Abstract
Background
Several evidences indicate that gut microbiota is involved in the control of host energy metabolism.
Objective
To evaluate the differences in the composition of gut microbiota in rat models under different nutritional status and physical activity and to identify their associations with serum leptin and ghrelin levels.
Methods
In a case control study, forty male rats were randomly assigned to one of these four experimental groups: ABA group with food restriction and free access to exercise; control ABA group with food restriction and no access to exercise; exercise group with free access to exercise and feed ad libitum and ad libitum group without access to exercise and feed ad libitum. The fecal bacteria composition was investigated by PCR-denaturing gradient gel electrophoresis and real-time qPCR.
Results
In restricted eaters, we have found a significant increase in the number of Proteobacteria, Bacteroides, Clostridium, Enterococcus, Prevotella and M. smithii and a significant decrease in the quantities of Actinobacteria, Firmicutes, Bacteroidetes, B. coccoides-E. rectale group, Lactobacillus and Bifidobacterium with respect to unrestricted eaters. Moreover, a significant increase in the number of Lactobacillus, Bifidobacterium and B. coccoides–E. rectale group was observed in exercise group with respect to the rest of groups. We also found a significant positive correlation between the quantity of Bifidobacterium and Lactobacillus and serum leptin levels, and a significant and negative correlation among the number of Clostridium, Bacteroides and Prevotella and serum leptin levels in all experimental groups. Furthermore, serum ghrelin levels were negatively correlated with the quantity of Bifidobacterium, Lactobacillus and B. coccoides–Eubacterium rectale group and positively correlated with the number of Bacteroides and Prevotella.
Conclusions
Nutritional status and physical activity alter gut microbiota composition affecting the diversity and similarity. This study highlights the associations between gut microbiota and appetite-regulating hormones that may be important in terms of satiety and host metabolism.
Introduction
Human eating disorders such as anorexia nervosa (AN) is an enormous public health problem in industrialized countries, and is characterized by extreme dietary restriction resulting in a sustained low weight [1]. Additionally, a significant proportion of AN patients shows evidence of abnormally high physical activity levels [2], [3]. It should be emphasized that AN is linked with a significant loss of adipose tissue and a decrease in energy metabolism [4]. On the other hand, there is no doubt that leptin plays an important role in AN, because leptin secretion is profoundly altered in this eating disorder [5]. Leptin is secreted by adipocytes and its primary role is to provide the central nervous system with a signal of the state of the body energy balance, which helps to control the appetite and food intake, and to maintain a stable body weight. Several studies have observed that serum leptin concentration is proportional to body fat mass during energy balance [6], and declines sharply in periods of energy deficit [7]. In addition, it has been found that patients with AN have higher fasting serum ghrelin levels than healthy subjects, which return to normal values after weight gain [8]. Ghrelin is mainly produced by the stomach but also by many other tissues [9]. Among the functions of ghrelin are the stimulation of the appetite and food intake, increasing fat mass deposition and weight gain and influencing glucose and lipid metabolism [10], [11]. The deregulation of all these mechanisms would be responsible for eating disorders like anorexia. Recently, it has been suggested that gut microbiota composition plays a role in the pathophysiology of eating disorders, since gut microbiota is able to partially mediate the appetite control regulating the level and type of autoantibodies targeting the appetite-regulating hormones [12], [13]. In addition, the host nutritional status may be markedly influenced by the composition and activities of the gut microbiota [14]. Moreover, several evidences indicate that the gut microbiota is involved in the control of host energy metabolism [15].
Activity-based anorexia (ABA) is a well established animal model used to study different aspects of AN and situations of undernutrition plus increased activity [16], [17]. In this rat model, anorexia is induced by restricting food intake to one daily feeding period (1–2 h) and permitting free access to a running wheel during the rest of the day (22–23 h). In this situation ABA rats exhibit a reduction in food intake, an increase in wheel activity and a progressive body weight decrease [16], [6].
The aim of the present study, therefore, was to characterize the composition of fecal microbiota in rat models under different nutritional status and physical activity in order to determine whether there were significant differences in the gut microbiota composition between these rat groups; if so, to quantify the differences and to identify the possible association of the gut microbiota found in these rat models with their serum leptin and ghrelin levels.
Materials and Methods
Ethics Statement
The animal studies were conducted in accordance with the ethical guidelines for the care and use of laboratory animals of the National Institutes of Health. All procedures were approved by the Animal Care ethics Committee of Santiago de Compostela University (Santiago de Compostela, Spain).
Animal models
Forty Male Sprague Dawley rats (160 g/5 weeks old) (Charles River Breeding Laboratory, Raleigh, NC) from the same bred were housed in a temperature-controlled room with a 12-h light/12-h dark cycles with free access to standard chow diet and water. A case-control study was performed. After 5 days acclimatization weight-matched animals were randomly assigned to one of four experimental groups (10 rats by group): (a) Activity based anorexia (ABA) group: rats starved by restricting food access to 23 hours per day and confined to running wheels except during a 60 min meal per day, (b) Control ABA group: rats submitted to the same food restriction schedule as ABA with no wheel access exercise, (c) Exercise group: rats feed ad libitum with free access to the activity wheel and (d) Ad libitum group: rats feed ad libitum but without access to the activity wheel. The ABA group was performed following previously established model by Routtenberg and Kuznesof [16]. For ethical reasons, ABA rats were not allowed to lose more than 20–30% of their initial body weight. All animals were housed individually in custom-designed, stainless steel cages, which in the running groups were connected to running wheels (16 cm in diameter) coupled to a turn counter (Harvard Apparatus, MA). Food intake, wheel running and body weight were measured daily. These rats were sacrificed by decapitation after 6 days of experiment and blood samples, tissues and fecal content were collected. The 6-day experimental length was fixed as it is the time where ABA rats reach an approximately 30% body weight reduction and some of them started to show gastric ulcers.
Hormone determination
Blood samples collected at the end of the study after the rats were sacrificed were immediately centrifuged and total ghrelin and leptin levels were determined in serum by a double antibody RIA using kits provided by Linco Research (St Charles, MI) as previously described [18], [19]. The limits of sensitivity of the assays were 100 pg/ml for ghrelin, 0.5 ng/ml for leptin, and 0.1 ng/ml for insulin.
DNA extraction from fecal samples
Fecal samples were immediately kept after collection at −80°C and stored until analyzed. DNA extraction from 200 mg of stools was performed using the QIAamp DNA stool Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The DNA concentration was determined by absorbance at 260 nm (A260), and the purity was estimated by determining the A260/A280 ratio with a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Analysis of fecal microbiota by PCR-DGGE
Fecal samples from each subject were examined by determining PCR-DGGE profiles as recently published by us [20]. The V2–V3 regions of the 16S rRNA genes (positions 339 to 539 in the Escherichia coli gene) of bacteria in the fecal samples were amplified by primers HDA1-GC (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG T-3′; (the GC clamp is in boldface) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) generating a 200 bp product. Aliquots (2 µL) of DNA were amplified by real-time PCR (20 µL final volume) in a 7500 Fast Real-Time PCR Systems instrument using Fast SYBR Green Master Mix and 200 nM of each of the universal primers HDA1-GC/HDA2 with the following amplification program: initial denaturation was at 95° for 20 s, amplification was carried out using 45 cycles including denaturation at 95°C for 3 s, annealing at 55°C for 30 s and extension at 72°C for 1 min.
After real time PCR 15 µL of products were mixed with 6 µL loading dye before loading. Electrophoresis was performed with a DCode ™ Universal Mutation Detection System instrument (Bio-Rad). 6% polyacrylamide gels were prepared and electrophoresed with 1× TAE buffer prepared from 50× TAE buffer (2 M Tris base, 1 M glacial acetic acid, 50 mM EDTA). The denaturing gradient was formed by using two 6% acrylamide (acrylamide/bisacrylamide ratio 37.5∶1) stock solutions (Bio-Rad). The gels contained a 20-80% gradient of urea and formamide that increase in the direction of electrophoresis. Electrophoretic runs were in a Tris-acetate-EDTA buffer (TAE 1x) (40 mmol/L Tris, 20 mmol/L acetic acid, and 1 mmol/L EDTA, pH 7.4) at 130 V and 60°C for 4.5 h. Electrophoresis was stopped when a xylene cyanol dye marker reached the bottom of a gel. Gels were stained with ethidium bromide (0.5 mg/L) for 5 min, rinsed with deionized water, viewed by UV transillumination and photographed with Gelcapture image acquisition software (DNR Bio-Imaging Systems Ltd). All the samples were analyzed on the same DGGE run to avoid the possible influence of variations in electrophoretic conditions between different runs. Similarities between banding patterns in the DGGE profile were calculated based on the presence and absence of bands and expressed as a similarity coefficient (Cs). Gels were analyzed using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). Normalized banding patterns were used for cluster analysis. The Dice similarity coefficient was used to calculate pairwise comparisons of the DGGE fingerprint profiles obtained. A Cs value of 100% indicates that DGGE profiles are identical while completely different profiles result in a Cs value of 0%. The UPGMA (unweighted pair group method with arithmetic mean) algorithm was used for construction of dendrograms.
Sequencing of selected bands from DGGE gels
Bands of specific interest were excised from DGGE gels with a sterile razor, placed in 40 µl sterile water and incubated at 4°C for diffusion of DNA into the water. DNA were used in a second PCR with HDA1/2 primers without GC-clamp (initial denaturation 95° for 20 s, followed 45 cycles including denaturation at 95°C for 3 s, annealing at 55°C for 15 s and extension at 72°C for 10 s). Subsequently, the PCR products will be directly cloned into pCR® 4-TOPO (Invitrogen, UK) according to the manufacturer's instructions. Plasmid DNA will be isolated from the cells using the Qiagen Mini Spin Prep kit (QIAGEN, Germany), and subjected to PCR (HDA1/2-GC) as earlier described. PCR products were diluted until 20 ng/μL, purified with ExoSAP-IT (USB corporation, Miles Road, Cleveland, Ohio, USA) and sequenced in an ABI 3130 (Applied Biosystems, USA) using the BigDie-Kit-Standard. Nucleotide sequence data obtained were analyzed using MicroSeqID v2.1.1 software (Applied Biosystems, USA).
Microbial quantification by real-time PCR
Specific primers targeting different bacterial genera were used to characterize the fecal microbiota by real-time qPCR (Table 1) [21]–[28]. Briefly, real-time qPCR experiments were performed with a LightCycler 2.0 PCR sequence detection system using the FastStart DNA Master SYBR Green kit (Roche Diagnostics, Indianapolis, IN). All PCR tests were carried out in duplicate with a final volume of 20 µL containing 1 µL of each fecal DNA preparation and 200 nM of each primer (Table 1). The thermal cycling conditions used were as follows: an initial DNA denaturation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10 s, primer annealing at optimal temperature (Table 1) for 20 s, extension at 72°C for 15 s. Finally, melt curve analysis was performed by slowly cooling the PCRs from 95 to 60°C (0.05°C per cycle) with simultaneous measurement of the SYBR Green I signal intensity. Melting-point-determination analysis allowed the confirmation of the specificity of the amplification products.
Table 1. Primers used for real-time PCR.
| Target group | Oligonucleotide sequence (5′–3′) | Reference | Amplicon size (bp) |
| Bacteroidetes | CATGTGGTTTAATTCGATGAT | (21) | 126 |
| AGCTGACGACAACCATGCAG | |||
| Bacteroides | GAGAGGAAGGTCCCCCAC | (21) | 106 |
| CGCTACTTGGCTGGTTCAG | |||
| Lactobacillus | GAGGCAGCAGTAGGGAATCTTC | (22) | 126 |
| GGCCAGTTACTACCTCTATCCTTCTTC | |||
| Fusobacteium | CCCTTCAGTGCCGCAGT | (23) | 273 |
| GTCGCAGGATGTCAAGAC | |||
| Firmicutes | ATGTGGTTTAATTCGAAGCA | (21) | 126 |
| AGCTGACGACAACCATGCAC | |||
| Actinobacteria | CGCGGCCTATCAGCTTGTTG | (24) | 600 |
| CCGTACTCCCCAGGCGGGG | |||
| Bifidobacterium | CTCCTGGAAACGGGTGG | (25) | 550 |
| GGTGTTCTTCCCGATATCTACA | |||
| Prevotella | GGTTCTGAGAGGAAGGTCCCC | (26) | 121 |
| TCCTGCACGCTACTTGGCTG | |||
| Enterococcus | CCCTTATTGTTAGTTGCCATCATT | (27) | 144 |
| ACTCGTTCTTCCCATGT | |||
| Proteobacteria | CATGACGTTACCCGCAGAAGAAG | (23) | 195 |
| CTCTACGAGACTCAAGCTTGC | |||
| Clostridium | GCACAAGCAGTGGAGT | (25) | 239 |
| Cluster IV | CTTCCTCCGTTTTGTCAA | ||
| Blautia coccoides–Eubacterium rectale group | CGGTACCTGACTAAGAAGC AGTTTCATTCTTGCGAACG | (27) | 429 |
| Methanobrevibacter smithii | CCGGGTATCTAATCCGGTTC CTCCCAGGGTAGAGGTGAAA | (28) | 123 |
The bacterial concentration from each fecal sample was calculated by comparing the Ct values obtained from the standard curves with the LightCycler 4.0 software. Standard curves were constructed for each experiment using serial tenfold dilutions of bacterial genomic DNA (of known concentration) from pure cultures, corresponding to 101–1010 16S rRNA gene copies/ gram of feces. The mass for one bacterial genome was calculated by using the Avogadro constant and assuming the mean molecular weight of a base pair to be 660 g/mol. Standard curves were normalized to the copy number of the 16S rRNA gene for each species. For the species whose copy number of 16S rRNA operon was not published, it was calculated by averaging the operon numbers of the closest bacterial taxa from the ribosomal RNA database rrnDB [29]. The 16S rRNA gene copies in each sample were normalized to gram of feces. The different strains used were obtained from the Spanish Collection of Type Cultures (CECT) (Bacteroides vulgatus NCTC 11154, Fusobacterium varium NCTC 10560, Enterococcus faecalis CECT 184, Enterobacter cloacae CECT 194, Clostridium perfringens CECT 376) and the American Type Culture Collection (ATCC) (Bifidobacterium bifidum ATCC 15696, Lactobacillus casei ATCC 334D-5, Prevotella intermedia ATCC 25611D-5, Ruminococus productus, ATCC 27340D-5, Methanobrevibacter smithii ATCC 35061).The data presented are the mean values of duplicate qPCR analyses.
Statistical analysis
Results are expressed as mean values and standard deviations. The statistical analysis was performed with SPSS 15.0 software (SPSS Inc., Chicago, IL). The 16S rRNA gene copies values were converted into logarithmic values before the statistical analysis. The Kruskal-Wallis test was used to check changes in bacterial number and the biochemical variables between the groups of rats. The Mann-Whitney U test with Bonferroni Post-hoc test was used to compare one group of rats to each other. The Spearman correlation coefficient was calculated to estimate the linear correlations between variables. A multivariate regression analysis was performed to identify independent predictors for serum leptin and ghrelin levels. In this analysis, bacterial group, weight, food intake and activity were selected as independent variables. Barnard's exact unconditional test was used to compare the proportions of one group of rats to each other. Statistical significance was set at a P value of <0.05. All data are presented in the text as the mean ± SD.
Results
Exercise, dietary aspects and appetite-regulating hormones levels
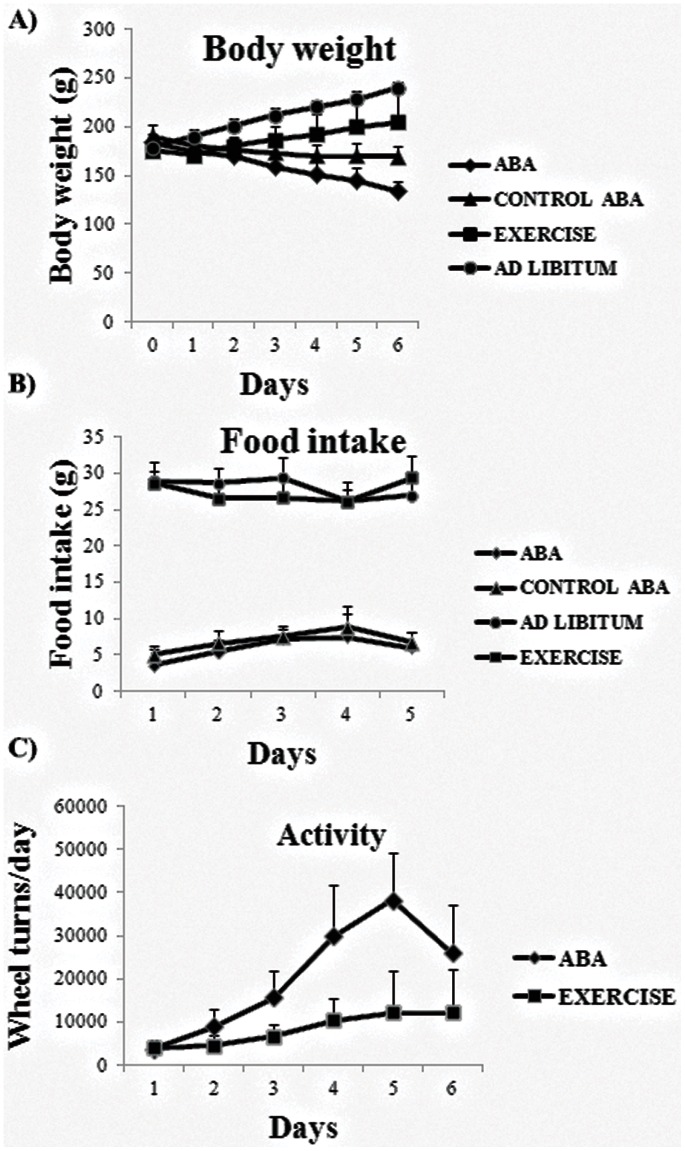
As figure 1A shows, our results confirmed that exercise and ad libitum groups of rats showed a progressive increase in body weight and significant differences were found between them. In contrast, feeding restriction to 1 h per day (control ABA group), and particularly the combination of this fasting regime with exercise (ABA group), significantly reduced body weight with respect to exercise and ad libitum groups from days 1 to 6. While, the control ABA group, showed a decrease in body weight that was stabilized from day 3 to 6 (figure 1A). With regard to food intake, we found no significant variations in daily food intake between exercise and ad libitum groups and between ABA and control ABA groups, respectively (figure 1B). Finally, when measuring the activity we found that the exercise group showed significantly less wheels turns than ABA group from days 3 to 5 (figure 1C).
Figure 1. Representation of the relative daily body weight variation (A) and daily food intake (B) in the all study groups (N = 10 rats per group).
The figure (C) shows a 23 h registered activity in ABA and exercise groups.
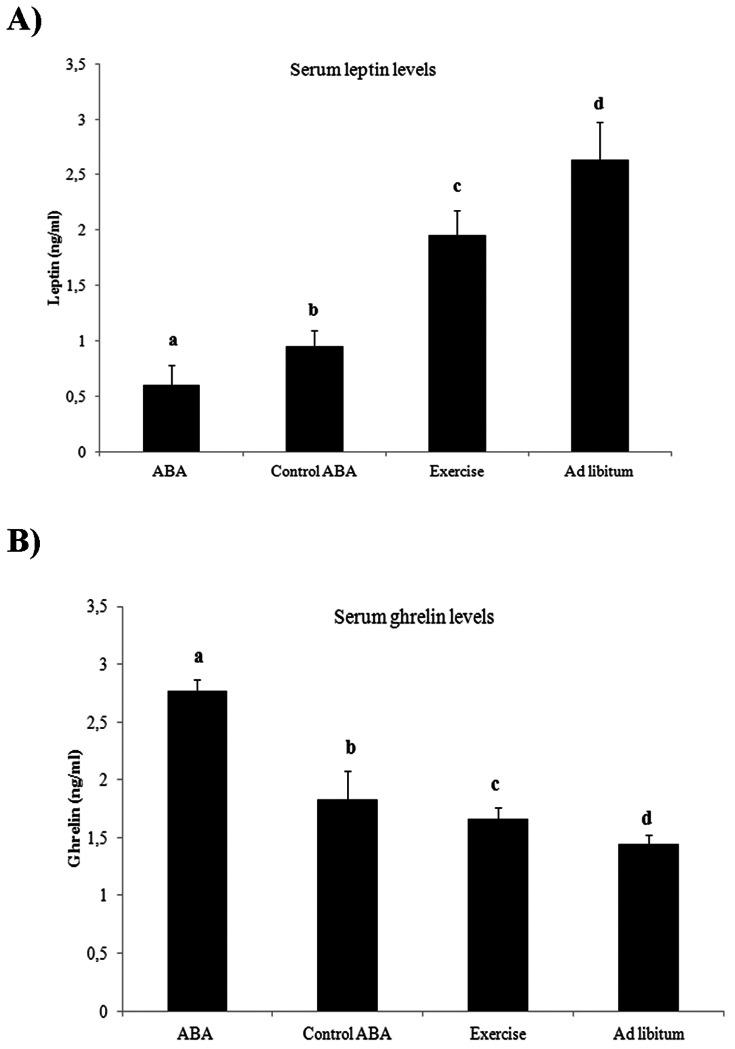
On the other hand, we observed a significant serum leptin decrease and ghrelin increase
in both ABA and control ABA compared to exercise and ad libitum. Moreover, we have found significant differences between the ABA and control ABA groups and between exercise and ad libitum groups in the serum leptin and ghrelin levels (figure 2A and 2B).
Figure 2. Serum leptin (A) and grhelin (B) levels (C) at day 6 of study in all rat groups (N = 10 rats per group).
The Mann-Whitney U test was used to compare a rat group to each other. Different superscript letters are significantly different P<0.05 (Bonferroni Post-hoc test).
PCR-DGGE fingerprint analysis and bacterial band identification in the fecal samples
Variations were found in the presence or absence (qualitative) and intensity (quantitative) of the bands between rat groups in the generated host-specific fingerprints. DGGE band profiles showed differences in band richness between the four groups. Analyzing the diversity of microbiota, we have found that the mean of DGGE bands was 11.16±2.0 for ABA, 13.50±1.04 for control ABA, 15.66±1.21 for exercise and 19.0±1.41 for ad libitum groups. Moreover, these differences in band richness were significant between all groups (P<0.05). On the other hand, some bands were seen in fingerprints from all the rats (in different lane but at the same position), indicating that specific species of the predominant microbiota were common to all rat groups.
The Dice similarity coefficient was used to calculate the similarity index of the DGGE band profiles for the four rat groups. The means of similarity index were 35.67%, 37.93%, 42.57% and 49.37% for ABA, control ABA, exercise and ad libitum groups respectively. The mean inter-group similarity index between ABA and control ABA groups was 25.75%, between exercise and ad libitum groups was 30.51%, between ABA and exercise group was 21.22% and between control ABA and ad libitum was 24.77%, which was fewer than the intra-group similarity index for the four rat groups above described. The intensity and position of DGGE detected bands were subjected to cluster analysis. The dendrogram shows four large clusters of feeding restriction plus exercise rats, feeding restriction rats, exercise rats and ad libitum rats, except for two exercise rats belonging to the cluster of ad libitum rats (Figure 3).
Figure 3. Dendrograms of electrophoretic band patterns obtained in the denaturing gel gradient electrophoresis experiment with universal primers in the fecal samples collected from ABA, control ABA (cABA), exercise and ad libitum rats.
A: Cluster analysis; B: DGGE profiles of fecal samples.
All the bands from all rat profiles in the different groups were cloned and sequenced to identify the dominant microbiota and the sequence similarity matches for bands were analyzed by MicroSeqID v2.1.1 software. Bacterial identification showed that the majority of bacteria represented in our fingerprints corresponded to four phyla (Table 2). Most of the sequences belonged to Firmicutes and Bacteroidetes, with the rest distributed among Actinobacteria and Proteobacteria. Nevertheless, we also observed important differences between all rat groups in the frequencies of different genus within these phyla. In the ABA and ad libitum groups, we have found an increase in the frequencies of Bacteroides, Prevotella, Clostridium and Helicobacter with respect to the control ABA and exercise groups respectively. Finally, with respect to the frequencies of Lactobacillus and Bifidobacterium, we observed a significant decrease in the ABA group with respect to exercise group (p = 0.008 and p = 0.05 respectively). In addition, a significant decrease was also found between exercise and ad libitum groups (p = 0.05) (Table 2).
Table 2. Bacterial identification after the sequencing of the bands cloned from the DGGE analysis of fecal samples from the all rat groups.
| Type bacteria genus(sequencing results ofthe bands) | ABA groupan = 45 | Control ABA groupan = 35 | Exercise groupan = 34 | Adlibitum groupan = 46 | Sequencesimilarity (%) |
| Phylum Bacteroidetes | |||||
| Genus Bacteroides | 13 (28.8%)a | 10 (28.6%)a | 8 (23.5%) a | 12 (26%) a | 99.86 |
| Genus Prevotella | 14 (28.6%) a | 10 (28.6%) a | 7 (20.58%)a | 13 (28.3%)a | 99.91 |
| Phylum Firmicutes | |||||
| Genus Lactobacillus | 0a, c | 1 (2.8%) a, b,c | 5 (14.7%)b | 1 (2.2%)a,c | 99.73 |
| Genus Clostridium | 15 (33.3%) | 11 (31.4%) | 9 (26.5%) | 16(34.8%) | 99.99 |
| Phylum Actinobacteria | |||||
| Genus Bifidobacterium | 0a | 1 (2.8%) a, b | 3(8.8%)b | 1 (2.2%)a,b | 99.92 |
| Phylum Proteobacteria | |||||
| Genus Helicobacter | 2 (4.4%) a | 1 (2.8%) a | 1 (2.9%) a | 2 (5.3%) a | 99.65 |
| Genus Campylobacter | 1 (2.2%) a | 1 (2.8%) a | 1 (2.9%) a | 1 (2.2%) a | 99.73 |
Refers to the frequency (and percent) of each unique bacteria genus in the ABA or Control ABA or exercise or ad libitum group.
"n" number of bands cloned, sequenced and identified in each rat group.
N = 10 rats per group. Barnard's exact unconditional test was used to compare the proportions of one group of rats to each other.
Values in a row with different superscript letters are significantly different P<0.05.
Comparative analysis of gut microbiota communities in the rat models under different nutritional status and physical activity
Changes in the bacterial population abundance were assessed in the fecal samples of all groups of rats both at phylum and genera levels (Tables 3). We found a significant decrease in the number of Actinobacteria phylum between the ABA group and the rest of the study groups. In addition, a significant increase in the number of Proteobacteria and a significant decrease in the quantity of Bacteroidetes and Firmicutes were observed in the ABA with respect to exercise and ad libitum groups. Finally, the quantity of Actinobacteria and Bacteroidetes was significantly increased while the number of Firmicutes was significantly decreased in exercise group compared to ad libitum group.
Table 3. Real-time PCR quantification of microbiota phyla, genera, groups and species in the study groups of rat.
| ABAgroup | Control ABAgroup | Exercisegroup | Ad libitum group | P | |
| Proteobacteria | 7.14±0.84a | 6.83±0.69a | 5.15±0.18c | 5.16±0.28 c | 0.001 |
| Fusobacteria | 6.42±0.26a | 6.31±0.16a | 6.40±0.49 a | 6.36±0.59 a | 0.180 |
| Actinobacteria | 5.91±0.99a | 7.41±0.45b | 8,33±0.01 c | 7.84±0.20 d | 0.013 |
| Bifidobacterium | 6.75±1.07a | 6.84±1.2a | 9.33±0.10 c | 7.97±0.37d | 0.003 |
| Bacteroidetes | 9.24±0.22a | 9.43±0.18a | 9.82±0.14 c | 9.68±0.24 c | 0.037 |
| Bacteroides | 7.95±0.13a | 7.64±0.48 b | 6.12±0.01 c | 7.10±0.38 d | 0.001 |
| Prevotella | 8.85±0.50a | 8.04±0.03b | 7.17±0.08 c | 7.45±0.13 d | 0.001 |
| Firmicutes | 7.24±0.33a | 7.33±0.15a | 7.63±0.22 c | 8.80±0.35 d | 0.007 |
| B. Coccoides- E. rectale group | 7.34±1.23a | 7.23±0.98a | 9.74±1.14c | 8.55±0.92 d | 0.001 |
| Enteroccocus | 8.41±1.73a | 8.35±1.8a | 4.08±0.66c | 6.55±1.7 d | 0.002 |
| Clostridium | 5.30±0.16a | 5.10±0.2b | 4.39±0.06 c | 4.87±0.14 d | 0.001 |
| Lactobacillus | 6.20±0.10a | 6.29±0.15a | 7.69±0.16 c | 6.91±0.21 d | 0.033 |
| Euryarchaeota M. smithii | 7.95±0.99 a | 7.89±1.07 a | 6.32±1.02 c | 6.43±1.10 c | 0.001 |
Values are presented as means ± SD and expressed as log10 16S rRNA gene copies/gram of feces. N = 10 rats per group. The P value in the last column was based on the Kruskal-Wallis test. The Mann-Whitney U test was used to compare a rat group to each other. Values in a row with different superscript letters are significantly different P<0.05 (Bonferroni Post-hoc test).
Within Firmicutes, the number of Clostridium was significantly increased in the ABA group with respect to the control ABA and the exercise and ad libitum groups. In addition, we found a significant increase in the number of Enteroccocus accompanied by a significant decrease in the quantities of B. Coccoides-E rectale group and Lactobacillus in the ABA and control ABA groups with respect to the exercise and ad libitum groups. On the other hand, the quantity of B. Coccoides-E rectale group and Lactobacillus showed a significant increase while the number of Clostridium and Enteroccocus presents a significant decrease in the exercise group with respect to ad libitum group.
Within Bacteroidetes, we have found a significantly higher quantity of Bacteroides and Prevotella in the ABA group with respect to control ABA, exercise and ad libitum group. In addition, a significant decrease in the number of Prevotella and Bacteroides was observed in the exercise group with respect to ad libitum group. Moreover, within Actinobacteria a significant decrease in the number of Bifidobacterium was observed in the ABA, control ABA and ad libitum groups with respect to exercise group. Finally, within Euryarchaeota a significant increase in the quantity of M. smithii in the ABA and control ABA with respect to exercise and ad libitum group has been found.
Relation between gut microbiota composition and serum leptin and ghrelin levels
Moreover, we found a significant univariate correlation between the quantity of specific bacterial groups and the serum leptin and ghrelin levels. The analysis showed a significant positive correlation between the quantity of Bifidobacterium (r = 0.429, P <0.05) and Lactobacillus (r = 0.466, P<0.05) and serum leptin levels, and a significant and negative correlation among the number of Clostridium (r = –0.677, P<0.001), Bacteroides (r = –0.531, P<0.001) and Prevotella (r = –0.885, P<0.001) and serum leptin levels in all the studied population. Furthermore, serum ghrelin levels were negatively correlated with the quantity of Bifidobacterium (r = –0.496, P<0.05), Lactobacillus (r = –0.499, P<0.05) and B. coccoides–Eubacterium rectale group (r = –0.628, P = 0.001) and positively correlated with the number of Bacteroides (r = 0.529, P<0.05) and Prevotella ( = 0.686, P<0.001). A multivariate regression analysis that included the weight, food intake, activity and all the bacterial groups analyzed as independent variables, showed that only the increase in the number of Lactobacillus and Bifidobacterium (R2 = 0.989; P<0.001, β = 0.563 and P<0.05, β = 0.979 respectively) and the decrease in Prevotella and Clostridium (R2 = 0.989;P<0.05, β = –0.241 and P<0.05, β = –0.448 respectively) were associated with the serum leptin level. While the serum ghrelin level was associated with the decrease in the quantity of Bifidobacterium, Lactobacillus and B. coccoides-E rectale group (R2 = 0.984; P = 0.001, β = –0.400; P<0.05, β = –0.247 and P<0.001, β = –0.578 respectively) and the increase in the number of Prevotella (R2 = 0.984; P<0.001, β = 0.446).
Discussion
In the present study we found that the nutritional status (food restriction and ad libitum regimens) and physical activity affected the composition of the gut microbiota. The food intake and activity present in our experimental rat models followed similar patterns as described previously [30], [31], however the gut microbiota composition of these animals has not been studied previously. In our study, to reduce variation in microbiota based in genetics, age and sex, male rats with similar weight from the same bred were acquired. Moreover, they were single housed to avoid competition for food as a source of variation. In this work, we have observed that rats with activity-based anorexia (ABA) develop hyperactivity and consume less food than rats with limited access to food and without access to running wheel (control ABA), since these rats adapt their feeding behavior to the shorter period of food availability. In presence of exercise, unrestricted eaters (exercise group) significantly increased the food intake compared to restricted eaters (ABA group), possibly because the exercise is more effective in creating a negative energy balance. In addition, the significant body weight reduction due to food restriction with access to exercise (ABA rats) was accompanied by a significant enhancement in the reduction of serum leptin concentration and a significant increase in the serum ghrelin levels, which reinforce the function of leptin as a starvation signal [32]. Moreover, this data suggest that low leptin levels could represent the key signal that triggers hyperactivity [33].
The analysis of the PCR-DGGE banding profiles showed that bacterial diversity was significantly lower in the two groups of rats with feeding restriction, especially in the ABA model. This lower bacterial composition reduced the set of ecosystem processes available in this bacterial community that may lead to less healthy animals [34]. In addition, the fecal DGGE analysis revealed a significantly lower intra-group similarity index in the restricted eaters groups (ABA and control ABA) than in the unrestricted eaters groups (exercise and ad libitum). Moreover, these intra-group similarity indexes of all rat groups were significantly higher than the inter-group similarity indexes, showing cluster of banding patterns characteristic for each rat group. These data suggest that the nutrition status and exercise may affect the diversity and similarity of the gut microbiota community present in this study models.
Sequencing results of the DGGE bands cloned also revealed differences in the microbiota composition between all the investigated groups. The sequence analysis of DGGE bands showed that the gut microbiota of the four rat groups was predominately composed of Firmicutes and Bacteroidetes. However, major differences were observed in the frequency of each genus of bacteria. We have observed differences in the frequency of bacteria in the ABA and ad libitum groups with respect to control ABA and exercise groups. These results suggest that the dominant microbiota genera could be modulated by both the feeding restriction and exercise.
Additional analyses with real-time qPCR were performed to obtain a quantitative estimation of the changes in the gut microbiota between the four experimental groups. We found significant differences between groups with or without feeding restriction at both phylum and genus level. Thus, we have detected a significant increase in the number of Proteobacteria, Bacteroides, Clostridium, Enteroccoco, Prevotella and M. smithii and a significant decrease in the quantity of Actinobacteria, Firmicutes, Bacteroidetes, B. coccoides-E. rectale group, Lactobacillus and Bifidobacterium in the restricted eaters with respect to the unrestricted eater groups. This distribution of gut microbiota among groups with and without feeding restriction may be linked to different nutrient availability in the gut. In accordance with us, Armougon et al. found that anorexic patients showed a significantly more quantity of M. smithii than lean patients. One explanation for these results may be associated with an adaptative attemp towards optimal exploitation of the very low caloric diet absorbed by these patients (35). Then, the increase of M. smithii in our restricted eaters may lead to optimization of food transformation in very low calorie diets.
It has been previously described that feeding restriction (fasting situation) stimulates the growth of the mucin degrading bacteria because these bacteria present a competitive advantage during nutrient deprivation [36], [37]. We have observed a significant increase in the number of Prevotella (which are responsible for the degradation of the intestinal mucin) in the ABA and control ABA with respect to exercise and ad libitum situations. This increase in Prevotella in the restricted eaters could involve an increase in the degradation and the lack of mucin (a glycoprotein produced by the host that maintains the integrity of the gut epithelium against pathogenic microorganisms as well as chemical, physical or enzymatic damage) on the epithelial layer of the gut, which would lead to a significant alteration in intestinal permeability. Increased mucosal permeability has previously reported by Sonoyama et al in fasted hamster due to the significant increase of Akkermansi muciniphila, a mucin degrading bacteria [38]. On the other hand, in these ABA and control ABA groups we have found a significant increase in the quantity of Bacteroides and Clostridium with respect to exercise and ad libitum groups. Based on cross-feeding mechanism among different bacterial groups, Clostridium spp. is able to utilize lactate and to convert it into acetate and propionate, and acetate is also produced by nearly all species of Bacteroides; however, these short fatty acids do not induce mucin synthesis [39]. Acetate was reported to protect against diet-induced obesity and had no acute effect on gut hormones while propionate was shown to induce gut hormones and reduce food intake [40]
Previous studies have described that weight reduction affects the composition of the gut microbial community in both mice and humans [41]–[43]. Moreover, it has been suggested that the effects of body weight changes on the gut microbiota may be mediated, in part, by changes in circulating leptin concentrations [32] since the level of this hormone may affect the composition of the gut microbiota by the stimulation of mucin production in the intestine [44], [45]. Accordingly, we have observed a connection between serum leptin concentrations and the composition of the gut microbiota. Thus, we have found that quantity of several bacterial genus is significantly correlated with serum leptin level. For instance, quantity of Bifidobacterium, Lactobacillus, and serum leptin concentrations is positively correlated, whereas the number of Clostridium, Bacteroides and Prevotella was negatively correlated with serum leptin level. Then, the significant decrease in body weight together with the significantly lower serum leptin levels found in the ABA and the control ABA animals with respect to exercise and ad libitum groups could be used to explain the significant changes found in the gut microbiota between these groups.
On the other hand, in our study we have observed a significant increase in the number of Lactobacillus, Bifidobacterium and Blautia coccoides–Eubacterium rectale group in the exercise group with respect to the ABA, control ABA and ad libitum groups. Both Bifidobacteria and Lactobacillus have the capacity to produce the organic acid lactate, which is converted into butyrate by butyrate-producing bacteria in the gut [46]. Barcenilla et al. showed that most of the butyrate-producing isolates from human fecal samples are related to the Blautia coccoides–Eubacterium rectale group [47]. Matsumoto et al. using exercise and sedentary Wistar rats suggested that exercise altered the composition of the microbiota in the cecum and increased the concentration of n-butyrate in the cecal content of the exercise rats [48]. Moreover, previous studies have shown that butyrate induces mucin synthesis [39], decreases bacterial transport across the epithelium [49], improves gut integrity by increasing tight junction assembly [50] and reduces serum ghrelin levels [40]. In our study we have found a significantly lower serum ghrelin level in the exercise and ad libitum groups with respect to ABA and control ABA groups. Furthermore, the serum ghrelin levels showed a significant negative correlation with the quantity of Bifidobacterium, Lactobacillus and Blautia coccoides–Eubacterium rectale group, and a significant positive correlation with the number of Bacteroides and Prevotella. Other authors have previously described the associations here found between fecal bacterial families and blood hormones, such as leptin and ghrelin in kittens under a protein/carbohydrate dietary intervention.They also found a positive association between Lactobacillus and blood leptin levels, but unlike us, the correlation between Bifidobacteriaceae and blood ghrelin levels was positive (51). These data suggest a role of the gut microbiota on satiety control.
In conclusion, these findings indicated that differences in the nutritional status and exercise alter the gut microbiota composition affecting the diversity and similarity of the bacterial community. In general, food restrictions and especially, situations of extreme food restriction plus increased activity (anorexia) seemed to have a potential negative impact on the quantity of health promoting bacteria as well as to enhance the growth of bacteria which may be related to the disruption of the gut mucosal barrier and the optimal exploitation of the very low caloric diet. On the other hand, this study highlights the associations between the gut microbiota and the appetite-regulating hormones such as leptin and ghrelin that may be important in terms of satiety control and host metabolism.
Acknowledgments
We gratefully acknowledge the help of Ian Johnstone for his expertise in preparing this manuscript. The research group belongs to the “Centros de Investigación en Red” (CIBEROBN, CB06/03/0018) of the “Instituto de Salud Carlos III. We gratefully acknowledge the very useful technical assistance provided by the Plataforma de Secuenciacion y Genotipado and the Unidad Central de Biología Molecular (Servicio de PCR a tiempo real) of the IBIMA.
Funding Statement
This work was supported in part by a grant from CIBER (Centro de Investigación en Red) CB06/03/0018 of the "Instituto de Salud Carlos III (ISCIII)", the Instituto de Salud Carlos III CP07/0095 Madrid (Spain), the Servicio Andaluz de Salud (PI0696/2010) and Fondo de Investigacion Sanitaria (FIS) PI12/02355 from the Instituto de Salud Carlos III (ISCIII), Madrid (Spain). The post-doctoral grant “Sara Borrell” (CD11/0030) from the Spanish Ministry of Economy and Competitiveness (MM) and the Miguel Servet Fellow from the Instituto de Salud Carlos III/SERGAS (MP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yager J, Anderson AE (2005) Anorexia nervosa. N Engl J Med 353(14): 1481–1488. [DOI] [PubMed] [Google Scholar]
- 2. Affenito SG, Dohm FA, Crawford PB, Daniels SR, Striegel-Moore RH (2002) Macronutrient intake in anorexia nervosa: The National Heart, Lung, and Blood Institute Growth and Health Study. J Pediatr 141(5): 701–705. [DOI] [PubMed] [Google Scholar]
- 3. Payne AN, Chassard C, Banz Y, Lacroix C (2012) The composition and metabolic activity of child gut microbiota demonstrate differential adaptation to varied nutrient loads in an in vitro model of colonic fermentation. FEMS Microbiol Ecol 80(3): 608–623. [DOI] [PubMed] [Google Scholar]
- 4. Kowalska I, Karczewska-Kupczewska M, Strączkowski M (2011) Adipocytokines, gut hormones and growth factors in anorexia nervosa. Clin Chim Acta 412(19–20): 1702–1711. [DOI] [PubMed] [Google Scholar]
- 5. Hebebrand J, Muller TD, Holtkamp K, Herpertz-Dahlmann B (2007) The role of leptin in anorexia nervosa: clinical implications. Mol Psychiatry 12(1): 23–35. [DOI] [PubMed] [Google Scholar]
- 6. Boakes RA (2007) Self-starvation in the rat: running versus eating. Span J Psychol 10(2): 251–257. [DOI] [PubMed] [Google Scholar]
- 7. Bulik CM, Berkman ND, Brownley KA, Sedway JA, Lohr KN (2007) Anorexia nervosa treatment: a systematic review of randomized controlled trials. Int J Eat Disord 40(4): 310–320. [DOI] [PubMed] [Google Scholar]
- 8. Nedvídková J, Krykorková I, Barták V, Papezová H, Gold PW, et al. (2003) Loss of meal-induced decrease in plasma ghrelin levels in patients with anorexia nervosa. J Clin Endocrinol Metab 88(4): 1678–1682. [DOI] [PubMed] [Google Scholar]
- 9. Castaneda TR, Tong J, Datta R, Culler M, Tschöp MH (2010) Ghrelin in the regulation of body weight and metabolism. Front Neuroendocrinol 31(1): 44–60. [DOI] [PubMed] [Google Scholar]
- 10. De Vriese C, Delporte C (2008) Ghrelin: a new peptide regulating growth hormone release and food intake. Int J Biochem Cell Biol 40(8): 1420–1424. [DOI] [PubMed] [Google Scholar]
- 11. Broglio F, Prodam F, Riganti F, Muccioli G, Ghigo E (2006) Ghrelin: from somatotrope secretion to new perspectives in the regulation of peripheral metabolic functions. Front Horm Res 35: 102–114. [DOI] [PubMed] [Google Scholar]
- 12. Ducrotte P, Hokfelt T, Dechelotte P (2008) Autoantibodies against appetite-regulating peptide hormones and neuropeptides: putative modulation by gut microflora. Nutrition 24(4): 348–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fetissov SO, Hamze Sinno M, Coquerel Q, Do Rego JC, Coeffier M, et al. (2008) Emerging role of autoantibodies against appetite-regulating neuropeptides in eating disorders. Nutrition 24(9): 854–859. [DOI] [PubMed] [Google Scholar]
- 14. Sekirov I, Russell SL, Antunes LC, Finlay BB (2010) Gut microbiota in health and disease. Physiol Rev 90(3): 859–904. [DOI] [PubMed] [Google Scholar]
- 15. Cani PD, Delzenne NM (2009) The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des 15(13): 1546–1558. [DOI] [PubMed] [Google Scholar]
- 16. Routtenberg A, Kuznesof AW (1967) Self-starvation of rats living in activity wheels on a restricted feeding schedule. J Comp Physiol Psychol 64(3): 414–421. [DOI] [PubMed] [Google Scholar]
- 17. Kinzing KP, Hargrave SL (2010) Adolescent activity-based anorexia increases Anxiety-like behavior in Adulthood. Physiol Behav 101(2): 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seoane LM, Al-Massadi O, Barreiro F, Dieguez C, Casanueva F (2007) Growth hormone and somatostatin directly inhibit gastric ghrelin secretion. An in vitro organ culture system. J Endocrinol Invest 30(9): RC22–25. [DOI] [PubMed] [Google Scholar]
- 19. Seoane LM, Al-Massadi O, Caminos JE, Tovar SA, Dieguez C, et al. (2007) Sensory stimuli directly acting at the central nervous system regulate gastric ghrelin secretion. An ex vivo organ culture study. Endocrinology 148(8): 3998–4006. [DOI] [PubMed] [Google Scholar]
- 20. Queipo-Ortuño MI, Boto-Ordoñez M, Murri M, Gomez-Zumaquero JM, Clemente-Postigo M, et al. (2012) Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am J Clin Nutr 95(6): 1323–1334. [DOI] [PubMed] [Google Scholar]
- 21. Guo X, Xia X, Tang R, Zhou J, Zhao H, et al. (2008) Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol 47(5): 367–373. [DOI] [PubMed] [Google Scholar]
- 22. Delroisse JM, Boulvin AL, Parmentier I, Dauphin RD, Vandenbol M (2008) Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol Res 163(6): 663–670. [DOI] [PubMed] [Google Scholar]
- 23. Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, et al. (2010) Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One 5(1): e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stach JE, Maldonado LA, Ward AC, Goodfellow M, Bull AT (2003) New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ Microbiol 5(10): 828–841. [DOI] [PubMed] [Google Scholar]
- 25. Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R (2004) Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol 70(12): 7220–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bekele AZ, Koike S, Kobayashi Y (2010) Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol Lett 305(1): 49–57. [DOI] [PubMed] [Google Scholar]
- 27. Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A (2004) Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol 97(6): 1166–1177. [DOI] [PubMed] [Google Scholar]
- 28. Dridi B, Henry M, El Khéchine A, Raoult D, Drancourt M (2009) High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One 4(9): e7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee ZMP, Bussema C, Schmidt TM (2009) rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res 37: D489–D493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boakes RA, Juraskova I (2001) The role of drinking in the suppression of food intake by recent activity. Behav Neurosci 115(3): 718–730. [DOI] [PubMed] [Google Scholar]
- 31. Pardo M, Roca-Rivada A, Al-Massadi O, Seoane LM, Camiña JP, et al. (2010) Peripheral leptin and ghrelin receptors are regulated in a tissue-specific manner in activity-based anorexia. Peptides 31(10): 1912–1919. [DOI] [PubMed] [Google Scholar]
- 32. Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, et al. (2012) Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity 20(4): 738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hebebrand J, Exner C, Hebebrand K, Holtkamp C, Casper RC, et al. (2003) Hyperactivity in patients with anorexia nervosa and in semistarved rats: evidence for a pivotal role of hypoleptinemia. Physiol Behav 79(1): 25–37. [DOI] [PubMed] [Google Scholar]
- 34. Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, et al. (2011) Toward defining the autoimmune microbiome for type 1 diabetes. Isme J 5(1): 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Armougom F, Henry M, Vialettes B, Raccah D, Raoult D (2009) Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS One 4(9): e7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deplancke B, Vidal O, Ganessunker D, Donovan SM, Mackie RI, et al. (2002) Selective growth of mucolytic bacteria including Clostridium perfringens in a neonatal piglet model of total parenteral nutrition. Am J Clin Nutr 76(5): 1117–1125. [DOI] [PubMed] [Google Scholar]
- 37. Miller RS, Hoskins LC (1981) Mucin degradation in human colon ecosystems. Fecal population densities of mucin-degrading bacteria estimated by a “most probable number” method. Gastroenterology 81(4): 759–765. [PubMed] [Google Scholar]
- 38. Sonoyama K, Fujiwara R, Takemura N, Ogasawara T, Watanabe J, et al. (2009) Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl Environ Microbiol 75(20): 6451–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J (2009) The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J 420(2): 211–219. [DOI] [PubMed] [Google Scholar]
- 40. Lin HV, Frassetto A, Kowalik EJ Jr, Nawrocki AR, Lu MM (2012) Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 7(4): e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ley RE (2010) Obesity and the human microbiome. Curr Opin Gastroenterol 26(1): 5–11. [DOI] [PubMed] [Google Scholar]
- 42. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI (2008) Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3(4): 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. (2009) A core gut microbiome in obese and lean twins. Nature 457(7228): 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. El Homsi M, Ducroc R, Claustre J, Jourdan G, Gertler A, et al. (2007) Leptin modulates the expression of secreted and membrane-associated mucins in colonic epithelial cells by targeting PKC, PI3K, and MAPK pathways. Am J Physiol Gastrointest Liver Physiol 293(1): 365–373. [DOI] [PubMed] [Google Scholar]
- 45. Plaisancie P, Ducroc R, El Homsi M, Tsocas A, Guilmeau S, et al. (2006) Luminal leptin activates mucinsecreting goblet cells in the large bowel. Am J Physiol Gastrointest Liver Physiol 290(4): 805–812. [DOI] [PubMed] [Google Scholar]
- 46. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, et al. (2011) Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One 6(10): e25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, et al. (2000) Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 66(4): 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsumoto M, Inoue R, Tsukahara T, Ushida K, Chiji H, et al. (2008) Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci Biotechnol Biochem 72(2): 572–576. [DOI] [PubMed] [Google Scholar]
- 49. Lewis K, Lutgendorff F, Phan V, Soderholm JD, Sherman PM, et al. (2010) Enhanced Translocation of Bacteria Across Metabolically Stressed Epithelia is Reduced by Butyrate. Inflamm Bowel Dis 16(7): 1138–1148. [DOI] [PubMed] [Google Scholar]
- 50. Peng LY, Li Z, Green RS, Holzman IR, Lin J (2009) Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr 139(9): 1619–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hooda S, Vester Boler BM, Kerr KR, Dowd SE, Swanson KS (2012) The gut microbiome of kittens is affected by dietary protein: carbohydrate ratio and associated with blood metabolite and hormone concentrations. Br J Nutr 31: 1–10. [DOI] [PubMed] [Google Scholar]