Abstract
Introduction
Prior studies identifying patients with inflammatory bowel disease (IBD) utilizing administrative codes have yielded inconsistent results. Our objective was to develop a robust electronic medical record (EMR) based model for classification of IBD leveraging the combination of codified data and information from clinical text notes using natural language processing (NLP).
Methods
Using the EMR of 2 large academic centers, we created data marts for Crohn’s disease (CD) and ulcerative colitis (UC) comprising patients with ≥ 1 ICD-9 code for each disease. We utilized codified (i.e. ICD9 codes, electronic prescriptions) and narrative data from clinical notes to develop our classification model. Model development and validation was performed in a training set of 600 randomly selected patients for each disease with medical record review as the gold standard. Logistic regression with the adaptive LASSO penalty was used to select informative variables.
Results
We confirmed 399 (67%) CD cases in the CD training set and 378 (63%) UC cases in the UC training set. For both, a combined model including narrative and codified data had better accuracy (area under the curve (AUC) for CD 0.95; UC 0.94) than models utilizing only disease ICD-9 codes (AUC 0.89 for CD; 0.86 for UC). Addition of NLP narrative terms to our final model resulted in classification of 6–12% more subjects with the same accuracy.
Conclusion
Inclusion of narrative concepts identified using NLP improves the accuracy of EMR case-definition for CD and UC while simultaneously identifying more subjects compared to models using codified data alone.
Keywords: Crohn’s disease, ulcerative colitis, disease cohort, natural language processing, informatics
INTRODUCTION
Electronic medical records (EMR) are increasingly being used in clinical practice and research allowing efficient development of cohorts, ascertainment of outcomes, and opportunities for translational research when linked to biospecimen repositories1–8. However, optimal use of EMR data requires accurate definition of diseases and outcomes, a major challenge for researchers. Thus far, a majority of models to define disease have relied solely on administrative billing codes. For example, a diagnosis of inflammatory bowel disease (IBD) using administrative datasets was defined by the presence of a single billing code for Crohn’s Disease (CD) or Ulcerative Colitis (UC), multiple billing codes, or a combination of billing codes, procedures, and medications9–12. However, the accuracy of these algorithms varies widely between 75–97% and is limited by the variations in coding practices, the fact that billing codes are often assigned by administrative non-clinical staff not directly involved in patient care, and incomplete medical history. Importantly, there are additional data in the EMR that do not have billing codes (i.e. endoscopic, pathologic, or radiologic findings) that provide important additional information for accuracy of disease definition. However, these data are often embedded within narrative text reports that typically require laborious manual medical record review for extraction.
Natural language processing (NLP) is a range of computational techniques for analyzing and representing naturally occurring written or oral texts for the purpose of achieving human-like language processing for a range of tasks or applications13. One such application is within the EMR where NLP has been used to define medication use, adverse events, complications, or response to treatment6, 7, 14–20. NLP has also been applied to aid in development of disease cohorts including prior work from our group7 that demonstrated that this informatics-based approach was accurate, improved sensitivity, and was portable to other institutions with distinct EMR systems1.
The goals of this study were to (1) develop and validate an algorithm for definition of Crohn’s disease (CD) and ulcerative colitis (UC) within a multi-institutional EMR; (2) compare the performance of case-definition models utilizing codified data alone to those incorporating narrative free text extracted using NLP, in particular focusing on the added contribution of NLP to improving sensitivity and accuracy; and (3) demonstrate disease associations that can uniquely be identified using NLP data.
METHODS
Data Source
We studied EMR data from Massachusetts General Hospital (MGH) and Brigham and Women’s Hospital (BWH), both tertiary referral hospitals serving over 3 million patients in the Boston metropolitan area. The EMR (Partners Longitudinal Medical Record (LMR)) has been in existence at MGH from October 1, 1994 and at BWH from October 3, 1996. We first created two datasets of all potential IBD patients – the “CD mart” comprised all patients with at least 1 International Classification of diseases, 9th edition (ICD-9) code for CD (555.x, n = 14,288), and the “UC mart,” which comprised patients with at least 1 ICD-9 diagnosis code for UC (556.X, n=14,335) (Figure 1). The ICD-9 codes for outpatient encounters, inpatient stays, and procedures are embedded within the dataset.
Figure 1.
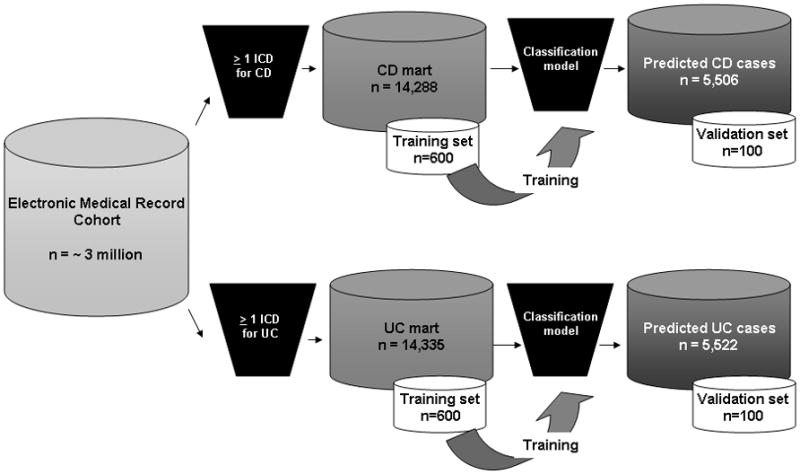
Classification model for defining inflammatory bowel disease cohorts in the electronic medical record cohort
Selection of codified variables
For each subject in the CD and UC mart, we identified the total number of ICD-9 codes for CD or UC. In addition, we identified the number of such codes that were assigned to an inpatient hospitalization, gastroenterologist visit, or associated with an endoscopic procedure. We also identified the number of codes for competing diagnoses with a similar clinical presentation (irritable bowel syndrome, ischemic colitis, diverticulitis), CD- or UC-related complications (intestinal fistulae, strictures, perianal fistulae or abscesses), or surgeries (small or large intestinal resection perirectal surgery) (Supplementary Table 1). Finally, we included whether a patient was prescribed or listed as being on a CD or UC-related medication by their physician using the EMR electronic prescription program at any point in their follow-up. These medications including 5-aminosalicylates (mesalamine, sulfasalazine, balsalazide), corticosteroids (prednisone, hydrocortisone, budesonide), immunomodulators (azathioprine, 6-mercaptopurine, methotrexate) and anti-tumor necrosis factor-α therapies (infliximab, adalimumab).
Narrative terms and NLP analysis
We used six different types of notes as source for our narrative terms – outpatient notes, discharge summaries, operative notes, radiology, endoscopy, and pathology reports. Routine inpatient progress notes are not available in electronic format and were not included. In addition to PDF format, endoscopy reports are available in text format and could be processed for NLP analysis. We processed the notes using the clinical Text Analysis and Knowledge Extraction System (cTAKES)21 (http://ohnlp.svn.sourceforge.net/viewvc/ohnlp/trunk/cTAKES/), which processes clinical text notes and identifies when the term is mentioned in the text, along with qualifying attributes (i.e., negated, non-negated, current, history of, family history of). We created an expert-defined list of terms we considered relevant to identifying subjects with IBD. The terms were then mapped to the Systemized Nomenclature of Medicine-Clinical Terms (SNOMED-CT), a hierarchically organized clinical healthcare terminology index with over 300,000 concepts, to allow for variations in language use, or the RxNorm, a normalized naming systemic for generic and branded drugs.
The selection of the relevant terms followed a structure similar to our codified data. First, we defined the number of times the terms were mentioned “Crohn’s disease/Crohn disease”, or “ulcerative colitis” in the narrative notes. Other terms extracted included those that were relatively specific for CD (“ileitis”), UC (“proctosigmoiditis”), common to both diagnoses, disease-related complications (“perianal abscess”) and surgeries (“ileocecal resection”). We categorized each as CD-specific, UC-specific, or common across both diseases. We also included the number of times the terms were mentioned in the clinical texts for each subject, for each of the potential competing diagnoses, supportive endoscopy, pathology, and radiology findings. For example, the colonoscopic findings that could support a diagnosis of IBD included “aphthous ulcer” “friable mucosa”, or “loss of vascularity” while pathology findings included “chronic active colitis”, “ileitis” and radiology findings included “bowel wall thickening” and “wall enhancement”.
To examine the accuracy of NLP in identifying the terms, 100 random sentences were selected for each of the main concepts of interest (“Crohn’s disease”, “ulcerative colitis”, medications). cTAKES was defined as having identified a term accurately if the sentence extracted contained a mention of the disease or medication. For the medication, an accurate mention included if the medication was currently being taken, had been taken in the past, was contemplated being initiated, or was being temporarily held. Negative mentions of the disease or medication terms (for example, “no evidence of Crohn’s disease”) were considered as accurate only if cTAKES was able to accurately identify that the term was negated. The precision of the identification of terms by NLP was defined as the number of sentences where the NLP output was confirmed by physician review/total number of sentences identified by NLP. Overall precision of NLP was high – Crohn’s disease – 100%; ulcerative colitis – 98%; anti-TNF agents – 98%; corticosteroids – 97%; and immunomodulators – 96%.
Development of the classification algorithm
A training set of 600 patients were selected at random from CD mart and another 600 were selected at random from the UC mart (Figure 1). A board certified gastroenterologist (A.N.A) reviewed the EMR of all patients and classified them as having CD, UC, or not having IBD. CD or UC was diagnosed based on the presence of typical symptoms, chronicity of presentation, and supportive endoscopic, histologic, or radiologic findings22–24. However, where primary data pertaining to the diagnosis was not available within our electronic medical record, we considered CD or UC as being present based on consistent mention within the medical record and use of an appropriate CD or UC-related medication without the presence of an alternate indication for that treatment. For patients who may have had pre-surgical UC and developed Crohn’s of the J-pouch following their surgery, or for those with IBDU, the IBD diagnosis type presumed during the majority of their clinical encounters was assigned. A penalized logistic regression with the adaptive LASSO procedure25 was used to select the informative variables for our final predictive model. The tuning parameter for the penalized regression was selected based on the Bayesian Information Criterion26. We constructed four separate models to predict a diagnosis of CD or UC in our EMR cohort – (1) model utilizing number of CD or UC ICD-9 codes alone (ICD-9 model); (2) model comprising all codified variables including disease complications (codified model); (3) model including narrative terms identified through NLP only (NLP model); and (4) a combined model including both codified and NLP variables (combined model). The regression model assigned each patient a probability of truly having a diagnosis of CD or UC on a continuous scale.
The accuracy of the models at various specificity levels were calculated non-parametrically27 and the overall prediction performance of each model evaluated based on the area under the receiver operating characteristic curve (AUC). To correct for over-fitting bias, the 0.632-bootstrap28 was used to estimate these accuracy measures. The standard error estimates were obtained via the bootstrap procedure with 1000 replicates. For all models, we selected a probability threshold corresponding to a specificity of 97% and classified patients with probability exceeding the threshold value as truly having the disease within the data mart. The accuracy of our classification rule was validated by reviewing the medical records of 100 additional patients each predicted by the final combined model to have CD or UC. Finally, we compared the performance of our final combined model to other published algorithms for defining CD or UC in an EMR cohort.
Histologic disease activity and risk of surgery
Findings such as histologic evidence of active disease are not available in routine non-research clinical datasets or administrative data and require laborious manual review of EMR for extraction. To further explore the utility of NLP in research, we examined the association between presence of histologic activity identified through narrative text extraction by NLP and risk of surgery in CD and UC. Relevant IBD-related surgeries were identified through ICD-9 codes as in previous studies29, 30. Patients with ≥1 ICD-9 for an IBD-related surgery were classified as having the outcome of surgery. In this exploratory analysis, for each patient we summed the number of NLP identified mentions of “cryptitis”, “crypt abscesses”, “chronic inflammation”, “chronic active colitis”, or “enteritis” to estimate cumulative burden of histologic disease activity, and divided patients into four strata based on the distribution of data – 0 mentions, 1–2 mentions (tertile 1), 2–6 mentions (tertile 2), and > 6 mentions (tertile 3). Logistic regression models adjusting for age, duration of follow-up, and intensity of healthcare utilization (number of facts) were used to examine the association between tertiles of cumulative burden of histologic disease activity and undergoing surgery during follow-up. Number of facts refers to number of distinct encounters with the medical system and is a marker of healthcare utilization. For example, an office visit, a laboratory test, and a colonoscopy each contribute 1 fact. The study was approved by the Institutional Review Board of Partners Healthcare.
RESULTS
Training set characteristics
The CD training set consisted of 600 patients with ≥ 1 ICD-9 code for CD (Figure 1); 399 patients (67.5%) were confirmed to have CD, 66 had UC (11.0%) and the remaining 135 did not have IBD (Table 1A). The mean number of ICD-9 codes for CD was greater in those with confirmed CD (34.7±2.8) compared to those with UC (6.1±1.5, p < 0.001) or without IBD (1.7±0.1, p < 0.001). Confirmed CD patients also had a greater number of narrative mentions of Crohn’s disease compared to UC or non-IBD patients.
Table 1A.
Characteristics of patients in the Crohn’s disease training set (n=600), stratified by physician-confirmed diagnosis of CD, UC, and non-IBD
| Physician assigned diagnosis by chart review | |||
|---|---|---|---|
| Parameter | Crohn’s disease (n = 399) | Ulcerative colitis (n = 66) | Non-IBD (n = 135) |
| Age at first diagnosis code [(mean (SD)) (in years) | 40 (19) | 40 (18) | 45 (20) |
| Female (%) | 59% | 67% | 61% |
| Total ICD-9 codes for CD [mean (SD)] | 34.7 (56) | 6.1 (12) | 1.7 (2) |
| Total ICD-9 codes for UC [mean (SD)] | 1.1 (3) | 26.2 (24) | 0.3 (1) |
| Inpatient ICD-9 codes for CD [mean (SD)] | 5.1 (16) | 0.4 (1) | 0.3 (1) |
| Narrative mentions of symptoms [mean (SD)] | 27.3 (78) | 24.4 (36) | 35.7 (66) |
| Narrative mentions of “Crohn’s disease” [mean (SD)] | 61.3 (94) | 7.7 (16) | 4.1 (7) |
| Narrative mentions of “ulcerative colitis” [mean (SD)] | 1.2 (1) | 33.5 (34) | 1.7 (2) |
| Number of facts [mean (SD)] † | 1758 (3236) | 1455 (1407) | 2273 (2800) |
| Duration of follow up (in days) [mean (SD)] ‡ | 3923 (2448) | 4559 (2428) | 4741 (2403) |
Note: Crohn’s disease training set ≥1 ICD-9 code for CD (555.x)
Number of facts refers to number of distinct encounters with the medical system and is a marker of healthcare utilization. For example, an office visit, a laboratory test, and a colonoscopy each contribute 1 fact.
Duration of follow-up is the difference between first and most recent clinical encounter (office visit, X-ray, laboratory test) within the Partners Healthcare system.
Among the UC training set of 600 patients with ≥ 1 ICD-9 code for UC, 378 (63%) were confirmed on chart review to have UC, 72 to have CD (12%) and 150 did not have IBD (25%). Those with confirmed UC had a greater number of total ICD-9 codes for UC (23.0±1.6) than those with CD (8.1±2.8) or non-IBD controls (1.8±0.2) (p < 0.001) (Table 1B), and a greater number of narrative mentions of ulcerative colitis. Thus, the PPV of a single ICD-9 code for CD or UC in the training sets were only 67.5% and 63% respectively.
Table 1B.
Characteristics of patients in the ulcerative colitis training set (n=600), stratified by physician-confirmed diagnosis of CD, UC and non-IBD
| Physician assigned diagnosis by chart review | ||||
|---|---|---|---|---|
| Parameter | Ulcerative colitis (n=378) | Crohn’s disease (n=72) | Non-IBD (n=150) | |
| Age at first diagnosis code [(mean (SD)) (in years) | 43 (18) | 37 (19) | 52 (21) | |
| Female (%) | 53 | 53 | 60 | |
| Total ICD-9 codes for UC [mean (SD)] | 23.0 (30) | 8.1 (23) | 1.8 (2) | |
| Total ICD-9 codes for CD [mean (SD)] | 3.5 (14) | 36.8 (41) | 0.1 | (0) |
| Inpatient ICD-9 codes for UC [mean (SD)] | 3.1 (7) | 1.3 (3) | 0.40 | (1) |
| Narrative mentions of symptoms [mean (SD)] | 20.8 (55) | 30.6 (45) | 31.4 (64) | |
| Narrative mentions of “Crohn’s disease” [mean (SD)] | 5.6 (23) | 62.4 (76) | 1.5 (8) | |
| Narrative mentions of “ulcerative colitis” [mean (SD)] | 32.9 (51) | 9.0 (23) | 1.1 (5) | |
| Number of facts [mean (SD)] † | 1602 (2466) | 1640 (2035) | 2564 (3641) | |
| Duration of follow up (in days) [mean (SD)] ‡ | 4362 (2544) | 4000 (2197) | 4791 (2543) | |
Note: Ulcerative colitis training set ≥1 ICD-9 code for UC (556.x)
Number of facts refers to number of distinct encounters with the medical system and is a marker of healthcare utilization. For example, an office visit, a laboratory test, and a colonoscopy each contribute 1 fact.
Duration of follow-up is the difference between first and most recent clinical encounter (office visit, X-ray, laboratory test) within the Partners Healthcare system.
Tables 2A and 2B present the frequency of various codified terms and corresponding NLP narrative mentions within the training sets, grouped by diagnoses of CD, UC or non-IBD assigned by chart review. We found that NLP identified narrative terms provided more information regarding current or past use of medications than codified mentions (Figures 2A and 2B). Less than one-fifth of the CD cohort had codified mentions of anti-TNF therapy; however, this proportion increased to 42% among those with narrative mentions of these agents. NLP was also useful in identifying supportive endoscopic and histologic features. A significantly greater proportion of those with CD or UC in both training sets had narrative mentions supportive of active inflammation on colonoscopy or histology than those classified as not having IBD.
Table 2A.
Distribution of codified variables and NLP mentions in the Crohn’s disease training set (n=600), stratified by physician-confirmed diagnoses of CD, UC and non-IBD.
| Parameter | Codified data | NLP narrative mentions | ||||
|---|---|---|---|---|---|---|
| Crohn’s disease (n = 399) % | UC (n = 66) % | Non-IBD (n = 135) % | Crohn’s disease (n = 399) % | UC (n = 66) % | Non-IBD (n = 135) % | |
| Fistulizing disease | 12 | 8 | 7 | 36 | 29 | 17 |
| Stricturing disease | 25 | 23 | 18 | 40 | 23 | 20 |
| Perianal disease | 13 | 11 | 3 | 11 | 0 | 1 |
| 5-ASA | 44 | 56 | 9 | 75 | 88 | 22 |
| Immunomodulator | 27 | 20 | 10 | 59 | 55 | 14 |
| Anti-TNF | 17 | 21 | 0 | 42 | 32 | 3 |
| Bowel surgery | 12 | 12 | 5 | 26 | 6 | 3 |
| Endoscopy* | - | - | - | 56 | 70 | 36 |
| Pathology* | - | - | - | 51 | 89 | 25 |
| Elevated CRP or ESR+ | 49 | 59 | 36 | - | - | - |
Note: Crohn’s disease training set ≥1 ICD-9 code for CD (555.x)
ASA – aminosalicylates, anti-TNF – tumor necrosis factor antibodies (infliximab, adalimumab, certolizumab), CRP – C-reactive protein, ESR – erythrocyte sedimentation rate, NA – not applicable
Endoscopy and pathology refer to the proportion of patients with at least 1 positive mention for a supportive endoscopic or histologic finding
CRP or ESR were assigned as being elevated if above the reference limit of normal for each hospital laboratory
Table 2B.
Distribution of codified variables and NLP mentions in the ulcerative colitis training set (n=600), stratified by physician-confirmed diagnoses of CD, UC and non-IBD.
| Parameter | Codified data | Data discovered through NLP (or NLP-based approach) | ||||
|---|---|---|---|---|---|---|
| UC (n=378) % | Crohn’s disease (n=72) % | Non-IBD (n=150) % | UC (n=378) % | Crohn’s disease (n=72) % | Non-IBD (n=150) % | |
| 5-ASA | 50 | 57 | 4 | 76 | 89 | 89 |
| Immunomodulator | 20 | 40 | 5 | 40 | 72 | 72 |
| Anti-TNF | 10 | 27 | 0 | 24 | 58 | 58 |
| Bowel Surgery | 13 | 24 | 11 | 24 | 40 | 50 |
| Endoscopy* | - | - | - | 56 | 60 | 60 |
| Pathology* | - | - | - | 67 | 68 | 68 |
| Elevated CRP or ESR+ | 46 | 68 | 43 | - | - | - |
Note: Ulcerative colitis training set ≥1 ICD-9 code for UC (556.x)
ASA – aminosalicylates, anti-TNF – tumor necrosis factor antibodies (infliximab, adalimumab, certolizumab), CRP – C-reactive protein, ESR – erythrocyte sedimentation rate, NA – not applicable
Endoscopy and pathology refer to the proportion of patients with at least 1 positive mention for a supportive endoscopic or histologic finding
CRP or ESR were assigned as being elevated if above the reference limit of normal for each hospital laboratory
Figure 2.
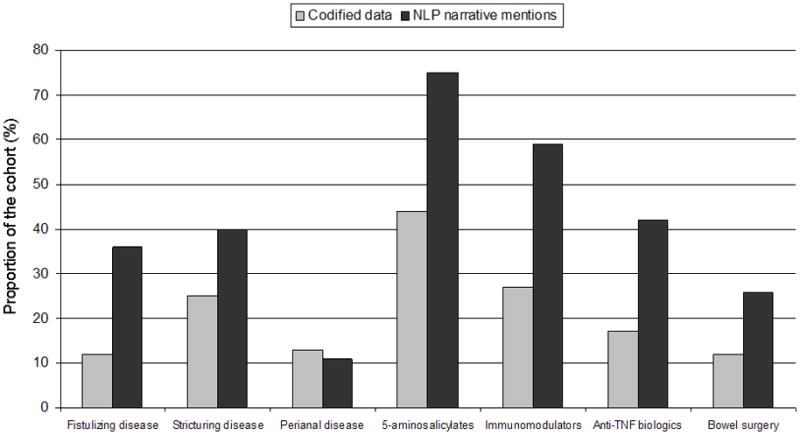
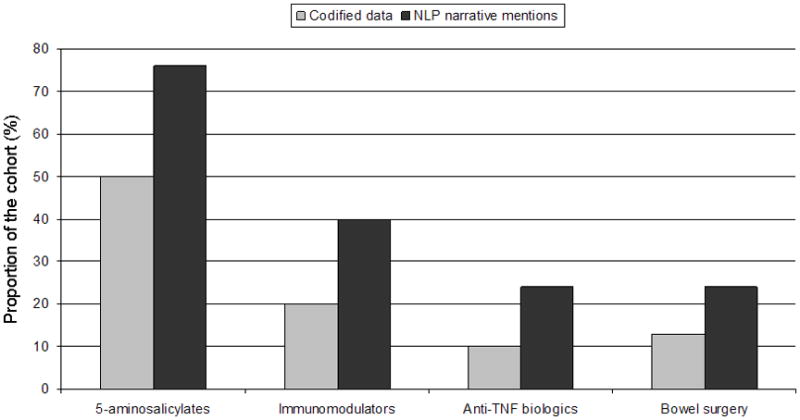
Figure 2a: Comparison of codified data and narrative mentions of disease complications, medications and outcomes in confirmed Crohn’s disease patients in the training set (n = 399)
Figure 2b: Comparison of codified data and narrative mentions of medications and outcomes in confirmed ulcerative colitis patients in the training set (n = 378)
Derivation of the classification algorithm
Figures 3A and 3B present the variables that were selected for inclusion in our final models to define CD and UC, respectively, in order of magnitude of the regression coefficients. The strongest variables for the prediction of CD were the number of ICD-9 codes for CD and the number of NLP mentions of CD. Other informative variables include IBD-related complications and medications. The presence of competing diagnosis codes including that for UC and NLP mentions for irritable bowel syndrome were negative predictors of CD.
Figure 3.
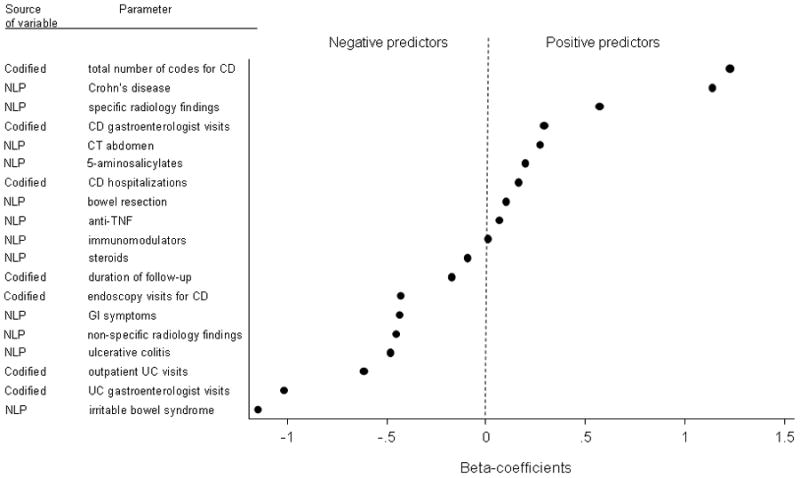
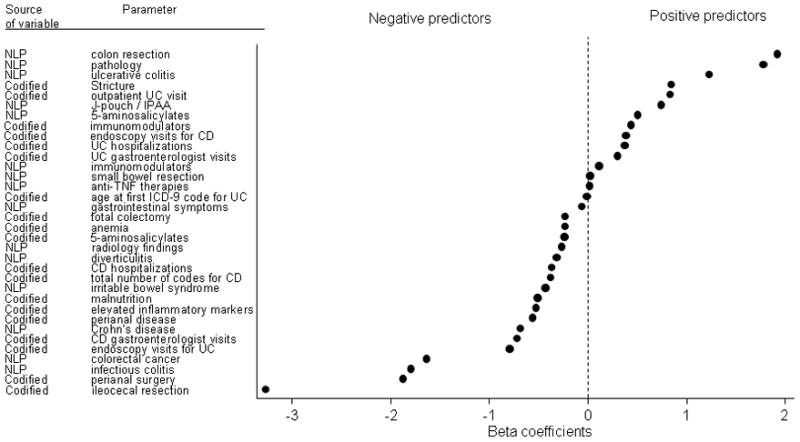
Figure 3a: Beta-coefficients of significant predictors included in the final combined model for Crohn’s disease
Figure 3b: Beta-coefficients of significant predictors including in the final model for ulcerative colitis
The number of NLP mentions for colon resection, presence of supportive findings on pathology, and UC were most predictive of UC diagnosis, while ICD-9 codes or NLP identified mentions of perianal disease (CD-related complication) and competing diagnoses were negative predictors of UC (Figure 3B).
Performance and Validation of the algorithm
The combined model incorporating both narrative and codified data had greater accuracy for identification of CD (AUC 0.95, 95% CI 0.93 – 0.97) than a model that contained only ICD-9 billing codes for CD (AUC 0.89, 95% CI 0.87 – 0.92) (Supplemental Figure 1). Similarly for UC, the combined model had better accuracy (AUC 0.94, 95% CI 0.93–0.96) than the ICD-9 model alone (AUC 0.86, 95% CI 0.83–0.89) or a model containing ICD-9 codes and disease complications (codified model) (Supplemental Figure 2).
The combined CD model classified 5,502 CD patients when applied to the CD mart, while the combined UC model classified 5,519 UC patients when applied to the UC mart. On review of medical records from an additional random set of 100 patients each predicted to have CD or UC by the combined model, 97 each were correctly identified as having this diagnosis through chart review resulting in a PPV of 97% for each algorithm.
Figure 4 demonstrates the proportion of patients in our IBD data mart who would be classified as having CD or UC at the same 97% specificity level from each of the 4 models: ICD-9, codified, NLP and combined model. Addition of NLP to a model containing codified data alone improved the sensitivity and therefore the proportion of EMR patients classified as having CD or UC. The improved sensitivity resulted in an additional 851 CD and 1887 UC patients who could be classified as truly having CD or UC when compared to the ICD-9 model, and 325–584 patients over the next best performing model. Compared to previously published algorithms, our present algorithm demonstrated significantly improved specificity and PPV as well without a decrease in sensitivity (Table 3).
Figure 4. Proportion of patients in the entire EMR data mart classified as having Crohn’s disease (CD) or ulcerative colitis (UC) with 97% specificity.
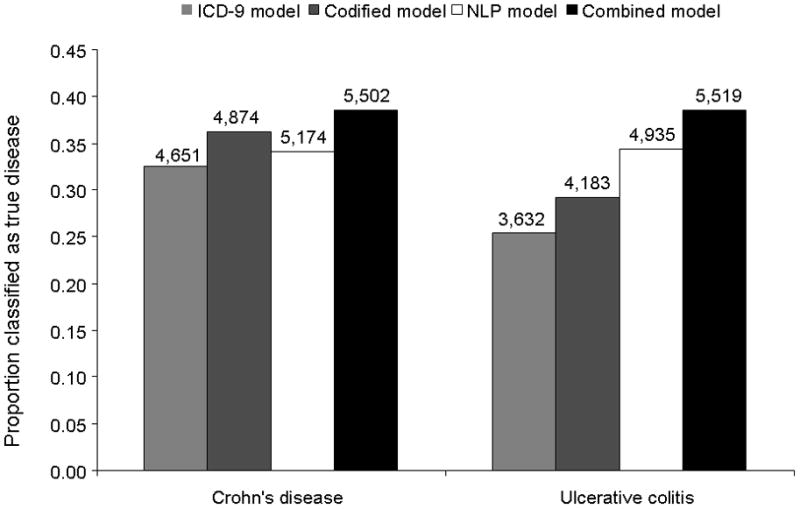
The numbers over the bar graph represent the estimated size of our EMR cohort for CD and UC using each of the four models
Table 3.
Comparison of performances of various published algorithms to define Crohn’s disease or ulcerative colitis in an electronic medical record cohort
| Algorithm | Positive Predictive value | Sensitivity | Specificity | Predicted number of patients in the IBD mart |
|---|---|---|---|---|
| Crohn’s disease | ||||
| Combined CD model | 98 (97 – 100) | 69 (65 – 74) | 97 (96 – 100) | 5,506 |
| 5 separate CD ICD-9 codes 10 | 91 (88 – 94) | 66 (61 – 71) | 88 (82 – 92) | 5,504 |
| 1 outpatient/inpatient CD ICD-9 code and 1 endoscopy12 | 85 (80 – 89) | 53 (48 – 58) | 81 (75 – 86) | 4,914 |
| 4 outpatient or 2 inpatient CD ICD-9 codes 9 | 92 (88 – 95) | 65 (60 – 70) | 88 (83 – 92) | 5,678 |
| Ulcerative colitis | ||||
| Combined UC model | 97 (97 – 100) | 79 (75 – 83) | 97 (95 – 100) | 5,522 |
| 5 separate UC ICD-9 codes10 | 90 (86– 94) | 67 (62 – 71) | 88 (83 – 92) | 4,893 |
| 1 outpatient/inpatient UC ICD-9 code and 1 endoscopy12 | 85 (80 – 89) | 51 (46 – 56) | 85 (79 – 89) | 4,489 |
| 4 outpatient or 2 inpatient UC ICD-9 codes 9 | 89 (85 – 93) | 66 (61 – 72) | 86 (81 – 91) | 5,070 |
Association between histologic activity and surgery
Among the cohort of UC patients, patients in the second and third tertiles of histologic activity had significantly greater odds of surgery with odds ratios (OR) of 3.20 (95% CI 2.43 – 4.21) and 6.37 (95% CI 5.02 – 8.07) respectively, compared to those with no mentions of histologic activity. Similarly increasing tertiles of histologic activity for CD were also associated with CD-related surgery with adjusted ORs of 1.41 (95%CI 1.05 – 1.89), 2.07 (95%CI 1.64 – 2.62), and 2.83 (95%CI 2.31 – 3.47) respectively.
DISCUSSION
We demonstrate that a CD or UC classification model incorporating clinical data extracted using NLP from narrative text has improved accuracy for identification of CD or UC patients in the EMR over a model utilizing CD or UC billing codes alone. The addition of NLP derived variables increased accuracy of CD identification by 6% compared to an algorithm containing billing codes alone (ICD-9 model), and resulted in a 7–15% increase in PPV compared to previously published algorithms9–12. Importantly, addition of NLP resulted in classifying a significantly greater proportion of patients in our EMR cohort as truly having the disease without loss of specificity and accuracy, increasing the size of our IBD cohorts by approximately 6–12%. Finally, we also demonstrate that NLP has the ability to contribute valuable clinical information not available in codified data; we observed an association between increasing NLP derived mentions of histologic disease and surgical outcome in CD and UC patients.
Accurate definition of diseases and outcomes is an important prerequisite in both administrative data and EMR based research. Prior disease algorithms relying on ICD-9 codes have yielded PPV of 75–97%9–12 in the published literature. In our study, the presence of a single ICD-9 code for CD or UC yielded a PPV of only 65–70%. Even without narrative data, addition of codes for disease complications (codified model) improved the PPV over the ICD-9 model, suggesting that such approaches should be considered in administrative database research. In particular, given the significant (~10%) misclassification of CD as UC or vice versa using only CD or UC ICD-9 codes, inclusion of disease specific complications (perianal disease, intestinal fistulae) may help improve the specificity of such algorithms.
The myriad modifications required in different EMR databases to achieve comparable accuracy in defining the disease of interest highlights the challenges in using billing codes alone to classify disease. This approach is also vulnerable to errors. First, there is the possibility of inaccurate coding as assignment of diagnosis codes is often performed by non-medical providers not involved in direct patient care. Second, several diseases (for example, primary sclerosing cholangitis) may lack distinct diagnosis codes31. In addition, reliance exclusively on billing codes ignores the wealth of information available as narrative free text within the medical record. We demonstrate that adding data extracted through NLP to models containing only billing data improved the accuracy substantially.
NLP is a range of computational techniques for analyzing the most ubiquitous human product, namely, language13. There are several benefits to incorporating NLP to analyze narrative text in EMR research. First, this allows for identifying not only disease terms but also supportive symptoms, laboratory tests, and investigations. Thus, by accurately identifying the results of a specific investigation rather than merely having a code for the test having been performed allows for significantly greater confidence in assigning a subject as truly having the disease. Indeed, a recent approach utilizing real-time NLP in the Veterans Affairs medical records revealed the ability to detect post-operative complications with superior sensitivity compared to patient safety indicators3, 6. Second, it contributes to increasing the confidence in disease diagnosis by ascertaining the presence of mentions for competing diagnoses that may mimic the disease in question. For example, colonoscopy revealing “erythema and ulceration in the splenic flexure watershed consistent with ischemic colitis” reduces the confidence in a diagnosis of ulcerative colitis though a common billing code may have been utilized. Third, NLP allows for identifying disease outcomes such as disease activity that are not available through billing codes. Fourth, addition of NLP to our case definition model resulted in a substantial increase in the size of our disease cohorts without compromising specificity. The utility of an EMR cohort for translational research relies on the ability to develop a sufficiently large cohort for genotype-phenotype studies, while not compromising on the specificity and accuracy of identifying true cases. Importantly, the 6–12% increase in cohort size in our study while maintaining a high level of accuracy for classification could significantly improve statistical power in genotype-phenotype correlation studies using biological samples linked to EMR data.
As EMR data is increasingly being used for research, and in particular translational research aimed at examining genotype-phenotype relationships, developing disease cohorts of adequate size to allow for power for genetic analyses is important. At the same time, it is important to preserve specificity of disease definition to ensure accuracy of genetic analysis. We demonstrate that addition of NLP is a valuable tool by allowing for classification of a greater number of patients as having CD or UC disease without increasing the false positive rate. Finally, we demonstrate that NLP can also be an invaluable source for mining the clinical narrative3 by defining an association between histologic disease activity through narrative searches for terms indicating active bowel inflammation, and requirement for surgery in both CD and UC, confirming prior findings from small studies32.
There are limitations to our study. First, it was restricted to the EMR from a single healthcare system, Partners Healthcare, which uses a common electronic record. Further studies are required to examine the portability of our IBD algorithm to different EMR systems. Notably, our group has recently demonstrated that an algorithm to define rheumatoid arthritis (RA) patients utilizing a combination of codified and NLP data developed in the Partners EMR7 was portable to other institutions using distinct EMR systems1. As an increasing number of institutions adopt electronic medical records, an approach utilizing the wealth of free text narrative information available within the EMR offers significant opportunities for efficient, cost-effective research and collaborative. Second, as our health system is comprised of referral hospitals and is not a ‘closed system’, a portion of our patients may receive part of their care at other hospitals. Our use of narrative free text mentions in addition to the billing codes allows a greater ability to ascertain such outcomes from the text within the medical notes. However, we acknowledge that this may still leave us with missing information.
Our findings have several implications. To our knowledge, this is one of the first studies to use NLP in addition to billing codes to improve on the predictive value of case definition models in the EMR for IBD. Validation of such an approach has the potential to allow for efficient development of multicenter cohorts to examine disease outcomes. In particular, develop of such multicenter cohorts will allow for study of uncommon phenotypes2 and complications such as primary sclerosing cholangitis that require large numbers of subjects. Second, several healthcare systems including ours have developed tools to allow for linkage of discarded or consented blood specimens to such EMR data2, 33, 34. This offers the exciting ability to define genotype-phenotype relationships for various outcomes8. Our group has already demonstrated the feasibility of such an approach for an RA cohort4 and we have begun collection and linkage of such biospecimens in our IBD cohort (457 unique plasma and buffy coat samples over 5 months). Once accrual of an adequate number of samples has occurred, our IBD cohort can be utilized to answer key and unique clinical questions that require narrative free text analysis and cannot be addressed using administrative datasets such as genetic prediction of treatment response or treatment related adverse events.
In conclusion, we demonstrate that incorporation of narrative free text data within the disease definition algorithm of an EMR cohort allows for superior accuracy and a higher positive predictive value than algorithms using billing codes alone or prior published studies. This novel methodology offers considerable promise towards multi-institution cohort development and efficient and cost-effective clinical and translational research.
Supplementary Material
Supplementary Figure 1: Performance of Crohn’s disease classification models in an electronic medical record cohort
Supplementary Figure 2: Performance of ulcerative colitis classification models in an electronic medical record cohort
Supplementary Table 1: List of International Classification of Diseases, 9th Edition (ICD-9) codes used in developing the disease classification model
Acknowledgments
Sources of Funding: The study was supported by NIH U54-LM008748. A.N.A is supported by funding from the American Gastroenterological Association. K.P.L. is supported by NIH K08 AR060257 and the Katherine Swan Ginsburg Fund. R.M.P. is supported by grants from the US National Institutes of Health (NIH) (R01-AR056768, U01-GM092691 and R01-AR059648) and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Footnotes
The work was presented in part at the Digestive Disease Week 2012, May 19-22, San Diego, CA.
References
- 1.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, Pacheco JA, Boomershine CS, Lasko TA, Xu H, Karlson EW, Perez RG, Gainer VS, Murphy SN, Ruderman EM, Pope RM, Plenge RM, Kho AN, Liao KP, Denny JC. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012 doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, Wang X, Pacheco JA, Kho AN, Hayes MG, Weston N, Matsumoto M, Kopp PA, Newton KM, Jarvik GP, Li R, Manolio TA, Kullo IJ, Chute CG, Chisholm RL, Larson EB, McCarty CA, Masys DR, Roden DM, de Andrade M. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jha AK. The promise of electronic records: around the corner or down the road? Jama. 2011;306:880–1. doi: 10.1001/jama.2011.1219. [DOI] [PubMed] [Google Scholar]
- 4.Kurreeman F, Liao K, Chibnik L, Hickey B, Stahl E, Gainer V, Li G, Bry L, Mahan S, Ardlie K, Thomson B, Szolovits P, Churchill S, Murphy SN, Cai T, Raychaudhuri S, Kohane I, Karlson E, Plenge RM. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet. 2011;88:57–69. doi: 10.1016/j.ajhg.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love TJ, Cai T, Karlson EW. Validation of psoriatic arthritis diagnoses in electronic medical records using natural language processing. Semin Arthritis Rheum. 2011;40:413–20. doi: 10.1016/j.semarthrit.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murff HJ, FitzHenry F, Matheny ME, Gentry N, Kotter KL, Crimin K, Dittus RS, Rosen AK, Elkin PL, Brown SH, Speroff T. Automated identification of postoperative complications within an electronic medical record using natural language processing. Jama. 2011;306:848–55. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- 7.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, Szolovits P, Churchill S, Murphy S, Kohane I, Karlson EW, Plenge RM. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–7. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–28. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 9.Benchimol EI, Guttmann A, Griffiths AM, Rabeneck L, Mack DR, Brill H, Howard J, Guan J, To T. Increasing incidence of paediatric inflammatory bowel disease in Ontario, Canada: evidence from health administrative data. Gut. 2009;58:1490–7. doi: 10.1136/gut.2009.188383. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein CN, Blanchard JF, Rawsthorne P, Wajda A. Epidemiology of Crohn’s disease and ulcerative colitis in a central Canadian province: a population-based study. Am J Epidemiol. 1999;149:916–24. doi: 10.1093/oxfordjournals.aje.a009735. [DOI] [PubMed] [Google Scholar]
- 11.Herrinton LJ, Liu L, Lafata JE, Allison JE, Andrade SE, Korner EJ, Chan KA, Platt R, Hiatt D, O’Connor S. Estimation of the period prevalence of inflammatory bowel disease among nine health plans using computerized diagnoses and outpatient pharmacy dispensings. Inflamm Bowel Dis. 2007;13:451–61. doi: 10.1002/ibd.20021. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Allison JE, Herrinton LJ. Validity of computerized diagnoses, procedures, and drugs for inflammatory bowel disease in a northern California managed care organization. Pharmacoepidemiol Drug Saf. 2009;18:1086–93. doi: 10.1002/pds.1824. [DOI] [PubMed] [Google Scholar]
- 13.Liddy ED, Turner AM, Bradley J. Modeling interventions to improve access to public health information. AMIA Annu Symp Proc. 2003:909. [PMC free article] [PubMed] [Google Scholar]
- 14.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med. 1998;37:1–7. [PubMed] [Google Scholar]
- 15.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc. 2005;12:448–57. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meystre S, Haug PJ. Evaluation of Medical Problem Extraction from Electronic Clinical Documents Using MetaMap Transfer (MMTx) Stud Health Technol Inform. 2005;116:823–8. [PubMed] [Google Scholar]
- 17.Savova GK, Olson JE, Murphy SP, Cafourek VL, Couch FJ, Goetz MP, Ingle JN, Suman VJ, Chute CG, Weinshilboum RM. Automated discovery of drug treatment patterns for endocrine therapy of breast cancer within an electronic medical record. J Am Med Inform Assoc. 2011 doi: 10.1136/amiajnl-2011-000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn S, Kocher JP, Chute CG, Savova GK. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc. 2011;18 (Suppl 1):i144–9. doi: 10.1136/amiajnl-2011-000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonager K, Sorensen HT, Rasmussen SN, Moller-Petersen J, Vyberg M. Assessment of the diagnoses of Crohn’s disease and ulcerative colitis in a Danish hospital information system. Scand J Gastroenterol. 1996;31:154–9. doi: 10.3109/00365529609031980. [DOI] [PubMed] [Google Scholar]
- 23.Loftus EV, Jr, Silverstein MD, Sandborn WJ, Tremaine WJ, Harmsen WS, Zinsmeister AR. Ulcerative colitis in Olmsted County, Minnesota, 1940–1993: incidence, prevalence, and survival. Gut. 2000;46:336–43. doi: 10.1136/gut.46.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loftus EV, Jr, Silverstein MD, Sandborn WJ, Tremaine WJ, Harmsen WS, Zinsmeister AR. Crohn’s disease in Olmsted County, Minnesota, 1940–1993: incidence, prevalence, and survival. Gastroenterology. 1998;114:1161–8. doi: 10.1016/s0016-5085(98)70421-4. [DOI] [PubMed] [Google Scholar]
- 25.Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006:101. [Google Scholar]
- 26.Friedman J, Hastie T, Tibshirani R. The elements of statistical learning. 2001. [Google Scholar]
- 27.Pepe MS. The statistical evaluation of medical tests for classification and prediction. Oxford University Press; 2004. [Google Scholar]
- 28.Efron B, Tibshirani R. An introduction to the bootstrap. Chapman & Hall/CRC; 1993. [Google Scholar]
- 29.Ananthakrishnan AN, McGinley EL, Binion DG, Saeian K. A nationwide analysis of changes in severity and outcomes of inflammatory bowel disease hospitalizations. J Gastrointest Surg. 15:267–76. doi: 10.1007/s11605-010-1396-3. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein CN, Nabalamba A. Hospitalization, surgery, and readmission rates of IBD in Canada: a population-based study. Am J Gastroenterol. 2006;101:110–8. doi: 10.1111/j.1572-0241.2006.00330.x. [DOI] [PubMed] [Google Scholar]
- 31.Molodecky NA, Myers RP, Barkema HW, Quan H, Kaplan GG. Validity of administrative data for the diagnosis of primary sclerosing cholangitis: a population-based study. Liver Int. 2011;31:712–20. doi: 10.1111/j.1478-3231.2011.02484.x. [DOI] [PubMed] [Google Scholar]
- 32.Riley SA, Mani V, Goodman MJ, Dutt S, Herd ME. Microscopic activity in ulcerative colitis: what does it mean? Gut. 1991;32:174–8. doi: 10.1136/gut.32.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schildcrout JS, Basford MA, Pulley JM, Masys DR, Roden DM, Wang D, Chute CG, Kullo IJ, Carrell D, Peissig P, Kho A, Denny JC. An analytical approach to characterize morbidity profile dissimilarity between distinct cohorts using electronic medical records. J Biomed Inform. 2010;43:914–23. doi: 10.1016/j.jbi.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Performance of Crohn’s disease classification models in an electronic medical record cohort
Supplementary Figure 2: Performance of ulcerative colitis classification models in an electronic medical record cohort
Supplementary Table 1: List of International Classification of Diseases, 9th Edition (ICD-9) codes used in developing the disease classification model


