneous cell populations within an organ or tissue further complicates the issue of conferring expression specificity. Gene expression can to an extent be regulated through vehicle choice. While lentivirus, AAV, and AV are able to infect dividing and non-dividing cells each virus type exhibits selectivity in the cell types they infect [52, 53]. In fact, cell type specificity can be conferred by viral serotype; for example, rAAV-2/5 is able to transduce both photoreceptors and retinal-pigmented epithelial (RPE) cells while rAAV-2/4 only transduces RPE cells [53]. Similarly, the nasal cavity possesses two populations of ciliated cells, OSNs and respiratory epithelial cells. Both cell types are exposed to virus upon intranasal gene delivery, but the use of lentivirus has shown preferential transduction of respiratory epithelial cells but not OSNs, although it should be noted that this specificity appears to depend on injection method [52, 54]. In contrast, AV can robustly transduce neurons in the OE and is also capable of infecting cells of the respiratory tract [55].
Utilization of tissue specific promoter elements currently offers a promising means of controlling tissue-specific gene therapeutic expression. For example, within the OE, the olfactory marker protein (OMP) is strongly and specifically expressed in mature OSNs [56, 57]. A functional OMP promoter element could therefore potentially confer expression in only this cell type. In the retina, promoters including rhodopsin and rhodopsin kinase have been successfully used in several studies for specific expression of genes delivered to photoreceptor cells [19, 20, 58-60]. However, in regards to most tissue types (and by extension most cell types) affected in ciliopathies, an appropriate promoter for driving gene therapeutic expression with high specificity has not been developed. Progress toward this end will require overcoming current size restrictions of our delivery vehicles and/or defining compact minimal promoter elements for each cell type and optimizing their expression.
A common phenotype in ciliopathies is the eventual degeneration of affected tissues. This is best characterized in the retina where photoreceptor cilia dysfunction leads to progressive tissue loss. In PCD patients, loss of motile cilia function in the airway results in bronchiectasis and chronic infections which contribute to lung destruction over time [21], and in various ciliopathies, the kidney and liver exhibit unabated epithelial cell proliferation that manifests cyst formation. Clearly, degeneration of the target tissue is a complicating challenge facing successful gene therapy treatment of ciliopathies. Initiating treatment in tissues prior to the onset of degeneration naturally offers the best hope of delaying or preventing tissue deterioration. However, in many cases (particularly with concern to the kidney), tissue defects begin to emerge prior to birth. This problem underscores the need to diagnose patients and develop personalized treatment as early as possible [61]. Additionally, because the retina lacks regenerative capabilities, there is a limited time frame in which photoreceptors can be treated before onset of cell death occurs. This necessitates the development of treatments that convey permanent gene expression changes or treatments that can be reapplied over time with relative ease.
In contrast to the retina, kidney and other tissues, in the olfactory system, ciliopathy gene mutations often reduce or ablate olfactory function, but the OE itself does not show tissue degeneration [6, 23, 24]. This has been shown across animal models representing mutations in a diverse array of genes including CEP290, BBS1, BBS4, BBS6, BBS8 and IFT88 [6, 23-25, 62]. One possibility to the difference in tissue degeneration between the retina and the OE is that the loss of sensory signaling does not appear to be significantly detrimental to OSN cell survival [24, 63-66]. In addition, the OE undergoes continuous cellular turnover with OSNs exhibiting natural planned cell death and replacement via proliferation and differentiation of a neuronal stem cell population. Potential differences in lifespan of OSNs with and without cilia have not been formally examined but are also not apparent from the aforementioned studies. Regardless of the mechanism, the limited lifespan of OSNs (typically between 30 and 90 days) makes the stable integration of genetic material into OSNs or optimization of gene therapies for extremely long OSN expression potentially unnecessary. Ectopic expression using AVs persists for a minimum of 4 weeks post-infection [6]. Although OE degeneration is not at issue in ciliopathies, natural tissue turnover will nevertheless cause gene therapy treatments to eventually “wear off” due to the replacement of rescued OSNs with new, mutant cells. This will in fact be an issue in any tissue that undergoes regular turnover, for example, the airway epithelium. Thus, regardless of the stability of integration, therapy would need to be repeatedly delivered to these types of tissues.
Conclusion
While the current state of gene therapy strategies presents challenges and side effects that will require further study (Text box 2), recent advances showing proof-of-concept demonstrate that gene therapy is a worthy approach to treating ciliopathies. However, the overarching hurdle of ciliopathic pleiotropy remains as the single most daunting challenge opposing the development of effective treatment. Not only do ciliopathy gene mutations show penetrance in multiple tissues, the degree of penetrance and severity of phenotypes can vary widely, based on an individual's genetic background as well as factors that are currently not understood [67]. To date, gene therapy has been successful in correcting ciliopathy defects of a single targeted tissue but has not been broadly applied to multiple affected tissues within an organism. It is possible that a therapy that shows promise in one tissue, such as the retina may not be effective in another. This raises the question of value in developing therapies that offer significant palliative care versus a global cure. It is our opinion that the value of these mollifying therapy options is significant. In fact, palliative care options provide a real opportunity to increase the quality of life for ciliopathy patients. Nonetheless, combining therapies or developing globally effective strategies will be key to implementing personalized care that improves both mortality and quality of life. Successful implementation of gene therapeutics in ciliopathies will likely require refining our current tools as well as developing novel strategies that overcome the unique problems posed by the complexity of ciliopathic disorders. Excitement, however, should be derived from proof-of-concept studies in ciliopathy models highlighted here as well as patient trials outside the ciliopathies where gene therapy has shown efficacy. As individualized medicine and gene therapy advances the treatment of ciliopathies may be a reality for diseases currently with limited therapeutic options.
Text box #2. Limitations and challenges to gene therapies.
While reports of successful gene therapy applications are growing, several challenges still exist that must be overcome. In the clinical setting, AAV mediated therapy is attractive due to the relative low immunogenic response to AAV exposure, although some serotypes can elicit immune responses in humans [75]. However as mentioned, AAVs have a limited capacity for DNA insertion. Emerging high capacity AV vectors, that are devoid of most viral genes, can incorporate 30kb of DNA; however, these vectors still illicit unwanted immune responses requiring the use of immune suppressing drugs [75]. As AVs only offer transient expression, reduced efficiency of repeated treatments is also predicted due to generation of neutralizing antibodies in the patient. In addition to potential vector induced immune responses, strategies such as partial gene delivery, modulating splice factors, RNA oligonucleotides and ZFNs have their own limitations that must be addressed. Expression of protein fragments must be thoroughly tested as the fragments could induce an immune response, or over-stimulate degradation pathways leading to cytotoxicity. Methods to alter RNA splicing must improve specificity to limit off-target effects on splicing of other transcripts. The delivery of antisense oligonucleotides to silence pathogenic alleles also presents challenges, such as activation of Toll-like receptors initiating immune responses leading to tissue degeneration [76, 77]. Finally ZFNs can induce cell toxicity through non-specific cleavage of DNA [76], necessitating their further study and development in animal models prior to clinical trials. It should also be noted that the efficiency of recombination with ZFNs is still quite low, and these therapies may be better suited for combination with stem cell therapies [78]. Altogether, optimization of these techniques for both their efficacy and safety in vivo must occur to ensure their use as viable solutions for treating in patients.
Highlights.
Gene therapies have been successfully used in several mammalian ciliopathy models.
Rescued defects include cilia mobility, vision and olfaction
Overcoming challenges presented by pleiotropy remain a goal for effective treatments.
Therapies providing palliative care offer substantial benefits to patients.
Glossary
- Adenovirus (AV)
A naked, dsDNA virus that is capable of infecting both dividing and non-dividing cells. Adenovirus can typically package 7.5kb of DNA, but certain variants can now contain 30kb. Adenovirus-delivered DNA does not incorporate into the genome thus allowing only transient expression.
- Adeno-associated virus (AAV)
A naked ssDNA virus capable of infecting both dividing and non-dividing cells. Due to their small genome, AAVs can only hold about 5Kb of DNA. These viruses can incorporate DNA into chromosome 19 of humans, or exist in an extrachromosomal state, providing long lasting expression.
- Autosomal dominant polycystic kidney disease (ADPKD)
Genetic disorder affecting the kidneys as well as other organs. The disease is caused primarily by mutations in genes encoding polycystin cation channels, PKD1 and PKD2.
- Bardet-Biedl syndrome (BBS)
A highly pleiotropic ciliopathy affecting many tissues resulting in a wide array phenotypes including renal cyst formation and failure, retinitis pigmentosa, obesity, polydactyly, and anosmia.
- Ciliopathy
Genetic disorder affecting the development, maintenance, or function of cilia.
- Intraflagellar Transport (IFT)
Process by which proteins are moved into and out of the cilium mediated by kinesin and dynein motors and accessory proteins.
- Lentivirus
Enveloped retrovirus that is capable of infecting both dividing and non-dividing cells. Lentivirus particles are able to hold approximately 10kb of DNA and incorporate into the host genome providing potentially long lasting expression.
- Mutational load
In ciliopathy patients, this refers to all the mutated alleles that may exist across multiple genes.
- Nephronophthisis (NPHP)
Ciliopathy affecting primarily the kidneys of children and associated with mutations in a group of proteins at the base of the cilium.
- Olfactory sensory neurons (OSNs)
Ciliated neurons responsible for detection of odors in the olfactory epithelium.
- Primary cilia dyskinesia (PCD)
disorder rendering motile cilia immotile, effecting sperm flagella and cilia lining the respiratory tract, fallopian tubes and brain ventricles.
- Unfolded protein response (UPR)
Cellular stress response to the accumulation of unfolded or misfolded proteins in the endoplasmic reticulum.
- Zinc-finger nucleases (ZFN)
Fusion of zinc-finger DNA binding domains with DNA cleavage domains creating artificial restriction enzymes that can be designed to target specific sequences in the genome. These enzymes can be used to induce double stranded DNA breaks. ZFN mediated DNA damage enables insertion of a single nucleotide or large pieces of DNA through homologous recombination with a DNA template.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for As a service to our customers we are providing this early version of the The manuscript will undergo copyediting, typesetting, and review of the resulting before it is published in its final citable form. Please note that during the process errors may be discovered which could affect the content, and all legal that apply to the journal pertain.
Literature cited
- 1.Berbari NF, et al. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda S, Narita K. Structure and function of vertebrate cilia, towards a new taxonomy. Differentiation. 2012;83:S4–11. doi: 10.1016/j.diff.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Chaki M, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150:533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Reeuwijk J, et al. Scrutinizing ciliopathies by unraveling ciliary interaction networks. Hum Mol Genet. 2011;20:R149–157. doi: 10.1093/hmg/ddr354. [DOI] [PubMed] [Google Scholar]
- 6.McIntyre JC, et al. Gene therapy rescues cilia defects and restores olfactory function in a mammalian ciliopathy model. Nat Med. 2012;18:1423–1428. doi: 10.1038/nm.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel V, et al. Advances in the pathogenesis and treatment of polycystic kidney disease. Curr Opin Nephrol Hypertens. 2009;18:99–106. doi: 10.1097/MNH.0b013e3283262ab0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang MY, Ong AC. Mechanism-based therapeutics for autosomal dominant polycystic kidney disease: recent progress and future prospects. Nephron Clin Pract. 2012;120:c25–34. doi: 10.1159/000334166. discussion c35. [DOI] [PubMed] [Google Scholar]
- 9.Leuenroth SJ, et al. Triptolide is a traditional Chinese medicine-derived inhibitor of polycystic kidney disease. Proc Natl Acad Sci U S A. 2007;104:4389–4394. doi: 10.1073/pnas.0700499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masyuk TV, et al. Octreotide inhibits hepatic cystogenesis in a rodent model of polycystic liver disease by reducing cholangiocyte adenosine 3′,5′-cyclic monophosphate. Gastroenterology. 2007;132:1104–1116. doi: 10.1053/j.gastro.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 11.Shillingford JM, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A. 2006;103:5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahl PR, et al. Inhibition of mTOR with sirolimus slows disease progression in Han:SPRD rats with autosomal dominant polycystic kidney disease (ADPKD). Nephrol Dial Transplant. 2006;21:598–604. doi: 10.1093/ndt/gfi181. [DOI] [PubMed] [Google Scholar]
- 13.Torres VE, et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2012;367:2407–2418. doi: 10.1056/NEJMoa1205511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mockel A, et al. Pharmacological modulation of the retinal unfolded protein response in Bardet-Biedl syndrome reduces apoptosis and preserves light detection ability. J Biol Chem. 2012;287:37483–37494. doi: 10.1074/jbc.M112.386821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chhin B, et al. Ciliary beating recovery in deficient human airway epithelial cells after lentivirus ex vivo gene therapy. PLoS Genet. 2009;5:e1000422. doi: 10.1371/journal.pgen.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simons DL, et al. Gene therapy prevents photoreceptor death and preserves retinal function in a Bardet-Biedl syndrome mouse model. Proc Natl Acad Sci U S A. 2011;108:6276–6281. doi: 10.1073/pnas.1019222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou J, et al. Whirlin replacement restores the formation of the USH2 protein complex in whirlin knockout photoreceptors. Invest Ophthalmol Vis Sci. 2011;52:2343–2351. doi: 10.1167/iovs.10-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acland GM, et al. Gene therapy restores vision in a canine model of childhood blindness. Nat Genet. 2001;28:92–95. doi: 10.1038/ng0501-92. [DOI] [PubMed] [Google Scholar]
- 19.Pawlyk BS, et al. Replacement gene therapy with a human RPGRIP1 sequence slows photoreceptor degeneration in a murine model of Leber congenital amaurosis. Hum Gene Ther. 2010;21:993–1004. doi: 10.1089/hum.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawlyk BS, et al. Gene replacement therapy rescues photoreceptor degeneration in a murine model of Leber congenital amaurosis lacking RPGRIP. Invest Ophthalmol Vis Sci. 2005;46:3039–3045. doi: 10.1167/iovs.05-0371. [DOI] [PubMed] [Google Scholar]
- 21.Bush A, Hogg C. Primary ciliary dyskinesia: recent advances in epidemiology, diagnosis, management and relationship with the expanding spectrum of ciliopathy. Expert Rev Respir Med. 2012;6:663–682. doi: 10.1586/ers.12.60. [DOI] [PubMed] [Google Scholar]
- 22.Lehman JM, et al. The Oak Ridge Polycystic Kidney mouse: modeling ciliopathies of mice and men. Dev Dyn. 2008;237:1960–1971. doi: 10.1002/dvdy.21515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulaga HM, et al. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 24.McEwen DP, et al. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc Natl Acad Sci U S A. 2007;104:15917–15922. doi: 10.1073/pnas.0704140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tadenev AL, et al. Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc Natl Acad Sci U S A. 2011;108:10320–10325. doi: 10.1073/pnas.1016531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berbari NF, et al. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domire JS, et al. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell Mol Life Sci. 2011;68:2951–2960. doi: 10.1007/s00018-010-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lechtreck KF, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin H, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura DY, et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci U S A. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphries MM, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- 32.Abd-El-Barr MM, et al. Impaired photoreceptor protein transport and synaptic transmission in a mouse model of Bardet-Biedl syndrome. Vision Res. 2007;47:3394–3407. doi: 10.1016/j.visres.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichers ER, et al. Phenotypic characterization of Bbs4 null mice reveals age-dependent penetrance and variable expressivity. Hum Genet. 2006;120:211–226. doi: 10.1007/s00439-006-0197-y. [DOI] [PubMed] [Google Scholar]
- 34.Peral B, et al. A stable, nonsense mutation associated with a case of infantile onset polycystic kidney disease 1 (PKD1). Hum Mol Genet. 1996;5:539–542. doi: 10.1093/hmg/5.4.539. [DOI] [PubMed] [Google Scholar]
- 35.Peral B, et al. Screening the 3′ region of the polycystic kidney disease 1 (PKD1) gene reveals six novel mutations. Am J Hum Genet. 1996;58:86–96. [PMC free article] [PubMed] [Google Scholar]
- 36.Hornef N, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baye LM, et al. The N-terminal region of centrosomal protein 290 (CEP290) restores vision in a zebrafish model of human blindness. Hum Mol Genet. 2011;20:1467–1477. doi: 10.1093/hmg/ddr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaghloul NA, et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc Natl Acad Sci U S A. 2010;107:10602–10607. doi: 10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masyukova SV, et al. Assessing the pathogenic potential of human Nephronophthisis disease-associated NPHP-4 missense mutations in C. elegans. Hum Mol Genet. 2011;20:2942–2954. doi: 10.1093/hmg/ddr198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janssen S, et al. Mutation analysis in Bardet-Biedl syndrome by DNA pooling and massively parallel resequencing in 105 individuals. Hum Genet. 2011;129:79–90. doi: 10.1007/s00439-010-0902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid F, et al. U1 snRNA-mediated gene therapeutic correction of splice defects caused by an exceptionally mild BBS mutation. Hum Mutat. 2011;32:815–824. doi: 10.1002/humu.21509. [DOI] [PubMed] [Google Scholar]
- 43.Hopp K, et al. B9D1 is revealed as a novel Meckel syndrome (MKS) gene by targeted exon-enriched next-generation sequencing and deletion analysis. Hum Mol Genet. 2011;20:2524–2534. doi: 10.1093/hmg/ddr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walczak-Sztulpa J, et al. Cranioectodermal Dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. Am J Hum Genet. 2010;86:949–956. doi: 10.1016/j.ajhg.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramprasad VL, et al. Identification of a novel splice-site mutation in the Lebercilin (LCA5) gene causing Leber congenital amaurosis. Mol Vis. 2008;14:481–486. [PMC free article] [PubMed] [Google Scholar]
- 46.Ashkinadze E, et al. Combining fetal sonography with genetic and allele pathogenicity studies to secure a neonatal diagnosis of Bardet-Biedl syndrome. Clin Genet. 2012 doi: 10.1111/cge.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanner G, et al. Therapeutic strategy to rescue mutation-induced exon skipping in rhodopsin by adaptation of U1 snRNA. Hum Mutat. 2009;30:255–263. doi: 10.1002/humu.20861. [DOI] [PubMed] [Google Scholar]
- 48.Schmid F, et al. A Gene Therapeutic Approach to Correct Splice Defects with Modified U1 and U6 snRNPs. Hum Gene Ther. 2012 doi: 10.1089/hum.2012.110. [DOI] [PubMed] [Google Scholar]
- 49.Urnov FD, et al. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 50.Overlack N, et al. Gene repair of an Usher syndrome causing mutation by zinc-finger nuclease mediated homologous recombination. Invest Ophthalmol Vis Sci. 2012;53:4140–4146. doi: 10.1167/iovs.12-9812. [DOI] [PubMed] [Google Scholar]
- 51.Chung DC, et al. Adeno-Associated Virus-Mediated Gene Transfer to Renal Tubule Cells via a Retrograde Ureteral Approach. Nephron Extra. 2011;1:217–223. doi: 10.1159/000333071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel M, et al. High efficiency gene transfer to airways of mice using influenza hemagglutinin pseudotyped lentiviral vectors. J Gene Med. 2013 doi: 10.1002/jgm.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber M, et al. Recombinant adeno-associated virus serotype 4 mediates unique and exclusive long-term transduction of retinal pigmented epithelium in rat, dog, and nonhuman primate after subretinal delivery. Mol Ther. 2003;7:774–781. doi: 10.1016/s1525-0016(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 54.Sadrian B, et al. Lentivirus-mediated genetic manipulation and visualization of olfactory sensory neurons in vivo. J Vis Exp. 2011 doi: 10.3791/2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gau P, et al. Air-assisted intranasal instillation enhances adenoviral delivery to the olfactory epithelium and respiratory tract. Gene Ther. 2011;18:432–436. doi: 10.1038/gt.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers KE, et al. Molecular cloning and sequencing of a cDNA for olfactory marker protein. Proc Natl Acad Sci U S A. 1987;84:1704–1708. doi: 10.1073/pnas.84.6.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danciger E, et al. Olfactory marker protein gene: its structure and olfactory neuron-specific expression in transgenic mice. Proc Natl Acad Sci U S A. 1989;86:8565–8569. doi: 10.1073/pnas.86.21.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khani SC, et al. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Invest Ophthalmol Vis Sci. 2007;48:3954–3961. doi: 10.1167/iovs.07-0257. [DOI] [PubMed] [Google Scholar]
- 59.Koch S, et al. Gene therapy restores vision and delays degeneration in the CNGB1(-/-) mouse model of retinitis pigmentosa. Hum Mol Genet. 2012;21:4486–4496. doi: 10.1093/hmg/dds290. [DOI] [PubMed] [Google Scholar]
- 60.Sun X, et al. Gene therapy with a promoter targeting both rods and cones rescues retinal degeneration caused by AIPL1 mutations. Gene Ther. 2010;17:117–131. doi: 10.1038/gt.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Helal I, et al. Emergent early markers of renal progression in autosomal-dominant polycystic kidney disease patients: implications for prevention and treatment. Am J Nephrol. 2012;36:162–167. doi: 10.1159/000341263. [DOI] [PubMed] [Google Scholar]
- 62.Rachel RA, et al. Combining Cep290 and Mkks ciliopathy alleles in mice rescues sensory defects and restores ciliogenesis. J Clin Invest. 2012;122:1233–1245. doi: 10.1172/JCI60981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brunet LJ, et al. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 64.Belluscio L, et al. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 65.McIntyre JC, et al. Emx2 stimulates odorant receptor gene expression. Chem Senses. 2008;33:825–837. doi: 10.1093/chemse/bjn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong ST, et al. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 67.Ware SM, et al. Spectrum of clinical diseases caused by disorders of primary cilia. Proc Am Thorac Soc. 2011;8:444–450. doi: 10.1513/pats.201103-025SD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsiokas L, et al. Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc Natl Acad Sci U S A. 1997;94:6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cole DG, et al. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kozminski KG, et al. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci U S A. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 72.Sang L, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dowdle WE, et al. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet. 2011;89:94–110. doi: 10.1016/j.ajhg.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis EE, Katsanis N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr Opin Genet Dev. 2012;22:290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goyenvalle A, et al. Therapeutic approaches to muscular dystrophy. Hum Mol Genet. 2011;20:R69–78. doi: 10.1093/hmg/ddr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossmiller B, et al. Gene therapy in animal models of autosomal dominant retinitis pigmentosa. Mol Vis. 2012;18:2479–2496. [PMC free article] [PubMed] [Google Scholar]
- 77.Kleinman ME, et al. Short-interfering RNAs induce retinal degeneration via TLR3 and IRF3. Mol Ther. 2012;20:101–108. doi: 10.1038/mt.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gilliam JC, et al. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029–1041. doi: 10.1016/j.cell.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]